Abstract
Recent isolation of Pseudomonas aeruginosa strains from the open ocean and subsequent pulsed-field gel electrophoresis analyses indicate that these strains have a unique genotype (N. H. Khan, Y. Ishii, N. Kimata-Kino, H. Esaki, T. Nishino, M. Nishimura, and K. Kogure, Microb. Ecol. 53:173-186, 2007). We hypothesized that ocean P. aeruginosa strains have a unique phylogenetic position relative to other strains. The objective of this study was to clarify the intraspecies phylogenetic relationship between marine strains and other strains from various geographical locations. Considering the advantages of using databases, multilocus sequence typing (MLST) was chosen for the typing and discrimination of ocean P. aeruginosa strains. Seven housekeeping genes (acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE) were analyzed, and the results were compared with data on the MLST website. These genes were also used for phylogenetic analysis of P. aeruginosa. Rooted and unrooted phylogenetic trees were generated for each gene locus and the concatenated gene fragments. MLST data showed that all the ocean strains were new. Trees constructed for individual and concatenated genes revealed that ocean P. aeruginosa strains have clusters distinct from those of other P. aeruginosa strains. These clusters roughly reflected the geographical locations of the isolates. These data support our previous findings that P. aeruginosa strains are present in the ocean. It can be concluded that the ocean P. aeruginosa strains have diverged from other isolates and form a distinct cluster based on MLST and phylogenetic analyses of seven housekeeping genes.
The ubiquitously distributed gammaproteobacterium Pseudomonas aeruginosa is capable of thriving in a large number of dissimilar ecological niches (12, 15, 16, 21, 22, 24, 33, 37, 38, 39, 41, 42, 44, 50, 52, 54, 55, 56). Our recent study documented its survival and existence in the open ocean (22, 24). Molecular fingerprinting techniques, such as pulsed-field gel electrophoresis, have demonstrated that oceanic P. aeruginosa strains harbor a unique genotype. Their growth and survival under nutrient-free, high-NaCl, and high-pH stress conditions in laboratory-based microcosms have been evaluated, and it was found that they could survive and thrive longer than clinical and freshwater strains (22). In light of these results, we hypothesized that marine P. aeruginosa possesses new sequence types and assumes unique positions in intraspecies phylogenetic trees.
The genome sizes of P. aeruginosa range from 5.2 to 7.1 Mbp (46). Recent work suggests that more than 80% of the genome of the sequenced strain (strain PAO1) is shared (with only 0.5% nucleotide divergence) with the isolate from cystic fibrosis patients and with environmental strains (47), indicating that the genome of this bacterium is characterized by a combination of relatively conserved core genes and some variable accessory genes. Therefore, depending on the genes analyzed and the methods used, apparently various genomic divergences are obtained.
Denamur et al. (6) and Picard et al. (40) argued that the species had a panmictic population structure, but Kiewitz and Tummler (23) proposed a net-like structure characterized by high frequencies of recombination. Lomholt et al. (29) favored an epidemic structure, and Pirnay et al. (41) combined sequencing-based techniques, such as sequencing of the outer membrane lipoprotein, with serotyping and pyoverdine type determination in a polyphasic approach to reveal extensive genetic mosaicism, particularly in the oprD gene.
Multilocus sequence typing (MLST) is a recently developed strain-typing system that focuses strictly on conserved housekeeping genes (5, 10, 31, 53). The choice of seven loci ensures adequate variability such that one can distinguish between the most closely related strains and still be able to track the global clonal history of the species with the highest possible accuracy. Because of these advantages, we used an MLST scheme to characterize strains of P. aeruginosa isolated from the open ocean, which enabled us to compare our data with those in the database generated from various clinical and environmental P. aeruginosa strains. In addition, we attempted to clarify the phylogenetic relationships of these isolates by concatenating seven genes used for MLST. These genes are generally conserved and less likely to undergo horizontal gene transfer and are therefore suitable for phylogenetic analysis.
In order to test the hypothesis that marine P. aeruginosa comprises a population uniquely adapted to the ocean environment, seven housekeeping genes (acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE) from 34 P. aeruginosa strains were sequenced, and the data were compared with those for MLST alleles in the existing database. The concatenated sequences were used for phylogenetic analysis to determine the position of ocean P. aeruginosa strains relative to other strains. To our knowledge, this is the first phylogenetic study of marine P. aeruginosa and will contribute significantly to a new ecological perspective on these well-known bacterial strains.
MATERIALS AND METHODS
Bacterial strains.
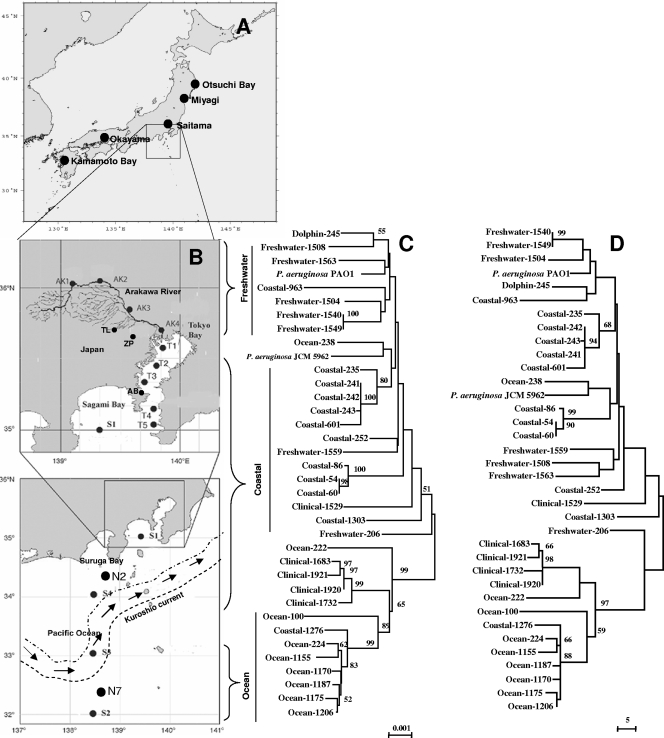
Thirty-four environmental and clinical strains were used. The environmental strains were isolated from the open ocean, coastal areas, and freshwater environments in Japan (Table 1 and Fig. 1). The coastal strains (12 strains) included those from Tokyo Bay, Sagami Bay (station S1), Suruga Bay (station N2), and station S4. Although stations N2 and S4 were at least 50 km away from the shore, both were north of the Kuroshio Current, and the possible influence of terrestrial environments was not completely rejected; thus, strains from these areas were considered coastal. Two strains each from Kumamoto Bay and Aburatsubo Bay were also included in the coastal category. Kumamoto Bay is far from Tokyo Bay, whereas Aburatsubo Bay is close to Tokyo Bay and located between Tokyo Bay and Sagami Bay. Ocean strains (9 strains) were isolated from the south of the Kuroshio Current (22), where the influence of human activities is negligible. Freshwater strains were isolated from ponds, lakes, and rivers near the greater Tokyo metropolitan area (22). The sampling locations for freshwater strains are shown in Fig. 1. Five strains were isolated from human blood, sputum, and urine samples and collected from different hospitals in Japan. One strain was isolated from a dead dolphin from Otsuchi Bay, Japan. The possible source of the P. aeruginosa strain isolated from the dolphin could not be clarified. As the strain was isolated from a dead dolphin, a possible secondary infection could not be rejected, as there are reports of fatal bronchopneumonia and dermatitis of bottlenose Atlantic dolphins induced by P. aeruginosa (7). The sources, origins, dates of isolation, and serotypes of the strains are given in Table 1. The origins and geographical locations of the sampling points are indicated in Fig. 1A and B. Except for the strains numbered Coastal-54 to Coastal-252 (Table 1), all strains were analyzed in our previous study (22).
TABLE 1.
Strain numbers and corresponding groups, origins, times of isolation, and serotypes for P. aeruginosa and other pseudomonads used for phylogenetic and MLST analyses
| Strain | Groupa | Origin (station, depth [m]) | Time of isolation | Serotype | Source or reference |
|---|---|---|---|---|---|
| P. aeruginosa | |||||
| Coastal-54 | Coastal | Suruga Bay (N2, 200) | March 2004 | I | This study |
| Coastal-60 | Coastal | Suruga Bay (N2, 200) | March 2004 | I | This study |
| Coastal-86 | Coastal | Suruga Bay (N2, 0) | March 2004 | I | This study |
| Ocean-100 | Ocean | Open ocean (N7, 0) | March 2004 | B | This study |
| Arakawa River-206 | Freshwater | Arakawa River, Saitama (AK1) | May 2004 | F | This study |
| Ocean-222 | Ocean | Open ocean (S2, 0) | May 2004 | I | This study |
| Ocean-224 | Ocean | Open ocean (S2, 0) | May 2004 | E | This study |
| Coastal-235 | Coastal | Sagami Bay (S1, 0) | May 2004 | G | This study |
| Ocean-238 | Ocean | Open ocean (S2, 0) | May 2004 | I | This study |
| Coastal-241 | Coastal | Sagami Bay (S1, 0) | May 2004 | G | This study |
| Coastal-242 | Coastal | Sagami Bay (S1, 0) | May 2004 | G | This study |
| Coastal-243 | Coastal | Sagami Bay (S1, 0) | May 2004 | G | This study |
| Dolphin-245 | Animal | Dolphin | May 2004 | E | This study |
| Coastal-252 | Coastal | Aburatsubo Bay, close to Tokyo Bay (0) | May 2004 | NDb | This study |
| Coastal-601 | Coastal | Kumamoto Bay (0) | December 2003 | ND | 22 |
| Coastal-963 | Coastal | Tokyo Bay (T1, 0) | May 2003 | F | 22 |
| Ocean-1155 | Ocean | Open ocean (S2, 0) | June 2003 | E | 22 |
| Ocean-1170 | Ocean | Open ocean (S2, 0) | June 2003 | E | 22 |
| Ocean-1175 | Ocean | Open ocean (S2, 0) | June 2003 | E | 22 |
| Ocean-1187 | Ocean | Open ocean (S2, 0) | June 2003 | E | 22 |
| Ocean-1206 | Ocean | Open ocean (S2, 0) | June 2003 | E | 22 |
| Coastal-1276 | Coastal | Tokyo Bay (0) | June 2003 | B | 22 |
| Coastal-1303 | Coastal | North of Kuroshio (S4, 0) | June 2003 | K | 22 |
| Arakawa River-1504 | Freshwater | Arakawa River, Tokyo (AK2) | November 2003 | K | 22 |
| Arakawa River-1508 | Freshwater | Arakawa River, Tokyo (AK4) | November 2003 | B | 22 |
| Clinical-1529 | Clinical | Urine, Tokyo | 2002 | M | 22 |
| Pond-1540 | Freshwater | Zenpukujii Pond, Tokyo | September 2003 | M | 22 |
| Pond-1549 | Freshwater | Zenpukujii Pond, Tokyo | September 2003 | G | 22 |
| Tama Lake-1559 | Freshwater | Tama Lake, Tokyo | September 2003 | G | 22 |
| Tama Lake-1563 | Freshwater | Tama Lake, Tokyo | September 2003 | G | 22 |
| Clinical-1683 | Clinical | Sputum, Okayama | 2002 | E | 22 |
| Clinical-1732 | Clinical | Urine, Myagi | 2002 | E | 22 |
| Clinical-1920 | Clinical | Blood, Saitama | 2002 | E | 22 |
| Clinical-1921 | Clinical | Blood, Saitama | 2002 | E | 22 |
| JCM 5962Tc,d | Unknown | Unknown | Unknown | ND | 8 |
| PAO1e | Clinical | Wound, Australia | 1955 | ND | 49 |
| Other species | |||||
| P. fluorescens JCM 5963Tc | Environmental | Prefilter tanks | 1961 | ND | 9 |
| P. putida KT2440e | Plant | Osaka, Japan | 1961 | ND | 34 |
| P. syringae B728ae | Plant | United States | 1987 | ND | 30 |
| P. fluorescens Pf-5e | Rhizosphere | United States | Unknown | ND | 17 |
The strains were assigned to different groups depending on their sources.
ND, not done.
These strains were used for DNA-DNA hybridization.
This strain was used for sequencing and comparative analyses.
These strains were used for comparative analyses, and the data were taken from the GenBank database.
FIG. 1.
Correlation between the geographical locations (sampling points) of P. aeruginosa strains and their positions in the phylogenetic trees. (A) Map showing the origins and sampling sites of clinical and coastal strains in Japan outside the Tokyo area. (B) Map showing the sampling sites in the Pacific Ocean, coastal, and freshwater environments in and around Tokyo, Japan. The figure shows the path of the Kuroshio Current as of the sampling time in 2003 during the Tansei Maru cruise in the Pacific Ocean. AB, Aburatsubo Bay; AK, Arakawa River; TL, Tama Lake; and ZP, Zenpukujii Pond. (C) Unrooted tree of the concatenated nucleotide sequences of seven housekeeping genes (acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE) of P. aeruginosa obtained using the NJ method with Kimura 2 correction for distance calculations. The total length of the concatenated group of nucleotide sequences was 5,700 bp. Bootstrap percentages retrieved in 1,000 replications are shown at the nodes. Clusters and bootstrap values that were 50% or greater are indicated. The scale bar (0.001) indicates the number of nucleotide substitutions per site. (D) The tree was constructed using the MP method with the same data set. Bootstrap percentages retrieved in 1,000 replications are shown at the nodes. Clusters and bootstrap values that were 50% or greater are indicated.
Isolation and identification of P. aeruginosa strains.
The isolation methods for P. aeruginosa were described previously (22). Several methods were applied for the identification of P. aeruginosa. Selective agar culture media were used for the identification of P. aeruginosa at the initial stage. Suspected colonies that showed the characteristic appearance and color on nalidixic acid cetrimide agar plates were inoculated onto cetrimide kanamycin nalidixic acid agar (26) and incubated at 42°C overnight. Isolates that showed growth at 42°C were primarily identified as P. aeruginosa. Colonies were transferred to other selective agar plates and incubated at 4°C for 3 to 4 days. No colonies appeared at 4°C. Sequences of the 16S rRNA gene were determined by direct sequencing using the primers 27f and 1492r (28). The PCR products were purified with an EXOSAP-IT kit (USB Corp., Cleveland, OH), sequenced using an ABI Prism BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster city, CA), and run on an ABI 3100 genetic analyzer (Applied Biosystems). BLAST programs were used to find the most closely related sequences (2). The isolates were also identified by the BD (Becton, Dickinson and Company, MD) Phoenix automated microbiology system according to the method previously described by Khan et al. (22). It is necessary to perform DNA-DNA hybridization to judge whether a novel isolate belongs to a species when the isolate and species representatives share more than 97% 16S rRNA gene sequence similarity (14, 48). Thus, two marine strains were randomly selected for DNA-DNA hybridization. P. aeruginosa JCM 5962T and P. fluorescence JCM 5963T were selected as the same species and a closely related species, respectively. DNA-DNA hybridization was determined by using photobiotin-labeled probes in a polystyrene 96-well plate (Nunc Maxisorp) as described by Ezaki et al. (11), utilizing Bacteroides sp. strain IAM 15343 as a negative control.
Culture of isolates and preparation of chromosomal DNA.
Bacterial strains were maintained at −80°C in 40% (vol/vol) glycerol in nutrient broth, streaked to single colonies, and cultured on Mueller-Hinton agar at 37°C under aerobic conditions. Chromosomal DNA was extracted from 1-ml cultures of strains grown overnight in nutrient broth by using a Qiagen DNeasy tissue extraction kit (Valencia, CA).
Locus selection.
The seven genes acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE (see Table 3) were selected according to the MLST scheme for P. aeruginosa (http://pubmlst.org/paeruginosa/). The criteria governing locus selection included biological role (comprising genes with diverse central housekeeping roles, such as mismatch repair, DNA replication, and amino acid biosynthesis), size (∼600 bp), location (i.e., a minimum of 6 kbp upstream or downstream from known virulence factors, lysogenic phages, or insertion sequence elements), and suitability for nested primer design and sequence diversity (5). All phylogenetic analyses were performed with the same criteria.
TABLE 3.
Functions, executable lengths, and numbers of variable sites for the PCR and MLST fragments of the genes analyzed
| Gene | Function | PCR fragment
|
MLST fragment
|
||
|---|---|---|---|---|---|
| Length (bp) | No. of variable sites | Length (bp) | No. of variable sites | ||
| acsA | Acetyl coenzyme A synthetase | 779 | 28 | 390 | 14 |
| aroE | Shikimate dehydrogenase | 755 | 28 | 495 | 15 |
| guaA | GMP synthase | 844 | 24 | 372 | 13 |
| mutL | DNA mismatch repair protein | 822 | 35 | 441 | 28 |
| nuoD | NADH dehydrogenase I chains C and D | 839 | 22 | 366 | 6 |
| ppsA | Phosphoenolpyruvate synthase | 901 | 25 | 369 | 11 |
| trpE | Anthralite synthetase component I | 760 | 31 | 441 | 12 |
| Total | 5,700 | 185 | 2,873 | 103 | |
Amplification of the loci.
The seven housekeeping genes were amplified with Z-Taq polymerase (Takara Bio, Inc., Shiga, Japan), using the primers described by Curran et al. (5). The primers had variable melting temperatures. The 25-μl amplification reaction mixture comprised 0.5 to 1.0 μl of chromosomal DNA (final concentration, 4 to 8 ng/μl), 0.5 μl each of the 10-pmol primers, 2.5 μl 10× Z-Taq buffer, 0.25 μl of Takara Z-Taq (2.5 units/μl), and 2 μl of deoxynucleoside triphosphate mixture (2.5 mM) and was brought up to the correct reaction volume with sterilized distilled water. The following PCR protocol was used: 1 s of denaturing at 98°C and 30 to 33 cycles (depending on the gene) of 1 s at 98°C (denaturing), 10 s at 57°C to 60°C (primer annealing), and 20 s at 72°C (polymerase extension). Reactions were performed with a PTC-100 thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA).
BigDye reaction and sequencing.
Two microliters of ExoSAP-IT was added directly to the 7-μl reaction products following PCR. ExoSAP-IT was inactivated at 80°C for 15 min, and the treated PCR products were ready for subsequent analysis in BigDye reactions, which required the DNA to be free of excess primers and nucleotides. BigDye reactions were carried out using an ABI Prism BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA). DNA sequencing was performed with an ABI 3100 genetic analyzer (Applied Biosystems) with a 50-cm array. Both forward and reverse sequences were obtained by using the PCR primer or internal sequence primers (specifically for MLST sequences) for each locus.
Sequence data analysis.
Both the forward and the reverse primer sequences of acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE were verified with the Chromas software program, version 2.32 (Technelysium Pty., Ltd.). Several representative DNA sequences were randomly selected from each group of isolates and converted into amino acid sequences, and multiple alignments of these sequences were calculated. These results and the published alignment data were used as templates to conduct multiple alignments of all DNA sequences determined (45). Positions in which gaps were present in any of the aligned sequences were excluded from the analysis. All sequences were confirmed by individual BLAST searches to determine whether the corresponding data matched well with the appropriate gene of P. aeruginosa. Both forward and reverse primer sequences were assembled by using ATGC, version 4.0 (Genetyx Corporation, Tokyo, Japan). The assembled sequences were saved with Genetyx-Win, version 5.0 (Genetyx Corporation, Tokyo, Japan) in Fasta format. The alignment was performed using a hierarchical method for multiple alignments implemented in the CLUSTAL-W tool in MEGA 3.1 (27, 51). Automatically aligned sequences were checked manually.
Allele and strain type assignment.
A number was assigned to each distinct allele within a locus according to the number available in the P. aeruginosa MLST database. Any allele that did not match with existing data was designated “new.” Each isolate was therefore given seven numbers that represented its strain type. Any strain type that did not match with the existing database was numbered as a “new” type. MLST analysis yielded alleles for individual loci of seven housekeeping genes. The alleles were numbered according to their entries in the MLST database. Alleles that did not exist in the database were termed “new.” A table was prepared (see Table 4), and all the alleles were provided separate numbers. Strains having the same allele were given the same number.
TABLE 4.
MLST allelic profiles and strain types of 34 P. aeruginosa strainsa
| P. aeruginosa strain | Strain groupb | Allelic type
|
Strain type | ||||||
|---|---|---|---|---|---|---|---|---|---|
| acsA | aroE | guaA | mutL | nuoD | ppsA | trpE | |||
| Ocean-100 | Ocean | New | 8 | 57 | New | 1 | 6 | 3 | 1 (new) |
| Ocean-222 | Ocean | 47 | 8 | 7 | New | 8 | 11 | 40 | 2 (new) |
| Ocean-224 | Ocean | 13 | 8 | 9 | 3 | 1 | 6 | 9 | 3 (new) |
| Ocean-238 | Ocean | 6 | 5 | 12 | New | 4 | 6 | 7 | 4 (new) |
| Ocean-1155 | Ocean | 13 | 8 | 9 | New | 1 | 6 | 9 | 5 (new) |
| Ocean-1170 | Ocean | 13 | 8 | 9 | 3 | 1 | 6 | 9 | 3 (new) |
| Ocean-1175 | Ocean | 13 | 8 | 9 | New | 1 | 6 | 9 | 6 (new) |
| Ocean-1187 | Ocean | 13 | 8 | 9 | New | 1 | 6 | 9 | 6 (new) |
| Ocean-1206 | Ocean | 13 | 8 | 9 | 3 | 1 | 6 | 9 | 3 (new) |
| Coastal-1303 | Coastal | 23 | 5 | 57 | 3 | 5 | 4 | 3 | 7 (new) |
| Coastal-54 | Coastal | 6 | 6 | 4 | 3 | 3 | 4 | 7 | 8 (9) |
| Coastal-60 | Coastal | 6 | 6 | 4 | 3 | 3 | 4 | 7 | 8 (9) |
| Coastal-86 | Coastal | 6 | 6 | 4 | 3 | 3 | 4 | 7 | 8 (9) |
| Coastal-235 | Coastal | 6 | 5 | 1 | New | 1 | 12 | 1 | 9 (new) |
| Coastal-241 | Coastal | 6 | 5 | 1 | 1 | 1 | 12 | 1 | 10 (new) |
| Coastal-242 | Coastal | 6 | 5 | 1 | 1 | 1 | 12 | 1 | 10 (new) |
| Coastal-243 | Coastal | 6 | 5 | 1 | 1 | 1 | 12 | 1 | 10 (new) |
| Coastal-252 | Coastal | 6 | 5 | 5 | 3 | 2 | 4 | 2 | 11 (new) |
| Coastal-601 | Coastal | 6 | 5 | 1 | 1 | 1 | 12 | 1 | 10 (new) |
| Coastal-963 | Coastal | 28 | 22 | 5 | New | 1 | 14 | 19 | 12 (new) |
| Coastal-1276 | Coastal | 13 | 8 | 9 | New | 1 | 6 | New | 13 (new) |
| Arakawa River-206 | Freshwater | 6 | 5 | 5 | 3 | 1 | 6 | 3 | 14 (new) |
| Arakawa River-1504 | Freshwater | 22 | 5 | New | 3 | 4 | 4 | 7 | 15 (new) |
| Arakawa River-1508 | Freshwater | 36 | 27 | 28 | 3 | 4 | 13 | 7 | 16 (179) |
| Pond-1540 | Freshwater | 17 | 5 | 11 | 5 | 4 | 4 | 7 | 17 (new) |
| Pond-1549 | Freshwater | 17 | 5 | 11 | 5 | 4 | 4 | 7 | 17 (new) |
| Tama Lake-1559 | Freshwater | New | 5 | 26 | 3 | 4 | 4 | 1 | 18 (new) |
| Tama Lake-1563 | Freshwater | 16 | 5 | 36 | 3 | New | New | 1 | 19 (new) |
| Clinical-1529 | Clinical | 40 | 5 | 11 | 3 | 1 | New | New | 20 (new) |
| Clinical-1683 | Clinical | 38 | 11 | 3 | New | 1 | 2 | 4 | 21 (new) |
| Clinical-1732 | Clinical | 38 | 11 | 3 | New | 1 | 2 | New | 22 (new) |
| Clinical-1920 | Clinical | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 23 (new) |
| Clinical-1921 | Clinical | 3 | 11 | 3 | New | 1 | 2 | 4 | 22 (new) |
| Dolphin-245 | Animal | 28 | 5 | 36 | 3 | 3 | 13 | 7 | 24 (155) |
| JCM 5962T | Type strain | 6 | 5 | 1 | 11 | 3 | 6 | 7 | (New) |
The number assigned to each gene corresponds to the allelic type available in the MLST database. Alleles that did not match with the existing database were designated “new.” Strains that had seven known alleles were further searched for strain type. The numbers assigned to the strain types outside parentheses correspond to the strain types in this study only. Known strain types available in the database are in parentheses. Types for strains having seven known alleles that did not match with any known strain type in the database were also designated “new.” The total numbers of allelic types were as follows: for acsA, 13; for aroE, 6; for guaA, 13; for mutL, 11; for nuoD, 6; and for ppsA, 11. There were 24 strain types.
Groups of strains were assigned depending on the source of the strain.
Phylogenetic analysis.
Similarity and evolutionary distance were calculated with different software programs. Gene distance matrices were calculated from nucleotide sequences by the Jukes-Cantor method (20). Mann-Whitney's U test was employed for within-group and between-group mean distances, and significance levels were calculated (32). Dendrograms were generated by neighbor joining (NJ), maximum parsimony (MP), minimum evolution, the unweighted-pair group method with arithmetic means, and split decomposition (SD) with bootstrap analysis (1,000 replications) using MEGA (27) and PAUP* (version 4; D. L. Swofford, Sinauer Associates, Sunderland, MA). Analyses were performed on individual gene sequences as well as on the concatenated data set. Rooted and unrooted NJ trees of the concatenated seven-gene sequence data were generated with MEGA, version 3.1 (27), using the Kimura 2 parameter model (25) with gamma correction and 1,000 bootstrap replicates for all sequences. In NJ analyses, closely related pseudomonads were used as an outgroup. The data for outgroups were collected from the NCBI GenBank database. An MP tree was generated from the concatenated gene sequence data from which bootstrap percentages were retrieved for 1,000 replicates. The number of polymorphic nucleotide sites and the ratio of the number of nonsynonymous substitutions to the number of synonymous substitutions (the dN/dS ratio) were calculated with the MEGA and START programs (18, 19, 27, 35).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study were submitted to GenBank. The accession numbers for the 16S rRNA gene, acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE are EU710848 to EU710881, EU561186 to EU561219, EU56084 to EU561117, EU561118 to EU561151, EU561152 to EU561185, EU590764 to EU590797, EU590730 to EU590763, and EU590696 to EU590729, respectively.
RESULTS
Identification of P. aeruginosa isolates.
The marine strains were confirmed as P. aeruginosa following observation of their physical properties (22) by BD Phoenix automated microbiology systems (22) and 16S rRNA sequence data. The 16S rRNA data revealed 99 to 100% similarity to the existing P. aeruginosa database in GenBank. When a dendrogram was prepared from the 16S rRNA sequence data and other P. aeruginosa strains were used as the outgroup, all P. aeruginosa strains formed a single cluster. The distance between P. aeruginosa and other Pseudomonas spp. was supported by a 99% bootstrap value (Fig. 2). All P. aeruginosa strains belonged to a single cluster, in which P. aeruginosa PAO1 and type strain P. aeruginosa JCM 5962T were included. Individual gene sequence data for each of the seven housekeeping genes also showed 98 to 100% similarity to the sequences in the P. aeruginosa database in GenBank when BLAST searches were carried out. For further confirmation of the P. aeruginosa strains, a standard DNA-DNA hybridization method was employed. The levels of DNA-DNA relatedness at the optimal temperature (51.6°C for P. aeruginosa, which was calculated based on G+C molecule contents) within each species were 89.8 to 93.2% (Table 2). All DNA-DNA relatedness values were above the threshold value that has been suggested as delineating the same bacterial species (approximately 70%) under optimal hybridization conditions (12). On the other hand, the values for DNA-DNA relatedness among the selected strains and Pseudomonas fluorescens were less than 70% (Table 2). This confirmed the view that strains Ocean-1187 and Coastal-54 represent the species P. aeruginosa.
FIG. 2.
Rooted tree of 16 rRNA gene sequence data of P. aeruginosa along with data on other Pseudomonas spp. obtained using the NJ method with Kimura 2 correction for distance calculations. Bootstrap percentages retrieved in 1,000 replications are shown at the nodes. Clusters and bootstrap values that were 50% or greater are indicated. The scale bar (0.005) indicates the number of nucleotide substitutions per site. Data for P. aeruginosa JCM 5962T were collected from the DDBJ database, and data for all outgroup strains were collected from the NCBI GenBank database.
TABLE 2.
DNA-DNA relatedness between strains Ocean-1187, Coastal-54, P. aeruginosa JCM 5962T, and P. fluorescens 5963T
| Strain | DNA-DNA relatedness (%) for indicated strain
|
|||
|---|---|---|---|---|
| Ocean-1187 | Coastal-54 | P. aeruginosa JCM 5962T | P. fluorescens JCM 5963T | |
| Ocean-1187 | 100 | 89.8 | 89.8 | 10.9 |
| Coastal-54 | 100 | 93.2 | 10.1 | |
| P. aeruginosa JCM 5962T | 100 | 12.7 | ||
| P. fluorescens JCM 5963T | 100 | |||
Nucleotide fragments and allelic profiles.
The fragment lengths for MLST data (obtained from MLST primers) and whole-gene sequence data (obtained from PCR amplification primers) for the seven housekeeping genes and their variable sites are given in Table 3. The allelic sequences obtained were searched for in the P. aeruginosa MLST database. For aroE, all profiles were found in the existing database, whereas one (guaA and nuoD) to eight (mutL) new profiles were found for other genes (Table 4). Most of the oceanic and clinical isolates produced new allelic types in mutL gene sequences. We stipulated that if a strain possessed at least one new profile among the seven genes examined, it produced a new sequence type; among 34 strains, 16 were deemed to be new. Furthermore, even if all seven alleles were known, the combination of each profile sometimes represented new sequence types; 10 strains belonged to this category. The remaining eight strains matched with existing types.
Of the 34 P. aeruginosa strains isolated from different sources, we found 24 distinct strain types. Among them, three (types 9, 155, and 179) existed in the MLST database (Table 4), while the other 21 were new. Three coastal strains (Coastal-54, Coastal-60, and Coastal-86) belonged to type 9. Three strains with this type had been deposited, one from an unknown source from France (strain 5757) and two from sputum samples from Canada (strains R1 and T1). The strain Dolphin-245, isolated from a dolphin in Otsuchi Bay, Iwate Prefecture, Japan, matched with type 155, which comprises two clinical strains from Canada (strains B5 and S3). The Arakawa River-1508 strain, from Arakawa River, matched with type 179, which includes five clinical strains from Canada (strains B4, E4SIB1, C1, W4, and C2). Other than for these three types, the strains did not match with any types available in the 409-strain database. Among ocean (9 strains) and coastal (12 strains) isolates, six and six new strain types, respectively, were found.
Genetic distances between the P. aeruginosa groups.
The average mean genetic distances between and within the designed P. aeruginosa groups were calculated by MEGA. Mann-Whitney's U test was carried out to see whether the differences within and between the group average means were statistically significant (32). In concatenated gene sequences (5,700 bp), the distances within groups were lower than the distances between groups (P < 0.05). The average mean distance between ocean-derived and other P. aeruginosa groups varied from 0.0075 to 0.0092, which was higher than the within-group average mean distance (P < 0.05) (Table 5).
TABLE 5.
Average genetic distances within and between P. aeruginosa groupsa
| Group | Distance within group | Distance between groups
|
||||||
|---|---|---|---|---|---|---|---|---|
| Ocean | Coastal | Freshwater | Clinical | Animal | PAO1 | Outgroup | ||
| Ocean | 0.0046 | 0 | 0.0077 | 0.0086 | 0.0075 | 0.0092 | 0.0078 | 0.2439 |
| Coastal | 0.0047 | 0 | 0.0052 | 0.0075 | 0.0053 | 0.0047 | 0.2434 | |
| Freshwater | 0.0047 | 0 | 0.0080 | 0.0046 | 0.0035 | 0.2438 | ||
| Clinical | 0.0042 | 0 | 0.0087 | 0.0076 | 0.2426 | |||
| Animal | NCb | 0 | 0.0031 | 0.2442 | ||||
| PAO1 | NCb | 0 | 0.2431 | |||||
| Outgroup | 0.1800 | 0 | ||||||
Distances were calculated for the concatenated PCR sequences (5,700 bp) by using MEGA. Rates among sites were uniform. The pattern among lineages was homogeneous. The total numbers of strains are as follows: open ocean, 9; coastal, 12; freshwater, 7; clinical, 5; animal, 1; PAO1, 1; and outgroup, 3.
NC, could not be calculated.
Phylogenetic analysis and comparison with the geographical locations of the origins of the strains.
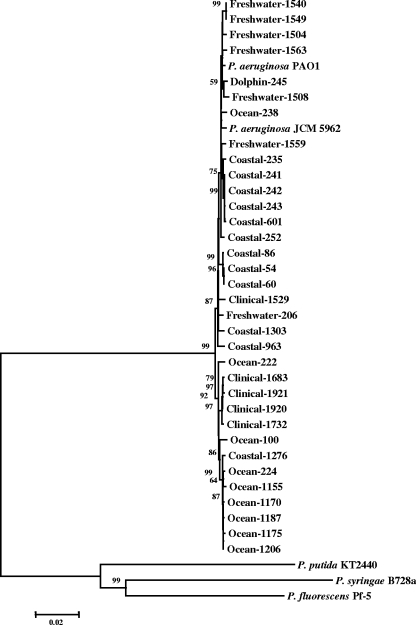
In order to assess the phylogenetic relationships between all the strains tested, PCR fragments for each gene, as well as the concatenated sequences of all PCR fragments, were analyzed to construct trees based on NJ, MP, minimum evolution, the unweighted-pair group method with arithmetic means, or SD. Thirty-nine strains were used, including P. aeruginosa PAO1, P. aeruginosa JCM 5962T, and three outgroup species strains (Pseudomonas putida KT2440, P. fluorescens Pf-5, and Pseudomonas syringae B728a) obtained from the NCBI database. Because the most conservative segments of the housekeeping genes were used in this study, the distance to the outgroups was too great to display in the tree. Only the unrooted trees are shown (Fig. 1 and 4). Figure 3 shows the rooted NJ tree of the concatenated data set of seven housekeeping genes with the outgroup.
FIG. 4.
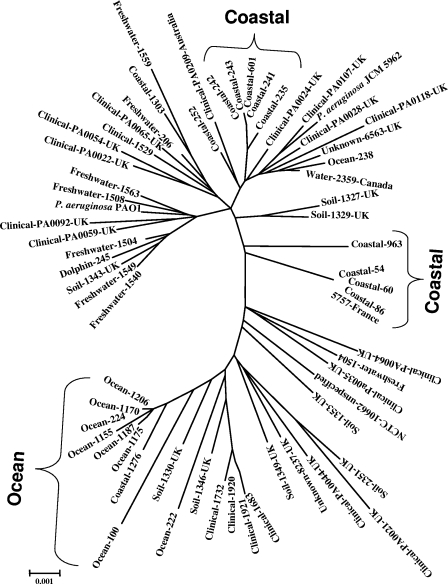
Unrooted and NJ radiation tree based on concatenated MLST sequences of seven gene loci. acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE gene sequences (2,873 bp) from 63 taxa (34 from the present study, 28 from the MLST database, and 1 P. aeruginosa PAO1 strain from the database) were concatenated and constructed by the MEGA 3.1 program. Sequence similarity was corrected for by using the Kimura 2 parameter. All nodes were supported by 1,000 bootstrap replications. The largest cluster was marked as open ocean, as all strains were from the ocean, and two other clusters were marked as coastal. The scale bar (0.001) indicates the number of nucleotide substitutions per site.
FIG. 3.
Rooted tree of the concatenated nucleotide sequences of seven housekeeping genes (acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE) of P. aeruginosa obtained using the NJ method with Kimura 2 correction for distance calculations. The total length of the concatenated group of nucleotide sequences was 5,700 bp. Bootstrap percentages retrieved in 1,000 replications are shown at the nodes. Clusters and bootstrap values that were 50% or greater are indicated. The scale bar (0.02) indicates the number of nucleotide substitutions per site.
In this study, we present only the data derived by the NJ and MP methods, as other methods showed similar patterns for the trees, so they were eliminated. Figure 1 shows two unrooted trees constructed by NJ and MP methods from concatenated gene sequence data together with a map identifying the geographical locations of the isolates. All open-ocean strains showed a trend of forming separate clusters in the trees except two ocean P. aeruginosa strains. Except for strains Ocean-222 and Ocean-238, all ocean strains formed a single cluster, with one coastal strain (Coastal-1276) included. Although the freshwater strains did not show a distinct cluster, they were mostly separated from the other strains, with the exception of strains Freshwater-206 and -1559. Strains P. aeruginosa PAO1, Dolphin-245, and Coastal-963 were included in the freshwater group. Mixing of coastal strains with freshwater strains might indicate the influence of freshwater outfalls on coastal systems. Although the coastal strains are rather scattered in the tree, they were mostly close to each other and formed two separate clusters (one from the strains of Sagami Bay and one from those of Suruga Bay) that were always prominent. Four of the five clinical isolates also clustered together.
Phylogenetic analysis of individual loci.
The relationships between the 34 strains were further analyzed by constructing unrooted trees using the NJ method based on the nucleotide sequences of PCR fragments for each gene (data not shown). Ocean alleles were clustered with respect to acsA, aroE, guaA, ppsA, and trpE. This clustering was supported by bootstrap values higher than 50%, indicating their distinct positions in the trees.
Phylogenetic analysis of the MLST sequences.
To clarify the phylogenetic positions of marine isolates among those from various sources, concatenated allelic data on 28 strains randomly selected from the MLST database were obtained. In addition, the corresponding sequence of P. aeruginosa PAO1 was taken from the NCBI database, and the sequence of type strain P. aeruginosa JCM 5962T was added. Unrooted trees were constructed for all strains through different methods. Regardless of the methodology used for designing phylogenetic trees, all open-ocean strains except two (Ocean-238 and Ocean-222) invariably formed clusters distinct from those of other strains. Here, we present a radiation tree prepared by the NJ method, using MEGA as the representative method (Fig. 4). The clinical and soil strains taken from the database tended to scatter throughout the entire tree. The freshwater strains showed loose association with some clinical strains. Three separate clusters were observed for marine strains, two for coastal isolates and one for ocean strains. Each coastal cluster was composed of isolates from different geographical areas. The biggest and most distinct cluster was represented by the P. aeruginosa strains from the ocean.
DISCUSSION
Our previous studies revealed that the oceanic P. aeruginosa strains have advantages over other strains with regard to their physiological adaptability to high-NaCl and pH stresses in laboratory-based microcosms. Molecular typing studies (pulsed-field gel electrophoresis) also indicated a clonal structure for oceanic P. aeruginosa strains that was distinct from those of other strains (22). Based on these results, we hypothesized that there are new sequence types of ocean P. aeruginosa strains not found in other strains, which confirms that the ocean P. aeruginosa strains have diverged from other isolates and form a distinct cluster, as determined by MLST and phylogenetic analyses of seven housekeeping genes.
In the present study, we applied MLST analysis to 34 strains and found that most marine strains represent new types. In addition, analysis using concatenated gene sequences indicated that ocean and coastal strains form their own phylogenetic clusters (Fig. 1, 3, and 4). Although freshwater strains tend to mix with soil or clinical strains (Fig. 4), the phylogenetic positions of ocean, coastal, and freshwater strains roughly reflect their geographic distributions, with some exceptions (Fig. 1). To our knowledge, this is the first report on phylogenetic analysis of marine P. aeruginosa and the first finding of a possible connection of the phylogeny of this bacterium to geographical distribution in aquatic environments.
We used MLST in this study primarily because it enabled us to compare our data with those on other strains, especially clinical isolates and environmental isolates from various areas. In addition, phylogenetic analysis of each gene and concatenated gene fragments clarifies the intraspecies phylogenetic positions of marine strains. The MLST database so far includes >409 types of P. aeruginosa (4) (http://www.mlst.org/) but does not yet include any marine or coastal P. aeruginosa strain. As this study is the first application to marine isolates, we used the analytical methods with care. Genes were sequenced using established primers (5), and only assembled sequences from forward and reverse primers were used.
In addition to the type determination, the genetic relatedness among strains was calculated from the MLST allelic profiles and the whole-gene sequence data by the phylogenetic analyses shown in Table 4 and Fig. 1, 3, and 4, which indicates that the relatedness is primarily dependent on the origins. The ocean isolates formed one cluster (except Ocean-222 and Ocean-238), which includes only one coastal strain (Coastal-1276) and none from other environments. The presence of such clusters is also implied by the trees constructed for each gene (data not shown), a result further supported by MEGA analysis. The unrooted phylogenetic trees shown in Fig. 1 demonstrate a rough relationship between the geographic distributions and phylogenetic positions of the strains. The unrooted tree constructed from concatenated MLST data further clarified the presence of three clusters unique to ocean and coastal environments (Fig. 4). These results confirmed our hypothesis regarding the presence of distinct marine types. As the possibility of mixing of freshwater with the coastal environments could not be rejected, it is assumed that the influence of freshwater and terrestrial environments reflected on the distributions of the coastal strains between clinical and freshwater environments and the separation of freshwater strains toward the other parts of the trees. On the other hand, the unique ocean environment retained a distinct group of P. aeruginosa strains, which always formed a single cluster, separated from the other isolates, with the exception of one coastal strain (Coastal-1276). A possible explanation for this kind of distribution is difficult to infer. We sampled seawater across the Kuroshio Current, primarily because the influence of freshwater inflow is hardly expected in the south part of the current. In other words, those strains are not recent contaminants of terrestrial origin. However, this does not necessarily mean that coastal strains should never be found in the open ocean or that those strains should never survive for a considerable time in the open ocean. The ocean is composed of one huge water mass without physical barriers. Although the physical process is unknown, coastal strains may be transferred to the open ocean and survive for years. As P. aeruginosa was regarded as a potential pathogen of marine mammals causing bronchopneumonia and dermatitis (7), it may be possible for the pathogen to travel with an ocean mammal from the open ocean to the coastal region or vice versa. If there are certain carriers, such as dolphins, they may release strains that are apparently of coastal origins. The distributions of ocean strains 222 and 238 may also be clarified in a similar way.
The Kuroshio Current is also a variable phenomenon in the South Pacific Ocean regarding its flow path. It always changes its path. The samples were collected twice from inside and outside the Kuroshio Current, which indicated a clear distinction among the strains inside and outside the Kuroshio Current. At this stage, we cannot conclude that Kuroshio is the only factor that differentiated coastal and open-ocean P. aeruginosa strains. Further studies are needed to make such a comment.
Our idea on the biogeography of P. aeruginosa is that the majority of strains from a certain environment are composed of one or a few particular groups that are unique to the environment. The opposite idea is “random” distribution regardless of environment. We believe that the data shown in Fig. 1 support the former idea but never support the latter.
Recently, the biogeography of free-living bacteria has drawn considerable attention because of the accumulation of data from various environments. Theoretically, biogeographical distributions may be explained either by environmental variations or by historical events. We believe that our present results indicate that environmental variations may be more important than the historical aspects. We also believe that our work is of considerable impact on such scientific issues because this study is based on multiple gene sequences of many isolates, whereas most works on biogeography are solely dependent on genetic information without any culture.
MLST has been also applied to other bacterial species for similar purposes. Sarkar and Guttman (45) carried out a study of the evolution of core genes of P. syringae, demonstrating the presence of a distinct group of strains from the SD of their combined phylogenetic tree. Cladera et al. (4) compared seven marine Pseudomonas stutzeri strains isolated from Barcelona, Spain, with strains from other sources by MLST. This analysis indicated that among the 25 total strains investigated, there were 24 alleles, which included seven marine types. The NJ tree drawn by the data indicated that marine P. stutzeri strains were distributed throughout the tree, suggesting that the divergence patterns of P. aeruginosa and P. stutzeri may be different. However, it should also be noted that the present MLST method was originally developed primarily to type clinical strains. Selecting a greater number of variable sites or genes, which increases the sensitivity toward aquatic strains, may offer better resolution and reliability as a typing method. For phylogenetic work, the analysis of more gene sequences will definitely be required.
An understanding of the community structure and genetic relatedness among strains of P. aeruginosa present in nature (sea, river, soil, plants, and animals) is essential for gaining a greater insight into the ecology and distribution of this bacterium. There have been debates on the extent to which they share common niches in the environment and the extent to which environmental and clinical strains are genetically similar. Recently, Pirnay et al. (42) aimed to determine the biodiversity among the P. aeruginosa population of a limited natural environment, the Woluwe River in Belgium, and compared it with the global biodiversity. They found that the population structure in that particular area, a small river, seemed to be comparable with that on the global scale. P. aeruginosa resides primarily in an aquatic environment, yet the organism is indeed a typical opportunistic bacterium and thrives well, eventually causing disease, independent of geographical location or host-specific niche. They concluded that P. aeruginosa is an autochthonous member of the bacterial community of the Woluwe River, which strongly suggests a worldwide spread of a homogeneously diverse population of this bacterium. In the present study, the strong support of the NJ tree by the MP tree regarding formation of a single cluster of the ocean strains and the correlation between phylogenetic position and geographical location give insight into the adaptation of these strains to a particular environment.
Aoi et al. (3) proposed that P. aeruginosa strains isolated from the Ushubetsu River in Japan originated primarily from environments of human activity. Many other scientists have observed that environmental P. aeruginosa isolates are genotypically, chemotaxonomically, and functionally indistinguishable from clinical isolates (1, 13, 36, 43, 44). Together, these works indicate that the populations present in different environments are highly intermingled or that high rates of genetic recombination lead to relatively homogeneous genetic structures among the strains.
Our results, however, indicate a divergence of the marine P. aeruginosa strains from the other strains. The open ocean is generally characterized by low nutrient levels, high salinity, and alkaline pH (∼8). Therefore, it seems reasonable to assume that these conditions select certain types of strains that differ from those in terrestrial or freshwater environments. In addition, it is difficult to imagine how terrestrial strains can be transported to the open ocean several hundred kilometers away from the shore, cross the Kuroshio Current (22), and mix with autochthonous populations. However, our results do not reject the possibility that further analysis of more genes of more strains or an entire genome may show high genetic similarities between marine strains and other strains. It is suggested that at least the core genes of P. aeruginosa are extremely conserved. Further work with a greater number of environmental strains from diversified geographical locations and with various genes is required to answer this question. Currently, we are sequencing the entire genome of one of the marine strains. The data will be published elsewhere soon.
In conclusion, the presence of marine P. aeruginosa strains that have unique genotypes and distinct clusters was confirmed by MLST and phylogenetic analysis for the first time. In addition, the geographic distributions of these marine strains have a roughly close relation to intraspecies phylogenetic position, which needs to be clarified by further analyses with more strains from various geographical locations and origins. These findings will have a considerable impact on the study of the origin, ecology, physiology, and genetic characteristics of this well-known bacterium.
Acknowledgments
We thank the Japanese Society for the Promotion of Sciences for sponsoring Nurul Huda Khan with a postdoctoral fellowship. The study was supported by Grants-in-Aid for Creative Basic Research, no. 12NP0201 (DOBIS) and no. 14208063, from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
We are grateful to Yoshikazu Ishii, Department of Microbiology and Infectious Diseases, Toho University School of Medicine, for providing clinical P. aeruginosa strains. We remain indebted to Rita R. Colwell, University of Maryland, for her valuable comments and inspiration regarding this study. This publication made use of the P. aeruginosa MLST website (http://pubmlst.org/paeruginosa/), developed by Keith Jolley and located at the University of Oxford (18). The development of this site has been funded by the Wellcome Trust.
Footnotes
Published ahead of print on 29 August 2008.
REFERENCES
- 1.Alonso, A., F. Rojo, and J. L. Martínez. 1999. Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environ. Microbiol. 1:421-430. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., L. M. Thomas, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST & PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoi, Y., H. Nakata, and H. Kida. 2000. Isolation of Pseudomonas aeruginosa from Ushubetsu River water in Hokkaido, Japan. Jpn. J. Vet. Res. 48:29-34. [PubMed] [Google Scholar]
- 4.Cladera, A. M., A. Bennasar, M. Barcelo, L. Lalucat, and E. Garcia-Valdes. 2004. Comparative genetic diversity of Pseudomonas stutzeri genomovars, clonal structure, and phylogeny of the species. J. Bacteriol. 186:5239-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curran, B., D. Jonas, H. Grundmann, T. Pitt, and C. G. Dowson. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 42:5644-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denamur, E., B. Picard, G. Decoux, J. B. Denis, and J. Elion. 1993. The absence of correlation between allozyme and rrn RFLP analysis indicates a high gene flow rate within human clinical Pseudomonas aeruginosa isolates. FEMS Microbiol. Lett. 110:275-280. [DOI] [PubMed] [Google Scholar]
- 7.Diamond, S. S., D. E. Ewing, and G. A. Cadwell. 1979. Fatal bronchopneumonia and dermatitis caused by Pseudomonas aeruginosa in an Atlantic bottle nosed dolphin. J. Am. Vet. Med. Assoc. 175:984-987. [PubMed] [Google Scholar]
- 8.Editorial Secretary for the Judicial Commission of the International Committee on Nomenclature of Bacteria. 1970. Opinion 36: designation of strain ATCC 10145 as the neotype strain of Pseudomonas aeruginosa (Schroeter) Migula. Int. J. Syst. Bacteriol. 20:15-16. [Google Scholar]
- 9.Editorial Secretary for the Judicial Commission of the International Committee on Nomenclature of Bacteria. 1970. Opinion 37: designation of strain ATCC 13525 as the neotype strain of Pseudomonas fluorescens Migula. Int. J. Syst. Bacteriol. 30:17-18. [Google Scholar]
- 10.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 11.Ezaki, T., Y. Hashimoto, and E. Yabuuchi. 1989. Fluorometric deoxyrinucleic acid-deoxyribonucleic acid hybridization in micro-dilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39:224-229. [Google Scholar]
- 12.Finnan, S., J. P. Morrissey, F. O'Gara, and E. F. Boyd. 2004. Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment. J. Clin. Microbiol. 42:5783-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foght, J. M., D. W. S. Westlake, W. M. Johnson, and H. F. Ridgway. 1996. Environmental gasoline-utilizing isolates of Pseudomonas aeruginosa are taxonomically indistinguishable by chemotaxonomic and molecular techniques. Microbiology 142:2333-2340. [DOI] [PubMed] [Google Scholar]
- 14.Grimont, P. A. D. 1999. Taxonomy and classification of bacteria, p. 249-259 In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, DC.
- 15.Hoadley, A. W. 1977. Potential health hazards associated with Pseudomonas aeruginosa in water, p. 80-114. In A. W. Hoadley and B. J. Dutka (ed.), Bacterial indicators/health hazards associated with water. American Society for Testing and Materials, Philadelphia, PA.
- 16.Hoadley, A. W. 1968. On the significance of Pseudomonas aeruginosa in surface waters. J. N. Engl. Water Works Assoc. 82:99-111. [Google Scholar]
- 17.Howell, C. R., and R. D. Stipanovic. 1979. Control of Rhizoctonia solani on cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology 69:480-482. [Google Scholar]
- 18.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinform. 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jolley, K. A., F. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 20.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, Inc., New York, NY.
- 21.Khan, A. A., and C. E. Cerniglia. 1994. Detection of Pseudomonas aeruginosa from clinical and environmental samples by amplification of the exotoxin A gene using PCR. Appl. Environ. Microbiol. 60:3739-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan, N. H., Y. Ishii, N. Kimata-Kino, H. Esaki, T. Nishino, M. Nishimura, and K. Kogure. 2007. Isolation of Pseudomonas aeruginosa from open ocean and comparison with freshwater, clinical, and animal isolates. Microb. Ecol. 53:173-186. [DOI] [PubMed] [Google Scholar]
- 23.Kiewitz, C., and B. Tummler. 2000. Sequence diversity of Pseudomonas aeruginosa: impact on population structure and genome evolution. J. Bacteriol. 182:3125-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimata, N., T. Nishino, S. Suzuki, and K. Kogure. 2004. Pseudomonas aeruginosa isolated from marine environments in Tokyo Bay. Microb. Ecol. 47:41-47. [DOI] [PubMed] [Google Scholar]
- 25.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 26.Kodaka, H., M. Iwata, S. Yumoto, and F. Kashitani. 2003. Evaluation of a new agar medium containing cetrimide, kanamycin and nalidixic acid for isolation and enhancement of pigment production of Pseudomonas aeruginosa in clinical samples. J. Basic Microbiol. 43:407-413. [DOI] [PubMed] [Google Scholar]
- 27.Kumar, S., K. Tamura, and M. Nei. 1994. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci. 10:189-191. [DOI] [PubMed] [Google Scholar]
- 28.Lane, D. J. 1991. 16S/23S rRNA sequencing in nucleic acid techniques, p. 115-148. In E. Stackebrandt and M. Goodfellow (ed.), Bacterial systematics. John Wiley and Sons, New York, NY.
- 29.Lomholt, J. A., K. Poulsen, and M. Kilian. 2001. Epidemic population structure of Pseudomonas aeruginosa: evidence for a clone that is pathogenic to the eye and that has a distinct combination of virulence factors. Infect. Immun. 69:6284-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loper, J. E., and S. E. Lindow. 1987. Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pv. syringae on bean leaf surfaces. Phytopathology 77:1449-1454. [Google Scholar]
- 31.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mann, H. B., and D. R. Whitney. 1947. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 18:50-60. [Google Scholar]
- 33.Mates, A. 1992. The significance of testing for Pseudomonas aeruginosa in recreational seawater beaches. Microbios 71:89-93. [PubMed] [Google Scholar]
- 34.Nakazawa, T. 2002. Travels of a Pseudomonas, from Japan around the world. Environ. Microbiol. 4:782-786. [DOI] [PubMed] [Google Scholar]
- 35.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 36.Nicas, T. I., and B. H. Iglewski. 1986. Production of elastase and other exoproducts by environmental isolates of Pseudomonas aeruginosa. J. Clin. Microbiol. 23:967-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panagea, S., C. Winstanley, M. J. Walshaw, M. J. Ledson, and C. A. Hart. 2005. Environmental contamination with an epidemic strain of Pseudomonas aeruginosa in a Liverpool cystic fibrosis centre, and study of its survival on dry surfaces. J. Hosp. Infect. 59:102-107. [DOI] [PubMed] [Google Scholar]
- 38.Papapetropourou, M., and G. Rodopoulou. 1994. Occurrence of enteric and non-enteric indicators in coastal waters of southern Greece. Bull. Mar. Sci. 54:63-70. [Google Scholar]
- 39.Pellett, S., D. V. Bigley, and D. J. Grimes. 1983. Distribution of Pseudomonas aeruginosa in a riverine ecosystem. Appl. Environ. Microbiol. 45:328-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Picard, B., E. Denamur, A. Barakat, J. Elion, and P. Goullet. 1994. Genetic heterogeneity of Pseudomonas aeruginosa clinical isolates revealed by esterase electrophoretic polymorphism and restriction fragment length polymorphism of the ribosomal RNA gene region. J. Med. Microbiol. 40:313-322. [DOI] [PubMed] [Google Scholar]
- 41.Pirnay, J. P., D. De Vos, C. Cochez, F. Bilocq, A. Vanderkelen, M. Zizi, B. Ghysels, and P. Cornelis. 2002. Pseudomonas aeruginosa displays an epidemic population structure. Environ. Microbiol. 4:898-911. [DOI] [PubMed] [Google Scholar]
- 42.Pirnay, J. P., S. Matthijs, H. Colak, P. Chablain, F. Bilocq, V. Eldere, D. D. Vos, M. Zizi, L Triest, and P. Cornelis. 2005. Global Pseudomonas aeruginosa biodiversity as reflected in a Belgian river. Environ. Microbiol. 7:969-980. [DOI] [PubMed] [Google Scholar]
- 43.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 44.Römling, U., J. Wingender, H. Müller, and B. Tümmler. 1994. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl. Environ. Microbiol. 60:1734-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarkar, S. F., and D. S. Guttman. 2004. Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 70:1999-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt, K. D., B. Tummler, and U. Romling. 1996. Comparative genome mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J. Bacteriol. 178:85-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spencer, D. H., A. Kas, E. E. Smith, C. K. Raymond, E. H. Sims, M. Hastings, J. L. Burns, R. Kaul, and M. V. Olson. 2003. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J. Bacteriol. 185:1316-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 49.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. F. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 50.Szoboszlay, S., B. Atzel, and B. Kriszt. 2003. Comparative biodegradation examination of Pseudomonas aeruginosa (ATCC 27853) and other oil degraders on hydrocarbon contaminated soil. Commun. Agric. Appl. Biol. Sci. 68:207-210. [PubMed] [Google Scholar]
- 51.Thompson, J., D. G. Huggins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Velammal, A., B. Aiyamperumal, V. K. Venugopalan, and S. Ajmalkhan. 1994. Distribution of Pseudomonas aeruginosa in Pondicherry coastal environments. Ind. J. Mar. Sci. 23:239-241. [Google Scholar]
- 53.Vernez, I., P. Hauser, M. V. Bernasconi, and D. S. Blanc. 2005. Population genetic analysis of Pseudomonas aeruginosa using multilocus sequence typing. FEMS Immunol. Med. Microbiol. 43:29-35. [DOI] [PubMed] [Google Scholar]
- 54.Walker, T. S., H. P. Bais, E. Deziel, H. P. Schweizer, L. G. Rahme, R. Fall, and J. M. Vivanco. 2004. Pseudomonas aeruginosa-plant root interactions. Pathogenicity, biofilm formation, and root exudation. Plant Physiol. 34:320-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshpe-Purer, Y., and S. Golderman. 1987. Occurrence of Staphylococcus aureus and Pseudomonas aeruginosa in Israeli coastal water. Appl. Environ. Microbiol. 53:1138-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Young, A. L., A. T. Leung, L. L. Cheng, R. W. Law, A. K. Wong, and D. S. Lam. 2004. Orthokeratology lens-related corneal ulcers in children: a case series. Ophthalmology 111:590-595. [DOI] [PubMed] [Google Scholar]