Abstract
New rRNA-targeting oligonucleotide probes permitted the fluorescence in situ hybridization (FISH) identification of freshwater fungi in an Austrian second-order alpine stream. Based on computer-assisted comparative sequence analysis, nine taxon-specific probes were designed and evaluated by whole-fungus hybridizations. Oligonucleotide probe MY1574, specific for a wide range of Eumycota, and the genus (Tetracladium)-specific probe TCLAD1395, as well as the species-specific probes ALacumi1698 (Alatospora acuminata), TRIang322 (Tricladium angulatum), and Alongi340 (Anguillospora longissima), are targeted against 18S rRNA, whereas probes TmarchB10, TmarchC1_1, TmarchC1_2, and AlongiB16 are targeted against the 28S rRNA of Tetracladium marchalianum and Anguillospora longissima, respectively. After 2 weeks and 3 months of exposure of polyethylene slides in the stream, attached germinating conidia and growing hyphae of freshwater fungi were accessible for FISH. Growing hyphae and germinating conidia on leaves and in membrane cages were also visualized by the new FISH probes.
Freshwater fungi are known to be intensively involved in the degradation and conversion of biopolymers in streams (5). Organisms from the heterogeneous phylogenetic group of freshwater fungi perform the initial breakdown of allochthonous leaf material in lotic and lentic systems (20). The enzymatic breakdown and the fungal biomass itself condition the leaves as a food source for invertebrate shredders, which further fragment the litter into fine organic particles (2). Traditionally, aquatic fungal communities are characterized by concentrating conidia from stream and river water or conidia developed from substrates on incubated filters (4, 41). These methods cover sporulating fungi but disregard nonreproducing mycelia. For the assessment of the phylogenetic diversity and the biomass of freshwater fungi, a range of chemical, serological, and molecular techniques is available. Chemical stains often show limited fungal specificity (11, 32, 37). The measurement of the fungal membrane component ergosterol (33) is used to estimate the biomass but gives no information about the species composition. PCR-based methods, including restriction fragment length polymorphism analysis (16), random amplified polymorphic DNA analysis (13, 35), and denaturing gradient gel electrophoresis (e.g., see reference 34), are tools for determining genetic variation in fungal populations. Immunostaining, employing monoclonal antibodies and enzyme-linked immunosorbent assays, provides a means of identifying aquatic hyphomycete species in situ (9). However, the spatial distribution of growing mycelia on or within colonized substrata is still poorly understood.
Combining both specific in situ detection and information about the physiological status of the targeted cells, fluorescence in situ hybridization (FISH) is an established method in bacterial ecology (1, 30). The fluorescence signal intensities of the rRNA-targeting FISH probes are correlated with the ribosome content of the target cell, which indicates the viability of the target organism before fixation (14). A first attempt to use the FISH method for fungi was the detection of Aureobasidium pullulans on leaves (27). More recently, Baker et al. (3) showed Eurotiomycetes to be more abundant than Dothideomycetes in an extremely acidic mine drainage system by using 18S rRNA-targeting FISH probes. Baschien et al. (6) and McArthur et al. (31) applied FISH to freshwater fungi in pure cultures and on leaves. The often discussed limitations of FISH, such as inherent autofluorescence conferred by fungal structures and substrates and the limited permeability of the fungal cell wall, were shown previously to be moderated by several methodological adaptations in the performance of the molecular tool (6).
The objective of this study was to detect actively growing freshwater fungi with new taxon-specific rRNA-targeting FISH probes. Aquatic hyphomycetes were isolated from various substrate samples obtained from the Austrian alpine stream Oberer Seebach. Pure cultures of aquatic hyphomycetes were obtained from the Czech Collection of Microorganisms (CCM). FISH with newly developed fungal probes was applied to leaves and polyethylene (PE) slides after exposure in the stream. Furthermore, the accessibility of germinating fungal conidia for FISH under natural conditions in the stream was evaluated. For this purpose, conidia were exposed in membrane-coated cages in the stream and germinating conidia were subsequently visualized by FISH with specific probes.
MATERIALS AND METHODS
Study site.
Sampling and exposure experiments were carried out in the alpine second-order limestone gravel stream Oberer Seebach at the Biological Station Lunz, Lunz, Austria, within the study area RITRODAT (10). The riparian vegetation is dominated by Fraxinus excelsior, Acer pseudoplatanus, Salix caprea, Fagus sylvatica, and Picea abies.
Sampling, isolation, and exposure.
Sampling and exposure experiments were conducted during two 3-week campaigns in autumn 2000 (26 September to 15 October) and summer 2001 (2 to 23 August). Fungal isolations from foam were performed directly in the field by streaking a loopful of foam onto object slides coated with 2% malt extract agar (MA) and incubating the slides at 15°C for 12 to 24 h. Single germinating conidia were transferred onto fresh MA. Leaves and twigs collected in the Oberer Seebach were transferred into sterile screw-cap jars or plastic bags and returned to the laboratory under cooling conditions. The leaves or twigs were washed under tap water, rinsed with distilled water, and incubated in petri dishes with distilled water or submerged and aerated in sterile 50-ml centrifuge tubes (Labcon, San Rafael, CA). The incubated samples were observed daily. Developing conidia were picked using a very fine glass needle and transferred onto MA.
High-density-PE slides were exposed short-term for 2 weeks (28 September to 11 October) and long-term for a 3-month period (28 September to 20 December) in autumn 2000, for a period during the winter of 2000 to 2001 (11 October 2000 to 15 February 2001), and for 2 weeks in summer 2001 (2 to 16 August). PE slides were placed at the RITRODAT sampling sites 1R1, with a slow current, and 17G2.2, with a faster current. Following the harvesting of four replicates, the PE slides were fixed immediately for examination by FISH.
Freshly fallen leaves of Acer pseudoplatanus, Acer campestre, Fagus sylvatica, and Salix caprea were collected from the Oberer Seebach in autumn 2000. All leaves were autoclaved and incubated in petri dishes. The leaves were examined for fungal growth after 2 and 7 days by using a dissecting microscope to exclude contamination prior to exposure. The exposure of 0.48 and 0.23 g of sterile leaf material was performed in sterile polyamide mesh bags (0.25-mm pore size) at the RITRODAT sampling sites 2C1.1 (slow current) and 16F3.3 (fast current). The leaf packs were harvested after 2 weeks and either fixed immediately or incubated in moist chambers for fungal isolations.
Culture conditions.
All fungal strains were grown on MA and oatmeal agar and in malt extract broth (17) at 16 to 18°C without agitation under a cycle of 12 h of daylight exposure or incubated at 20°C on a shaker. Aquabacterium commune was grown on modified R2A medium (26).
Germination experiments.
To induce sporulation, pieces of pure fungal colonies were cut out and submerged in a tube with aerated distilled water or tap water. Conidia were harvested with a sterile Pasteur pipette 2 to 5 days after air bubbles had formed foam at the tube wall. The conidia were used immediately as inocula for subsequent germination experiments. Samples of 10 to 1,000 conidia ml−1 were put into cages between two cellulose acetate membranes (5-μm pore dimension; Millipore GmbH, Eschborn, Germany). The cage membranes were clamped between two hose connections which were screwed together and bound to a stiff metal wire for fixation at the sampling site. The cages were exposed in 5 cm of water at slow- and fast-current sites for 2 days (29 September to 1 October 2000) and 5 days (1 to 6 October 2000). After being harvested, germinating conidia on the cage membranes were counted directly.
Sequencing of aquatic hyphomycete strains.
Pure cultures were harvested after 8 weeks. Genomic DNA was isolated by using the FastDNA SPIN kit for soil in conjunction with the FastPrep FP120 instrument (Qbiogene, Heidelberg, Germany). 18S rRNA genes were amplified with the primer pair NS1/NS8 (44). The first two-thirds of the 28S rRNA gene (rDNA) were amplified with the primer pair 5.8SR/LR5 (42). PCR mixtures contained PCR buffer (10 mM Tris-HCl, 50 mM KCl, pH 8), 200 μM (each) deoxynucleotides, 1.5 mM magnesium chloride, 20 pM (each) primers, 50 to 200 ng of genomic DNA, and 2.5 U of Taq polymerase (Boehringer, Mannheim, Germany). The PCR was performed in a personal cycler (Primus 25 HE; MWG-Biotech, Ebersberg, Germany) with an initial denaturation step for 2 min at 95°C, followed by 35 cycles of denaturation for 30 s at 94°C, primer annealing for 1 min 30 s at 55°C (56°C for primers 5.8SR/LR5), and extension for 1 min 45 s at 72°C; final extension was for 10 min at 72°C. The PCR products were purified with the QIAquick purification kit (Qiagen, Hilden, Germany). Automatic sequencing was done with the BigDye Terminator ready reaction kit (PerkinElmer Applied Biosystems, Weiterstadt, Germany). Primers used for the sequencing of the 18S rDNA and 28S rDNA were NS1 and NS8 (44); nu-SSU-402-5′, nu-SSU-819-5′, nu-SSU-852-3′, and nu-SSU-305-3′ (18); and LR1, LROR, LR21, LR2R, LR3, and LR5 (42).
Sequences were generated with an ABI 373 sequencer (PerkinElmer Applied Biosystems, Foster City, CA) and analyzed with the sequence analysis software version 3.3 at Services in Microbiology (Berlin, Germany) by Martin Meixner. The resulting sequences were assembled, corrected, and exported as consensus text with Auto Assembler version 2.1 (PerkinElmer).
Modification of ARB software settings.
For sequence alignment, probe design, and probe checking, the ARB software package (28) (free to download at http://www.arb-home.de/) was used. As the original sequence database contains mainly bacterial 16S and 23S rRNA gene sequences and, to a much lesser extent, fungal gene sequence data, the database was extended with fungal rDNA sequences obtained from GenBank (http://www.ncbi.nlm.nih.gov/). The alignment settings were adapted to the work with fungi as follows. The ARB Fast Aligner tool incorporates secondary-structure information on the rRNA molecule by evaluating base pairing at secondary-structure positions. In ARB, secondary-structure information is controlled by the sequence-administrated information (SAI; SAI: HELIX and SAI: HELIX_NR) laid down for the reference organism within the ARB EDITOR (version 4). In order to adapt the alignment features of ARB to the work with fungal sequences, the bacterial reference organism Escherichia coli was exchanged for baker's yeast Saccharomyces cerevisiae. The 18S rDNA sequence of Saccharomyces cerevisiae (GenBank accession number JO1353) was copied to SAI and saved as that of a new reference organism. Helix information was manually adjusted by following the secondary-structure model for the nuclear small-subunit (SSU) rRNA of Saccharomyces cerevisiae (http://www.psb.ugent.be/rRNA/) (45). The same procedure was performed for alignments of 28S rDNA sequences by employing the data from and the secondary-structure model of the large-subunit (LSU) rRNA of Saccharomyces cerevisiae (accession number JO1355) (8).
Fixation.
Following harvesting, cultures and conidia were fixed with freshly prepared 3.7% paraformaldehyde solution in phosphate-buffered saline (PBS) for 1 to 4 h at 4°C in the dark. The paraformaldehyde-fixed thalli were washed twice with PBS and stored at −20°C in a storage buffer consisting of a 1:1 (vol/vol) mixture of PBS and 96% ethanol. Immediately after recovery, PE slides, leaves, and cage membranes were submerged in 3.7% paraformaldehyde solution and incubated for 1 h at 4°C. The biofilms were washed twice with PBS, air dried, and stored at room temperature.
Probe design and evaluation.
All sequences were aligned with various other sequences by using Fast Aligner version 1.03 of the ARB software package, involving the secondary structures of helices, loops, and bulges. All alignments were examined and manually optimized according to primary- and secondary-structure similarities.
Potential signatures for fungal in situ probes at different phylogenetic levels were evaluated using the Probe_Design tool and Probe_Match tool of the ARB software package. The specificity check was supplemented by the analysis of additional ribosomal gene sequences through searches in GenBank. To complement the computer-aided probe design, the specificity of the newly developed probes was verified under in situ conditions by the reproducible and discriminative staining of target and nontarget species. The probe EUB338 (Bacteria probe) (1) served as a negative control for nonspecific binding. The universal probe EUK516 (Eucarya probe) (1) was used as a positive control in fungal hybridizations. In tests for the nonspecific binding of the probe MY1574 with nontarget organisms, the probes EUK516, EUB338, and ARCH915 (Archaea probe) (40) were used as positive controls. For the construction of specific oligonucleotide probes for Tetracladium species, Alatospora acuminata, Anguillospora longissima, and Tricladium angulatum, the database was scanned to evaluate probe sequences; other fungi should be distinguished by at least one mismatch in the target region. Additionally, the target region of the probe should be located in a region of the 18S or the 28S rRNA molecule suitable for in situ hybridizations. In situ discrimination testing was performed with fungi displaying one or two mismatches within the target region, if available. The probe EUK516 was used as a positive control.
The target sites of newly designed probes were checked for accessibility using the prediction maps based on the 18S and the 28S rRNA of Saccharomyces cerevisiae (7, 25).
All hybridization probes were labeled at the 5′ terminus with a fluorescent marker, either the indocarbocyanine dye Cy3 or Oregon Green 488. All probes were synthesized and labeled by Metabion (Planegg, Germany) and stored in sterile distilled water at −20°C.
Determination of hybridization stringencies.
Hybridizations were performed at the recommended stringencies, 20 to 40% formamide for probes EUB338 and EUK516 and 20% formamide for probe ARCH915.
Hybridization stringencies for the newly designed division-, genus-, and species-specific probes were adjusted by the stepwise addition of formamide in hybridizations against selected reference strains displaying one, two, or more mismatches within the target region. All fungal probes were labeled with Cy3 for the determination of hybridization stringencies.
Probe MY1524 was evaluated against Verticillium lecanii (CBS 126.27), Syncephalastrum racemosum (CBS 557.81), Aquabacterium commune (DSMZ11901), and Methanosarcina barkeri (DSMZ800). Probes specific for the genus Tetracladium and for the species Tetracladium marchalianum were tested against Epicoccum nigrum, Tricladium angulatum strain CCM F-10200, and Tetracladium setigerum (CCM F-20987), Tetracladium furcatum (CCM F-11883), Tetracladium apiense (CCM F-23199), and Tetracladium maxilliforme (CCM F-14286). Probes for Tricladium angulatum were checked against Tricladium splendens (CCM F-12386 and CCM F-19087). Probes for Alatospora acuminata strains were tested against Alternaria alternata (CBS 154.31) and Geomyces pannorum and against the other Alatospora acuminata strains corresponding to the probe set. Anguillospora longissima probes were tested against Leptosphaeria bicolor (ATCC 42642) and Pleospora herbarum (CBS 191.86). For the evaluation of nonspecific staining and autofluorescent cells, control hybridizations with Cy3-labeled probe EUB338 were performed.
Hybridization procedure.
Fixed fungal cultures, conidia, PE slides, cage membranes, and leaf samples were washed twice with autoclaved distilled water and air dried. In order to decrease inherent autofluorescence signals, leaves were subjected to treatment with 100 mM Tris-HCl buffer (pH 8) with 50 mM sodium borohydride (NaBH4) for 30 min.
The samples were subsequently covered with permeabilization buffer (1× PBS, 1% sodium dodecyl sulfate [SDS], pH 5.5) and chitinase solution (Sigma-Aldrich) at a final concentration of 1 mg of chitinase per ml of buffer. The samples were incubated at 30°C for 10 min, rinsed gently with distilled water, and air dried. Following permeabilization, the samples were subjected to treatment with increasing concentrations of ethanol (50, 80, and 96%, for 3 min each).
Working solutions of probes had a concentration of 50 ng of DNA per μl. The hybridization buffer, containing 0.9 M NaCl, 20 mM Tris-HCl (pH 7.2), 0.03% SDS, and 20, 35, 40, or 50% formamide, and the fluorescent probe were gently mixed in a ratio of 10:1 (vol/vol) to get a final oligonucleotide concentration of 5 ng per μl. Small amounts of cultures or conidia were immobilized on a slide and covered with 10 μl of probe-containing hybridization buffer. The PE slides were partitioned into two sections with strong adhesive tape, enabling hybridizations with different probes on one slide. Fifty microliters of hybridization solution was placed onto each section. Leaves and cage membranes were mounted onto glass slides with 20 μl of hybridization solution and overlaid with 50 μl of hybridization solution. For hybridization, slides were placed in sampling tubes and incubated at 46°C in the dark for 1.5 to 16 h.
Following hybridization, the slides were washed with prewarmed washing buffer (20 mM Tris-HCl, 0.01% SDS, and 250, 88, 62.4, or 31.4 mM NaCl, corresponding to 20, 35, 40, and 50% formamide stringencies) for 20 min at 46°C, rinsed with double-distilled water, air dried, and mounted with the antifading reagent Citifluor AF 2 (Citifluor Ltd., London, United Kingdom).
Calcofluor white staining.
Fixed PE slides, leaves, and cellulose cage membranes were stained with an 0.5 mM solution of calcofluor white M2R {4,4′-bis[4-anilino-6-bis(2-ethyl)amino-s-triazin-2-ylamino]-2,2′-disulfonic acid; Molecular Probes Europe, Leiden, The Netherlands}. The targets of calcofluor white M2R are chitin, cellulose, and carboxylated polysaccharides. Following incubation in the dark for 5 min, the PE slides, leaves, and cellulose cage membranes were observed by confocal laser scanning microscopy (CLSM).
Microscopy and documentation.
CLSM was performed using a TCS 4D microscope and a TCS SP/MP microscope (Leica, Heidelberg, Germany). The TCS 4D microscope was attached to an inverted microscope and equipped with an argon-krypton and UV laser. Fungi were observed through 20× 0.6 NA (numerical aperture), 40× 0.75 NA, 63× 1.2 NA, and 100× 1.4 NA lenses. The microscope was used in the single- or multichannel mode to record the reflection signal, stained and hybridized fungi, environmental samples, and the autofluorescence of fungi and phototrophic organisms. For the presentation of micrographs, the standard software ScanWare version 5.1A (Leica) was used. The images were printed with Photoshop version 5.5 (Adobe, San Jose, CA).
The TCS SP/MP instrument (Leica) was set up with three visible lasers, Ar (458, 476, 488, and 514 nm), Kr (568 nm), and He/Ne (633 nm) (Omnichrome, Chino, CA). The SP (spectral photometer) feature allowed the adjustment of sliders on the detector side. Images were obtained with 40× 0.8 NA, 63× 0.9 NA, and 63× 1.2 NA water-immersible lenses (Leica). Numerical apertures of 40 by 0.8, 63 by 0.9, and 63 by 1.2. Image processing and three-dimensional reconstructions were done using the standard software package TCS NT version 1.6.587 (Leica, Heidelberg, Germany) delivered with the instrument and IMARIS version 3.06 (Bitplane, Zurich, Switzerland).
To evaluate the inherent autofluorescence signals emitted from the different fungal species, hybridization fluorescence intensities and autofluorescence intensities were measured simultaneously by performing a scan over the visible light spectrum (300- to 600-nm wavelength) using CLSM. Fungal samples not subjected to the FISH assay but treated with hybridization buffer and/or with permeabilization buffer and sodium borohydride were used as negative controls.
Nucleotide sequence accession numbers.
The sequences of the different regions of the rRNA genes of 39 aquatic hyphomycete strains were deposited in GenBank under accession numbers AY204573 to AY204578, AY204583 to AY204586, AY204597 to AY204605, AY204607, AY204612 to AY204619, AY204626 to AY204630, and AY204632. GenBank accession numbers are indicated in Table 1.
TABLE 1.
Names and provenances of aquatic hyphomycetes sequenced in this study and corresponding GenBank accession numbers
| Species | Strain no. | Origina | GenBank accession no. for:
|
|
|---|---|---|---|---|
| SSU | LSU | |||
| Alatospora acuminata sensu stricto | CCM F-02383 | UK | AY204583 | |
| CCM F-13089 | CZ | AY204584s | ||
| Alatospora acuminata sensu lato | CCM F-12186 | SK | ||
| CCM F-37194 | CA | AY204585 | ||
| L8 | AT | AY204586 | ||
| Anguillospora crassa | CCM F-07082 | SK | AY204578 | |
| CCM F-13483 | SK | AY204576 | ||
| CCM F-15583 | SK | AY204577 | ||
| CCM F-05584 | SK | AY204574 | ||
| CCM F-15283 | CZ | AY204575 | ||
| Anguillospora furtiva | L16 | AT | AY204601 | |
| Anguillospora longissima | CCM F-00980 | DE | AY204598 | AY204597 |
| CCM F-11891 | CZ | AY204599 | ||
| CCM F-10691 | CZ | |||
| L22 | AT | AY204600 | ||
| CCM F-11791 | CZ | |||
| Heliscus lugdunensis | CCM F-245 | SK | AY204603 | |
| Elbe 98 | DE | AY204604 | ||
| L5 | AT | AY204602 | ||
| Lemonniera aquatica | CCM F-04480 | CZ | AY204605 | |
| Lemonniera terrestris | CCM F-125 | CZ | AY204607 | |
| CCM F-11486 | SK | AY204607 | ||
| Tetracladium marchalianum | CCM F-19399 | CZ | AY204613 | |
| CCM F-26199 | CZ | AY204614 | AY204612 | |
| CCM F-26299 | CZ | AY204615 | ||
| CCM F-26399 | CZ | AY204616 | ||
| Elbe 50 | DE | AY204618 | ||
| Elbe 90 | DE | AY204617 | ||
| L27 | AT | AY204619 | ||
| Tricladium angulatum | CCM F-139 | CZ | AY204627 | |
| CCM F- 01380 | DE | AY204573 | ||
| CCM F-14186 | CZ | AY204628 | ||
| CCM F-10200 | CZ | AY204626 | ||
| Tricladium splendens | CCM F-12386 | CZ | AY204630 | |
| CCM F-19087 | CA | AY204632 | ||
| CCM F-11989 | CZ | AY204629 | ||
| CCM F-16599 | CZ | AY204631 | ||
| Varicosporium elodeae | CCM F-04276 | SK | AY425613 | |
| CCM F-11783 | CZ | AY425614 | ||
UK, United Kingdom; CZ, Czech Republic; SK, Slovakia; CA, Canada; AT, Austria; DE, Germany.
RESULTS
Development of rRNA-targeting oligonucleotide probes.
During this study, nine fungus-specific probes were developed. Computer-aided sequence analysis revealed at least one centrally located mismatch in all accessible 18S and 28S rRNA sequences of nontarget organisms for all probes except TRIang322. The last sequence data analysis was performed 11 August 2008. For Tetracladium species, Alatospora acuminata, Anguillospora longissima, and Tricladium angulatum, FISH probes targeting either the 18S or the 28S rRNA could be designed and evaluated on a genus- or species-specific level. FISH analysis of pure cultures revealed no significant differences among themselves in probe-conferred fluorescence signal intensities. Sequences of new oligonucleotide probes, target regions, probe binding sites, and formamide concentrations within the hybridization buffer are summarized in Table 2.
TABLE 2.
Fungal oligonucleotide probes and their target organisms, sequences, and target positions
| Probe | Hybridization stringencya (% formamide) | Target organism(s) | Probe sequence (5′ to 3′) | rRNA subunit, helix,b and binding positionc |
|---|---|---|---|---|
| MY1574 | 20 | Eumycota | TCCTCGTTGAAGAGC | 18S; 47; 1474-1489 |
| TCLAD1395 | 35 | Members of the genus Tetracladium | CGCACTTCCATTTGCTTG | 18S; 44; 1395-1413 |
| TmarchB10 | 40 | Tetracladium marchalianum | GCTTAAGGTCAGGGGTAT | 28S; B10; 182-200 |
| TmarchC1_1 | 20 | Tetracladium marchalianum | TCACCGGATGATCAACTG | 28S; C1_1; 605-623 |
| TmarchC1_2 | 20 | Tetracladium marchalianum | GCCCATTCCCAGGCCTTT | 28S; C1_2; 679-697 |
| ALacumi1698 | 40 | Alatospora acuminata | CCGCGCTAGGTGGTCGTT | 18S; 49; 1698-1716 |
| TRIang322 | 35 | Tricladium angulatum | GCCCACTACGCTACAATC | 18S; 12; 322-340 |
| Alongi340 | 40 | Anguillospora longissima | CCCGTTGAAACCATAGGT | 18S; 12; 340-358 |
| AlongiB16 | 35 | Anguillospora longissima | GCGCTGTTCCAAGGGTCT | 28S; B16; 345-363 |
Design and evaluation of specific oligonucleotide probes. (i) Fungal probe MY1574.
A comparative sequence analysis of MY1574 using GenBank and ARB databases resulted in matches with fungal 18S rDNA sequences from all divisions of the Eumycota. No matching sequences other than fungal rRNA sequences were observed. Under in situ conditions, probe MY1574 showed no nonspecific binding to the nontarget organisms Aquabacterium commune, Methanosarcina barkeri, Scenedesmus quadricauda, Verticillium lecanii, and Syncephalastrum racemosum. Verticillium lecanii and Syncephalastrum racemosum each displayed one centrally located mismatch within the probe target region. By adjusting the in situ hybridization stringency by the addition of 20% formamide to the hybridization buffer, Verticillium lecanii and Syncephalastrum racemosum could be unequivocally separated from the target fungi tested. Fig. 1A and E to J and L shows successful in situ hybridizations of target fungi using MY1574.
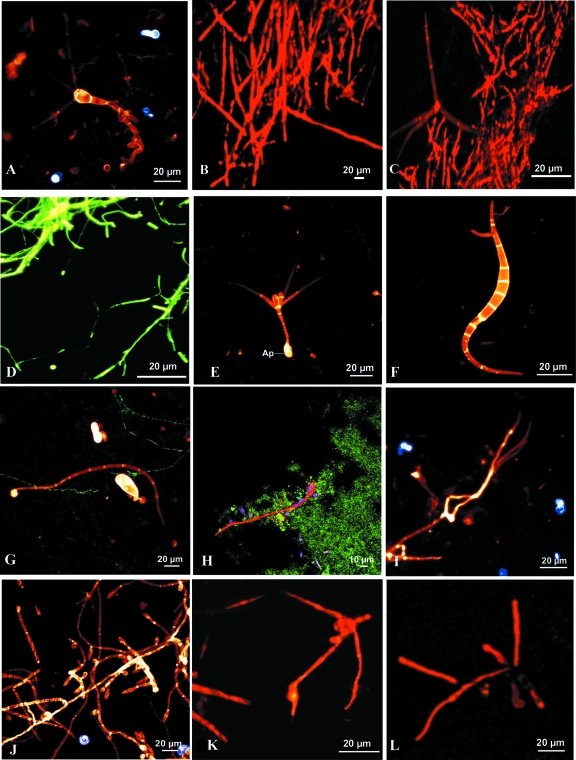
FIG. 1.
FISH with new specific oligonucleotide probes. (A) CLSM photomicrograph of a conidium and conidiophore of Clavariopsis aquatica growing out of a Fagus sylvatica leaf collected from the Oberer Seebach, shown after hybridization with Cy3-labeled probe MY1574. (B) CLSM photomicrograph showing FISH of Tetracladium marchalianum with specific 28S rRNA-targeting Cy3-labeled probe TmarchC1_1. (C) CLSM photomicrograph of Alatospora acuminata hybridized with 18S rRNA-targeting Cy3-labeled probe ALacumi1698. (D) Epifluorescence photomicrograph of Anguillospora longissima after FISH with 18S rRNA-targeting Cy3-labeled probe Alongi340. (E) Conidium of Tetracladium marchalianum attached to a PE slide exposed for 2 weeks within the Oberer Seebach (Lunz; autumn 2000) and then hybridized with Cy3-labeled probe MY1574 and simultaneously stained with calcofluor white; Ap, appressorium. (F) Germinating conidium of Anguillospora cf. crassa (similar to that of A. crassa or to the broad conidium of A. longissima) attached to a PE slide (after a 2-week exposure in autumn 2000), shown after FISH with Cy3-labeled probe MY1574 and simultaneous staining with calcofluor white. (G) Conidium of Anguillospora cf. filiformis attached to a PE slide exposed for 2 weeks within the Oberer Seebach (Lunz; autumn 2000) and then hybridized with Cy3-labeled probe MY1574 and simultaneously stained with calcofluor white, along with bacteria hybridized with Oregon green-labeled probe EUB338. (H) Biofilm obtained on a PE slide exposed for 3 months (September to November 2000) within the Oberer Seebach and analyzed with Oregon green-labeled probe EUB338 (green) and Cy3-labeled probe MY1574 (red). Also visible are autofluorescing diatoms. (I) CLSM photomicrograph of an unidentified fungus producing filiform phialoconidia on a leaf of Fagus sylvatica exposed for 2 weeks in the Oberer Seebach (autumn 2000), shown after hybridization with Cy3-labeled probe MY1574. (J) CLSM photomicrograph of fungal hyphae and phialids on a Fagus sylvatica leaf exposed for 2 weeks in the Oberer Seebach (autumn 2000), shown after hybridization with Cy3-labeled probe MY1574 and simultaneous staining with calcofluor white. (K) Germinating conidium of Tetracladium marchalianum after 2 days of exposure, probed with TmarchB10. (L) Germinating conidium of Varicosporium elodeae after 2 days of exposure, probed with MY1574.
(ii) Probe specific for the genus Tetracladium.
Probe TCLAD1395 showed at least one mismatch relative to all accessible non-Tetracladium 18S rRNA sequences. In situ hybridizations done with pure and mixed cultures of Tetracladium strains using probe TCLAD1395 showed clear FISH signals. Epicoccum nigrum as a nontarget organism displayed two mismatches within the target region of TCLAD1395. It could be clearly distinguished by adjusting the hybridization stringency to 35%.
(iii) Probes specific for Tetracladium marchalianum.
By using the available 18S rRNA sequence data, it was not possible to construct a probe on the intended species level for Tetracladium marchalianum. Among the Tetracladium species, the similarities of the 18S rRNA sequences are too high. Species-specific probes for Tetracladium marchalianum needed to be designed by targeting the 28S rRNA subunit. Three different probes, termed TmarchB10, TmarchC1_1, and TmarchC1_2, showed no nonspecific matches with bacterial and fungal sequences. In situ testing of the three probes against Tetracladium setigerum, Tetracladium furcatum, Tetracladium apiense, and Tetracladium maxilliforme revealed no nonspecific binding when the hybridization stringency was adjusted to 20% for probes TmarchC1_1 and TmarchC1_2 and to 40% for probe TmarchB10. FISH of Tetracladium marchalianum with any of the 28S rRNA-targeting probes TmarchB10, TmarchC1_1, or TmarchC1_2 resulted in clear hybridization signals in pure and mixed cultures and environmental samples (Fig. 1B).
(iv) Probe specific for Alatospora acuminata.
The 18S rRNA-targeting probe ALacumi1698 showed at least two mismatches relative to rRNA sequences from nontarget bacterial, fungal, or algal species. Sequences from four fungi of the order Lecanorales (GenBank accession numbers U86695, U86696, U86697, and AF241543) and from two fungi of the order Pertusariales (GenBank accession numbers AF274110 and AF274114) displayed two mismatches at positions 14 and 17 relative to the target region of probe ALacumi1698. Since none of the reference organisms were available for in situ testing, the hybridization stringency was adjusted to 40%. Alatospora acuminata strains CCM F-2380, CCM F-13089, CCM F-37194, CCM F-12186, CCM F-18799, and L8 in pure cultures and environmental samples showed clear FISH signals when hybridized with probe ALacumi1698 (Fig. 1C).
(v) Probe specific for Tricladium angulatum.
Probe TRIang322 targets the 18S rRNA and fits to all four Tricladium angulatum strains tested without mismatches. BLAST searches with the probe TRIang322 also yielded matches with the sequences of the lichens Lecanora achariana (GenBank accession number DQ973004) and Rhizoplaca peltata (GenBank accession number AY530887). FISH signals exclusively from Tricladium angulatum strains were obtained at a hybridization stringency level of 35% during in situ hybridizations testing Tricladium angulatum strains, Tricladium splendens strains, and Tetracladium marchalianum strains (data not shown).
(vi) Probes specific for Anguillospora longissima.
The 18S rRNA-targeting probe Alongi340 showed at least three mismatches at the 3′ terminus relative to the target regions of organisms other than Anguillospora longissima strains. With a 40% formamide concentration in the hybridization buffer, Anguillospora longissima could be clearly distinguished from Leptosphaeria bicolor. By performing FISH with the probe Alongi340, all strains of Anguillospora longissima could be unambiguously detected. The new 28S rRNA-targeting probe AlongiB16 showed at least two mismatches in its target region with all other sequences available in the databases. In situ testing of probe AlongiB16 against Anguillospora longissima strains CCM F-00980, CCM F-11891, CCM F-10691, CCM F-11791, and L22 and the nontarget organisms Leptosphaeria bicolor and Clavariopsis aquatica showed exclusive staining of the target organism when the stringency was adjusted to 35% (Fig. 1D).
FISH of freshwater fungi within environmental samples.
In order to check the applicability of the FISH method to the in situ detection of freshwater fungi in environmental samples, the exposed surfaces of PE slides and leaves were examined.
Biofilms on PE slides.
After 2 weeks of exposure in autumn 2000 and August 2001, many conidia of freshwater fungi were attached to the PE slides. They were accessible for FISH as soon as they formed appressoria or were germinating (Fig. 1E to G). Appressoria were formed by tetraradiate and by sigmoid conidia. In contrast to the samples exposed for 2 weeks, the PE slides exposed for 3 months in September, October, and November 2000 could not be shown to have conidia. Here, among thick layers of bacteria, hyphae were detected by FISH with the probe MY1574 (Fig. 1H). On the PE slides exposed during the winter (October 2000 to February 2001), no conidia were observed nor could hyphae be detected by FISH. No difference in the accessibility of fungal structures for FISH was observed between the PE slides exposed at sampling sites with slow and fast currents.
As in pure-culture experiments, probe-conferred signal intensities varied in different parts of the mycelia on the PE slides. Fungal structures that were suspected to have high-level growth activities, for example, appressoria, showed very bright FISH signals. In contrast, hyphae often showed both cells with FISH signals and cells without FISH signals.
Fungus-directed FISH on leaves.
Prior to FISH application, the leaves had to be subjected to treatment with sodium borohydride in order to lower the leaves' inherent autofluorescence. On all types of leaves, growing hyphae and germinating conidia could be detected by FISH. Most FISH signals were obtained by using the Eumycota probe MY1574 and, to a lesser extent, by using specific probes. Analyzing the leaves with probe MY1574 showed FISH signals for Clavariopsis aquatica (Fig. 1A) and an unidentified fungus producing filiform phialoconidia (Fig. 1I). Remarkably, the fungi detected by FISH signals of probe MY1574 often showed Acremonium- and Phialophora-like phialidic structures (Fig. 1J). About one-third of hyphae detected by calcofluor white staining were not accessible for FISH using the fungal probes available in this study. As for the PE slides, no difference in the accessibility of fungal structures for FISH between leaves exposed at the RITRODAT sampling sites 2C1.1 (slow current) and 16F3.3 (fast current) was observed.
Accessibility of germinating conidia for FISH within the Oberer Seebach.
The water temperature of the Oberer Seebach was stable at 11°C over the entire time of the germination experiments. The accessibility of the exposed conidia for FISH was tested by using the probes MY1574 and TmarchB10. All conidia in the stage of germination were accessible for FISH (Fig. 1K and L). After 2 days of exposure, 80% of the total number of exposed conidia were still visible on the cage membranes. In contrast, the longer exposure time resulted in increased losses of conidia (Table 3). It could not be determined clearly if the missing parts were washed out or somehow decayed. The germination of Varicosporium elodeae conidia was observed at slow-current exposure sites only.
TABLE 3.
Species, numbers of conidia exposed, exposition times and sites, and germination rates as percentages of total numbers originally exposed within the Oberer Seebach in autumn 2000
| Species | No. of conidia or spores exposed (ml−1) | Exposition site (current speed) | Exposition time (days) | Germination rate (%) |
|---|---|---|---|---|
| Varicosporium elodeae | 320 | 2C1.1 (slow) | 2 | 9-10 |
| 320 | 15F3.3 (fast) | 2 | 0 | |
| Tetracladium marchalianum | 1,000 | 2C1.1 (slow) | 2 | 5 |
| 1,000 | 15F3.3 (fast) | 2 | 0 | |
| Anguillospora crassa | 10 | 2C1.1 (slow) | 2 | 10 |
| 10 | 15F3.3 (fast) | 2 | 20 | |
| Varicosporium elodeae | 400 | 2C1.1 (slow) | 5 | 16-20 |
| 400 | 15F3.3 (fast) | 5 | 0 | |
| Tetracladium marchalianum | 1,000 | 2C1.1 (slow) | 5 | 8-10 |
| 1,000 | 15F3.3 (fast) | 5 | 0 | |
| Anguillospora crassa | 10 | 2C1.1 (slow) | 5 | 10 |
| 10 | 15F3.3 (fast) | 5 | Cage was gone |
DISCUSSION
For the reliable characterization of microorganisms within their microenvironments by FISH, it is critical to know the degrees of specificity of the oligonucleotide probes employed. Of approximately 100,000 currently described fungi and an estimated 1.5 million fungal species worldwide (22), only 18,424 species are presently listed in the GenBank databases (GenBank Taxonomy Browser statistics, August 2008). Of the 143,016 fungal rRNA sequences presently listed in GenBank, many represent internal transcribed spacer (ITS) data. In the present study, it was sometimes impossible to find sequences in GenBank displaying only one mismatch in the target region. This finding does not mean that the specificities of our probes are reliably restricted to certain species, because one does not know whether organisms which coincidentally have the same target sequences are present in a particular environmental sample. For that reason, theoretical and practical testing of probes over short time intervals should be performed (30). No 18S rRNA-targeting Tetracladium marchalianum-specific probe could be designed because the sequences of other Tetracladium species were too similar. This issue leads to the general question of the suitability of fungal rRNA genes for the design of specific FISH probes. The 18S rDNA is known to be useful for examining ancient evolutionary events (12, 23, 24). This approach allows the construction of group-specific oligonucleotides, such as the widely used fungal primers of White et al. (44). Consequently, the design of the probe MY1574, specific for a wide range of Eumycota, was easily achieved. It is noteworthy that probe MY1574 was designed on the basis of a much larger database than the fungal probe FUN1429 (6). While the phylogenetic target range of probe FUN1429 seems to be restricted to members of the subphylum Pezizomycotina (29), probe MY1574 also matches members of the Saccharomycotina and Taphrinomycotina and the Basidiomycetes and Zygomycetes. The binding sites of FUN1429 and MY1574 are located in regions known to be conserved in most fungi, but not in all eukaryotes (45). Thus, probe MY1574 may be specific for a large number of members of the kingdom Fungi. The genus-specific probe TCLAD1395 and the species-specific probes ALacumi1698 and Alongi340 bind to regions with high degrees of variability. The probe specific for Alatospora acuminata has a binding site in a hypervariable region at helix 49. The slightly less variable probe signatures found in the SSU sequences of Tricladium angulatum and Anguillospora longissima were sufficient to construct a species-specific probe only in the case of Anguillospora longissma. According to data from GenBank searches, the probe TRIang322 also matches two lichens. Nevertheless, the probe TRIang322 may be used in aquatic environments, as both lichens are terrestrial and furthermore should easily be distinguished from Tricladium angulatum by morphology. By using current knowledge of fungal 18S sequences, the highly variable regions of the SSU rRNA appear to be sufficient to design species-specific probes, with limitations. Most likely, the highly variable helical regions E23_1, E23_2, E23_5, 10, 29, and 43 provide signature sites for specific probe design. The fungal 18S rRNA is more likely than the bacterial 16S rRNA to be a useful marker molecule for the design of probes with specificity on higher phylogenetic levels, such as orders and classes. This utility should become evident as more fungal SSU sequences become available.
The 28S rRNA gene provides more variations in rates of evolution than the 18S rDNA (15, 24), but fungal 28S rRNA sequences are currently less available in public databases. The species-specific probes for Anguillospora longissima and Tetracladium marchalianum both bind to different helical loci within the variable V2 region (15) of the LSU rRNA. Remarkably, a hybridization stringency of only 20% was sufficient to discriminate successfully among the closely related nontarget fungi Tetracladium setigerum, Tetracladium furcatum, Tetracladium apiense, and Tetracladium maxilliforme by using the Tetracladium marchalianum-specific LSU-targeting probes TmarchC1_1 and TmarchC1_2. The LSU-targeting probe AlongiB16, specific for Anguillospora longissima, could be tested only against fungi that are presumed to be present in the same habitat. The suitability of the LSU rRNA for the construction of species-specific probes is difficult to estimate. However, the LSU rRNA molecule provides at least enough variable sequence stretches for the design of in situ probes specific to the genus level. This conclusion will probably be corroborated as soon as more LSU rDNA sequence data become available.
On the basis of nucleotide variability, the ITS regions seem to be the most appropriate target for the design of species-specific FISH probes. Because ITS-derived RNA is not located in the cytoplasm, FISH cannot be applied.
FISH with the new probes gave evidence of actively growing fungi on substrates exposed in the Oberer Seebach stream. Active mycelia could be detected by FISH on all leaves sampled from the Oberer Seebach, as well on the PE slides exposed in the Oberer Seebach during the summer and autumn. By using the newly developed set of probes specific for different aquatic hyphomycetes, only Alatospora acuminata was detected, by the probe ALacumi1698, on PE slides. However, by using probe MY1574, aquatic hyphomycetes other than Alatospora acuminata (including Clavariopsis aquatica, Tetracladium marchalianum, and an undetermined species having scolecoform conidia) could be successfully detected. Remarkably, many Acremonium-, Chalara-, and Phialophora-like fungi were detected on collected and exposed leaves. Strong hybridization signals of conidiogenous cells indicated that they were capable of sporulation under water. Terrestrial, epiphytic, and endophytic fungi, already present on the leaf before abscission, are understood to be of limited importance in leaf decomposition in streams and are generally replaced by aquatic hyphomycetes as the decomposition process develops (19, 39). The observation of active epiphytic fungi by FISH may be due to the short period of exposure of the leaves (2 weeks), and aquatic hyphomycetes might not yet have been representing the major component of the leaf fungal community. This assumption is also supported by the observation of many nongerminated conidia attached to the leaves and by the previous observations of Sridhar and Bärlocher (39), who also observed low levels of aquatic hyphomycete diversity and biomass on maple leaves in a small Canadian stream after 2 weeks. On the other hand, the detection of many terrestrial fungi on the leaves may indicate that the contribution of these fungi to leaf litter decomposition in streams may be underestimated. In traditional sporulation analyses, the small conidia (e.g., 1 to 5 μm) of terrestrial fungi are easily overlooked due to the pore sizes of filters used with this method. By the sporulation method, the conidial production on aerated leaves under laboratory conditions can be assessed. The FISH method allows the detection of freshly attached and germinating conidia in situ. This approach allows the characterization of the upcoming active fungal population.
On PE slides exposed for 2 weeks, the majority of aquatic hyphomycetes detected by FISH were in the process of attachment, showing appressorium formation and germ tubes. Developed mycelia could be detected only on PE slides exposed for longer periods in the autumn. These PE slides also showed a dense layer of bacteria and algae which may have provided more nutrients for fungal growth than naked PE slides which were exposed for only a short time. The lack of freshly attached conidia after 3 months of exposure in autumn (September to November) indicates that no conidial inoculum was available in the last days of November (winter). This result is in conjunction with the general assumption that the conidial inoculation of substrates in streams in moderate climates occurs mainly during leaf fall events in late summer and autumn. On slides exposed during the winter (November to February), no fungi were detected, indicating that no fungal activity occurs at very low water temperatures (1 to 4°C) (43).
About one-third of fungal mycelia on leaves and PE slides observed by calcofluor white staining could not be hybridized using the fungal probes. In part, these structures might have been nongrowing mycelia not accessible by FISH. However, the probe MY1574 with broad phylogenetic specificity also has a limited phylogenetic range. Another explanation may be the low rRNA content of mycelia exhibiting low-level growth activities, which would make the detection by FISH difficult. Furthermore, the lack of sufficient permeabilization may prevent the penetration of fungal cell walls by FISH probes. First attempts to use a more sensitive FISH protocol, such as catalyzed reported deposition-FISH (36), did not result in satisfying probe-conferred fluorescence signals from freshwater fungi on leaves (unpublished data). These limitations indicate that one must be cautious with diversity and biomass estimations when using FISH as the only method.
Germinating conidia of aquatic hyphomycetes could be successfully hybridized on the membranes exposed within the cages in the Oberer Seebach RITRODAT area. Hybridizations yielded strong fluorescence signals for highly active germ tubes and young hyphae. The sample size was small; consequently, one has to be careful in drawing conclusions from the germination rates. However, the germination rates of Tetracladium marchalianum, Varicosporium elodeae, and Anguillospora crassa of 5 to 20% after 2 days can be regarded as rather low. In the Oberer Seebach, the limiting factor for the efficient germination of the exposed species may be the small amount of nitrate and, especially, phosphate present in the stream water. Conidium production and fungal growth are stimulated by inorganic N and P (21, 38). The two cage membranes may have been exposed to less water circulation than the submerged leaves, and thus, the availability of oxygen and nutrients may have been too low for more efficient germination.
In conclusion, FISH is a reliable tool for the detection of growing stages and the spatial distribution of aquatic hyphomycetes in lotic systems. By the application of FISH, we could show the high-level growth activity of freshly attached and germinating conidia indicated by strong probe-conferred fluorescence signals on leaves, PE slides, and cellulose acetate membranes. Furthermore, the application of the new FISH probes provided evidence for the first time of the in situ presence of metabolically active terrestrial fungi on submerged leaves. One main issue of increasing the validity of FISH is the thorough control of the specificity of existing probes and the continuous design of new specific probes in synchronization with growing sequence databases. Also, FISH should not be used as the sole tool to characterize the freshwater fungal community, as estimates of the total living fungal biomass, conidia production, and in situ enzymatic activities give additional important information.
Acknowledgments
We thank Felix Bärlocher (Mount Allison University, Canada) for providing cultures of Tetracladium marchalianum CCM F-11391, Tetracladium setigerum CCM F-20987, Tetracladium apiense, Tetracladium maxilliforme CCM F-14286, and Tetracladium furcatum CCM F-11883. The expert technical assistance of Ute Kuhlicke is gratefully acknowledged.
This work was supported by Czech Ministry of Education grant MSM0021622416 (to L.M.) and by UFZ grant UFZ-03/99.
Footnotes
Published ahead of print on 5 September 2008.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microb. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arsuffi, T. I., and K. Suberkropp. 1989. Selective feeding by shredders on leaf-colonizing stream fungi: comparison on macroinvertebrate taxa. Oecologia 79:30-37. [DOI] [PubMed] [Google Scholar]
- 3.Baker, B. J., M. A. Lutz, S. C. Dawson, P. L. Bond, and J. F. Banfield. 2004. Metabolically active eukaryotic communities in extremely acidic mine drainage. Appl. Environ. Microbiol. 70:6264-6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bärlocher, F. 1982. Conidium production from leaves and needles in four streams. Can. J. Bot. 60:1487-1494. [Google Scholar]
- 5.Bärlocher, F. 1992. Community organisation, p. 38-69. In F. Bärlocher (ed.), The ecology of aquatic hyphomycetes. Springer Verlag, Berlin, Germany.
- 6.Baschien, C., W. Manz, T. R. Neu, and U. Szewzyk. 2001. Fluorescence in situ hybridization of freshwater fungi. Int. Rev. Hydrobiol. 86:371-384. [Google Scholar]
- 7.Behrens, S., C. Rühland, J. Inácio, H. Huber, Á. Fonseca, I. Spencer-Martins, B. M. Fuchs, and R. Amann. 2003. In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes. Appl. Environ. Microbiol. 69:1748-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben Ali, A., J. Wuyts, R. De Wachter, A. Meyer, and Y. Van de Peer. 1999. Construction of a variability map for eukaryotic large subunit ribosomal RNA. Nucleic Acids Res. 27:2825-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bermingham, S., L. Maltby, and F. M. Dewey. 1997. Use of immunoassays for the study of natural assemblages of aquatic hyphomycetes. Microb. Ecol. 33:223-229. [DOI] [PubMed] [Google Scholar]
- 10.Bretschko, G. 1991. The limnology of a low order alpine gravel stream (Ritrodat-Lunz study area, Austria). Int. Assoc. Theor. Appl. Limnol. 24:1908-1912. [Google Scholar]
- 11.Brul, S., J. Nussbaum, and S. K. Dielbandhoesing. 1997. Fluorescent probes for cell wall porosity and membrane integrity in filamentous fungi. J. Microbiol. Methods 28:169-178. [Google Scholar]
- 12.Bruns, T. D., T. J. White, and J. W. Taylor. 1991. Fungal molecular systematics. Annu. Rev. Ecol. Syst. 22:525-564. [Google Scholar]
- 13.Charcosset, J.-Y., and M. Gardes. 1999. Infraspecific genetic diversity and substrate preference in the aquatic hyphomycete species, Tetrachaetum elegans. Mycol. Res. 103:736-742. [Google Scholar]
- 14.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 15.De Rijk, P., Y. Van de Peer, S. Chapelle, and R. De Wachter. 1994. Database on the structure of large ribosomal subunit RNA. Nucleic Acids Res. 22:3495-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fargues, J., M.-C. Bon, S. Manguin, and Y. Couteaudier. 2002. Genetic variability among Paecilomyces fumosoroseus isolates from various geographical and host insect origins based on the rDNA-ITS regions. Mycol. Res. 106:1066-1074. [Google Scholar]
- 17.Gams, W., E. S. Hoekstra, and A. Aptroot. (ed.). 1998. CBS course of mycology, 4th ed., p. 1-165. Centraalbureau voor Schimmelcultures, Baarn, The Netherlands.
- 18.Gargas, A., and P. T. DePriest. 1996. A nomenclature for fungal PCR primers with examples from intron-containing SSU rDNA. Mycologia 88:745-748. [Google Scholar]
- 19.Garnett, H., F. Bärlocher, and D. Giberson. 2000. Aquatic hyphomycetes in Catamaran Brook: colonization dynamics, seasonal patterns, and logging effects. Mycologia 92:29-41. [Google Scholar]
- 20.Gessner, M. O., V. Gulis, K. A. Kuehn, E. Chauvet, and K. Suberkropp. 2007. Fungal decomposers of plant litter in aquatic ecosystems, p. 301-324. In C. P. Kubicek and I. S. Druzhinina (ed.), Environmental and microbial relationships, 2nd ed., vol. IV. The Mycota: a comprehensive treatise on fungi as experimental systems for basic and applied research. Springer Verlag, Berlin, Germany. [Google Scholar]
- 21.Gulis, V., and K. Suberkropp. 2004. Effects of whole-stream nutrient enrichment on the concentration and abundance of aquatic hyphomycete conidia in transport. Mycologia 96:57-65. [PubMed] [Google Scholar]
- 22.Hawksworth, D. L. 2004. Fungal diversity and its implications for genetic resource collections. Stud. Mycol. 50:9-18. [Google Scholar]
- 23.Hibbett, D. S. 1992. Ribosomal RNA and fungal systematics. Trans. Mycol. Soc. Japan 33:533-556. [Google Scholar]
- 24.Hillis, D. M., and M. T. Dixon. 1991. Ribosomal DNA: molecular evolution and phylogenetic inference. Q. Rev. Biol. 66:411-452. [DOI] [PubMed] [Google Scholar]
- 25.Inácio, J., S. Behrens, B. M. Fuchs, Á. Fonseca, I. Spencer-Martins, and R. Amann. 2003. In situ accessibility of Saccharomyces cerevisiae 26S rRNA to Cy3-labeled oligonucleotide probes comprising the D1 and D2 domains. Appl. Environ. Microbiol. 69:2899-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalmbach, S. 1998. Polyphasic characterization of the microbial population of drinking water biofilms. Ph.D. thesis. Technische Universitaet, Berlin, Germany. www.dissertation.de.
- 27.Li, S., R. N. Spear, and J. H. Andrews. 1997. Quantitative fluorescence in situ hybridization of Aureobasidium pullulans on microscope slides and leaf surfaces. Appl. Environ. Microbiol. 63:3261-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüssmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lumbsch, H. T., and S. M. Huhndorf (ed.). 2007. Outline of Ascomycota—2007. Myconet 13:1-58. [Google Scholar]
- 30.Manz, W. 1999. In situ analysis of microbial biofilms by rRNA-targeted oligonucleotide probes. Methods Enzymol. 310:79-91. [DOI] [PubMed] [Google Scholar]
- 31.McArthur, F. A., M. O. Baerlocher, N. A. B. McLean, M. D. Hiltz, and F. Bärlocher. 2001. Asking probing questions: can fluorescent in situ hybridization identify and localise aquatic hyphomycetes on leaf litter? Int. Rev. Hydrobiol. 86:429-438. [Google Scholar]
- 32.Müller, U., and P. von Sengbusch. 1983. Visualization of aquatic fungi (Chytridiales) parasitizing on algae by means of induced fluorescence. Arch. Hydrobiol. 94:471-485. [Google Scholar]
- 33.Newell, S. Y. 1992. Estimating fungal biomass and productivity in decomposing litter, p. 521-562. In G. C. Carroll and D. T. Wicklow (ed.), The fungal community. Its organization and role in the ecosystem, 2nd ed. Marcel Dekker, Inc., New York, NY.
- 34.Nikolcheva, L. G., A. M. Cockshutt, and F. Bärlocher. 2003. Determining diversity of freshwater fungi on decaying leaves: comparison of traditional and molecular approaches. Appl. Environ. Microbiol. 69:2548-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peláez, F., G. Platas, J. Collado, and M. T. Díez. 1996. Infraspecific variation in two species of aquatic hyphomycetes assessed by RAPD analysis. Mycol. Res. 100:831-837. [Google Scholar]
- 36.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robin, B. J., R. C. Arffa, I. Avni, and N. A. Rao. 1986. Rapid visualization of three common fungi using fluorescein-conjugated lectins. Investig. Ophthalmol. Vis. Sci. 27:500-506. [PubMed] [Google Scholar]
- 38.Robinson, C. T., and M. O. Gessner. 2000. Nutrient addition accelerates leaf breakdown in an alpine springbrook. Oecologia 122:258-263. [DOI] [PubMed] [Google Scholar]
- 39.Sridhar, K. R., and F. Bärlocher. 2000. Initial colonization, nutrient supply, and fungal activity on leaves decaying in streams. Appl. Environ. Microbiol. 66:1114-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 41.Suberkropp, K., and M. J. Klug. 1976. Fungi and bacteria associated with leaves during processing in a woodland stream. Ecology 57:707-719. [Google Scholar]
- 42.Vilgalys, R., and M. Hester. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172:4238-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner, F. H., and M. Leichtfried. 2003. Endbericht des Langzeit-Forschungsprogramms RITRODAT. Institute for Limnology, Austrian Academy of Science, Mondsee, Austria.
- 44.White, T. J., T. Bruns, S. Lee, and J. W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gefland, J. J. Sninsky, and T. J. White (ed.), PCR protocols. A guide to methods and applications. Academic Press, San Diego, CA.
- 45.Wuyts, J., P. De Rijk, Y. Van de Peer, G. Pison, P. Rousseeuw, and R. De Wachter. 2000. Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eucaryotic small subunit ribosomal RNA. Nucleic Acids Res. 28:4698-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]