Abstract
During a previous study on the molecular interaction between commensal bacteria and host gut immunity, two novel bacterial strains, A911T and G707T, were isolated from the gut of Drosophila melanogaster. In this study, these strains were characterized in a polyphasic taxonomic study using phenotypic, genetic, and chemotaxonomic analyses. We show that the strains represent novel species in the family Acetobacteraceae. Strain G707T, a highly pathogenic organism, represents a new species in the genus Gluconobacter, “Gluconobacter morbifer” sp. nov. (type strain G707 = KCTC 22116T = JCM 15512T). Strain A911T, dominantly present in the normal Drosphila gut community, represents a novel genus and species, designated “Commensalibacter intestini” gen. nov., sp. nov. (type strain A911 = KCTC 22117T = JCM 15511T).
Life on Earth emerged at least 3.8 billion years ago (32); Earth has been inhibited by bacteria for at least 2.5 billion years and by eukaryotes for 0.5 to 1 billion years (2). All life forms containing mucosal epithelia have had to adapt to the presence of commensal microbiota in their gastrointestinal tract; this has been achieved via interactions such as symbiosis, commensalism, and pathogenicity (20). Research on ubiquitous bacterial symbionts has defined the mechanisms by which these organisms infect their host cells, prevent the host's immune responses, and control host physiology in insects (7). The in-depth analysis of gut-microbe interactions presents difficulties because most symbiotic bacteria cannot be cultured ex vivo and genetically amenable animal models are not diverse; however, Drosophila melanogaster has been successfully used as a genetically tractable model organism for the investigation of pathogen-initiated immunity in the gut (11, 17, 18, 37, 47, 50).
We have previously investigated the molecular interactions between commensal bacteria and host gut immunity. The Drosophila gut maintains the appropriate antimicrobial peptide (AMP) levels for preservation of the normal flora community structure by recruiting the developmental master control gene caudal (cad) (36). We showed that a deregulated immune genotype (i.e., AMP overexpression due to the loss of cad) was linked to a chronic gut inflammation phenotype, not because it was a direct cause of the disease but because it altered the commensal microbiota community structure in which the pathogenic commensal strain G707T became dominant. Two strains were isolated from wild-type Drosophila midguts: strain A911T was a predominant commensal member (more than 105 bacteria per gut), and strain G707T was a minor member. When caudal expression was disrupted by RNA interference (RNAi) (caudal RNAi), the population of strain A911T was significantly diminished and was maintained at a low level (less than 103 bacteria per gut) whereas strain G707T became a dominant commensal member (to more than 104 bacteria per gut). The characteristics of strain G707T (highly resistant to AMP) and strain A911T (sensitive to AMP) suggest that G707T could be dominant in the gut of cad RNAi flies as a result of the overexpression of AMPs. A high level of apoptosis in epithelial cells was also detected in the guts of G707T-challenged germ-free wild-type flies, but not in G707T-challenged, conventionally reared wild-type flies; however, it is still unclear how this change in bacterial population induces apoptosis in Drosophila. Thus, it is necessary to explore the fundamental characteristics of these two novel bacterial strains in order to elucidate the mechanism leading to apoptosis.
Phylogenetics is the study of evolutionary relationships among various groups of organisms. In accordance with the accumulated phylogenetic knowledge database, when comparing novel isolates with evolutionally closely related strains, researchers can determine which taxon they belong to, whether new isolates are novel, and which enzyme activities or metabolites are crucial for their ecological roles in the environment.
The primary goal of this study was to phylogenetically characterize the novel strains G707T and A911T through phenotypic, genetic, and chemotaxonomic analyses. Strain G707T is a highly pathogenic commensal organism, and strain A911T is predominant in the normal gut community structure. Here we show that these two novel isolates from the microbiota community in Drosophila midguts are affiliated with the family Acetobacteraceae; strain G707T belongs to a novel species in the previously established genus Gluconobacter, and strain A911T belongs to a novel species in the novel genus Commensalibacter.
MATERIALS AND METHODS
Bacterial strains.
Drosophila gut homogenates were inoculated on mannitol-agar plates (25 g/liter n-mannitol, 5 g/liter yeast extract, 3 g/liter peptone, and 15 g/liter agar), a medium selective for members of the family Acetobacteraceae (35), and incubated at 25°C. The colonies were repeatedly restreaked on agar plates in order to obtain a pure culture of novel strain candidates, as described previously (36). Bacterial cultures of the isolates were stored at −80°C in the presence of 20% (vol/vol) glycerol.
Morphology and physiological characteristics.
Cell morphology was examined by light microscopy (ECLIPSE 80i; Nikon, Japan) and electron microscopy. Motility was observed by the wet-mount method (33). The Gram type was determined using the Gram staining method (16) and the nonstaining method using KOH (3), and spore formation was examined using the Schaeffer-Fulton staining method (41). Cells were grown on mannitol-agar plates to determine the optimum growth conditions at various temperatures, and mannitol broth media were used to determine the optimum pH, adjusted with HCl or NaOH. Tolerance of NaCl was determined using mannitol broth media supplemented with the appropriate concentrations of NaCl. Oxidase activity was determined using an oxidase reagent (BioMerieux), and catalase activity was determined by bubble production in 3% (vol/vol) hydrogen peroxide solution. Growth on R2A agar (Difco), tryptic soy agar (TSA; Difco), marine agar (MA; Difco), nutrient agar (NA; Difco), and Luria agar (LA; Difco) was also determined. Anaerobic growth on mannitol-agar plates was analyzed with the BBL GasPak pouch system. Acid production from carbohydrate, utilization of sole carbon sources, and enzyme activities were determined using API20NE, API50CH, and API ZYM test strips (BioMerieux) and Biolog gram-negative (GN) plates with GN/gram-positive (GP) inoculating fluid.
16S rRNA gene sequencing, DNA-DNA hybridization, and phylogenetic analysis.
Chromosomal DNA was extracted using a G-spin DNA extraction kit (iNtRON Biotechnology, Republic of Korea), and the 16S rRNA gene was PCR amplified from chromosomal DNA using PCR premix (Solgent, Republic of Korea). Two universal primers were used for amplification: forward primer 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and reverse primer 1492r (5′-TACGGYTACCTTGTTACGACTT-3′). PCR was initiated by denaturation at 94°C for 2 min. Reaction products were amplified for 30 cycles as follows: denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1.5 min. A final extension was performed at 72°C for 10 min. PCR products were purified and sequenced using a PCR purification kit (Cosmo Genetech, Republic of Korea) and a BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems), respectively, according to the manufacturers' instructions. A Prism 3730XL DNA analyzer (Applied Biosystems) was used to analyze the resulting reaction mixtures, and full-length 16S rRNA gene sequences were assembled using SeqMan software (DNASTAR). Pairwise 16S rRNA gene sequence similarity was determined using the EzTaxon server (http://www.eztaxon.org/) (5) to determine phylogenetic neighbors, and sequences from the novel strains and related taxa (NCBI database) were aligned using the multiple sequence alignment program CLUSTAL X, version 1.8 (46). A DNA-DNA homology experiment was performed as described previously (34). Phylogenetic relationships between representative species in the family Acetobacteraceae were determined using the MEGA3 software program (25). Distance matrices were determined (23) and used to elaborate dendrograms by the neighbor-joining (39), minimum-evolution (38), and maximum-parsimony (24) methods. To evaluate stability, a bootstrap analysis was performed using a consensus tree based on 1,000 randomly generated trees (10).
Genomic analysis.
The G+C content was determined using high-performance liquid chromatography, as described by Mesbah and Whitman (30). The genome sizes were analyzed by pulsed-field gel electrophoresis (42, 43).
Chemotaxonomy.
The cell biomass for cellular fatty acid composition analysis was collected from mannitol-agar plates after incubation at 25°C for 2 days. Cells were harvested, and the cellular fatty acids were saponified, methylated, and extracted by following the instructions in the manual for the Sherlock microbial identification system. Fatty acids were analyzed by gas chromatography (Hewlett-Packard; 6890) and identified using the Microbial Identification software package (40).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of strains G707T and A911T are available from the GenBank nucleotide database at the NCBI website (http://www.ncbi.nlm.nih.gov) under accession numbers EU409602 and EU409601, respectively.
RESULTS
Morphology and physiological characteristics.
The cells from strains G707T and A911T are aerobic, non-spore forming, nonmotile, and rod shaped. Strain G707T cells are gram variable. Strain G707T colonies are pink pigmented and circular after 2 days of growth on mannitol-agar at 25°C. This strain can grow on conventional media such as TSA and R2A agar plates. Growth occurs on mannitol-agar plates containing up to 2% NaCl, but not at 3% NaCl. Strain G707T cannot reduce nitrate to nitrite or nitrogen. Strain A911T cells are gram negative, and its colonies are cream pigmented. It can also grow on conventional media such as TSA, R2A agar, and LA plates. Growth occurs on mannitol-agar plates containing up to 3% NaCl, but not at 4% NaCl. Strain A911T can reduce nitrate to nitrite. Both strains are oxidase negative and catalase positive. The optimal temperature range for growth is 25 to 30°C, and the optimal pH range is 3.6 to 7.8 for both strains. Additional descriptions of the strains appear in the species description. Tables 1 and 2 show a comparison between the characteristics of closely related strains and G707T and A911T, respectively.
TABLE 1.
Taxonomic characteristics of G. morbifer sp. nov., strain G707T and type strains of the other species in the genus Gluconobacter
| Characteristic | Result for straina:
|
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| Growth at 37°C | + | − | NR | +c | −c | −c | wc |
| Growth at pH 3.0 | − | NR | NR | +c | −c | +c | +c |
| Acid production from: | |||||||
| d-Mannose | + | NR | +d | wc | +c | +c | −c |
| d-Sorbitol | − | w | +d | −c | +c | −c | −c |
| Melibiose | + | + | +d | wc | +c | +c | −c |
| Sucrose | + | + | +d | −c | −c | +c | −c |
| d-Arabitol | + | NR | NR | −b | +b | +b | NR |
| l-Arabitol | + | NR | NR | −b | −b | +b | NR |
| Growth on: | |||||||
| d-Glucose | − | + | w | + | + | + | + |
| d-Fructose | + | + | w or − | + | + | + | + |
| d-Mannitol | + | + | w or − | + | + | + | + |
| d-Sorbitol | + | vw | vw or − | + | + | + | − |
| Glycerol | + | + | w or − | + | + | + | + |
| Sucrose | + | + | w | − | w | + | + |
| d-Arabitol | + | + | vw or − | vw/−e | + | + | + |
| DNA G+C content (mol%) | 54.1 | 59.8 | 60d | 60.3d | 55.9d | 55.1d | 55.8c |
Strains: 1, Gluconobacter morbifer, sp. nov., G707T; 2, G. kondonii NBRC 3266T; 3, G. albidus NBRC 3250T; 4, G. oxydans NBRC 14819T; 5, G. cerinus NBRC 3267T; 6, G. frateurii NBRC 3264T; 7, G. thailandicus F149-1T. Data for reference strains are from Malimas et al. (28). +, positive; w, weakly positive; vw, very weakly positive; −, negative; NR, not reported.
Cited from Yamada et al. (54).
Cited from Tanasupawat et al. (45).
Cited from Yukphan et al. (56).
Negative datum from Tanasupawat et al. (45).
TABLE 2.
Differential characteristics of gen. nov. strain A911T and related other genera in the family Acetobacteraceae
| Characteristic | Result for genusa:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| Flagellation | Nonmotile | Peritrichous or nonmotile | Nonmotile | Peritrichous or nonmotile | Peritrichous or nonmotile | Polar or nonmotile | Nonmotile | Nonmotile |
| Pigmentation | + | − | − | +/− | − | + | − | − |
| Production of water-soluble brown pigment(s) | − | − | − | − | +/− | −/+ | − | − |
| Growth on: | ||||||||
| 1% (wt/vol) glutamate agar | + | + | − | + | + | − | NR | − |
| Mannitol-agar | + | w | + | + | + | + | NR | + |
| Acid production from: | ||||||||
| l-Arabinose | − | +/− | + | + | +/− | + | − | + |
| d-Arabinose | − | − | − | + | − | + | NR | +/− |
| d-Xylose | + | +/− | + | + | +/− | + | − | + |
| l-Rhamnose | − | − | − | +/− | − | − | NR | − |
| d-Glucose | + | +/− | + | + | + | + | + | + |
| d-Galactose | − | +/− | + | + | + | + | − | + |
| d-Mannose | − | +/− | + | + | +/− | + | − | + |
| Melibiose | − | − | + | + | − | + | NR | + |
| Sucrose | + | − | + | + | − | + | − | +/− |
| Dulcitol | − | − | − | + | − | − | NR | − |
| Glycerol | + | − | − | + | + | + | − | + |
| DNA G+C content (mol%) | 37 | 52-60 | 52-53 | 59-61 | 55-66 | 54-63 | 63-66 | 56-57 |
Genera: 1, Commensalibacter gen. nov.; 2. Acetobacter; 3, Saccharibacter; 4, Asaia; 5, Gluconacetobacter; 6, Gluconobacter; 7, Acidomonas; 8, Kozakia. Data in columns 2 and 4 to 7 from the left for reference genera are from Urakami et al. (48), those in column 3 are from Jojima et al. (21), and those in column 8 are from Lisdiyanti et al. (26). +, positive; −, negative; NR, not reported.
Anticompetition between two novel strains.
An in vitro experiment to identify antagonism between two novel strains on a mannitol-agar plate revealed that their populations were able to coexist well together. One strain cannot exclude the other strain in a static plate environment. No antagonism, such as antibiotic-mediated competition, between two strains of Escherichia coli that produce an antibacterial toxin (colicin) and are sensitive to colicin (4) was observed, even though antagonism between two novel strains in the guts of G707T-challenged conventionally reared wild-type flies was reported (36).
Phylogenetic analysis.
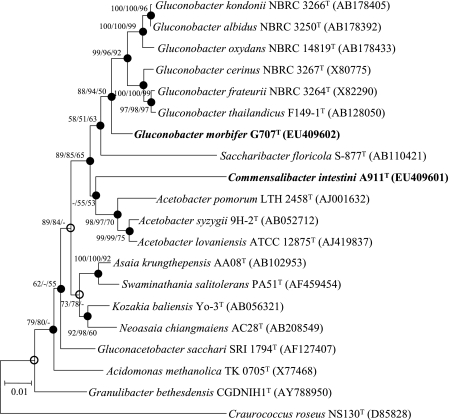
The nearly complete 16S rRNA gene sequences of strains G707T (1,367 bp) and A911T (1,482 bp) were obtained. The 16S rRNA gene sequence similarity showed that these two strains are associated with the family Acetobacteraceae. Strain G707T exhibited the highest 16S rRNA gene similarity to Gluconobacter albidus BCC 14434T (97.9%), Gluconobacter cerinus ATCC 19441T (97.6%), Gluconobacter oxydans ATCC 19357T (97.5%), Gluconobacter frateurii ATCC 49207T (97.4%), and Gluconobacter thailandicus F149-1T (97.4%), whereas strain A911T exhibited the greatest similarity to type strains Acetobacter syzygii 9H-2 (93.5%), Acetobacter lovaniensis ATCC 12875 (93.4%), Acetobacter pomorum LTH 2458 (93.3%), and Gluconacetobacter sacchari SRI 1794 (93.2%) and strains of other species belonging to the family Acetobacteraceae (≤93.1%). The sequence similarity observed for strain A911T implies that this strain must be classified as a new species, if not a new genus. Construction of phylogenetic trees based on the 16S rRNA gene sequences confirmed that both strains are also associated with the family Acetobacteraceae (Fig. 1); strain G707T clustered within a clade of the genus Gluconobacter (Fig. 1). The sequence of strain A911T was phylogenetically distinct, implying a relatively long subline of descent separate from other species of the family Acetobacteraceae (Fig. 1). Despite the low bootstrap values (approximate 50%) of the three treeing methods used, supporting a branching point between the major Acetobacter cluster and strain A911T, most of the branching points remained unchanged regardless of the treeing methods, supporting the affiliation of the strain A911T with the family Acetobacteraceae. A DNA-DNA homology study showed an average level of DNA-DNA relatedness below the 70% threshold value (51) for strain G707T and each of its closest relatives: G. albidus BCC 14434T (35.4%), G. cerinus ATCC 19441T (30.9%), G. oxydans ATCC 19357T (41.7%), G. frateurii ATCC 49207T (30.7%), G. thailandicus F149-1T (28.9%), and Gluconobacter kondonii NBRC 3266T (32.9%). Both strains can therefore be considered to represent a distinct genospecies.
FIG. 1.
Neighbor-joining tree showing the phylogenetic positions of two novel strains and related species based on 16S rRNA gene sequences. Filled circles and open circles indicate generic branches that were also recovered using the minimum-evolution and maximum-parsimony algorithms and the minimum-evolution algorithm, respectively. Numbers at nodes indicate bootstrap values as calculated on the basis of neighbor-joining/minimum-evolution/maximum-parsimony probabilities expressed as percentages of 1,000 replications. Bootstrap values greater than 50% are shown at the branch points. Bar, 0.01 accumulated change per nucleotide.
Chemotaxonomic characteristics.
The predominant fatty acids in strain G707T are C18:1 ω7c (48.64%), C16:0 (17.60%), C16:0 2-OH (15.76%), C18:0 3-OH (3.18%), C18:0 (2.68%), C16:0 3-OH (2.64%), C14:0 2-OH (2.33%), C14:0 (1.76%), summed feature 2 comprising C14:0 3-OH and/or C16:1 iso I (2.08%), and other fatty acids (less than 1.00%). The predominant fatty acids in strain A911T are C18:1 ω7c (65.29%), C18:1 2-OH (14.08%), C16:0 (10.39%), C18:0 (3.91%), C16:0 2-OH (2.69%), C16:0 3-OH (1.06%), summed feature 2 comprising C14:0 3-OH and/or C16:1 iso I (1.34%), and other fatty acids (less than 1.00%). The characteristic composition profile of the genus Gluconobacter exhibits a large amount of straight-chain unsaturated C18:1, straight-chain saturated C16:0 and 2-hydroxy fatty acid, and C16:0 2-OH as major fatty acids, with a minor presence of C18:0, C14:0 3-OH, C16:0 3-OH, C18:0 3-OH, and others (48). A predominant fatty acid profile with C18:1 ω7c as the major fatty acid is characteristic of members of the family Acetobacteraceae (12, 55). The fatty acid profile of strain A911T is roughly in agreement with those of validly described species of other genera in the family Acetobacteraceae.
Genomic analysis.
The genomic DNA G+C contents of strains G707T and A911T were 54.1 mol% and 37.0 mol%, respectively. The genomic G+C contents of six currently validated type species found in the genus Gluconobacter are within the 55.1- to 60.3-mol% range (28, 45, 56); this is similar to the genomic G+C content of novel strain G707T. These data confirm that strain G707T belongs to the genus Gluconobacter. In contrast, the genomic G+C content of A911T is significantly lower than that (more than at least 50 mol%) of any other validated species in the family Acetobacteraceae. The value of the genomic G+C content, in addition to the 16S rRNA gene sequence similarity and fatty acid profile, also supports the idea that strain A911T could be a distinct genospecies.
The sizes of the genomes of strains G707T and A911T were estimated by pulsed-field gel electrophoresis to be 1,900 kb and 2,100 kb, respectively. These two novel strains have rather smaller genome sizes than other host-associated or free-living archaea and bacteria whose genomes were obtained from a compilation of 244 complete published genomes (14). The genome sizes of novel strains are smaller than the sizes determined for strains of the species Gluconobacter oxydans subsp. oxydans (2,240 to 3,787 kb) (49).
DISCUSSION
It is known that the human gastrointestinal tract is made up of more than 500 microbial phylotypes comprising an estimated 1014 microbes; most of them are uncultivated species and novel microorganisms revealed by contemporary 16S rRNA gene sequencing techniques (9). Compared with the diversity of the human gastrointestinal tract, the microbial consortium in Drosophila is extremely simple (6). In the gastrointestinal consortium of Drosophila, 9 to 13 microbial phylotypes were found in the entire bodies of laboratory-reared Drosophila and at most 25 phylotypes were found in captured wild Drosophila.
A previous study (36) identified 10 phylotypes from control and cad RNAi Drosophila midguts including Lactobacillus plantarum, Lactobacillus brevis, Acetobacter pomorum, Acetobacter malorum, Gluconacetobacter intermedius, Gluconacetobacter rhaeticus, Gluconacetobacter europaeus, Wolbachia pipientis, and two novel strains (G707T and A911T). This study used culture-independent methods to perform the first analysis of the midgut commensal consortium in Drosophila; these techniques included PCR-denaturing gradient gel electrophoresis and the use of 16S rRNA gene clone libraries. The microbial diversity in the midgut was significantly lower than that in the entire Drosophila body (31). Two novel candidates that were isolated from the Drosophila midguts under adjusted conditions using mannitol-agar plates to focus on the culture of species in the family Acetobacteraceae were characterized in this study.
Since the genus Gluconobacter, belonging to the family Acetobacteraceae, was first proposed by Asai (the type strain was isolated from fruits in 1935 [1]), six species of this genus have been validated: G. albidus (56), G. cerinus (53), G. frateurii (29), G. oxydans (8), G. kondonii (28), and G. thailandicus (45). Among Gluconobacter species, one strain, Gluconobacter asaii NBRC 3276 (Mason and Claus 1989) is regarded as a heterotypic synonym of G. cerinus, due to the high degree of DNA-DNA relatedness to the type strain of G. cerinus, according to Katsura et al. (22). Many strains belonging to the genus Gluconobacter have been isolated from various sources including flowers, bees, various fruits, beer, cider, sour porridge, lemonade, vinegar, and sewage on soil (15). Members of this genus are gram negative or variable and motile or nonmotile; if motile, the cells have three to eight polar flagella, are obligately aerobic, have ellipsoidal to rod-shaped cells, have strong catalase activity, and do not form endospores. Some strains can cause pink diseases and rot in fruits (19). Results from the 16S rRNA gene sequence similarity, fatty acid profile, and genomic DNA G+C content assessments of strain G707T, which is dominant in the gut of cad RNAi flies, are consistent with properties of the genus Gluconobacter; the phenotypic differences between strain G707T and other Gluconobacter species are illustrated in Table 1. Thus, on the basis of phenotypic, genetic, and chemotaxonomic comparisons to previously described taxa, strain G707T can be classified as a novel species in the genus Gluconobacter, for which the name “Gluconobacter morbifer” sp. nov. is proposed.
The family Acetobacteraceae, belonging to the class Alphaproteobacteria, was first introduced by Beijerinck in 1898; the name of the family was subsequently validated in 1980 (13). At the time of writing, this family is currently assigned 26 genera, including the genera Acetobacter, Gluconacetobacter, Gluconobacter, Acidomonas, Asaia, and Kozakia. Bacteria in this family are often referred to as the acetic acid bacteria because a typical feature is their ability to produce acetic acid from ethanol. Within the acetic acid bacteria, whole cells and enzymes associated with the genera Acetobacter and Gluconobacter are widely utilized for biosensor construction in the biotechnology industry (27, 44). Among 26 genera belonging to the family Acetobacteraceae, most currently contain only a single species; also, many unclassified 16S rRNA sequences that can be assigned to the family Acetobacteraceae were registered in the NCBI database. These data mean that there are still a lot of strains in this family that can be isolated and validated from the environment. Phylogenetic analysis based on 16S rRNA gene sequences indicated that strain A911T is associated with the family Acetobacteraceae. Within the phylogenetic trees, this novel strain shares a branching point with the genus Acetobacter and a low 16S rRNA gene sequence similarity to the closely related type strain of Acetobacter syzygii, 9H-2. On the bases of the large phylogenetic distance from its closest relatives shown in Table 2 and its significantly lower genomic G+C content and different phenotypic characteristics, strain A911T differs from those of all known validly described genera in the family Acetobacteraceae. Thus, it can be concluded that strain A911T should be classified as a novel genus in the family Acetobacteraceae, for which the name “Commensalibacter intestini” gen. nov., sp. nov., is proposed. The nomenclature and description of the two strains are given below.
In this present study, two novel species belonging to the genus Gluconobacter and the new genus Commensalibacter in the family Acetobacteraceae were microbiologically characterized through a polyphasic taxonomic study. It was recently reported that the molecular pathogenesis of chronic inflammatory bowel diseases is connected to deregulation of the NF-κB and AMP signaling pathways in mammalian gut epithelia (52). By using a genetically amenable model organism harboring an extremely simple gut commensal microbiota, we have established the molecular links among immune genotype, commensal microbiota structure, and disease phenotype at the organism level. In this model, the dominance of a pathogenic commensal microbe Gluconobacter morbifer G707T due to the immune-deregulated cad RNAi genotype has been shown to induce chronic gut pathology (36). Thus, Gluconobacter morbifer G707T in the Drosophila gut should play an important role in gut pathogenesis. This study should provide fundamental information for future investigations to characterize the metabolites of these novel strains and confirm the relationship between the immune genotype-based commensal community structure and host physiology.
Description of Gluconobacter morbifer sp. nov.
Gluconobacter morbifer (mor′bi.fer. L. masc. adj. morbifer, that brings disease).
Cells are aerobic, non-spore forming, nonmotile, and rod shaped (0.7 by 1.8 to 2.0 μm), and the Gram stain is variable, with some cells staining positive and others negative. Colonies are pink-pigmented and circular, measuring approximately 1.0 to 1.5 mm in diameter after 2 days of growth on mannitol-agar at 25°C in aerobic conditions. Growth also occurs on TSA and R2A agar, but not on LA, MA, or NA. The temperature range for growth is 13 to 37°C (optimum, 25 to 30°C), but no growth occurs at 12 and 38°C. No growth occurs in NaCl concentrations greater than 3%. The pH range for growth is 3.6 to 7.8. Strain G707 is oxidase negative and catalase positive and cannot reduce nitrate to nitrite or nitrogen. Indole is not produced, and d-glucose fermentation occurs. Cells are l-arginine dihydrolase and urease negative. Esculin, gelatin, and p-nitrophenyl-β-d-galactopyranoside hydrolysis does not occur. In Biolog GN plates the strain is positive for the substrates d-arabitol, i-erythritol, d-fructose, d-mannitol, d-sorbitol, sucrose, xylitol, d-gluconic acid, dl-lactic acid, and glycerol. In API50CH test strips for acid production the strain was positive for glycerol, erythritol, d-arabinose, l-arabinose, d-ribose, d-xylose, d-galactose, d-glucose, d-mannose, methyl-α-d-glucoside, amygdalin, d-maltose, d-lactose, d-melibiose, sucrose, d-trehalose, inulin, d-raffinose, starch, xylitol, gentiobiose, d-lyxose, d-fucose, d-arabitol, l-arabitol, and gluconate. Enzyme activities determined using API ZYM test strips indicated the strain was positive for esterase (C4), esterase lipase (C8), leucine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, and N-acetyl-β-glucosaminidase. In contrast, alkaline phosphatase, lipase (C14), valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, α-mannosidase, and α-fucosidase activities were not observed. The major fatty acids are C18:1 ω7c (48.64%), C16:0 (17.60%), C16:0 2-OH (15.76%), and others (less than 5%). The G+C content of the genomic DNA and the size of the genome of the type strain are 54.1 mol% and 1,900 kb, respectively. The type strain is G707, which was isolated from Drosophila midguts. Strain G707T was deposited with the KCTC (Korean Collection for Type Cultures) and JCM (Japan Collection of Microorganisms) under accession numbers KCTC 22116T and JCM 15512T, respectively.
Description of Commensalibacter gen. nov.
Commensalibacter (com.men.sa.li.bac′ter. M.L. adj. commensalis, commensal; N.L. masc. n. bacter, a rod; N.L. masc. n. Commensalibacter, a commensal bacterium).
Cells are aerobic, non-spore-forming, nonmotile rods and are gram negative, oxidase negative, and catalase positive. The major fatty acids are C18:1 ω7c, C18:1 2-OH, and C16:0. The DNA G+C content of the type species is 37.0 mol%. The genus is a member of the family Acetobacteraceae. The type species is Commensalibacter intestini.
Description of Commensalibacter intestini sp. nov.
Commensalibacter intestini (in.tes.ti′ni. L. gen. n. intestini, of the gut).
The description is as for the genus with the following additional properties. Cells are 0.5 μm wide and 2.0 to 2.5 μm long. Colonies are cream colored and circular, measuring approximately 1.0 to 2.0 mm in diameter after 2 days of growth on mannitol-agar at 25°C in aerobic conditions. Growth also occurs on TSA, R2A agar, and LA but not on MA or NA. The temperature range for growth is 18 to 34°C (optimum, 25 to 30°C), but no growth occurs at 17 and 35°C. No growth occurs in NaCl concentrations greater than 4%. The pH range for growth is 3.6 to 7.8. Strain A911T is oxidase negative and catalase positive and can reduce nitrate to nitrite. Indole is not produced, and d-glucose fermentation does not occur. Cells are l-arginine dihydrolase and urease negative. Esculin and p-nitrophenyl-β-d-galactopyranoside hydrolysis occurs, but gelatin hydrolysis does not occur. In Biolog GN plates the strain is positive for the substrates adonitol, d-arabitol, i-erythritol, d-fructose, gentiobiose, α-d-glucose, maltose, d-mannitol, d-mannose, β-methyl-d-glucoside, d-sorbitol, xylitol, and glycerol. In API50CH test strips for acid production the strain was positive for glycerol, erythritol, d-ribose, d-xylose, d-glucose, d-fructose, d-mannitol, d-sorbitol, amygdalin, esculin, salicin, d-maltose, d-lactose, sucrose, d-trehalose, starch, glycogen, xylitol, gentiobiose, d-arabitol, and 5-ketogluconate. Enzyme activities determined using API ZYM test strips indicated the train was positive for alkaline phosphatase, esterase (C4), esterase lipase (C8), acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-galactosidase, and β-glucosidase. In contrast, lipase (C14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, β-glucuronidase, α-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase activities were not observed. The major fatty acids are C18:1 ω7c (65.29%), C18:1 2-OH (14.08%), C16:0 (10.39%), and others (less than 5%). The G+C content of the genomic DNA and the size of the genome of the type strain are 37.0 mol% and 2,100 kb, respectively. The type strain is A911, which was isolated from Drosophila midguts. Strain A911T was deposited with the KCTC and JCM under accession numbers KCTC 22117T and JCM 15511T, respectively.
Acknowledgments
We thank J. P. Euzéby (Ecole Nationale Vétérinaire, France) for etymological advice.
This work was supported by the KRIBB Research Initiative Program, the Environmental Biotechnology National Core Research Center (KOSEF: R15-2003-012-02002-0), the National Creative Research Initiative Program, and the 21C Frontier Microbial Genomics and Application Center Program.
Footnotes
Published ahead of print on 22 August 2008.
REFERENCES
- 1.Asai, T. 1935. Taxonomic studies on acetic acid bacteria isolated from and allied oxidative bacteria isolated from fruits. A new classification of the oxidative bacteria. J. Agric. Chem. Soc. Jpn. 11:499-513, 610-620, 674-708. [Google Scholar]
- 2.Brocks, J. J., G. A. Logan, R. Buick, and R. E. Summons. 1999. Archean molecular fossils and the early rise of eukaryotes. Science 285:1033-1036. [DOI] [PubMed] [Google Scholar]
- 3.Buck, J. D. 1982. Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl. Environ. Microbiol. 44:992-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao, L., and B. R. Levin. 1981. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc. Natl. Acad. Sci. USA 78:6324-6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun, J., J. H. Lee, Y. Jung, M. Kim, S. Kim, B. K. Kim, and Y. W. Lim. 2007. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57:2259-2261. [DOI] [PubMed] [Google Scholar]
- 6.Cox, C. R., and M. S. Gilmore. 2007. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect. Immun. 75:1565-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dale, C., and N. A. Moran. 2006. Molecular interactions between bacterial symbionts and their hosts. Cell 126:453-465. [DOI] [PubMed] [Google Scholar]
- 8.De Ley, J. 1961. Comparative carbohydrate metabolism and a proposal for a phylogenetic relationship of the acetic acid bacteria. J. Gen. Microbiol. 24:31-50. [DOI] [PubMed] [Google Scholar]
- 9.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 11.Ferrandon, D., A. C. Jung, M. Criqui, B. Lemaitre, S. Uttenweiler-Joseph, L. Michaut, J. Reichhart, and J. A. Hoffmann. 1998. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J. 17:1217-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franke, I. H., M. Fegan, C. Hayward, G. Leonard, E. Stackebrandt, and L. I. Sly. 1999. Description of Gluconacetobacter sacchari sp. nov., a new species of acetic acid bacterium isolated from the leaf sheath of sugar cane and from the pink sugar-cane mealy bug. Int. J. Syst. Bacteriol. 49:1681-1693. [DOI] [PubMed] [Google Scholar]
- 13.Gillis, M., and J. De Ley. 1980. Intra- and intergeneric similarities of the ribosomal ribonucleic acid cistrons of Acetobacter and Gluconobacter. Int. J. Syst. Bacteriol. 30:7-27. [Google Scholar]
- 14.Giovannoni, S. J., H. J. Tripp, S. Givan, M. Podar, K. L. Vergin, D. Baptista, L. Bibbs, J. Eads, T. H. Richardson, M. Noordewier, M. S. Rappe, J. M. Short, J. C. Carrington, and E. J. Mathur. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242-1245. [DOI] [PubMed] [Google Scholar]
- 15.Gossele, F., J. Swings, K. Kersters, and J. De Ley. 1983. Numerical analysis of phenotypic features and protein gel electropherograms of Gluconobacter asai 1935 emend. mut. char. Asai, Iizuka, and Komagata 1964. Int. J. Syst. Bacteriol. 33:65-81. [Google Scholar]
- 16.Gram, H. 1884. Über die isolierte Färbung der Schizomyceten in Schnitt- und Trockenpräparaten. Fortschritte Med. 2:185-189. [Google Scholar]
- 17.Ha, E. M., C. T. Oh, Y. S. Bae, and W. J. Lee. 2005. A direct role for dual oxidase in Drosophila gut immunity. Science 310:847-850. [DOI] [PubMed] [Google Scholar]
- 18.Ha, E. M., C. T. Oh, J. H. Ryu, Y. S. Bae, S. W. Kang, I. H. Jang, P. T. Brey, and W. J. Lee. 2005. An antioxidant system required for host protection against gut infection in Drosophila. Dev. Cell 8:125-132. [DOI] [PubMed] [Google Scholar]
- 19.Holt, J. G., N. R. Krieg, P. H. A. Sneath, J. T. Staley, and S. T. Williams (ed.). 1994. Bergey's manual of determinative bacteriology, 9th ed. Lippincott Williams & Wilkins, Baltimore, MD.
- 20.Hooper, L. V., and J. I. Gordon. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115-1118. [DOI] [PubMed] [Google Scholar]
- 21.Jojima, Y., Y. Mihara, S. Suzuki, K. Yokozeki, S. Yamanaka, and R. Fudou. 2004. Saccharibacter floricola gen. nov., sp. nov., a novel osmophilic acetic acid bacterium isolated from pollen. Int. J. Syst. Evol. Microbiol. 54:2263-2267. [DOI] [PubMed] [Google Scholar]
- 22.Katsura, K., Y. Yamada, T. Uchimura, and K. Komagata. 2002. Gluconobacter asaii Mason and Claus 1989 is a junior subjective synonym of Gluconobacter cerinus Yamada and Akita 1984. Int. J. Syst. Evol. Microbiol. 52:1635-1640. [DOI] [PubMed] [Google Scholar]
- 23.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 24.Kluge, A. G., and F. S. Farris. 1969. Quantitative phyletics and the evolution of anurans. Syst. Zool. 18:1-32. [Google Scholar]
- 25.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 26.Lisdiyanti, P., H. Kawasaki, Y. Widyastuti, S. Saono, T. Seki, Y. Yamada, T. Uchimura, and K. Komagata. 2002. Kozakia baliensis gen. nov., sp. nov., a novel acetic acid bacterium in the α-Proteobacteria. Int. J. Syst. Evol. Microbiol. 52:813-818. [DOI] [PubMed] [Google Scholar]
- 27.Macauley, S., B. McNeil, and L. M. Harvey. 2001. The genus Gluconobacter and its applications in biotechnology. Crit. Rev. Biotechnol. 21:1-25. [DOI] [PubMed] [Google Scholar]
- 28.Malimas, T., P. Yukphan, M. Takahashi, M. Kaneyasu, W. Potacharoen, S. Tanasupawat, Y. Nakagawa, M. Tanticharoen, and Y. Yamada. 2007. Gluconobacter kondonii sp. nov., an acetic acid bacterium in the α-Proteobacteria. J. Gen. Appl. Microbiol. 53:301-307. [DOI] [PubMed] [Google Scholar]
- 29.Mason, L. M., and W. Claus. 1989. Phenotypic characteristics correlated with deoxyribonucleic acid sequence similarities for three species of Gluconobacter: G. oxydans (Henneberg 1897) De Ley 1961, G. frateurii sp. nov., and G. asaii sp. nov. Int. J. Syst. Bacteriol. 39:174-184. [Google Scholar]
- 30.Mesbah, M., and W. B. Whitman. 1989. Measurement of deoxyguanosine/thymidine ratios in complex mixtures by high-performance liquid chromatography for determination of the mole percentage guanine + cytosine of DNA. J. Chromatogr. 479:297-306. [DOI] [PubMed] [Google Scholar]
- 31.Mlodzik, M., and W. J. Gehring. 1987. Expression of the caudal gene in the germ line of Drosophila: formation of an RNA and protein gradient during early embryogenesis. Cell 48:465-478. [DOI] [PubMed] [Google Scholar]
- 32.Mojzsis, S. J., G. Arrhenius, K. D. McKeegan, T. M. Harrison, A. P. Nutman, and C. R. L. Friend. 1996. Evidence for life on Earth before 3,800 million years ago. Nature 384:55-59. [DOI] [PubMed] [Google Scholar]
- 33.Murray, R. G. E., R. N. Doetsch, and C. F. Robinow. 1994. Determinative and cytological light microscopy, p. 21-41. In R. Gerhardt (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC.
- 34.Roh, S. W., Y. Sung, Y. D. Nam, H. W. Chang, K. H. Kim, J. H. Yoon, C. O. Jeon, H. M. Oh, and J. W. Bae. 2008. Arthrobacter soli sp. nov., a novel bacterium isolated from wastewater reservoir sediment. J. Microbiol. 46:40-44. [DOI] [PubMed] [Google Scholar]
- 35.Rojo-Bezares, B., Y. Saenz, M. Zarazaga, C. Torres, and F. Ruiz-Larrea. 2007. Antimicrobial activity of nisin against Oenococcus oeni and other wine bacteria. Int. J. Food Microbiol. 116:32-36. [DOI] [PubMed] [Google Scholar]
- 36.Ryu, J.-H., S.-H. Kim, H.-Y. Lee, J. Y. Bai, Y.-D. Nam, J.-W. Bae, D. G. Lee, S. C. Shin, E.-M. Ha, and W.-J. Lee. 2008. Innate immune homeostasis by the homeobox gene Caudal and commensal-gut mutualism in Drosophila. Science 319:777-782. [DOI] [PubMed] [Google Scholar]
- 37.Ryu, J. H., E. M. Ha, C. T. Oh, J. H. Seol, P. T. Brey, I. Jin, D. G. Lee, J. Kim, D. Lee, and W. J. Lee. 2006. An essential complementary role of NF-κB pathway to microbicidal oxidants in Drosophila gut immunity. EMBO J. 25:3693-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rzhetsky, A., and M. Nei. 1992. A simple method for estimating and testing minimum-evolution trees. Mol. Biol. Evol. 9:945-967. [Google Scholar]
- 39.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 40.Sasser, M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Inc., Newark, DE.
- 41.Schaeffer, A. B., and M. D. Fulton. 1933. A simplified method of staining endospores. Science 77:194. [DOI] [PubMed] [Google Scholar]
- 42.Schwartz, D. C., and C. R. Cantor. 1984. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 37:67-75. [DOI] [PubMed] [Google Scholar]
- 43.Smith, C. L., J. G. Econome, and A. Schutt. 1987. A physical map of the Escherichia coli K12 genome. Science 236:1448-1453. [DOI] [PubMed] [Google Scholar]
- 44.Svitel, J., J. Tkac, I. Vostiar, M. Navratil, V. Stefuca, M. Bucko, and P. Gemeiner. 2006. Gluconobacter in biosensors: applications of whole cells and enzymes isolated from Gluconobacter and Acetobacter to biosensor construction. Biotechnol. Lett. 28:2003-2010. [DOI] [PubMed] [Google Scholar]
- 45.Tanasupawat, S., C. Thawai, P. Yukphan, D. Moonmangmee, T. Itoh, O. Adachi, and Y. Yamada. 2004. Gluconobacter thailandicus sp. nov., an acetic acid bacterium in the α-Proteobacteria. J. Gen. Appl. Microbiol. 50:159-167. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tzou, P., S. Ohresser, D. Ferrandon, M. Capovilla, J. M. Reichhart, B. Lemaitre, J. A. Hoffmann, and J. L. Imler. 2000. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity 13:737-748. [DOI] [PubMed] [Google Scholar]
- 48.Urakami, T., J. Tamaoka, K. I. Suzuki, and K. Komagata. 1989. Acidomonas gen. nov., incorporating Acetobacter methanolicus as Acidomonas methanolica comb. nov. Int. J. Syst. Bacteriol. 39:50-55. [Google Scholar]
- 49.Verma, V., P. Qazi, J. Cullum, and G. N. Qazi. 1997. Genetic heterogeneity among keto-acid-producing strains of Gluconobacter oxydans. World J. Microbiol. Biotechnol. 13:289-294. [Google Scholar]
- 50.Vodovar, N., M. Vinals, P. Liehl, A. Basset, J. Degrouard, P. Spellman, F. Boccard, and B. Lemaitre. 2005. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc. Natl. Acad. Sci. USA 102:11414-11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wayne, L. G., D. J. Brenner, and R. R. Colwell. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 52.Xavier, R. J., and D. K. Podolsky. 2007. Unravelling the pathogenesis of inflammatory bowel disease. Nature 448:427-434. [DOI] [PubMed] [Google Scholar]
- 53.Yamada, Y., and M. Akita. 1984. An electrophoretic comparison of enzymes in strains of Gluconobacter species. J. Gen. Appl. Microbiol. 30:115-126. [Google Scholar]
- 54.Yamada, Y., R. Hosono, P. Lisdyanti, Y. Widyastuti, S. Saono, T. Uchimura, and K. Komagata. 1999. Identification of acetic acid bacteria isolated from Indonesian sources, especially of isolates classified in the genus Gluconobacter. J. Gen. Appl. Microbiol. 45:23-28. [DOI] [PubMed] [Google Scholar]
- 55.Yamada, Y., M. Nunoda, T. Ishikawa, and Y. Tahara. 1981. The cellular fatty acid composition in acetic acid bacteria. J. Gen. Appl. Microbiol. 27:405-417. [Google Scholar]
- 56.Yukphan, P., M. Takahashi, W. Potacharoen, S. Tanasupawat, Y. Nakagawa, M. Tanticharoen, and Y. Yamada. 2004. Gluconobacter albidus (ex Kondo and Ameyama 1958) sp. nov., nom. rev., an acetic acid bacterium in the α-Proteobacteria. J. Gen. Appl. Microbiol. 50:235-242. [DOI] [PubMed] [Google Scholar]