Abstract
Two methyl coenzyme M reductases (MCRs) encoded by the mcr and mrt operons of the hydrogenotrophic methanogen Methanothermobacter thermautotrophicus ΔH are expressed in response to H2 availability. In the present study, cis elements and trans-acting factors responsible for the gene expression of MCRs were investigated by using electrophoretic mobility shift assay (EMSA) and affinity particle purification. A survey of their operator regions by EMSA with protein extracts from mrt-expressing cultures restricted them to 46- and 41-bp-long mcr and mrt upstream regions, respectively. Affinity particle purification of DNA-binding proteins conjugated with putative operator regions resulted in the retrieval of a protein attributed to IMP dehydrogenase-related protein VII (IMPDH VII). IMPDH VII is predicted to have a winged helix-turn-helix DNA-binding motif and two cystathionine β-synthase domains, and it has been suspected to be an energy-sensing module. EMSA with oligonucleotide probes with unusual sequences showed that the binding site of IMPDH VII mostly overlaps the factor B-responsible element-TATA box of the mcr operon. The results presented here suggest that IMPDH VII encoded by MTH126 is a plausible candidate for the transcriptional regulator of the mcr operon in this methanogen.
Hydrogenotrophic methanogenic archaea, which live on methane formation by CO2 reduction with H2, are found in various anoxic environments such as soil, aquatic sediments, or animal intestines (32). Hydrogenotrophic methanogens are indispensable microbes for the complete degradation of organic matter in environments where electron acceptors other than CO2 are scarce. Low-molecular-weight amines, alcohols, and organic acids, which are formed from the decomposition of organic matter in anoxic environments, are difficult to degrade by fermentative microbes, since anaerobic oxidation of these molecules accompanied by H2 production is energetically unfavorable, unless a very low H2 partial pressure is maintained (26). Therefore, H2 consumption by hydrogenotrophic methanogens could promote the degradation of organic matter in anoxic environments and maintain the balance between H2 production and consumption, a balance essential for the continuous anaerobic degradation of organic compounds. In this context, the ability of methanogens to deal with fluctuating H2 availability is important for methanogenic ecosystems.
The methanogenesis pathway of hydrogenotrophic methanogens has been extensively studied in the thermophile Methanothermobacter thermautotrophicus ΔH (formerly Methanobacterium thermoautotrophicum ΔH) (30). Methane formation in M. thermautotrophicus ΔH occurs by the reduction of CO2 with H2 as an electron donor via seven reaction steps by the methanogenesis pathway. In this pathway, several sets of isofunctional enzymes are involved in the same reaction steps. For example, two formylmethanofuran dehydrogenases (FWD and FMD), F420-dependent and H2-dependent N5,N10-methylene tetrahydromethanopterin dehydrogenases (MTD and HMD), and methyl coenzyme M reductases (MCRI and MCRII) are involved in the first, fourth, and final steps of the reaction, respectively (25). In addition, at least two types of hydrogenases are known to be present in Methanothermobacter species (1, 6, 24, 33). The actual reason why M. thermautotrophicus ΔH retains more than one enzyme responsible for the same reaction step of the methanogenesis pathway on its small genome (1.75 Mbp; reference 29) has not yet been adequately addressed; however, gene expression of these enzymes is known to depend on growth conditions such as medium composition, growth phase, temperature, gassing rate (H2 in the input gas or the impeller speed of the fermentor), and syntrophic growth (16, 17, 19, 20, 22, 25).
Among methanogenesis-related enzymes, MCRI and MCRII, encoded by the operons mcrBDCGA and mrtBDGA, alter their expression levels depending particularly on H2 availability. Briefly, MCRI is predominantly expressed under H2-limited conditions provided by, for instance, a low gassing rate or impeller speed, whereas MCRII is dominated when H2 is sufficiently supplied by means of a high rate of gassing or stirring (17, 19). On the other hand, enzymatic differences in MCRs have been documented, such as the facts that MCRI is superior to MCRII in substrate specificity (Km) while MCRII has a higher maximum turnover rate (Vmax) than MCRI (5).
Considering the kinetic features of MCRs, their manner of expression, depending on H2 availability, is reasonable from the aspect of efficient use of fluctuating H2 in a natural environment; however, regardless of the drastic expression change of MCRs, their regulatory scheme is still unclear and nothing is known about their relevant transcriptional factors. In addition, the H2-sensing mechanism of M. thermautotrophicus ΔΗ has not been elucidated, although it is proposed that H2 availability sensing somehow occurs directly or indirectly. Information about transcriptional factors directly regulating the gene expression of MCRs would contribute to a better understanding of the regulatory scheme of the methanogenesis pathway and also provide important clues to clarify the H2-sensing mechanism of hydrogenotrophic methanogens.
In the present study, we attempted to identify a candidate for the transcriptional regulator that directs the gene expression of MCRs by using cells predominantly expressing either MCRI or MCRII, and we found a protein with DNA-binding activity specific to the promoter region of the mcr operon, implying its gene-specific transcriptional regulatory function. Furthermore, we discuss the H2-sensing mechanism of M. thermautotrophicus ΔΗ based on the structural feature of this candidate protein.
MATERIALS AND METHODS
Organism and growth conditions.
M. thermautotrophicus ΔH (DSM1053) was purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). M. thermautotrophicus ΔH was grown in a 1.3-liter serum bottle containing the minimal salts medium described previously (16) under an atmosphere of H2-CO2 (80:20 [vol/vol]) at 0.2 MPa. The medium was reduced by adding 1% (vol/vol) Na2S-9H2O and cysteine-HCl solutions (0.3 g liter−1 [final concentration]) before inoculation. All cultivations were carried out at 55°C, and stirring was done with a 40-mm-long Teflon-coated stirrer bar at 700 rpm.
Measurements of hydrogen and methane.
Hydrogen and methane in the headspace of serum bottles were determined with a gas chromatograph (GC8-AIT; Shimadzu, Kyoto, Japan) equipped with a 60/80 mesh column (Unibeads C; Shimadzu) and a thermal conductivity detector. Argon was used as the carrier gas. The column and detector temperatures were kept at 145 and 150°C, respectively.
Northern blot analysis.
DNA probes used for Northern blot analysis were prepared from DNA fragments amplified by PCR. Four milliliters of a full-growth culture of M. thermautotrophicus ΔH was harvested by centrifugation at 8,000 × g and 4°C for 10 min. The harvested cells were suspended in 0.1 ml of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA), and then the suspension was processed with a bead-beating device (Fast-Prep; Bio 101, Irvine, CA) at maximum speed for 1 min twice. Genomic DNA was prepared from the resulting lysate with a Fast DNA preparation kit (Bio 101) according to the manufacturer's protocol, followed by phenol-chloroform extraction. DNA probes were designed to target mRNAs encoding the subunits of MCR isozymes (mcrC and mrtD). Partial sequences of target genes were amplified with the primer sets listed in Table 1. PCR amplification was performed with a 50-μl reaction mixture (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2, 50 mM each deoxynucleoside triphosphate, 10 pmol of each primer, 2.5 U of Taq DNA polymerase [AmpliTaq Gold DNA polymerase; Applied Biosystems, Foster City, CA]) under the following temperature cycling conditions: initial denaturation at 96°C for 9 min; 35 cycles of 96°C for 30 s, 52°C for 30 s, and 72°C for 1.5 min; and a final extension at 72°C for 10 min. The PCR products were labeled with a PCR DIG Probe synthesis kit (Roche Diagnostics, Basel, Switzerland) according to the manufacturer's instructions.
TABLE 1.
Synthetic oligonucleotides used in this study as primers or probes
| Name | Sequence (5′-3′) | Experiment(s)a |
|---|---|---|
| mcrCf | ATGATCGGAAAGTGCACT | Northern |
| mcrCr | TCATGCACCTCCTAATGC | Northern |
| mrtDf | ATGATGTCAGAAACAGGA | Northern |
| mrtD52r | CCGATTACCCTTGGATC | Northern |
| 1132-01f | CAGATGCAGGATTCCAGTTA | EMSA |
| 1132-01r | CCGCTTATGCTTCAGTTTTTT | EMSA |
| 1132-02b | GCATAAGCGGTTATATTCAGATTAATACTGGATAATAACGAGATGTAC | EMSA |
| 1132-03f | CGAGATGTACAGTATGGCTT | EMSA |
| 1132-03r | ACTGATGGGGTTATGGGATT | EMSA |
| 1132-04b | CCCCATCAGTTTTATTAATAAAATAGTAAATTTTATTAATAAATAAATAAAACAAGAGGT | EMSA |
| 1132-05b | CGAGATGTACAGTATGGCTTGTTCTATAATGAGCTGTGCTCTGGAG | EMSA |
| 1132-06b | TGCTCTGGAGGTTGGAGGATGAACTTCGGTATAACCTGAAGGTCCATTTCG | EMSA |
| 1132-07b | GTCCATTTCGAGACATTGGTGATGGATAGCTCCTTCATGAGGTGACCATTT | EMSA |
| 1132-08b | GTGACCATTTCCATGGATTATCGCTGGCAATCCCATAACCCCATCAGT | EMSA |
| 1132-09rc | AAATGGTCACCTATGAAGGAG | EMSA |
| 1132-10rc | CGAAATGGACCTTCAGGTTAT | EMSA |
| 1132-11b | TAATGAGCTGTGCTCTGGAGGTTGGAGGATGAACTTCGGT | EMSA |
| 1132-12b | GAACTTCGGTATAACCTGAAGGTCCATTTCGAGACATTGGT | EMSA, affinity |
| 1168-01f | GTCACTCTCTCTGTGGAGA | EMSA |
| 1168-01r | AGTATCATCAGCAGGGAGTA | EMSA |
| 1168-02f | TGATGATACTCTGCTTTGGG | EMSA |
| 1168-02r | TCTGAATCAAGTCGAAAGCTT | EMSA |
| 1168-03b | TTGATTCAGAAAGAAATCTTAATTAATTATAATCAAGTTAAATATGCATCGTATACTAA | EMSA |
| 1168-04f | GTATACTAAGACGGGTGTTTT | EMSA |
| 1168-04r | TTCCGTATACTAAACATCCAG | EMSA |
| 1168-05b | TTGATTCAGAAAGAAATCTTAATTAATTATAATCAAGTTAAATATG | EMSA, affinity |
| 1168-06b | CTTAATTAATTATAATCAAGTTAAATATGCATCGTATACTAAG | EMSA |
| 1168-07b | AGAAAGAAATCTTAATTAATTATAATCAAG | EMSA |
| 1168-08b | AGATTCTAATCTTAATTAATTATAATCAAG | EMSA |
| 1168-11b | AGAAAGAGGTCTTAATTAATTATAATCAAG | EMSA |
| 1168-12b | AGAAAGAAATCTTGGTTAATTATAATCAAG | EMSA |
| 1168-13b | AGAAAGAAATCTTAATTGGTTATAATCAAG | EMSA |
| 1168-14b | AGAAAGAAATCTTAATTAATTATGGTCAAG | EMSA |
| 1168-15b | AGAAAGAAATCTTAATTAATTATAATCGGG | EMSA |
| 126exp10f | ATTCATAATACATATGCATAATGGATCTGCAGGTGd | Expression |
| 126exp10r | GTTAAAAATAGGATCCTCATTCAAGTCCGGCGATGCe | Expression |
Northern, Northern blot analysis; affinity, affinity particle purification of DNA-binding proteins; expression, cloning and expression of IMPDH VII.
Oligonucleotides were annealed with their complement counterparts to prepare double-stranded EMSA probes.
Oligonucleotide used for PCR with forward primer 1132-03f.
The underlined sequence is an NdeI site.
The underlined sequence is a BamHI site.
To evaluate mRNA expression of the mcr and mrt operons under different culture volume conditions, total RNA was prepared from cultures grown with various culture volumes (100, 200, 300, 400, and 500 ml). The cultures were pregrown as described above until the optical density (600 nm) reached 0.1. The headspace of the pregrown culture was replaced by 30 min of gassing with H2-CO2 (80:20 [vol/vol]) and filled with the same gas mixture at 0.2 MPa. The headspace-refreshed culture was harvested after 2 h of continued cultivation with stirring at 700 rpm and 55°C. Total RNA preparation and subsequent Northern blot analysis were carried out according to the procedure described previously (16).
Preparation of crude protein extracts.
Crude protein extracts were prepared from 100- and 500-ml cultures grown by the same procedure as that used for RNA preparation. Cells were harvested by centrifugation at 8,000 × g and 4°C for 10 min and then suspended in 3.5 ml of 10 mM HEPES (pH 8.0) containing 5 mM dithiothreitol (DTT) and 5 μg ml−1 protease inhibitor mixture for bacteria (Wako Pure Chemical, Osaka, Japan). The cell suspension was passed three times through a French pressure cell (FA-003; Thermo Fisher Scientific, Waltham, MA) at about 138 MPa. The resulting lysate was centrifuged at 20,000 × g and 4°C for 20 min with an ultracentrifuge (L-70 ultracentrifuge with a type 70 Ti rotor; Beckman Coulter, Fullerton, CA). The supernatant solution was dispensed into 1.5-ml microtubes and stored at −20°C until use. The protein concentration of the solution was determined by the Folin method with a DC protein assay kit according to the procedure used with thiols (Bio-Rad Laboratories, Hercules, CA).
EMSA.
Electrophoretic mobility shift assay (EMSA) was performed with digoxigenin (DIG)-labeled PCR fragments or synthetic oligonucleotides as probes. For the probe regions and primer sets for making probes, see Fig. 3 and Table 1, respectively. Nucleotide probes longer than 60 bp were prepared from DNA fragments amplified by PCR from the M. thermautotrophicus ΔH genome. The PCR mixture was subjected to a subsequent probe-labeling reaction without further purification. Nucleotide probes less than 61 bp in length were made from synthetic sense and antisense oligonucleotides purchased from the manufacturer (Tsukuba Oligo Service, Tsukuba, Japan). The oligonucleotides were annealed by 30 min of slow cooling to 4°C after boiling for 3 min. The amplified DNA fragments and annealed synthetic oligonucleotides were labeled with DIG with a DIG Gel Shift Kit, 2nd generation (Roche Diagnostics), according to the manufacturer's protocol.
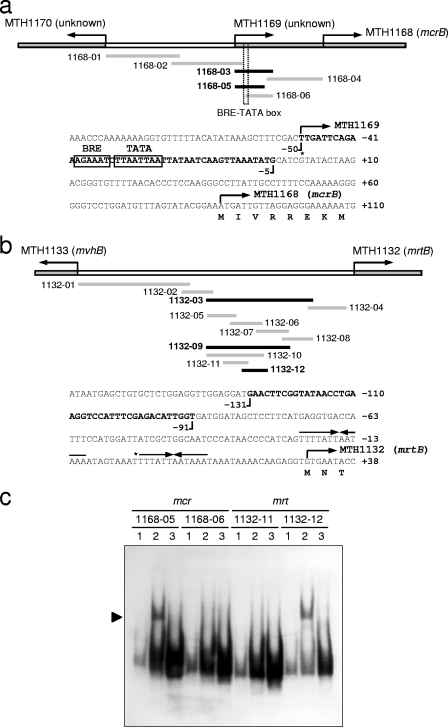
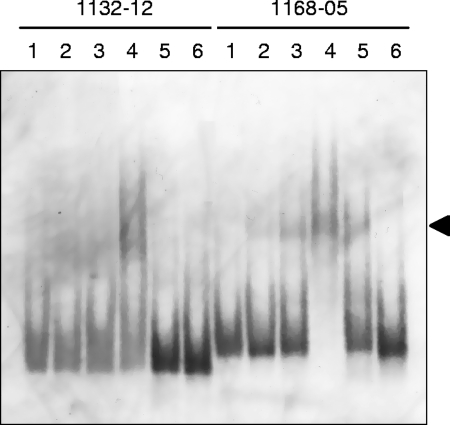
FIG. 3.
Survey for operator sites of the mcr and mrt operons by EMSA. The probes used in the survey and the sequence structures of putative operator sites of the mcr (a) and mrt (b) upstream regions are shown. Black and gray lines show the regions of shifted and nonshifted probes, respectively. The sequences in bold are the final restricted regions expected to contain operator sites. Transcriptional start sites and palindromic sequences previously reported (22) are represented by asterisks and arrows, respectively. The TATA box and the BRE are boxed. Electropherogram of an EMSA with fragmented upstream regions of the mcr and mrt operons (c). Lane 1, unbound probes (60 fg); lane 2, probes reacted with protein extract prepared from an mrt-expressing culture (20 μg); lane 3, probes reacted with protein extract and unlabeled probes (60 pg). All reactions were performed at 4°C for 2 h. Mobility shifts are clearly observed in reaction mixtures with probes 1168-05 and 1132-12, as indicated by the arrowhead.
EMSA was also performed with the DIG Gel Shift Kit, 2nd generation (Roche Diagnostics), essentially according to the manufacturer's instructions, unless otherwise stated. Extracted or purified protein was incubated with 45 or 60 fg of end-labeled double-stranded DNA fragments in a final volume of 25 μl. Incubations were carried out at 4°C for 2 h in a solution of 10 mM Tris-HCl (pH 7.6), 50 mM NaCl, 5% (vol/vol) glycerol, 1 mM EDTA, and 1 mM DTT. The reaction mixtures were loaded onto low-ionic-strength 5% (wt/vol) polyacrylamide gels (acrylamide-to-bisacrylamide weight ratio of 37.5:1) that were preelectrophoresed at 100 V for 1 h in 1× Tris-borate-EDTA buffer consisting of 8.9 mM Tris-HCl (pH 8.0), 8.9 mM boric acid, and 2 mM EDTA. Polyacrylamide gels were electrophoresed at 100 V at ambient temperature until bromophenol blue reached the bottom. The probes were transferred onto a positively charged nylon membrane (Hybond-N+; GE Healthcare, Uppsala, Sweden) with a blotting apparatus (Trans-Blot SD semidry transfer cell, Bio-Rad, Hercules, CA) and then detected according to the kit protocol.
Purification of DNA-binding protein with affinity particles.
DNA-binding proteins were purified with magnetic affinity particles conjugated with the putative mcr and mrt operator regions. DNA fragments harboring the putative regulatory sites were prepared by annealing sense and antisense strands of synthetic oligonucleotides as described for the preparation of EMSA probes; however, the 3′ end of the annealed oligonucleotides was phosphorylated and then concatenated by overnight ligation at 16°C before affinity particle preparation. The concatenated nucleotides were conjugated with magnetic particles with a DNA-Binding Protein Purification Kit (Roche Diagnostics). One-half milligram of magnetic affinity particles was reacted with 80 μg of crude protein extracts, which were prepared from 100-ml cultures at ambient temperature for 2 h. To collect a detectable amount of DNA-binding proteins, the same purification process was repeated 10 times and 0.8 mg of total proteins was subjected to affinity purification. The collected proteins were concentrated by acetone precipitation and then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein bands stained with Coomassie brilliant blue were excised and subsequently subjected to protein identification by peptide mass fingerprinting (PMF).
Identification of proteins by PMF.
Tryptic in-gel digestion of the protein band was carried out according to a previously published procedure (13). Protein bands excised from acrylamide gel were destained by one charge of 50% acetonitrile and 25 mM ammonium bicarbonate for 10 min with vigorous shaking; this mixture was subsequently removed. After destaining, 10 mM DTT in 100 mM ammonium bicarbonate was added and proteins were reduced for 1 h at 56°C. After cooling to room temperature, the solution was replaced with the same volume of 55 mM iodoacetamide in 100 mM ammonium bicarbonate. After 45 min incubation at room temperature in the dark with occasional vortexing, the gel pieces were washed with 100 mM ammonium bicarbonate for 10 min; they were subsequently washed several times with 50% methanol-10% acetic acid with vigorous shaking. The liquid phase was removed, and the gel pieces were soaked in 100 mM ammonium bicarbonate for 5 min with shaking. The mixture was then shaken with 100% acetonitrile for 5 min and completely dried in a SpeedVac evaporator (CVE-100D; EYELA, Tokyo, Japan). The dried gel pieces were swollen on ice for 20 min in a trypsin solution composed of 100 mM ammonium bicarbonate, 0.1% (vol/vol) n-octyl glucoside, and 0.1 μg μl−1 modified trypsin (Promega, Madison, WI). To the gels was added a small amount of the same buffer without trypsin, and the gels were incubated at 37°C for 4 h. The digested peptides were extracted with 75% acetonitrile in 0.1% trifluoroacetic acid (TFA). The extracts were concentrated in a SpeedVac evaporator.
The resulting solution was subjected to matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry analysis. A saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile-0.1% TFA and a solution of 2,5-dihydroxybenzoic acid in 33% acetonitrile-0.1% TFA (40 mg ml−1) were prepared as the matrix solutions. Either α-cyano-4-hydroxycinnamic acid or 2,5-dihydroxybenzoic acid solution was mixed with an equal volume (0.5 μl) of the peptide solution and spotted onto a MALDI target plate. MALDI-TOF mass spectrometry analyses were carried out with a Voyager DE-PRO TOF mass spectrometer operating in the positive reflectron mode (Applied Biosystems, Foster City, CA). Internal mass calibration was performed by using trypsin autolysis peaks (m/z 842.5099, 2,211.1045, and 2,807.3145). The peptide mass list was searched with the Mascot search engine (http://www.matrixscience.com/) against the NCBI nonredundant database.
Cloning, expression, and purification of DNA-binding protein.
Recombinant IMPDH VII was prepared with a pET expression system. The gene encoding IMPDH VII, MTH126, was amplified by PCR with primers 126exp10f and 126exp10r (Table 1). PCR was performed by the same procedure as that used for making the probes for the Northern blot analysis, except that the annealing temperature was 60°C. The PCR product was cloned into plasmid vector pCR2.1 with a TOPO TA cloning kit (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The cloned gene was cut out from the plasmid vector by digestion with NdeI and BamHI at 37°C for 2 h. The resulting digests were purified by electrophoresis on 1% agarose gel, followed by extraction with a QIAquick gel extraction kit (Qiagen, Hilden, Germany). The purified fragments were cloned into expression vector pET19b (EMD Chemicals, Gibbstown, NJ), and they were introduced into Escherichia coli BL21-CodonPlus(DE3)-RIL (Stratagene, La Jolla, CA). Protein expression of the transformants was induced by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the cells were harvested after 3 h of cultivation. Protein preparation was carried out by the same procedure as that used for the preparation of crude protein extracts of M. thermautotrophicus ΔH. His-tagged protein was purified in an Ni-Sepharose column (HisTrap HP kit; GE Healthcare) in accordance with the manufacturer's instructions. The N-terminal His tag was eliminated by enterokinase cleavage (Enterokinase cleavage capture kit; EMD Chemicals). The protein concentration was quantified by the Folin method, as used for the preparation of crude protein extracts.
RESULTS
Growth and gene expression of M. thermautotrophicus ΔH grown in various culture volumes.
M. thermautotrophicus ΔH was grown in a batch culture in a 1.3-liter glass vial filled with 0.2 MPa of H2-CO2 (80:20 [vol/vol]). To control H2 availability to the methanogen cells, we examined the batch culture volumes to alter the efficiency of gas diffusion into the culture. For this, the methanogen cells were pregrown at 55°C with stirring at 700 rpm to a cell optical density of 0.1 with different culture volumes ranging from 100 to 500 ml, and the cultures were further cultivated for 2 h after refreshing the headspace gas with H2-CO2 (80:20 [vol/vol]) at 0.2 MPa. In this case, gas diffusion into the medium would be limited, depending on the culture volume, if other factors such as cell density, gas composition and pressure, agitation, and temperature were mostly kept constant.
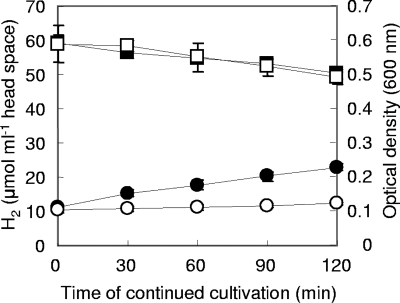
The time course of growth and H2 consumption in 100- and 500-ml cultures is shown in Fig. 1 as an example of the growth profiles obtained with cultures grown in different volumes. Hydrogen in the headspace of both cultures decreased at similar constant rates, which could be approximated by linear equations that yield H2 consumption rates of 310.1 and 247.4 nmol min−1 for 100- and 500-ml cultures, respectively. On the other hand, the average wet weights of cells harvested from 100- and 500-ml cultures after 2 h of continued cultivation were 71.1 and 125.3 mg, respectively. Accordingly, the H2 consumption rates per unit of cell wet weight of 100- and 500-ml cultures at this time were 4.3 and 2.0 μmol g−1 min−1, respectively, indicating that cells grown in 100-ml cultures consumed H2 about 2.2-fold faster than those grown in 500-ml cultures. The difference in the H2 consumption rate between cultures seemed to result from the different H2 availability in each culture.
FIG. 1.
Time course of optical density (circles) and H2 concentration (squares) during 2 h of continued cultivation of M. thermautotrophicus ΔH in 100-ml (closed) and 500-ml (open) cultures. The optical density (600 nm) of the cultures at the starting point (0 min) was 0.1, and the headspace was refreshed with H2-CO2 (80:20) at 0.2 MPa before continued cultivation. Data were obtained from triplicate cultures, and error bars indicate standard deviations. Detailed cultivation conditions are described in Materials and Methods.
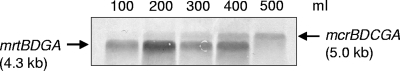
The gene expression of the mcr and mrt operons in cells grown in various culture volumes was evaluated by Northern blot analysis simultaneously with each transcript-specific probe. As shown in Fig. 2, a 5.0-kb mRNA fragment of the mcr operon increased gradually along with an increase in the culture volume, while 4.3 kb of the mrt operon was firmly expressed in all of the cultures, except for the 500-ml culture. Northern blot analysis also indicated that the cells grown in a 100-ml culture predominantly expressed the mrt operon, and conversely, those retrieved from the 500-ml culture exclusively expressed the mcr operon. In the following studies, 100- and 500-ml cultures were used as mrt-expressing (mcr-repressed) and mcr-expressing (mrt-repressed) cultures, respectively. In addition, the time required to reach an optical density of 0.1 was the same, about 12 h for both cultures, which suggested that the cells in both cultures grew under comparable H2 availability conditions until that time. Therefore, the difference in MCR expression was likely due to the 2 h of continued cultivation.
FIG. 2.
Northern blot analysis of transcripts from the mcr and mrt operons of cells grown in different culture volumes (100 to 500 ml). Twenty micrograms of total RNA was used for the analysis. Transcripts were simultaneously detected with each operon-specific probe prepared with the primers listed in Table 1. Only the cells grown in 100- and 500-ml cultures were used for the following studies as mrt-expressing (mcr-repressed) and mcr-expressing (mrt-repressed) cultures, respectively.
Survey of the operator regions of the mcr and mrt operons.
The operator regions of the mcr and mrt operons were evaluated by EMSA with crude protein extracts prepared from cultures expressing both mcr and mrt. Operator regions are expected to interact with any transcriptional factors when relevant genes are being expressed and/or repressed. In the present study, operator regions were surveyed by EMSA with various fragments derived from the upstream regions of each gene, as shown in Fig. 3. As a result, distinct mobility retardation patterns were observed in both the mcr and mrt upstream regions when proteins extracted from mrt-expressing cultures were examined. In contrast to mrt-expressing cultures, protein extracts prepared from mcr-expressing cultures showed no or weak mobility shifts when the same probe sets were used (data not shown). These results indicated that some protein factors with DNA-binding activity in the cell extracts of mrt-expressing cultures are associated with their upstream regions, which were deduced to be putative operator regions of the mcr and mrt operons. EMSA with various probes showed that the putative operator sites for the mcr and mrt operons located from base position −5 to base position −50 (46 bp long; probe 1168-05) and from base position −91 to base position −131 (41 bp long; probe 1132-12), respectively (Fig. 3a and b). Figure 3c shows the electropherogram of an EMSA with probes 1168-05 and 1132-12 for the mcr and mrt operons, respectively. In the putative operator site for the mcr operon, a potential factor B-responsible element (BRE)-TATA box sequence was found in the middle (22). On the other hand, no typical promoter sequence was identified in the putative mrt operator region.
Purification of DNA-binding proteins.
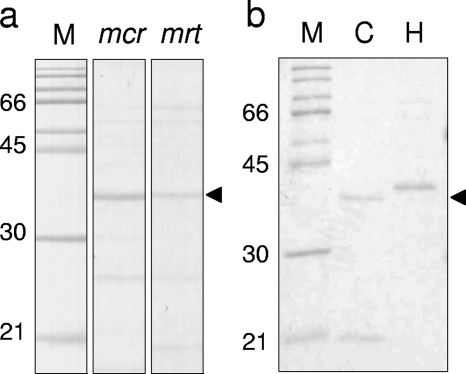
Purification of DNA-binding proteins was attempted by using affinity particles conjugated with the putative mcr and mrt operator regions (1132-12 and 1168-05, respectively, shown in Table 1). Crude proteins prepared from mrt-expressing cultures were used for the purification process, in which 0.8 mg crude protein in total was employed. The resulting protein solutions, further purified by acetone precipitation, were separated by SDS-PAGE as shown in Fig. 4. On the Coomassie brilliant blue-stained gel, a single protein band with an estimated molecular mass of about 32 kDa appeared in both reaction mixtures, each of which employed one of the putative operator regions; however, the band intensity of the protein obtained from the mrt operator region was relatively weak compared with that for the reaction mixture with the mcr operator site.
FIG. 4.
DNA-binding protein purified from mrt-expressing cultures and its recombinant protein. (a) SDS-PAGE of purified proteins with affinity particles conjugated with the putative operator regions of the mcr and mrt operons. (b) SDS-PAGE of His-tagged (H) and His-tagged and cleaved (C) recombinant IMPDH VII (0.5 μg each). M indicates the molecular mass standard. Gels were stained with Coomassie brilliant blue. The molecular mass of the purified protein is consistent with that of recombinant IMPDH VII (32.4 kDa).
The 32-kDa protein bands and some other weak bands were excised from the gels, which were then subjected to protein identification by PMF. As a result, only two dense peptide bands with a molecular mass of 32 kDa could be assigned. The other weak protein bands could not be clarified, probably because of the insufficient amount of protein. Seven and 11 peptide fragments of trypsin digests of the 32-kDa proteins retrieved from the mcr and mrt operator sites agreed with in silico-digested peptides of a protein encoded by MTH126 in the M. thermautotrophicus ΔH genome (40 and 30% coverage, 50 and 30 ppm mass tolerance, respectively). This protein is annotated as IMP dehydrogenase-related protein VII (IMPDH VII) in the original database (29) and in the Kyoto Encyclopedia of Genes and Genomes (12). The protein is predicted to have a winged helix-turn-helix-type DNA-binding motif at the N terminus and two cystathionine β-synthase (CBS) domains on opposite sides. The CBS domain is widely found in all domains of life; however, its function is still unclear.
DNA-binding activity of IMPDH VII at the putative mcr operator region.
The DNA-binding activity of IMPDH VII at the putative mcr and mrt operator regions was evaluated by EMSA with its recombinant protein. The IMPDH VII coding sequence, MTH126, was cloned and expressed in E. coli cells with the pET system. After Ni-Sepharose purification and enterokinase cleavage, the purified recombinant IMPDH VII protein was subjected to EMSA with the putative mcr and mrt operator regions. As a result, recombinant IMPDH VII was revealed to be associated with the putative mcr operator region, as shown in Fig. 5 (1168-05, lanes 2 to 5). The DNA-binding activity of IMPDH VII at the mcr operator site seemed to be strong, because the shifted band was found even in the presence of a nonspecific competitor [1 μg of poly(dI-dC), lane 5 of 1168-05]. On the other hand, DNA binding at the mrt operator region was observed only when a relatively high protein concentration (2 μg) was used (lane 4 of 1132-12); furthermore, this mobility shift disappeared in the presence of 1 μg of nonspecific competitors (lane 5 of 1132-12), and even at lower concentrations [0.1 μg of poly(dI-dC); data not shown]. Therefore, we concluded that the interaction between IMPDH VII and the putative mrt operator site was a nonspecific, sequence-independent reaction.
FIG. 5.
Specific binding of IMPDH VII to the putative mcr and mrt operator regions. Lane 1, free probe; lanes 2 to 4, 0.5, 1, and 2 μg of IMPDH VII, respectively; lane 5, 2 μg of IMPDH VII and 1 μg of poly(dI-dC); lane 6, 2 μg of IMPDH VII and 60 pg of unlabeled probes. Forty-five femtograms of DIG-labeled probes was used for the reactions. All reactions were carried out at 4°C for 2 h.
Binding site of IMPDH VII in the putative mcr promoter region.
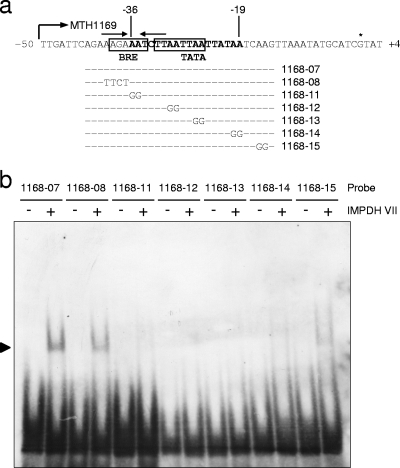
To specify the binding site of IMPDH VII in the mcr upstream region in more detail, we surveyed the sequence region important for this interaction with probes with unnatural sequences, as indicated in Fig. 6a. In this region, the TATA box sequence and BRE, which was identified by a consensus sequence of RNWAAW reported in Sulfolobus species (2), and one short palindromic sequence were found. Therefore, of the seven probes examined, one was used for a positive control with the original sequence (1168-07), one was designed to break the palindrome formation (1168-08), and the remaining five (1168-11 to -15) were constructed to alter the double adenine sequences located regularly in this region (two- and four-base intervals). As a result of evaluations with recombinant IMPDH VII, mobility shift was hindered by altering the double adenine sequences in the region from base position −19 to base position −36, as shown in Fig. 6. On the other hand, the loss of palindrome formation did not affect IMPDH VII binding to this region. These results indicated that the binding site of IMPDH VII mostly overlaps the BRE-TATA box of the mcr promoter region, also suggesting its possible function of interfering with the formation of the preinitiation complex.
FIG. 6.
Influence of base changes in the putative mcr operator region to unnatural sequences. (a) Probes and their corresponding sites in the putative mcr operator region. The BRE, TATA box, and transcriptional start site are indicated by the boxes and the asterisk, respectively. Palindromic sequences are indicated by arrows. (b) Electropherogram of an EMSA with probes harboring unnatural sequences. A minus or a plus sign indicates the absence or presence, respectively, of recombinant IMPDH VII. All EMSA reactions were carried out at 4°C for 2 h.
DISCUSSION
In the present study, cis elements and trans-acting factors of the genes for MCRs of the hydrogenotrophic methanogen M. thermautotrophicus ΔH were investigated and IMPDH VII, encoded by MTH126, was found to bind specifically to the mcr promoter region. By combining the results obtained in the experiments with crude protein extracts, IMPDH VII was speculated to function as a repressor of the mcr operon. The present study is the first report to present a candidate mcr regulator that evidently interacted directly with its promoter region.
Gene and protein expressions of methanogenesis-related enzymes have been examined to some extent in several studies, and it has been reported that H2 availability is a significant factor for directing the expression of mcr genes and other genes (16, 17, 19, 21, 22); however, almost nothing is known about the mechanism of H2 availability sensing, the existence of which has been speculated in the past (25). We therefore assumed that the identification of cis elements (operator sites) and trans-acting factors (transcriptional regulators) of regulated genes should provide clues to elucidate the mechanism of H2 sensing and its downstream gene regulation.
Upon starting this study, we prepared cells expressing either MCRI or MCRII by using a simple batch culture experiment, although most studies on the H2 response of methanogen cells have been performed with anaerobic reactors (17-19). In our batch culture experiments, precultivation and headspace refreshment enabled the creation of cultures with similar conditions of cell density and H2 partial pressure at the start point. In this case, a change in the culture volume led to a distinct total cell amount. Since H2 flow from the gas phase (headspace) to the liquid phase (medium) is basically defined by the proportion of medium surface area and the number of H2-consuming cells, different culture volumes should provide different levels of H2 availability to methanogen cells. Needless to say, controlling gas diffusion into a liquid culture should be applied to carbon dioxide, and this would affect the buffer effect of the medium. However, since the solubility of hydrogen (0.0175 liter liter water−1 at 25°C) is significantly lower than that of carbon dioxide (1.0535 liters liter water−1), the culture volume would have a greater influence on the hydrogen concentration. Moreover, considering the manner of expression of MCRs depending on H2 availability reported to date, the clear difference in mRNA expression in our studies indicated that H2 availability for methanogen cells is obviously regulated by the batch culture volume (Fig. 2). As described, this modified batch cultivation technique is simple and reliable, and changes in the gene expression of MCRs were successfully and repeatedly achieved.
A survey of operator regions by EMSA with protein extracts from mrt-expressing cultures prepared by batch cultivation resulted in narrowing down the regions to 46- and 41-bp lengths for the mcr and mrt operons, respectively. Within the putative mcr operator regions, an obvious BRE-TATA box sequence was located in the middle, suggesting its regulatory function for the mcr operon. Further EMSA with recombinant IMPDH VII also showed that the region neighboring the BRE-TATA box is essential for the interaction between IMPDH VII and the mcr upstream region. Considering that mobility retardation patterns were obtained from mrt-expressing (mcr-repressed) cultures but not mcr-expressing cells, it was speculated that IMPDH VII works as a transcriptional repressor by interfering with the formation of the preinitiation complex, which consists of the TATA-binding protein, transcriptional factor B, and RNA polymerase (23).
H2 sensing and its signal transduction system in prokaryotic cells have been extensively studied in the lithoautotrophic bacterium Ralstonia eutropha. In this bacterium, H2 directly interacts with a signal-sensing cytoplasmic [Ni-Fe]-hydrogenase and its signal is transmitted to a two-component signal transduction system consisting of a sensor kinase and a response regulator. This results in the expression of energy-generating hydrogenases (3, 15); however, such a complex and the sophisticated direct H2-sensing mechanism do not seem to be essentially applicable to all hydrogenotrophic microbes. In contrast to R. eutropha, which is also capable of heterotrophic growth, in strictly hydrogenotrophic species like M. thermautotrophicus ΔH, whose energy source is highly restricted, fluctuating H2 availability could influence not only methanogenesis but also the cellular energy status and other metabolisms. This suggests that small intracellular compounds could be good indicators of H2 availability. In earlier reports, cofactors such as F420 or F390, or C1 intermediates have been speculated to be molecules that signal the cellular energy status of M. thermautotrophicus ΔH (17, 19, 21, 22, 25, 31).
IMPDH VII is a functionally uncharacterized protein that possesses one winged helix-turn-helix-type DNA-binding domain and a pair of CBS domains. Such HTH-type DNA-binding proteins accompanied by CBS domains are found only in the archaeal domain, and their functions have not been assigned. However, CBS domains are widely distributed in a variety of proteins in all domains of life. Because mutations in the CBS domain cause several human hereditary diseases, CBS (28), IMPDH (14), AMP-activated protein kinase (4), and chloride channels (9) have been well investigated. It was recently revealed that CBS domains are associated with adenosyl-containing ligands such as AMP, ATP, or S-adenosylmethionine, and there has been speculation about their sensory role in intracellular metabolites (11). In the present study, the effect of adenosyl derivatives (AMP, ADP, ATP, and S-adenosylmethionine) and several nucleotides (UTP, GTP, CTP, and IMP) on the DNA-binding activity of IMPDH VII was examined by EMSA. However, unfortunately, no influence of these compounds was found in our mobility shift assay (data not shown).
Although ATP binding to CBS domains in IMPDH VII was not evident in our study and the intracellular ATP level of M. thermautotrophicus has not been reported, an arginine residue corresponding to base position 224 of human IMPDH II, which is indispensable for ATP binding (27), was conserved in IMPDH VII (data not shown). This structural feature implied the possible interaction between IMPDH VII and ATP. In addition, this conserved arginine residue was also found in IMPDH VII homologs, the hypothetical proteins encoded by MJ1232 and MMP0052 of Methanothermobacter jannaschii (7) and Methanocaldococcus maripaludis (formerly Methanococcus maripaludis) (10), respectively. This suggests that these homologs have similar functions in such hydrogenotrophic methanogens.
Our results and recent findings concerning CBS domains led to the hypothesis that IMPDH VII functions as an intracellular energy status sensor by sensing adenosyl derivatives, although the key signaling molecule remains to be clarified. To confirm the function of IMPDH VII in gene regulation, its transcriptional regulatory activity must be examined with a transcription system. At present, the only available platform for M. thermautotrophicus ΔH is an in vitro transcription system consisting of TATA-binding protein, transcriptional factor B, and RNA polymerase (8), since genetic tools have not been developed for this methanogen. Therefore, we are currently conducting further experiments to study this topic. We are also surveying the genes regulated by IMPDH VII. Further investigation of IMPDH VII could provide useful information that might clarify the gene regulation of M. thermautotrophicus ΔH based on H2 availability; this subject has not yet been adequately addressed.
Acknowledgments
We thank Tomoyuki Kosaka, Shun-ichi Ishii (National Institute of Advanced Industrial Science and Technology), and Kazuya Watanabe (Marine Biotechnology Institute) for valuable discussions and helpful advices. Thanks also to Yasuo Igarashi (The University of Tokyo) for continuous encouragement.
This work was supported by the New Energy and Industrial Technology Development Organization of Japan.
Footnotes
Published ahead of print on 29 August 2008.
REFERENCES
- 1.Alex, L. A., J. N. Reeve, W. H. Orme-Johnson, and C. T. Walsh. 1990. Cloning, sequence determination, and expression of the genes encoding the subunits of the nickel-containing 8-hydroxy-5-deazaflavin reducing hydrogenase from Methanobacterium thermoautotrophicum ΔH. Biochemistry 29:7237-7244. [DOI] [PubMed] [Google Scholar]
- 2.Bell, S. D., P. L. Kosa, P. B. Sigler, and S. P. Jackson. 1999. Orientation of the transcription preinitiation complex in archaea. Proc. Natl. Acad. Sci. USA 96:13662-13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernhard, M., T. Buhrke, B. Bleijlevens, A. L. De Lacey, V. M. Fernandez, S. P. Albracht, and B. Friedrich. 2001. The H2 sensor of Ralstonia eutropha. Biochemical characteristics, spectroscopic properties, and its interaction with a histidine protein kinase. J. Biol. Chem. 276:15592-15597. [DOI] [PubMed] [Google Scholar]
- 4.Blair, E., C. Redwood, H. Ashrafian, M. Oliveira, J. Broxholme, B. Kerr, A. Salmon, I. Östman-Smith, and H. Watkins. 2001. Mutations in the γ2 subunit of AMP-activated protein kinase cause familial hypertrophic cardiomyopathy: evidence for the central role of energy compromise in disease pathogenesis. Hum. Mol. Genet. 10:1215-1220. [DOI] [PubMed] [Google Scholar]
- 5.Bonacker, L. G., S. Baudner, E. Morschel, R. Bocher, and R. K. Thauer. 1993. Properties of the two isoenzymes of methyl-coenzyme M reductase in Methanobacterium thermoautotrophicum. Eur. J. Biochem. 217:587-595. [DOI] [PubMed] [Google Scholar]
- 6.Braks, I. J., M. Hoppert, S. Roge, and F. Mayer. 1994. Structural aspects and immunolocalization of the F420-reducing and non-F420-reducing hydrogenases from Methanobacterium thermoautotrophicum Marburg. J. Bacteriol. 176:7677-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. Geoghagen, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 8.Darcy, T. J., W. Hausner, D. E. Awery, A. M. Edwards, M. Thomm, and J. N. Reeve. 1999. Methanobacterium thermoautotrophicum RNA polymerase and transcription in vitro. J. Bacteriol. 181:4424-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estévez, R., M. Pusch, C. Ferrer-Costa, M. Orozco, and T. J. Jentsch. 2004. Functional and structural conservation of CBS domains from CLC chloride channels. J. Physiol. 557:363-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendrickson, E. L., R. Kaul, Y. Zhou, D. Bovee, P. Chapman, J. Chung, E. Conway de Macario, J. A. Dodsworth, W. Gillett, D. E. Graham, M. Hackett, A. K. Haydock, A. Kang, M. L. Land, R. Levy, T. J. Lie, T. A. Major, B. C. Moore, I. Porat, A. Palmeiri, G. Rouse, C. Saenphimmachak, D. Soll, S. Van Dien, T. Wang, W. B. Whitman, Q. Xia, Y. Zhang, F. W. Larimer, M. V. Olson, and J. A. Leigh. 2004. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 186:6956-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ignoul, S., and J. Eggermont. 2005. CBS domains: structure, function, and pathology in human proteins. Am. J. Physiol. Cell Physiol. 289:C1369-C1378. [DOI] [PubMed] [Google Scholar]
- 12.Kanehisa, M., and S. Goto. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katayama, H., T. Nagasu, and Y. Oda. 2001. Improvement of in-gel digestion protocol for peptide mass fingerprinting by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 15:1416-1421. [DOI] [PubMed] [Google Scholar]
- 14.Kennan, A., A. Aherne, A. Palfi, M. Humphries, A. McKee, A. Stitt, D. A. Simpson, K. Demtroder, T. Orntoft, C. Ayuso, P. F. Kenna, G. J. Farrar, and P. Humphries. 2002. Identification of an IMPDH1 mutation in autosomal dominant retinitis pigmentosa (RP10) revealed following comparative microarray analysis of transcripts derived from retinas of wild-type and Rho−/− mice. Hum. Mol. Genet. 11:547-557. [DOI] [PubMed] [Google Scholar]
- 15.Lenz, O., and B. Friedrich. 1998. A novel multicomponent regulatory system mediates H2 sensing in Alcaligenes eutrophus. Proc. Natl. Acad. Sci. USA 95:12474-12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo, H. W., H. Zhang, T. Suzuki, S. Hattori, and Y. Kamagata. 2002. Differential expression of methanogenesis genes of Methanothermobacter thermoautotrophicus (formerly Methanobacterium thermoautotrophicum) in pure culture and in cocultures with fatty acid-oxidizing syntrophs. Appl. Environ. Microbiol. 68:1173-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan, R. M., T. D. Pihl, J. Nolling, and J. N. Reeve. 1997. Hydrogen regulation of growth, growth yields, and methane gene transcription in Methanobacterium thermoautotrophicum ΔH. J. Bacteriol. 179:889-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukhopadhyay, B., E. F. Johnson, and R. S. Wolfe. 2000. A novel pH2 control on the expression of flagella in the hyperthermophilic strictly hydrogenotrophic methanarchaeaon Methanococcus jannaschii. Proc. Natl. Acad. Sci. USA 97:11522-11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nölling, J., T. D. Pihl, A. Vriesema, and J. N. Reeve. 1995. Organization and growth phase-dependent transcription of methane genes in two regions of the Methanobacterium thermoautotrophicum genome. J. Bacteriol. 177:2460-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pennings, J. L., J. L. de Wijs, J. T. Keltjens, and C. van der Drift. 1997. Medium-reductant directed expression of methyl coenzyme M reductase isoenzymes in Methanobacterium thermoautotrophicum (strain ΔH). FEBS Lett. 410:235-237. [DOI] [PubMed] [Google Scholar]
- 21.Pennings, J. L., J. T. Keltjens, and G. D. Vogels. 1998. Isolation and characterization of Methanobacterium thermoautotrophicum ΔH mutants unable to grow under hydrogen-deprived conditions. J. Bacteriol. 180:2676-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pihl, T. D., S. Sharma, and J. N. Reeve. 1994. Growth phase-dependent transcription of the genes that encode the two methyl coenzyme M reductase isoenzymes and N5-methyltetrahydromethanopterin:coenzyme M methyltransferase in Methanobacterium thermoautotrophicum ΔH. J. Bacteriol. 176:6384-6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qureshi, S. A., S. D. Bell, and S. P. Jackson. 1997. Factor requirements for transcription in the archaeon Sulfolobus shibatae. EMBO J. 16:2927-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reeve, J. N., G. S. Beckler, D. S. Cram, P. T. Hamilton, J. W. Brown, J. A. Krzycki, A. F. Kolodziej, L. Alex, W. H. Orme-Johnson, and C. T. Walsh. 1989. A hydrogenase-linked gene in Methanobacterium thermoautotrophicum strain ΔH encodes a polyferredoxin. Proc. Natl. Acad. Sci. USA 86:3031-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reeve, J. N., J. Nolling, R. M. Morgan, and D. R. Smith. 1997. Methanogenesis: genes, genomes, and who's on first? J. Bacteriol. 179:5975-5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schink, B. 1997. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 61:262-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott, J. W., S. A. Hawley, K. A. Green, M. Anis, G. Stewart, G. A. Scullion, D. G. Norman, and D. G. Hardie. 2004. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J. Clin. Investig. 113:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan, X., R. L. Dunbrack, Jr., S. A. Christopher, and W. D. Kruger. 2001. Mutations in the regulatory domain of cystathionine β-synthase can functionally suppress patient-derived mutations in cis. Hum. Mol. Genet. 10:635-643. [DOI] [PubMed] [Google Scholar]
- 29.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. McDougall, G. Shimer, A. Goyal, S. Pietrokovski, G. M. Church, C. J. Daniels, J.-I. Mao, P. Rice, J. Nölling, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thauer, R. K. 1998. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. 1998 Marjory Stephenson Prize lecture. Microbiology 144:2377-2406. [DOI] [PubMed] [Google Scholar]
- 31.Vermeij, P., E. Vinke, J. T. Keltjens, and C. Van der Drift. 1995. Purification and properties of coenzyme F390 hydrolase from Methanobacterium thermoautotrophicum (strain Marburg). Eur. J. Biochem. 234:592-597. [DOI] [PubMed] [Google Scholar]
- 32.Zinder, S. 2001. Ecology, physiology, biochemistry and genetics, p. 128-206. In J. Ferry (ed.), Methanogenesis. Chapman & Hall, New York, NY.
- 33.Zirngibl, C., W. Van Dongen, B. Schworer, R. Von Bunau, M. Richter, A. Klein, and R. K. Thauer. 1992. H2-forming methylenetetrahydromethanopterin dehydrogenase, a novel type of hydrogenase without iron-sulfur clusters in methanogenic archaea. Eur. J. Biochem. 208:511-520. [DOI] [PubMed] [Google Scholar]