Abstract
Motility is a critical function needed for nutrient acquisition, biofilm formation, and the avoidance of harmful chemicals and predators. Flagellar motility is one of the most pressure-sensitive cellular processes in mesophilic bacteria; therefore, it is ecologically relevant to determine how deep-sea microbes have adapted their motility systems for functionality at depth. In this study, the motility of the deep-sea piezophilic bacterium Photobacterium profundum SS9 was investigated and compared with that of the related shallow-water piezosensitive strain Photobacterium profundum 3TCK, as well as that of the well-studied piezosensitive bacterium Escherichia coli. The SS9 genome contains two flagellar gene clusters: a polar flagellum gene cluster (PF) and a putative lateral flagellum gene cluster (LF). In-frame deletions were constructed in the two flagellin genes located within the PF cluster (flaA and flaC), the one flagellin gene located within the LF cluster (flaB), a component of a putative sodium-driven flagellar motor (motA2), and a component of a putative proton-driven flagellar motor (motA1). SS9 PF flaA, flaC, and motA2 mutants were defective in motility under all conditions tested. In contrast, the flaB and motA1 mutants were defective only under conditions of high pressure and high viscosity. flaB and motA1 gene expression was strongly induced by elevated pressure plus increased viscosity. Direct swimming velocity measurements were obtained using a high-pressure microscopic chamber, where increases in pressure resulted in a striking decrease in swimming velocity for E. coli and a gradual reduction for 3TCK which proceeded up to 120 MPa, while SS9 increased swimming velocity at 30 MPa and maintained motility up to a maximum pressure of 150 MPa. Our results indicate that P. profundum SS9 possesses two distinct flagellar systems, both of which have acquired dramatic adaptations for optimal functionality under high-pressure conditions.
The deep sea constitutes the largest habitat in the biosphere, where physiochemical parameters such as low temperature, refractory organic carbon, and high hydrostatic pressure structure diverse communities. It has been hypothesized that the evolutionary modalities of autochthonous microbial residents are significantly affected by hydrostatic pressure at depths greater than 2,000 m (45). Hydrostatic pressure alters the free energy of equilibria, macromolecular packing, and hydration via influences on system volume changes (39). Piezophilic (“pressure-loving”) microorganisms harbor unique adaptations to cope with deep-sea high-pressure conditions (reviewed in reference 4). A bacterial strain with a Pkmax (pressure at which the growth rate is maximal) of >0.1 MPa and <60 MPa is termed a piezophile (46); a strain with a Pkmax of <30 MPa might be termed a “moderate” piezophile and one with a Pkmax of >60 MPa a “hyperpiezophile.”
Motility is considered one of the most pressure-sensitive cellular processes in nonpiezophilic microorganisms (4, 31). In Escherichia coli, increased hydrostatic pressure acts as an inhibitor of the formation of new flagella and of the functioning of previously assembled filaments (31). The bacterial flagellum is a helical filament of approximately 15 μm consisting of ∼30,000 flagellin monomers that polymerize to form a functional filament (24). High-resolution structures obtained from X-ray crystallography and electron cryomicroscopy of the Salmonella enterica serovar Typhimurium flagellin F41 fragment reveal four linearly connected domains (D0, D1, D2, and D3) with a 3-dimensional structure in the shape of a uppercase Greek gamma (Γ) (36, 48). The flagellar filament is composed of 11 protofilaments that form two conformations based on supercoiling: a left-handed (L type, or “normal”) and a right-handed (R type, or “curly”) symmetry. The N-terminal and C-terminal regions form an α-helical coiled-coil (D0-D1 domains) that constitutes a densely packed filament core and are highly conserved among eubacterial flagellins (6). Domains D2 and D3, on the other hand, are hypervariable regions projecting out from the filament core. These two regions vary both in sequence and in length and are thought to be important for folded flagellin conformation stability (26). In vitro studies of Salmonella serovar Typhimurium flagellin show an increase in partial molar volume of 340 cm3/mol upon polymerization, where filaments irreversibly depolymerize at approximately 340 MPa (40). It would therefore appear that the assembly and functioning of the flagellar apparatus under high hydrostatic pressure in deep-sea bacteria would necessitate adaptation.
The psychrotolerant, moderately piezophilic organism Photobacterium profundum SS9 has been extensively studied using genetic, genomic, and functional genomic approaches (12, 23, 41). Two flagellar motility gene clusters have been identified: a polar flagellum (PF) gene cluster and a potential lateral flagellum (LF) gene cluster (12, 41). Microarray hybridization experiments indicate that one of the most notable differences between SS9 and its shallow-water, pressure-sensitive relative Photobacterium profundum strain 3TCK is that the latter lacks the putative LF cluster found in SS9 (12). Based on its higher GC content, the LF cluster could have been horizontally transferred to SS9 (12).
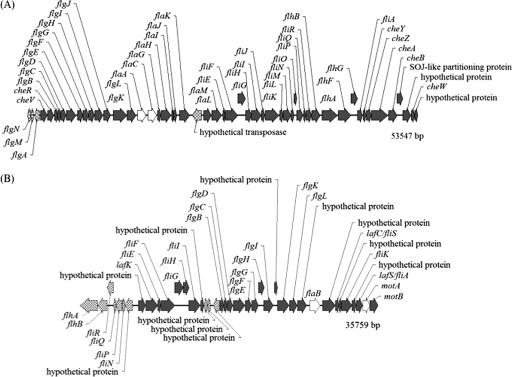
The organization of the P. profundum SS9 PF and LF clusters (Fig. 1) is almost identical to that of the major flagellar loci in Vibrio parahaemolyticus BB22 (28). The genes found in the PF cluster appear to be organized into nine operons based on close or overlapping sequences and the absence of transcriptional terminators. Only three flagellin genes are present in SS9, compared to the six polar flagellin genes of Vibrio parahaemolyticus and Vibrio fischeri and the five polar flagellin genes of Vibrio cholerae and Vibrio anguillarum (21, 29, 30, 32). flaA and flaC reside within the PF gene cluster, while flaB is the single flagellin gene present within the LF gene cluster. Interestingly, P. profundum SS9 flaA is differentially upregulated at atmospheric pressure compared to 28 MPa (12). Furthermore, flaC is expressed at a higher level than flaA based on microarray mean fluorescent values and is presumably incorporated into the filament more extensively.
FIG. 1.
Polar (A) and lateral (B) flagellar gene clusters in P. profundum SS9. Additional genes not found in the main clusters include the putative sodium motor genes motAB (PBPRA0808 and PBPRA0809, chromosome 1, positions 895239 to 896941), motX (PBPRA3344, chromosome 1, positions 3792907 to 3793536), and motY (PBPRA2571, chromosome 1, positions 2976252 to 2977142). Four of the five genes deleted via in-frame mutagenesis are represented by open arrows.
The LF cluster in P. profundum SS9 contains a number of sequences related to genes present in the V. parahaemolyticus LF system, with the notable exception that the lateral cluster in P. profundum SS9 is present on chromosome 1 instead of chromosome 2 and is not similarly organized into two regions (28). The first 15 genes from V. parahaemolyticus region 2 are inverted and reside upstream of the region 1 genes in the P. profundum SS9 LF system. Many of the genes in the SS9 LF cluster are annotated as hypothetical proteins due to low amino acid identity and sequence similarity to known flagellar components. P. profundum SS9 flaB is highly divergent and only weakly resembles a lateral flagellin. Instead, it is most closely related at the amino acid level to a Pseudomonas fluorescens flagellin (corresponding to GenBank accession number AAC63947; 41% identity).
Consistent with the V. parahaemolyticus motility system, P. profundum SS9 appears to possess two kinds of rotary motors for propulsion: a sodium-driven complex associated with PF rotation and components for a proton-driven motor found in the LF cluster. Four genes are linked to the sodium type polar motor, with motAB2 residing upstream of the PF cluster: motA (GenBank locus tag PBPRA0808), motB (PBPRA0809), motX (PBPRA3344), and motY (PBPRA2571). The putative proton-driven components motAB1 (PBPRA0048, PBPRA0049) reside in the LF cluster. Chemotaxis genes and numerous methyl-accepting chemotaxis protein genes are distributed throughout the genome.
In this study, the first examination of motility as a function of pressure in a deep-sea microbial species is reported. The results indicate that the SS9 PF and LF systems are fully functional and are adapted for swimming and swarming, respectively, at depth.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this work are described in Table 1. P. profundum SS9 strains were cultured aerobically in 2216 medium (28 g/liter; Difco Laboratories) at 16°C. E. coli strains were grown aerobically in Luria-Bertani (LB) medium at 37°C (37). High-pressure growth of SS9 strains was performed anaerobically at 16°C in 2216 medium supplemented with 20 mM glucose and 100 mM HEPES buffer (pH 7.5) (Sigma). Late-exponential-phase cultures were diluted 500-fold into fresh medium and used to fill 4.5-ml polyethylene transfer pipettes (Samco). Transfer pipettes were heat sealed with a handheld heat-sealing clamp (Nalgene) and incubated at 0.1 MPa and 30 MPa in stainless-steel pressure vessels (47). Antibiotics (Sigma) were used at the following concentrations: chloramphenicol, 30 μg ml−1; kanamycin, 75 μg ml−1 for E. coli and 200 μg ml−1 for SS9; rifampin, 100 μg ml−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| P. profundum | ||
| SS9R | Rifr SS9 derivative | 13 |
| EAE1 | ΔflaA SS9R; Rifr | This study |
| EAE2 | ΔflaB SS9R; Rifr | This study |
| EAE3 | ΔflaC SS9R; Rifr | This study |
| EAE4 | ΔmotA1 SS9R; Rifr (H+ driven) | This study |
| EAE5 | ΔmotA2 SS9R; Rifr (Na+ driven) | This study |
| E. coli | ||
| ED8654 | pRK2073 maintenance | 33 |
| DH5α | RecA−; cloning | 18 |
| XL1-Blue | RecA−; cloning | Stratagene, La Jolla, CA |
| TOP10 | RecA−; cloning | Invitrogen, Carlsbad, CA |
| Plasmids | ||
| pRK2073 | tra genes for conjugal transfer | 9 |
| pRL271 | sacB-containing suicide plasmid; Cmr | 10 |
| pMB2190 | pBR327 derivative; Knr | 2 |
| pEAE1k | flaA deletion construct::pRL271; Knr | This study |
| pEAE2k | flaB deletion construct::pRL271; Knr | This study |
| pEAE3k | flaC deletion construct::pRL271; Knr | This study |
| pEAE4k | motA1 deletion construct::pRL271; Knr | This study |
| pEAE5k | motA2 deletion construct::pRL271; Knr | This study |
Knr, kanamycin resistance; Cmr, chloramphenicol resistance; Rifr, rifampin resistance.
Measurement of swimming speed under high hydrostatic pressure.
The HPDS (Hartmann, Pfeifer, Dornheim, Sommer) high-pressure cell (Technische Universität München, Freising-Weihenstephan, Germany) was used to examine and analyze the swimming behavior of P. profundum strain 3TCK, Escherichia coli strain W3110, the parental strain SS9R, and its motility mutants under high hydrostatic pressure (16, 19). The HPDS high-pressure cell was fixed to an inverted microscope (DM-IRB; Leica, Germany) and examined using a 40× phase-contrast objective giving 0.5-μm resolution. The temperature and pressure were measured through a transducer and sent, via a DAQ (data acquisition hardware) device custom developed for use with the HPDS cell, to a computer for monitoring with a custom-developed program made in LabView, version 5.1 (National Instruments) (for details, see reference 19). Mid-exponential-phase (optical density at 600 nm [OD600], 0.3) cultures were diluted 1:10, and 3.5 μl (∼3.5 × 103 cells) was loaded into the HPDS cell for viewing. Samples were maintained at atmospheric pressure (0.1 MPa) for 2 min before the pressure was increased. Pressure treatment involved a stepwise increase by 10 MPa (up to 150 MPa) every 30 s and a final decompression. A 2/3-in charge-coupled device digital camera (Basler, Germany) (19) displayed live output feed, which was directly recorded to iMovie format and subsequently converted into MPEG image sequence files for analysis. Image analysis software (NIH ImageJ, version 1.36 for Mac; http://rsb.info.nih.gov/ij/) (1) was used to measure the swimming speeds of individual moving bacteria manually in 25 successive frames corresponding to 1 s. Between 50 and 100 individual swimming tracks were measured for each condition with approximately 20 to 30 cells/frame.
Generation of flagellin and motor protein mutants.
Marker exchange-eviction mutagenesis was performed using the sacB-containing suicide vector pRL271 (10, 35). Plasmids were constructed as described by Welch and Bartlett (43). First, a 2,642-bp region upstream (primers, FlaAUPF [5′-AGTCTCGAGTGATCGGCAGTGGGCATACC-3′] and FlaAUPR [5′-GATGCGGCCGCTTTGCTCTCCTTTGACTTTTCAC-3′]) and a 2,564-bp region downstream (primers, FlaADNF [5′-GATGCGGCCGCACACAGCTACAGTAAATATTG-3′] and FlaADNR [5′-Phos-GCGAGCGCCATTATCATCTTTG-3′]) of flaA were PCR amplified from wild-type SS9 using the Expand Long-Template PCR system (Roche Applied Science). The amplified flanking regions were digested with NotI, ligated using Quick-stick ligase (Bioline), and reamplified using primers FlaAUPF and FlaADNR. The resulting deletion construct was digested with XhoI and cloned into the XhoI-NaeI sites of the suicide plasmid pRL271, creating pEAE1. A SalI-digested fragment containing the kanamycin resistance gene from pMB2190 was cloned into pEAE1, creating plasmid pEAE1k. pEAE1k was then conjugated into SS9R, a rifampin-resistant derivative of wild-type SS9, by triparental conjugations using helper plasmid pRK2073 as described by Chi and Bartlett (13), with kanamycin selection to identify exconjugants with pEAE1k integrated into the flaA gene. pEAE1k contains the sacB gene, the product of which is lethal in gram-negative bacteria in the presence of sucrose (35). When exconjugants were plated onto 2216 medium with 5% sucrose, only clones that had undergone a second recombination event that excises sacB grew. A flaA deletion mutant was identified from the two possible recombination events by PCR with primers FlaACTRLF (5′-CCAAAGTGACGGTAACCCCAAAAA-3′) and FlaACTRLR (5′-TGTTTTGCTCACCGGTTTTATCTG-3′) and was designated EAE1. In-frame deletions of the flaB, flaC, motA1, and motA2 genes were similarly constructed and verified by PCR. Primer sequences used for the construction of pEAE1k to pEAE5k are listed in Table S1 in the supplemental material.
Microscopy.
A 0.5-μl aliquot of a stock solution of the fluorescent protein stain NanoOrange (Molecular Probes, Invitrogen) was added to 10-μl mid-exponential-phase (OD600, 0.3) liquid cultures on a microscope slide (17). Slides were incubated in the dark for 10 min to allow staining and were then examined on an inverted epifluorescence microscope (model IX71; Olympus) at a magnification of ×60 with a blue filter (excitation wavelength, 490 nm; emission wavelength, 520 nm). Images were captured with a MicroFire 2/3-in charge-coupled device digital camera and examined with the PictureFrame imaging application system (Optronics).
Motility assay.
Motility phenotypes for the flagellin and motor protein mutants were investigated qualitatively using 2216 medium-0.3% agar polyethylene transfer pipette bulbs. Late-exponential-phase cultures were inoculated into the bulb in a straight line by using a thin inoculation rod, and the bulb was subsequently heat sealed. The parental strain, SS9R, and its mutants were incubated for 48 h at 16°C and 0.1 MPa or 30 MPa in stainless-steel pressure vessels. Polyvinylpyrrolidone, with an average molecular weight of 360,000 (PVP-360; Sigma), was used as a viscosity-increasing agent in the 2216 medium-0.3% agar bulbs at increments of 1.25%, 2.5%, 5%, and 10% to assay for lateral motility (7). pFL185 (a plasmid expressing β-galactosidase) was conjugated into SS9R and motility mutants by using helper plasmid pRK2073. Plasmid-containing strains were inoculated into 2216 medium-0.3% agar bulbs with added S-Gal (3,4-cyclohexenoesculetin-β-d-galactopyranoside; Sigma) and ferric ammonium citrate (20).
RT-PCR expression analysis.
Reverse transcription-PCR (RT-PCR) was used to assess differential expression of flagellin and motor-protein genes from the parental strain, SS9R, under conditions of varying viscosity and high pressure. Mid-exponential-phase (OD600, 0.3) 30-ml cultures grown with or without 5% PVP-360 (Sigma) in 2216 medium (20 mM glucose and 100 mM HEPES buffer) at 30 MPa and 16°C were harvested for RNA extraction. Cell pellets were resuspended in 4 ml Trizol reagent (Invitrogen), and 800 μl chloroform was added. The solution was thoroughly mixed, incubated on ice for 5 min, and centrifuged at 9,000 × g for 15 min. Two milliliters of the aqueous phase was transferred to a new tube, and 2 ml isopropanol was added and then incubated at room temperature for 10 min. Tubes were centrifuged at 12,000 × g for 5 min, washed with 4 ml 75% ethanol, and centrifuged again at 9,000 × g for 5 min. RNeasy columns (Qiagen) were used for the second RNA cleanup with additional DNase treatment according to the manufacturer's instructions. RT-PCR was carried out using a Qiagen One-Step RT-PCR kit according to instructions with an added RNase inhibitor (Ambion). An internal fragment of flaB was amplified using primers flaBRNAF (5′-TGGCGGTTCAGTCTAAAAAT-3′) and flaBRNAR (5′-AATACCAGTACCGGCATCCTCAGT-3′). Uridine phosphorylase (PBPRA1431) (primers, udpF [5′-GTGCACCGTCAGCCATTATCG-3′] and udpR [5′-CGCCCAGCACGCCTTTCT-3′]) was used as a control due to low expression as determined from microarray data (12).
RESULTS
Agar matrix motility assays at high hydrostatic pressure.
Qualitative analyses of microbial motility are typically performed by inoculating cell cultures into plates with a low percentage of agar and visually inspecting the diameter of the growing cell population and the associated chemotaxis patterns (32). This method, however, is impractical at high hydrostatic pressure. Instead, pressurizable plastic pipette bulbs containing a low-percentage agar medium and a β-galactosidase indicator dye, and lacking significant air space, were utilized (see Materials and Methods). Inoculation into the bulbs of bacteria genetically modified to produce high levels of β-galactosidase activity, and their subsequent incubation at appropriate temperatures and pressures, provided a convenient 3-dimensional view of cell growth and movement over time.
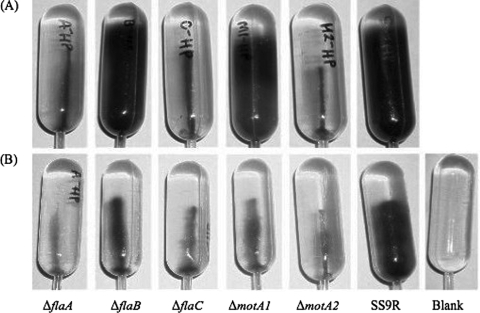
Motility bulb assay characteristics were obtained for derivatives of the P. profundum piezosensitive strain 3TCK and the P. profundum piezophilic strain SS9, along with its isogenic fla and mot mutants (Fig. 2 and 3). Flagellin and motor component deletion mutants were checked for growth defects to ensure that there were no pressure effects on the growth rate that would affect the interpretation of the growth-based motility agar assays. All five flagellin and motor component mutants were found to have growth rates comparable to that of the parental strain, SS9R (data not shown). 3TCK displayed vigorous swimming ability at 0.1 MPa, with cells moving out to the sides of the 140-mm bulb after 24 h at 16°C, while little swimming ability was evident at 30 MPa. The opposite was true for SS9, which filled the entire bulb after 48 h at 16°C and 30 MPa. The PF flaA, flaC, and motA2 mutants were nonmotile both at 0.1 MPa and at 30 MPa, whereas the LF flaB mutant and the motA1 mutant appeared to possess wild-type motility at 30 MPa (Fig. 3A). NanoOrange fluorescent microscopic visualization of these cells indicated that they all are monotrichously flagellated except for the flaA and flaC mutants (Fig. 4). Taken together, the results indicate that P. profundum possesses a single polar flagellum, which in the case of 3TCK functions optimally at atmospheric pressure and in the case of SS9 functions best at elevated pressure. SS9 FlaA and FlaC are required components of the polar flagellum, which is most likely driven by sodium motive force based on the sequence similarity of the motAB gene products to confirmed sodium-driven motors in V. parahaemolyticus (3).
FIG. 2.
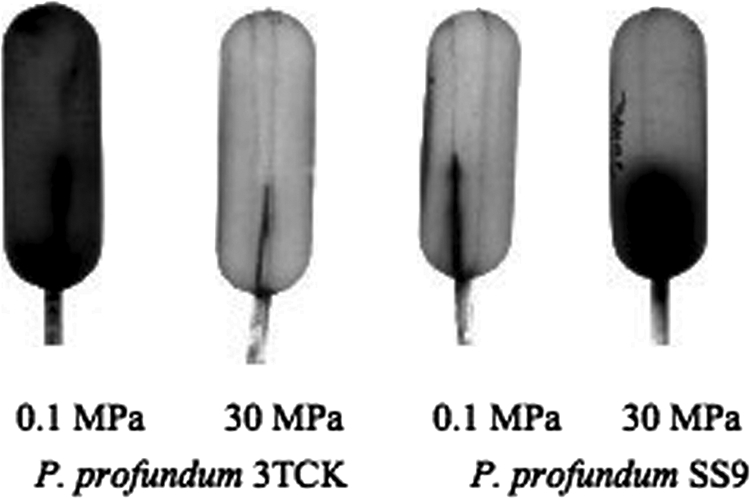
Growth-based 0.3% agar bulb assay to qualitatively assess motility for P. profundum strains 3TCK and SS9 at atmospheric pressure (0.1 MPa) and high pressure (30 MPa).
FIG. 3.
Growth-based agar bulb assay to qualitatively assess motility under high-hydrostatic-pressure and high-viscosity conditions. (A) Swimming motility bulbs under high-pressure conditions (30 MPa) with 0.3% agar. (B) Swarming motility bulbs under high-pressure (30 MPa) and increased-viscosity (0.3% agar with 2.5% PVP-360) conditions. The strains examined were SS9R (the parental strain) and the ΔflaA, ΔflaB, ΔflaC, ΔmotA1, and ΔmotA2 mutants.
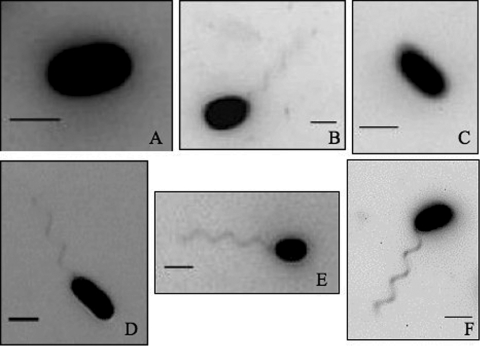
FIG. 4.
Flagella visualized with NanoOrange staining for P. profundum SS9 and deletion mutants. The ΔflaA (A) and ΔflaC (C) mutants do not produce a filament. The ΔflaB (B) and proton-motor (ΔmotA1) (D) mutants were seen to produce a polar flagellum comparable to that of the parental strain, SS9R (F). The sodium motor (ΔmotA2) (E) mutant was found to produce an intact flagellum yet was nonmotile. Scale bars, 2 μm.
SS9 swarming motility.
Lateral flagellar motility has been studied on 0.6 to 0.8% agar plates containing growth media with reduced iron levels and in similar liquid media containing viscosity-increasing agents (7). Viscosity-increasing agents such as polyvinylpyrrolidone, polyethylene glycol, and Ficoll effectively induce swarmer cell differentiation in Vibrio parahaemolyticus (7). The possible presence of a lateral flagellar motility system in SS9 was examined by modifying the bulb motility assays described above to include PVP-360 (Sigma) in increments of 1.25%, 2.5%, 5%, and 10%. Under these conditions, all of the SS9R-derived strains were nonmotile, except for the parental strain, SS9R. While the flaB and motA1 mutants were motile at high pressure under standard bulb assay conditions, they displayed nonmotile phenotypes when grown under conditions of high pressure and increased viscosity (Fig. 3B). These results provide the first phenotypic evidence for a functional lateral flagellar motility apparatus in SS9, a feature that is evident only under conditions of both high pressure and high viscosity. However, both NanoOrange fluorescence microscopy and scanning electron microscopy failed to reveal lateral flagella on SS9 cells, although these structures were clearly visible in control cultures of V. parahaemolyticus (data not shown).
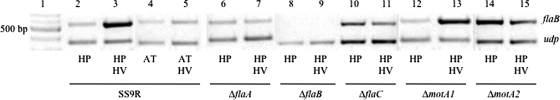
The results from the motility assays suggested induction of a lateral flagellar system under high-pressure and high-viscosity conditions, and therefore the possibility of upregulation of the transcription of the components of the lateral flagella and their associated motor proteins. The transcript abundances of two genes associated with putative lateral flagellar function, flaB and motA1, were assessed under high-pressure and high-viscosity conditions by semiquantitative RT-PCR, with uridine phosphorylase (udp; PBPRA1431) as a control (Fig. 5). Uridine phosphorylase was used as a reference due to its low-level constitutive expression, based on transcriptome data (13), which was comparable to the low expression levels of flaB. The results indicated that mid-log-phase cultures of SS9R grown under high-pressure conditions induce flaB at least fivefold under increased-viscosity conditions (5% PVP-360), based on the ratio of gene amplification under high-pressure conditions to that under high-pressure plus high-viscosity conditions relative to udp control expression. flaB was constitutively expressed at the same level in flaA, flaC, and motA2 mutants under conditions of high pressure with or without increased viscosity, suggesting that the upregulation of flaB and the production of lateral flagella are dependent on a functional polar flagellum.
FIG. 5.
Flagellin B levels increase under conditions of high pressure and increased viscosity. Shown are results of semiquantitative RT-PCR analysis of flaB from cells grown under conditions of high pressure (HP) or atmospheric pressure (AT), with or without increased viscosity (HV) (5% PVP-360). Lane 1, 2-log ladder (New England Biolabs); all other lanes are as marked. The upper band is an internal fragment of flaB; the lower band is a constitutively expressed control gene, uridine phosphorylase (udp; PBPRA1431).
Direct visualization of motility as a function of pressure.
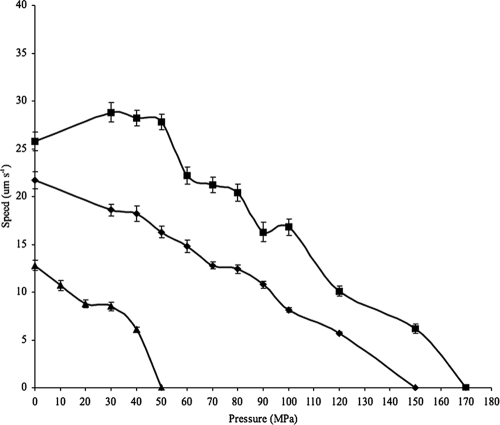
The HPDS high-pressure cell was used to examine and analyze the swimming behavior of Escherichia coli strain W3110, Photobacterium profundum strains 3TCK and SS9R, and the SS9R motility mutants under high hydrostatic pressure (16, 19). The temperature used for these experiments was 20°C. This system has been used previously to examine Spirogyra algae and human B cells, but this was the first application to measure swimming speeds under high hydrostatic pressure. E. coli was used as a reference because the swimming behavior of this species as a function of pressure has been investigated previously by capillary tube assays (31). Consistent with these prior results, E. coli strain W3110 in the HPDS high-pressure cell displayed its highest average swimming speed at atmospheric pressure (12.8 μm s−1), and its motility was completely abolished at 50 MPa (Fig. 6; see also Table S2 in the supplemental material). In contrast to E. coli, both 3TCK and SS9R displayed an extremely broad pressure range for swimming motility. 3TCK and SS9R displayed the highest average swimming speeds at their corresponding optimal growth pressures (21.7 μm s−1 for 3TCK at 0.1 MPa; 28.2 μm s−1 for SS9R at 30 MPa) (Fig. 6). Stepwise increases in hydrostatic pressure resulted in a striking decrease in swimming velocity for E. coli and a much more gradual reduction for 3TCK, which proceeded up to 120 MPa, while SS9R showed increased swimming velocity at 30 MPa and maintained motility up to a maximum pressure of 150 MPa (Fig. 6).
FIG. 6.
Swimming velocity as a function of increasing hydrostatic pressure for the piezophile P. profundum SS9 (squares), the pressure-sensitive strain P. profundum 3TCK (diamonds), and E. coli W3110 (triangles). For pressures up to 100 MPa, data are mean velocity measurements from 50 individual cells; beyond 100 MPa, velocity measurements for 3TCK and SS9 were taken for 10 to 20 individual cells. Error bars, standard errors.
DISCUSSION
In this study the PF and LF systems of a deep-sea bacterium have been explored using genetics in concert with novel phenotypic screening as a function of hydrostatic pressure. The results suggest that while motility is one of the most pressure-sensitive cellular processes in mesophilic microorganisms, piezophilic bacteria possess uniquely adapted motility systems to maintain movement under the high pressures found in the deep ocean. The combination of culture-based agar bulb assays and culture-independent short-term microscopic visualizations made it possible to examine motility under conditions requiring the assembly of new flagella (the bulb assay) or only the function of preexisting flagella (high-pressure microscopy). The former assay could have been addressed by using a variation of the Dorayaki plate method, in which colonial growth under high-pressure conditions is achieved by sandwiching cells between slabs of agar growth medium (34). However, while this method is useful for screening the growth of many strains on a solid medium under high-pressure conditions, bacterial growth tends to spread out in the layer between the two agar sections, which is especially problematic in media with a low percentage of agar. In our hands the bulb assay provided more-consistent results.
Bacterial motility in a liquid environment under high-pressure conditions has been investigated previously. Meganathan and Marquis utilized a capillary assay to examine E. coli swimming behavior under high-pressure conditions over periods as long as 48 h (31). Although they utilized a different method and a different E. coli strain, their results were very similar to those reported here using the high-pressure microscope. High-pressure microscopy provides a window into the behavior of individual cells as well as cell populations. In the future it would be useful to use the HPDS system to further examine motility as a function of pressure, temperature, viscosity, and time.
The results of motility assays for the P. profundum strains were particularly revealing. Not surprisingly, the swimming of SS9, like that of other marine bacteria (22, 25, 38), appears to be sodium powered, and its swarming motility, like that of V. parahaemolyticus (3), appears to be proton powered, based on the phenotypic characterization of the two motA deletion mutants and the deduced MotA primary structures. However, in contrast to the observation that because of redundancy none of the six polar flagellin genes are required for swimming motility in V. parahaemolyticus, flaA and flaC in SS9 are essential for swimming. Another major surprise is that both 3TCK and SS9 are capable of short-term swimming under pressures well above the known upper pressure limit for microbial life (45). Their pressure optima for swimming matched those for growth, suggesting that swimming analyses are a good proxy for the overall pressure adaptation of the microbe under examination. The swimming speeds measured for P. profundum in this study are comparable to those of Vibrio species (20 to 65 μm s−1) and reflect the high-velocity swimming characteristics of marine bacteria compared to the lower velocities of E. coli (8, 22, 25, 38).
While we were unable to visualize lateral flagella, phenotypic characterizations of flaB and motA1 mutants, as well as flaB expression analysis, indicate a functional lateral motility system under conditions of high pressure and increased viscosity. V. parahaemolyticus lateral flagella have been visualized using transmission electron microscopy under careful handling conditions with phosphotungstic acid staining (as opposed to uranyl acetate staining, since uranyl acetate destroys lateral flagella) (29). Lateral flagella are extremely fragile structures, so one explanation for our inability to visualize lateral flagella might be filament loss or damage following decompression and subsequent sample processing.
The functionality of the SS9 LF system is particularly noteworthy when SS9 is compared to its deep-sea relative P. profundum DSJ4, which also possesses an LF gene cluster, and the recently characterized dual motility system of Shewanella piezotolerans WP3 (12, 42). In the case of S. piezotolerans WP3, the two sets of flagellar systems were found to be inversely regulated: the LF system is upregulated at low temperatures, and the PF system is upregulated under high-pressure conditions (42). Our results suggest that P. profundum SS9 regulates its LF genes differently from S. piezotolerans WP3, since the lateral system is expressed only under conditions of high pressure and increased viscosity.
The SS9 lateral flagellum is a complex organelle encoded by almost 40 genes, which were potentially acquired via lateral gene transfer (12). Although it is not known how the LF block of 35 kbp was obtained, recently, within the family Vibrionaceae, Vibrio cholerae was demonstrated to be capable of chitin-mediated transformation of as much as 42 kbp of DNA, resulting in lipopolysaccharide serogroup conversion (11). Presumably, SS9 LF development requires a functional PF, since induction occurs only under conditions of high pressure and increased viscosity, and the regulation of LF flagellin (flaB) by viscosity is dependent on the expression of flaA, flaC, and motA2. In V. parahaemolyticus, the functionality of the polar flagellum is coupled to the transcription of laf genes, where physical or genetic disruption of the polar flagellum results in induction of lateral flagella (27). More work will be needed to sort out the details of the SS9 PF signal transduction process under high-pressure and increased-viscosity conditions, including more quantitative studies of laf gene expression and gene product abundance.
These results provide the first phenotypic evidence for a functional LF motility apparatus in SS9, a feature that is evident only under conditions of high pressure and increased viscosity. The ecological significance of having a dual motility system in the deep sea could stem from the particular lifestyle of P. profundum SS9, which was isolated from a deep-sea scavenging amphipod (5, 14). While the deep sea is generally described as an oligotrophic environment with limited utilizable carbon (44), the ability to attach to and colonize particles or animals would enable deep-sea bacteria to access a more dependable source of organic matter. Indeed, surface-adapted motility systems could assume greater significance at depth. Additional deep-sea bacteria have been found to contain LF gene components (42; also our unpublished results), and environmental genomic surveys suggest a potentially greater role for a surface-attached lifestyle in deeper-water microbial communities (15). It will be intriguing in the future to compare the motility systems of other piezophilic bacteria in order to further substantiate the ecological relevance and mechanisms of motility in the deep sea.
Supplementary Material
Acknowledgments
We thank Debbie Millikan for initial work on this project, Verena Pfund for work on the two motA deletion constructs, and Kai Linke and Markus Hartmann for assistance with the HPDS high-pressure system.
This work was supported by NSF grants MCB02-37059, MCB04-009, and MCB05-44524 to D.H.B.
Footnotes
Published ahead of print on 22 August 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Arps, P. J., and M. E. Winkler. 1987. Structural analysis of the Escherichia coli K-12 hisT operon by using a kanamycin resistance cassette. J. Bacteriol. 169:1061-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atsumi, T., L. McCarter, and Y. Imae. 1992. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature 355:182-184. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett, D. H. 2002. Pressure effects on in vivo microbial processes. Biochim. Biophys. Acta 1595:367-381. [DOI] [PubMed] [Google Scholar]
- 5.Bartlett, D. H., G. Ferguson, and G. Valle. 2008. Adaptations of the psychrotolerant piezophile Photobacterium profundum strain SS9, p. 319-337. In C. Michiels, D. H. Bartlett, and A. Aertsen (ed.), High-pressure microbiology. ASM Press, Washington, DC.
- 6.Beatson, S. A., T. Minamino, and M. J. Pallen. 2006. Variation in bacterial flagellins: from sequence to structure. Trends Microbiol. 14:151-155. [DOI] [PubMed] [Google Scholar]
- 7.Belas, M. R., M. Simon, and M. Silverman. 1986. Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J. Bacteriol. 167:210-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg, H. C., and D. A. Brown. 1972. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature 239:500-504. [DOI] [PubMed] [Google Scholar]
- 9.Better, M., and D. R. Helinski. 1983. Isolation and characterization of the recA gene of Rhizobium meliloti. J. Bacteriol. 155:311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. [DOI] [PubMed] [Google Scholar]
- 11.Blokesch, M., and G. S. Schoolnik. 2007. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog. 3:733-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campanaro, S. A., A. Vezzi, N. Vitulo, F. M. Lauro, M. D'Angelo, F. A. Simonato, A. G. Cestaro, G. Malacrida, G. Bertoloni, G. Valle, and D. H. Bartlett. 2005. Laterally transferred elements and high pressure adaptation in Photobacterium profundum strains. BMC Genomics 6:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi, E., and D. H. Bartlett. 1993. Use of a reporter gene to follow high-pressure signal transduction in the deep-sea bacterium Photobacterium sp. strain SS9. J. Bacteriol. 175:7533-7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLong, E. F. 1986. Adaptations of deep-sea bacteria to the abyssal environment. Ph.D. dissertation. University of California, San Diego.
- 15.DeLong, E. F., C. M. Preston, T. Mincer, V. Rich, S. J. Hallam, N.-U. Frigaard, A. Martinez, M. B. Sullivan, R. Edwards, B. R. Brito, S. W. Chisholm, and D. M. Karl. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311:496-503. [DOI] [PubMed] [Google Scholar]
- 16.Frey, B., M. Hartmann, M. Herrmann, R. Meyer-Pittroff, K. Sommer, and G. Bluemelhuber. 2006. Microscopy under pressure—an optical chamber system for fluorescence microscopic analysis of living cells under high hydrostatic pressure. Microsc. Res. Tech. 69:65-72. [DOI] [PubMed] [Google Scholar]
- 17.Grossart, H. P., G. F. Steward, J. Martinez, and F. Azam. 2000. A simple, rapid method for demonstrating bacterial flagella. Appl. Environ. Microbiol. 66:3632-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 19.Hartmann, M., M. Kreuss, and K. Sommer. 2004. High pressure microscopy—a powerful tool for monitoring cells and macromolecules under high hydrostatic pressure. Cell. Mol. Biol. 50:479-484. [PubMed] [Google Scholar]
- 20.Heuermann, K., and J. Cosgrove. 2001. S-Gal: an autoclavable dye for color selection of cloned DNA inserts. BioTechniques 30:1142-1147. [DOI] [PubMed] [Google Scholar]
- 21.Klose, K. E., and J. J. Mekalanos. 1998. Differential regulation of multiple flagellins in Vibrio cholerae. J. Bacteriol. 180:303-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen, M. H., N. Blackburn, J. L. Larsen, and J. E. Olsen. 2004. Influences of temperature, salinity and starvation on the motility and chemotactic response of Vibrio anguillarum. Microbiology 150:1283-1290. [DOI] [PubMed] [Google Scholar]
- 23.Lauro, F. M., K. Tran, A. Vezzi, N. Vitulo, G. Valle, and D. H. Bartlett. 2008. Large-scale transposon mutagenesis of Photobacterium profundum SS9 reveals new genetic loci important for growth at low temperature and high pressure. J. Bacteriol. 190:1699-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macnab, R. M. 1996. Flagella and motility. ASM Press, Washington, DC.
- 25.Magariyama, Y., S. Masuda, Y. Takano, T. Ohtani, and S. Kudo. 2001. Difference between forward and backward swimming speeds of the single polar-flagellated bacterium, Vibrio alginolyticus. FEMS Microbiol. Lett. 205:343-347. [DOI] [PubMed] [Google Scholar]
- 26.Malapaka, R. V., L. O. Adebayo, and B. C. Tripp. 2007. A deletion variant study of the functional role of the Salmonella flagellin hypervariable domain region in motility. J. Mol. Biol. 365:1102-1116. [DOI] [PubMed] [Google Scholar]
- 27.McCarter, L., M. Hilman, and M. Silverman. 1988. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54:345-351. [DOI] [PubMed] [Google Scholar]
- 28.McCarter, L. L. 2004. Dual flagellar systems enable motility under different circumstances. J. Mol. Microbiol. Biotechnol. 7:18-29. [DOI] [PubMed] [Google Scholar]
- 29.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGee, K., P. Hörstedt, and D. L. Milton. 1996. Identification and characterization of additional flagellin genes from Vibrio anguillarum. J. Bacteriol. 178:5188-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meganathan, R., and R. E. Marquis. 1973. Loss of bacterial motility under pressure. Nature 246:525-527. [DOI] [PubMed] [Google Scholar]
- 32.Millikan, D. S., and E. G. Ruby. 2004. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J. Bacteriol. 186:4315-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray, N. E., W. J. Brammar, and K. Murray. 1977. Lambdoid phages that simplify the recovery of in vitro recombinants. Genet. Genomics 150:53-61. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama, A., Y. Yano, and K. Yoshida. 1994. New method for isolating barophiles from intestinal contents of deep-sea fishes retrieved from the abyssal zone. Appl. Environ. Microbiol. 60:4210-4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ried, J. L., and A. Collmer. 1987. An nptl-sacB-sacR cartridge for constructing directed, unmarked mutations in Gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57:239-246. [DOI] [PubMed] [Google Scholar]
- 36.Samatey, F. A., K. Imada, S. Nagashima, F. Vonderviszt, T. Kumasaka, M. Yamamoto, and K. Namba. 2001. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410:331-337. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Sen, A., R. K. Nandi, and A. N. Ghosh. 2005. Ion-swimming speed variation of Vibrio cholerae cells. J. Biosci. 30:465-467. [DOI] [PubMed] [Google Scholar]
- 39.Somero, G. N. 1990. Life at low volume change: hydrostatic pressure as a selective factor in the aquatic environment. Am. Zoolog. 30:125-135. [Google Scholar]
- 40.Tamura, Y., K. Gekko, K. Yoshioka, F. Vonderviszt, and K. Namba. 1997. Adiabatic compressibility of flagellin and flagellar filament of Salmonella typhimurium. Biochim. Biophys. Acta 1335:120-126. [DOI] [PubMed] [Google Scholar]
- 41.Vezzi, A., S. Campanaro, M. D'Angelo, F. Simonato, N. Vitulo, F. M. Lauro, A. Cestaro, G. Malacrida, B. Simionati, N. Cannata, C. Romualdi, D. H. Bartlett, and G. Valle. 2005. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307:1459-1461. [DOI] [PubMed] [Google Scholar]
- 42.Wang, F., J. Wang, H. Jian, B. Zhang, S. Li, F. Wang, X. Zeng, L. Gao, D. H. Bartlett, J. Yu, S. Hu, and X. Xiao. 2008. Environmental adaptation: genomic analysis of the piezotolerant and psychrotolerant deep-sea iron reducing bacterium Shewanella piezotolerans WP3. PLoS One 3:e1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welch, T. J., and D. H. Bartlett. 1998. Identification of a regulatory protein required for pressure-responsive gene expression in the deep-sea bacterium Photobacterium species strain SS9. Mol. Microbiol. 27:977-985. [DOI] [PubMed] [Google Scholar]
- 44.Wirsen, C. O., and S. J. Molyneaux. 1999. A study of deep-sea natural microbial populations and barophilic pure cultures using a high-pressure chemostat. Appl. Environ. Microbiol. 65:5314-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yayanos, A. A. 1986. Evolution and ecological implications of the properties of deep-sea barophilic bacteria. Proc. Natl. Acad. Sci. USA 83:9542-9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yayanos, A. A. 1995. Microbiology to 10,500 meters in the deep sea. Annu. Rev. Microbiol. 49:777-805. [DOI] [PubMed] [Google Scholar]
- 47.Yayanos, A. A., and R. Van Boxtel. 1982. Coupling device for quick high pressure connections to 100 MPa. Rev. Sci. Instrum. 53:704-705. [Google Scholar]
- 48.Yonekura, K., S. Maki-Yonekura, and K. Namba. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424:643-650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.