Abstract
The apuB gene of Bifidobacterium breve UCC2003 was shown to encode an extracellular amylopullulanase. ApuB is composed of a distinct N-terminally located α-amylase-containing domain which hydrolyzes α-1,4-glucosidic linkages in starch and related polysaccharides and a C-terminally located pullulanase-containing domain which hydrolyzes α-1,6 linkages in pullulan, allowing the classification of this enzyme as a bifunctional class II pullulanase. A knockout mutation of the apuB gene in B. breve UCC2003 rendered the resulting mutant incapable of growth in medium containing starch, amylopectin, glycogen, or pullulan as the sole carbon and energy source, confirming the crucial physiological role of this gene in starch metabolism.
The study of the human gut microbiota in the context of health maintenance or improvement has in recent years enjoyed an exponentially increasing interest (reviewed in references 35 and 58). It is known that several pathogenic bacteria can cause acute gastroenteritis, while representatives of other bacterial genera, such as the lactobacilli, have the potential to provide protection against infection (11). It has also become apparent that elements of the colonic microbiota are capable of influencing the incidence and severity of gastrointestinal diseases and disorders, such as ulcerative colitis, bowel cancer, and pseudomembranous colitis (41). There have also been many studies focusing on prebiotics, which are “selectively fermented food ingredients that allow specific changes, both in the composition and/or activity in the gastrointestinal microbiota that confer benefits upon host well being and health” (33; see also references 28 and 42 for reviews). Carbohydrates that have been shown to exert prebiotic effects include those from whole-grain wheat, fructooligosaccharides, galactooligosaccharides, and lactulose (12, 17, 20, 33). The development of functional foods containing prebiotics and/or probiotics which can change the composition or activity of the microbiota such that it prevents or ameliorates gastrointestinal disorders is a key goal for both the pharma and food industries. Achieving this goal may be facilitated by the availability of an increasing number of genome sequences of gastrointestinal microbes (1, 9, 10, 26, 40, 48, 49), since the genomic data should allow the selection of novel, perhaps more-selective prebiotics and would also be pivotal in attaining a fundamental understanding of the probiotic effect.
Although starch and related compounds are digested and absorbed in the stomach and small intestine, a significant fraction that is resistant to digestion may represent valuable carbon and energy sources for colonic bacteria, such as bifidobacteria, and which thus may act as prebiotics (57). Plant cells and seeds are a rich source of starch where it is deposited as granules in the cytoplasm. Starch is composed of two high-molecular-weight components: amylose, which comprises 15 to 25% of starch, is a linear polymer consisting of α-1,4-linked glucopyranose, while the predominant component, amylopectin, is a branched polymer containing, in addition to α-1,4-glycosidic linkages, α-1,6-linked branch points occurring every 17 to 26 glucose units (18).
Glycogen is the storage form of glucose in animals and humans and plays a role which is analogous to the function of starch/amylopectin in plants. Glycogen is synthesized and stored mainly in the liver and muscles. Structurally, glycogen is a branching polymer consisting of chains of glucose units connected by α-1,4 linkages with branch points that are formed by α-1,6 linkages that occur at intervals of 10 to 13 glucose units.
Pullulan is a fermentation product of the yeast Aureobasidium pullulans that has a starch-like structure in that it is an α-glucan. Pullulan has a relatively simple structure of three α-1,4-linked glucose molecules that act as the repeated subunit and create a linear polymer through α-1,6 linkages on the terminal glucose of each subunit (31).
Starch degradation in most organisms proceeds via the combined action of amylases (EC 3.2.1.1, EC 3.2.1.2, and EC 3.2.1.3) and amylopullulanases (APU EC 3.2.1.41). Many of these amylolytic enzymes are industrially important for the liquefaction of starch and in saccharification processes. At present there is only limited knowledge available on the metabolism of starch and related α-glucans by bifidobacteria. Wang et al. (55) observed that bifidobacteria could efficiently utilize high-amylose maize starch granules and that bifidobacteria produced several starch-degrading enzymes of various molecular weights. Ji et al. (23) purified and characterized an extracellular amylase (AmyB) from Bifidobacterium adolescentis INT57, while Rhim et al. (43) have heterologously expressed AmyB from B. adolescentis INT57 in Bifidobacterium longum MG1. Ryan et al. (46) have reported on the screening of various bifidobacteria for α-amylase and/or pullulanase activity by investigating their ability to utilize starch, amylopectin, and pullulan. Of the bifidobacterial strains examined, five B. breve strains were identified that could utilize starch and pullulan as primary carbohydrate sources. These activities were found to be both inducible and extracellular, as well as consistent with pullulanase type II (amylopullulanase) activity.
This research reports on the characterization and mutagenesis of apuB, encoding an extracellular amylopullulanase, that was identified on the genome of Bifidobacterium breve UCC2003.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bifidobacterium breve UCC2003 was routinely cultured in reinforced clostridial medium (RCM; Oxoid Ltd., Basingstoke, Hampshire, United Kingdom). Carbohydrate utilization by B. breve was examined in de Man Rogosa and Sharpe medium (MRS) prepared from first principles (13). Prior to inoculation, the MRS was supplemented with cysteine HCl (0.05%) and a carbohydrate source (1%). The carbohydrates used (purchased from Sigma-Aldrich Ireland Ltd., Dublin, Ireland) were starch (derived from potato), amylopectin (derived from potato), glycogen (derived from oyster), pullulan (from Aureobasidium pullulans), and glucose. Bifidobacteria cultures were incubated at 37°C (unless otherwise stated) under anaerobic conditions which were maintained by using an Anaerocult oxygen-depleting system (Merck, Darmstadt, Germany) in an aerobic chamber. Escherichia coli was cultured in Luria Bertani broth (LB) (47) at 37°C with agitation. Lactococcus lactis strains were cultivated in M17 broth containing 0.5% glucose (50) at 30°C. The growth medium contained chloramphenicol (Cm; 5 μg ml−1 for L. lactis and 2 μg ml−1 for B. breve), erythromycin (Em; 100 μg ml−1 for E. coli and 1 μg ml−1 for B. breve), or kanamycin (50 μg ml−1 for E. coli) when required for plasmid maintenance. Recombinant E. coli cells containing pORI19 were selected on LB agar containing Em and supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 40 μg ml−1) and 1 mM IPTG (isopropyl-β-d-galactopyranoside).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli EC101 | Cloning host; repA+ Kanr | 30 |
| L. lactis strains | ||
| NZ9000 | MG1363, nisin-inducible overexpression host; pepN::nisRK | 14 |
| NZ9000-pNZ-αamy | NZ9000 containing pNZ-αamy | This study |
| NZ9000-pNZ-pull | NZ9000 containing pNZ-pull | This study |
| B. breve strains | ||
| UCC2003 | Isolate from nursling stool | 37 |
| UCC2003-apuB | pORI19-apuB insertion mutant of UCC2003 | This study |
| Plasmids | ||
| pNZ8048 | Cmr; nisin-inducible translational fusion vector | 14 |
| pNZ-αamy | Cmr; pNZ8048 derivative containing translational fusion of α-amylase encoding DNA fragment to nisin-inducible promoter | This study |
| pNZ-pull | Cmr; pNZ8048 derivative containing translational fusion of pullulanase encoding DNA fragment to nisin-inducible promoter on pNZ8048 | This study |
| pTGB019 | Cmr; 6.5-kb derivative of pVE6007 harboring temperature-sensitive repA gene | L. Steidler and S. Neirinck, unpublished results |
| pORI19 | Emr RepA− Ori+; cloning vector | 30 |
| pORI19-apuB | Internal 1-kb fragment of apuB cloned in pORI19 | This study |
Nucleotide sequence analysis.
Sequence data were obtained from the Artemis-mediated (45) genome annotations of the B. breve UCC2003 sequencing project (S. Leahy, M. O'Connell Motherway, J. Moreno Munoz, G. F. Fitzgerald, D. Higgins, and D. van Sinderen, unpublished data). Database searches were performed by using nonredundant sequences accessible at the National Center for Biotechnology Information internet site (http://www.ncbi.nlm.nih.gov) using BLAST (2, 3). Sequence alignments were performed by using the Clustal method of the MEGALIGN program of the DNASTAR software package (DNASTAR, Madison, WI). The biological software program SignalP (http://www.cbs.dtu.dk/services/SignalP) was used to predict the presence and precise location of the secretion signal in the ApuB protein (6). Screening for the cell wall anchoring motif LPXTG was performed manually.
DNA manipulations.
Chromosomal DNA was isolated from B. breve UCC2003 as previously described (38). Minipreparation of plasmid DNA from E. coli or L. lactis was achieved by using a Qiaprep spin plasmid miniprep kit (Qiagen GmBH, Hilden, Germany). For L. lactis, an initial lysis step was incorporated into the plasmid isolation procedure, and cells were resuspended in lysis buffer supplemented with lysozyme (30 mg ml−1) and incubated at 37°C for 30 min. The procedures for DNA manipulations were performed essentially as described by Sambrook et al. (47). Restriction endonucleases, shrimp alkaline phosphatase, and T4 DNA ligase were obtained from Roche Diagnostics and used according to the supplier's instructions. (Roche Diagnostics, Bell Lane, East Sussex, United Kingdom). The synthetic single-stranded oligonucleotide primers used in this study were synthesized by MWG Biotech AG (Ebersberg, Germany). Standard PCRs were performed using Taq PCR mastermix (Qiagen), while high-fidelity PCR was achieved by using KOD polymerase (Novagen, Darmstadt, Germany). B. breve colony PCRs were performed according to standard procedures with the addition of 2 units of mutanolysin to each PCR, while an initial cell lysis step of 37°C for 30 min was incorporated into the PCR conditions. PCR fragments were purified by using a Qiagen PCR purification kit (Qiagen). Electroporation of plasmid DNA into E. coli was performed as described by Sambrook et al. (47) and into L. lactis as described by Wells et al. (56). Electrotransformation of B. breve UCC2003 with pTGB019 and, subsequently, pORI19-apuB was performed as described by Mazé et al. (37). Southern transfer and hybridization were performed according to standard procedures using an ECL gene hybridization and detection system (GE Healthcare, United Kingdom).
Cloning of the α-amylase- and pullulanase-encoding domains of apuB in pNZ8048.
DNA fragments encompassing the α-amylase- or pullulanase-encoding domain of apuB were generated by PCR amplification from chromosomal DNA of B. breve UCC2003 using KOD DNA polymerase and primer combinations ApuAf and ApuAr or ApuPf and ApuPr (Table 2), respectively. NcoI and XbaI restriction sites were incorporated at the 5′ ends of the forward and reverse primers, respectively. In addition, an in-frame His10-encoding sequence was incorporated into each of the forward primers to facilitate protein purification using a Ni-nitrilotriacetic acid system (Qiagen). The amplicons, specifying Amy-HisApuB and Pull-HisApuB, were digested with NcoI and XbaI and ligated into the similarly digested nisin-inducible translational fusion plasmid pNZ8048 (Table 1). The ligation mixtures were introduced into L. lactis NZ9000 by electrotransformation, and transformants selected based on Cm resistance. The plasmid content of a number of Cmr transformants was screened by restriction analysis, and the integrity of positively identified clones was verified by sequencing.
TABLE 2.
Oligonucleotide primers used in this study
| Purpose | Primer | Sequencea |
|---|---|---|
| Cloning of α-amylase fragment of apuB in pNZ8048 | ApuAf | TGCATCCCATGGCCCATCACCATCACCATCACCATCACCATCACACCGAGGCCAATGCACAG |
| ApuAr | CTCGAATCTAGATTAGTAGGAGTCCTCGCCGGTG | |
| Cloning of pullulanase-encoding fragment of apuB in pNZ8048 | ApuPf | TGCACGCCATGGCCCATCACCATCACCATCACCATCACCATCACGCCGACACGAGCCGCACCATC |
| ApuPr | TGCGCATCTAGACTACTGTTCGGAGCCCTC | |
| Cloning of internal fragment of apuB | ApuBf | CTTGCGAAGCTTGGCGGCGACGGCTGGGTGAG |
| in pORI19 | ApuBr | GCGACGTCTAGACCTTCGGCGGGCTTGTAGTGC |
| Amplification of 5.1-kb apuB | ApuF | ATGGCCGTACGACGATTCAGCTC |
| fragment for use as probe in Southern hybridization | ApuR | CTGTTCGGAGCCCTCCTTACTG |
| Colony PCR primer pair | Apu1 | ATCGCCATCGCATTCCAAACCAAC |
| pORI19r | GATTAAGTTGGGTAAGCCG |
The sequences in bold correspond to restriction sites.
Protein overproduction and purification.
An amount of 400 ml of M17 broth supplemented with 0.5% glucose was inoculated with a 2% inoculum of a particular L. lactis strain, followed by incubation at 30°C until an optical density at 600 nm of 0.5 was reached, at which point protein overexpression was induced by the addition of purified nisin (5 ng ml−1) and cultures were incubated at 30°C for 90 min. Cells were harvested by centrifugation, washed, and concentrated 40-fold in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0]). Cell extracts were prepared by using 106-μm glass beads and a Mini BeadBeater-8 cell disrupter (Biospec Products, Bartlesville, Oklahoma). After homogenization, the glass beads and cell debris were sedimented by centrifugation and the supernatant containing the cytoplasmic fractions retained. Protein purification from the cytoplasmic fraction was performed by using Ni-nitrilotriacetic acid matrices in accordance with the manufacturer's instructions (Qiagen). Elution fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, as described by Laemmli (29), on a 12.5% polyacrylamide gel. After electrophoresis, the gels were fixed and stained with Coomassie brilliant blue to identify fractions containing the purified protein. Rainbow-prestained low-molecular-weight protein markers (New England Biolabs, Herdfordshire, United Kingdom) were used to estimate the molecular weight of the purified proteins.
HPTLC analysis.
High-performance thin-layer chromatography (HPTLC) allowed the qualitative determination of the breakdown products of starch, amylopectin, or pullulan following hydrolysis by the purified α-amylase, pullulanase, or cell extract of B. breve UCC2003. The purified α-amylase, pullulanase, cells, or cell extracts of B. breve UCC2003 were incubated with starch, amylopectin, glycogen, or pullulan in 50 mM phosphate buffer, pH 6.0, at 37°C for 72 h. An aliquot of the reaction mixture was spotted onto a silica gel 60 plate (10 by 10 cm; Merck) with a Nanomat 4 (Camag, Switzerland). The chromatogram was developed with a butanol-acetic acid-water (5:4:1, vol/vol/vol) solvent system in a horizontal developing chamber. Ascending development was repeated twice at room temperature. The plate was allowed to dry in a fume hood and then developed by spraying evenly with 20% (vol/vol) sulfuric acid in ethanol. The plate was dried and heated to 120°C for 10 min to visualize sugar-representing spots. Glucose, maltose, maltotriose, maltotetraose, maltopentaose, maltohexaose, and maltoheptaose were purchased from Sigma, and a mixture of these sugars was used as a standard marker for the HPTLC experiments.
Construction of pORI19-apuB.
An internal 1-kb fragment of apuB was amplified by PCR using B. breve UCC2003 chromosomal DNA as template and the oligonucleotide primers ApuBf and ApuBr (Table 2). The 1-kb PCR product generated was cloned into pORI19, an Ori+ RepA− integration plasmid (30), by using the unique HindIII and XbaI restriction sites that were incorporated into the primers and introduced into E. coli EC101 by electroporation. The expected structure of the recombinant plasmid, pORI19-apuB, was confirmed by restriction mapping prior to its introduction into B. breve harboring pTGB019 by electrotransformation and subsequent selection on RCA plates supplemented with Em and Cm.
RESULTS
Identification and analysis of a B. breve UCC2003 amylopullulanase-encoding gene.
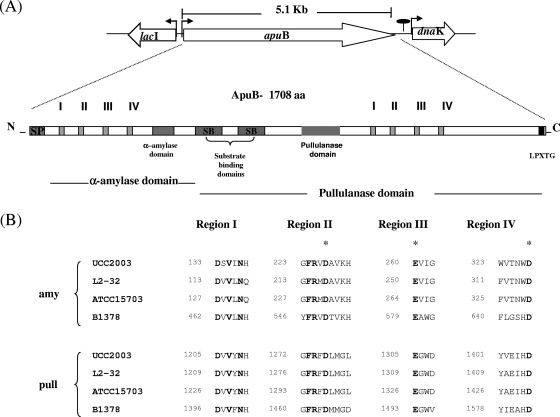
A gene designated apuB, predicted to encode amylopullulanase activity, was identified from the annotation of the genome sequence of B. breve UCC2003. (S. Leahy, M. O'Connell Motherway, J. Moreno Munoz, G. F. Fitzgerald, D. Higgins, and D. van Sinderen, unpublished results). apuB is located upstream of dnaK (53) and downstream from and in the opposite orientation to a gene predicted to encode an LacI-type transcriptional regulator (Fig. 1A). At the protein level, the predicted product of apuB displays significant similarity (67%) to predicted amylopullulanases encoded on the genomes of Bifidobacterium adolescentis strains L2-32 (gi:154487452; GenBank accession number AAXD0000000) and ATCC 15703 (gi:119025726; GenBank accession number AP009256). The apuB gene is 5,127 bp in length and corresponded to a deduced protein of 1,708 amino acids with an estimated molecular mass of 182.36 kDa. Based on the presence of a signal sequence of 34 amino acids with a corresponding cleavage site located between Ala34 and Asp35, ApuB is predicted to represent a secreted protein. The C-terminally located LPNTG sequence suggests that ApuB can be covalently linked to the peptidoglycan at the carboxy terminus via the LPXTG motif (51). The molecular mass of the mature extracellular, cell wall-anchored ApuB (from Asp35 to Thr1670) is expected to be 174.98 kDa. Individual domains representing α-amylase and pullulanase activity are predicted to be located in the amino-terminal portion and carboxy-terminal portion, respectively, of ApuB. Highly conserved regions were found when the amino acid sequence of ApuB was aligned with sequences of other characterized amylopullulanases (Fig. 1A) (22). For example, two copies of four highly conserved sequences designated I, II, III, and IV characteristic of the active site of amylolytic enzymes (7) were evident (Fig. 1B). The first of these is located between amino acids Asp133 and Asp328 in the predicted α-amylase domain, while the second set is located between amino acid positions Asp1205 and Asp1406 of the putative pullulanase domain. Based on the similarity with characterized α-amylases, pullulanases, and amylopullulanases, the catalytic residues of the presumed α-amylase domain (and of the pullulanase domain) of ApuB were predicted to be Glu260 (Glu1305) in conserved region III, Asp227 (Asp1276) in conserved region II, and Asp328 (Asp1406) in conserved region IV. Between the predicted α-amylase and pullulanase domains, two copies of a conserved substrate binding domain were identified; the first extends from Thr632 to Asp736 and the second from Gln744 to Asn857, and both are rich in aromatic amino acids (34). The identification of amylase and pullulanase domains, together with the presence of two apparently independently operating active sites, would indicate that ApuB is a so-called type II bifunctional amylopullulanase (21) that functions in cell wall-associated/extracellular metabolism of starch and related polysaccharides by B. breve UCC2003.
FIG. 1.
(A) Schematic representation of apuB and surrounding genes. ApuB is encoded by a single open reading frame of 5,127 bp, producing a protein of 1,708 amino acids which includes a signal sequence of 34 amino acids (SP). The α-amylase and pullulanase domains are located in the amino-terminal and carboxy-terminal portion, respectively. Within the protein, four regions highly conserved in α-amylase-like proteins were identified. In addition, specific α-amylase and pullulanase domains were identified. Two copies of a domain (SB) rich in aromatic amino acids were identified between the α-amylase and pullulanase domains. These domains are believed to be involved in substrate binding. aa, amino acids. (B) Two copies of the four regions highly conserved among α-amylases, pullulanases, and amylopullulanases were identified in ApuB and in amylopullulanases from B. adolescentis strains L2-32 and ATCC 15703. The amino acids in bold are conserved among all amylolytic enzymes, while the putative catalytic amino acids are denoted by asterisks. The sequence of the well-characterized alkaline amylopullulanase from Bacillus sp. KSM-1378 (GenBank accession no. D78258) is included.
ApuB represents a bifidobacterial amylopullulanase with dual specificity.
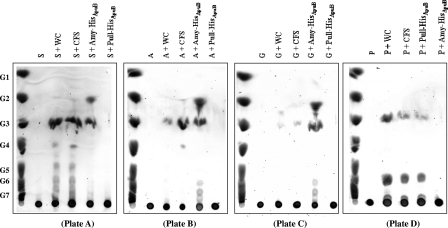
In order to verify that apuB encodes a type II bifunctional amylopullulanase, the predicted α-amylase- and pullulanase-encoding domains of apuB were individually amplified by PCR and cloned in the nisin-inducible expression vector pNZ8048 to generate pNZ-αamy and pNZ-pull, respectively (See Materials and Methods). The prospective α-amylase- and pullulanase-encoding gene products were each overexpressed and purified from the soluble cell extract fraction of L. lactis NZ9000 harboring the recombinant plasmid pNZ-αamy or pNZ-pull by using metal chelate affinity chromatography. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of Amy-HisApuB and Pull-HisApuB revealed, for each protein, a single band at an apparent molecular mass of approximately 70 kDa and 110 kDa, respectively, which is in agreement with the expected size calculated from the recombinant apuB gene sequence (results not shown). The end products formed by the hydrolysis of starch, amylopectin, glycogen, or pullulan following incubation with cells or cell-free supernatant of B. breve UCC2003 (representing the positive controls) or purified α-amylase (Amy-HisApuB) or pullulanase (Pull-HisApuB) were analyzed by HPTLC (Fig. 2). The results show that purified Amy-HisApuB can liberate maltooligosaccharides, predominantly maltotriose and maltose, from carbohydrates such as starch, amylopectin, and glycogen (Fig. 2, plates A, B, and C). Pull-HisApuB liberates predominantly maltotriose and polymers of maltotriose from pullulan (Fig. 2, plate D). Pull-HisApuB did not hydrolyze starch, amylopectin, or glycogen to short-chain maltooligosaccharides. In addition, no disaccharide or monosaccharide hydrolysis products, indicative of α-1,4-glucosidic activity, were detected after the incubation of Pull-HisApuB with pullulan. Similar end products were produced with all substrates when a cell-free supernatant or cells of B. breve UCC2003 were used, thereby indicating that some of the protein is cell bound while some is released from the cell into the culture medium. Long-chain maltooligosaccharides may have been produced by the hydrolysis of the α-1,6-glucosidic bonds in amylopectin, glycogen, and pullulan by Pull-HisApuB; however, the HPTLC system employed in this study does not allow the visualization of such long oligosaccharides. Collectively, these results demonstrate that Amy-HisApuB is an α-amylase exhibiting α-1,4-glucosidase activity while Pull-HisApuB is a true pullulanase, only hydrolyzing the α-1,6-glucosidic linkages in pullulan. Furthermore, these data illustrate that the extracellular amylopullulanase, encoded by apuB of B. breve UCC2003, is capable of hydrolyzing α-1,4 and α-1,6 linkages at different active sites and can therefore be classified as a so-called type II bifunctional amylopullulanase (15, 21).
FIG. 2.
HPTLC analysis of the reaction products generated by washed cells (WC) of B. breve UCC2003, cell-free supernatant (CFS) of B. breve UCC2003, or the purified α-amylase (Amy-HisApuB) or pullulanase (Pull-HisApuB) of ApuB following incubation with starch (S) (plate A), amylopectin (A) (plate B), glycogen (G) (plate C) or pullulan (P) (plate D). The standards glucose (G1), maltose (G2), maltotriose (G3), maltotetraose (G4), maltopentaose (G5), maltohexaose (G6), and maltoheptaose (G7) are included in each HPTLC.
Disruption of the apuB gene in B. breve UCC2003.
In order to determine whether ApuB is essential for the metabolism of starch and related polysaccharides by B. breve UCC2003, we tested the functionality of the plasmid integration strategy described by Law et al. (30) for the creation of an apuB gene knockout. The apuB disruption through plasmid integration in B. breve UCC2003 was achieved by using a strategy essentially as described for L. lactis (30), although with some modifications. pTGB019, a 6.5-kb derivative of the lactococcal temperature-sensitive plasmid pVE6007, was used as it exhibited a higher level of segregational instability compared to the smaller pVE6007 plasmid (M. O'Connell Motherway, unpublished data). The loss of pTGB019 from B. breve upon growth at elevated temperature is attributed to the combination of the temperature sensitivity and segregational instability of this plasmid. B. breve UCC2003 harboring pTGB019 was transformed with pORI19-apuB (see Materials and Methods) and plated on RCA supplemented with Em and Cm followed by incubation at 33°C under anaerobic conditions. Emr Cmr transformants carrying both pTGB019 and pORI19-apuB were cultured once overnight at 33°C in RCM broth. Subsequently, cells were passaged for 50 generations at 42°C, with selection for pORI19-apuB only, thus allowing integration into the chromosome upon the loss of pTGB019. Following this temperature-induced enrichment, serial dilutions of the culture were plated onto RCA with Em selection and incubated overnight at 42°C.
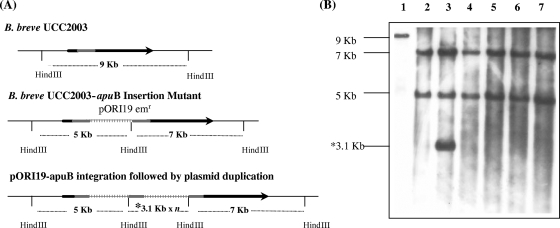
Screening for the loss of pTGB019 (Cms colonies) was performed by replica plating individual colonies onto RCA-Em and RCA-Cm with overnight incubation at 33°C. Potential apuB gene disruption isolates exhibited an Emr Cms phenotype and were verified by colony PCR analysis using a forward primer upstream of the region of integration and a reverse primer based on pORI19 (Apu1 and pORI19r) (Table 2). An expected PCR product of 1.7 kb was obtained in some cases, indicating that integration had occurred (data not shown). In order to unequivocally prove that the disruption of apuB was the result of the integration of pORI19-apuB, Southern hybridization was performed using HindIII-digested genomic DNA and the 5.1-kb PCR fragment encompassing apuB as a probe. HindIII was selected for the genomic digests as there are no corresponding restriction sites within the apuB gene sequence (Fig. 3A). The apuB fragment probe hybridized to a 9-kb fragment of strain UCC2003 genomic DNA, while in the suspected UCC2003-apuB mutant strains, this band was absent and instead two hybridization signals of 5 kb and 7 kb were observed. For one of the obtained UCC2003-apuB insertion mutants (Fig. 3B, lane 3), the apuB probe also hybridized to a 3.1-kb HindIII fragment. This hybridization signal indicated that duplication of pORI19-apuB had occurred after integration of the plasmid into the bacterial chromosome in this mutant strain. However, this hybridization profile does not identify the number of duplicated copies of the plasmid represented in the culture. The duplication of plasmid copies after the insertion of pORI-type plasmids has been reported previously for L. lactis and Lactobacillus acidophilus and has been attributed to recombinatory activity between flanking DNA regions of homology that result from Campbell-like integration (32, 44). The plasmid duplication is influenced by a number of factors that include the antibiotic selection and the nature and location of the insertion event.
FIG. 3.
(A) Schematic representation of the relevant regions of the B. breve UCC2003 and UCC2003-apuB chromosomes. Chromosomal DNA is represented by a thin line, the apuB gene is represented by a black arrow, the internal apuB fragment is indicated by a solid gray line, and pORI19 is indicated by a boxed line. HindIII sites relevant to the Southern hybridization analysis are indicated. (B) Southern hybridization analysis of HindIII chromosomal DNAs of B. breve UCC2003 (lane 1) and six representative B. breve UCC2003-apuB insertion mutants (lanes 2 to 7) obtained with pORI19-apuB integration. The sizes of the hybridizing fragments are indicated to the left of the panel. A PCR product of 5.1-kb encompassing apuB was used as a probe for the hybridization. “*3.1 Kb × n” represents the duplication of pORI19-apuB, present in an unknown number of copies (n) on the chromosome.
Analysis of the B. breve UCC2003-apuB insertion mutant phenotype.
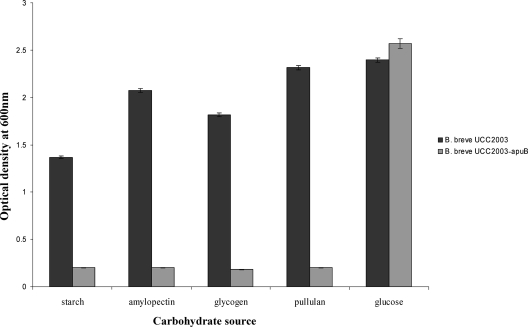
B. breve UCC2003 and B. breve UCC2003-apuB were analyzed for the ability to grow on starch, amylopectin, glycogen, or pullulan as the sole carbohydrate source (Fig. 4). B. breve UCC2003-apuB failed to grow on starch, amylopectin, glycogen, or pullulan in comparison to the growth of the parent strain, while comparable growth of the parent and apuB mutant strains was observed when glucose was the sole carbohydrate source. These results indicate that the extracellular enzyme specified by apuB is the sole enzyme responsible for the breakdown of starch, amylopectin, glycogen, and pullulan by B. breve UCC2003.
FIG. 4.
Growth profile analysis of B. breve UCC2003 and B. breve UCC2003-apuB in modified Rogosa broth supplemented with starch, amylopectin, glycogen, pullulan, or glucose as indicated. Error bars show standard deviations.
DISCUSSION
A gene, apuB, encoding an extracellular amylopullulanase, was identified on the genome of B. breve UCC2003. ApuB was shown to contain four highly conserved regions in two separate domains of the enzyme, each of which is proposed to contain an active center and a substrate binding site, a characteristic shared by other amylopullulanases (15, 16, 21). The N-terminally located α-amylase and C-terminally located pullulanase domains were independently overexpressed and purified. The α-amylase enzyme domain was shown to cleave α-1,4-glucosidic linkages in starch, amylopectin, and glycogen to produce maltooligosaccharides, while the pullulanase enzyme domain was shown to hydrolyze α-1,6-glucosidic linkages in pullulan to produce maltotriose and polymers of maltotriose (Fig. 2). Based on the end products formed following the hydrolysis of starch, amylopectin, glycogen, and pullulan by the enzymatic activity of the purified ApuB domains, the product of apuB can be classified as an amylopullulanase (4, 21), the first to be identified in the genus Bifidobacterium. Recent investigations of bifunctional type II pullulanases have led to a division of these enzymes into two distinct groups: those that hydrolyze both α-1,4- and α-1,6-glucosidic bonds using a single active center and substrate binding site and those, such as ApuB, that perform the two catalytic activities at two different sites within the same protein (5, 25).
The alkaline amylopullulanase from Bacillus sp. KSM-1378 exhibits different activities based on thermal stability, the pH stability profile, and inhibition by metals, indicating that the dual catalytic activities of the enzyme involved different active sites (25). The amylopullulanases from Thermoanaerobium sp. strain Tok-B1 (39) and Bacillus circulans F-2 (25) have also been suggested to contain separate active sites for the individual activities on the basis of the results of competitive kinetics studies with mixed substrates, namely, amylose and pullulan. In contrast, other reports unequivocally showed that a single active site is responsible for the hydrolyzing activities of certain amylopullulanases (24, 36). For example, in the amylopullulanase of Thermoanaerobacter ethanolicus 39E, the modification of one of the two conserved Asp residues by using site-directed mutagenesis was shown to lead to the loss of both α-amylase and pullulanase activities (36).
To establish if ApuB was the sole enzyme responsible for the metabolism of starch and related polysaccharides by B. breve UCC2003, an apuB gene disruption strain was created. To the best of our knowledge, gene inactivation via homologous recombination has not been reported yet for the genus Bifidobacterium. The ability to create gene disruptions/knockouts in Bifidobacterium has been a fundamental obstacle in attaining a full understanding of the probiotic effect (54). Here we successfully exploited and adapted the well-established lactococcal mutagenesis system as described by Law et al. (30). The B. breve UCC2003-apuB insertion mutants generated in this study were no longer capable of growth on starch, amylopectin, glycogen, or pullulan due to the disruption of apuB. This research thus illustrates that ApuB is essential in the metabolism of starch and starch-like polysaccharides by B. breve UCC2003.
Ryan et al. (46) demonstrated that all B. breve strains screened appeared to possess amylopullulanase activity. From these results, it can be suggested that this enzyme may be characteristic of B. breve strains, and if so, this activity must have some relevance to the organism's activity in the gut. It has been reported that after the first week of life of a newborn baby, a flora rich in Bifidobacterium spp. is established in the baby's gut (19, 52). The ApuB enzyme may play an important ecological role by allowing this B. breve strain to remain competitive in an environment where food sources change. During weaning, nonmilk foods are added to the diet and infants are exposed for the first time to different complex carbohydrates. A significant proportion of the carbohydrate, e.g., starch, will escape digestion and enter the colon because of the infant's lack of chewing ability and low levels of salivary amylase activity and the immaturity of the infant's intestine (8, 27). Such resistant starch sources therefore represent excellent carbohydrate sources for those bacteria that can produce amylolytic enzymes.
This extracellular bifunctional enzyme encoded by apuB may be one of the first crucial players in a carbohydrate metabolic pathway in B. breve, hydrolyzing extracellular starch or long-chain maltooligosaccharides to produce shorter (chain length ranging between 2 and 6) maltooligosaccharides. Analogous to other starch-degrading microorganisms, the short-chain maltooligosaccharides produced are expected to be taken up by the bifidobacterial cell where they are further degraded to glucose. It will be interesting to determine how B. breve UCC2003 performs the latter functions, and future research will focus on this aspect of starch metabolism.
Acknowledgments
This research was financially supported by the Science Foundation Ireland Alimentary Pharmabiotic Centre located at University College Cork.
Footnotes
Published ahead of print on 8 August 2008.
REFERENCES
- 1.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 102:3906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Ara, K., K. Igarashi, H. Hagihara, K. Sawada, T. Kobayashi, and S. Ito. 1996. Separation of functional domains for the alpha-1,4 and alpha-1,6 hydrolytic activities of a Bacillus amylopullulanase by limited proteolysis with papain. Biosci. Biotechnol. Biochem. 60:634-639. [DOI] [PubMed] [Google Scholar]
- 5.Ara, K., K. Saeki, K. Igarashi, M. Takaiwa, T. Uemura, H. Hagihara, S. Kawai, and S. Ito. 1995. Purification and characterization of an alkaline amylopullulanase with both alpha-1,4 and alpha-1,6 hydrolytic activity from alkalophilic Bacillus sp. KSM-1378. Biochim. Biophys. Acta 1243:315-324. [DOI] [PubMed] [Google Scholar]
- 6.Bendtsen, J. D., H. Nielsen, D. Widdick, T. Palmer, and S. Brunak. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buisson, G., E. Duée, R. Haser, and F. Payan. 1987. Three dimensional structure of porcine pancreatic alpha-amylase at 2.9 Å resolution. Role of calcium in structure and activity. EMBO J. 6:3909-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullen, C. L., P. V. Tearle, and A. T. Willis. 1976. Bifidobacteria in the intestinal tract of infants: an in-vivo study. J. Med. Microbiol. 9:325-333. [DOI] [PubMed] [Google Scholar]
- 9.Cerdeño-Tárraga, A. M., S. Patrick, L. C. Crossman, G. Blakely, V. Abratt, N. Lennard, I. Poxton, B. Duerden, B. Harris, M. A. Quail, A. Barron, L. Clark, C. Corton, J. Doggett, M. T. Holden, N. Larke, A. Line, A. Lord, H. Norbertczak, D. Ormond, C. Price, E. Rabbinowitsch, J. Woodward, B. Barrell, and J. Parkhill. 2005. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307:1463-1465. [DOI] [PubMed] [Google Scholar]
- 10.Claesson, M. J., Y. Li, S. Leahy, C. Canchaya, J. P. van Pijkeren, A. M. Cerdeño-Tárraga, J. Parkhill, S. Flynn, G. C. O'Sullivan, J. K. Collins, D. Higgins, F. Shanahan, G. F. Fitzgerald, D. van Sinderen, and P. W. O'Toole. 2006. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. USA 103:6718-6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corr, S. C., Y. Li, C. U. Riedel, P. W. O'Toole, C. Hill, and C. G. Gahan. 2007. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. USA 104:7617-7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costabile, A., A. Klinder, F. Fava, A. Napolitano, V. Fogliano, C. Leonard, G. R. Gibson, and K. M. Tuohy. 2008. Whole-grain wheat breakfast cereal has a prebiotic effect on the human gut microbiota: a double-blind, placebo-controlled, crossover study. Br. J. Nutr. 99:110-120. [DOI] [PubMed] [Google Scholar]
- 13.de Man, J., M. Rogosa, and M. E. Sharpe. 1960. A medium for the culture of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 14.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doman-Pytka, M., and J. Bardowski. 2004. Pullulan degrading enzymes of bacterial origin. Crit. Rev. Microbiol. 30:107-121. [DOI] [PubMed] [Google Scholar]
- 16.Dong, G., C. Vieille, and J. G. Zeikus. 1997. Cloning, sequencing, and expression of the gene encoding amylopullulanase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Appl. Environ. Microbiol. 63:3577-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dughera, L., C. Elia, M. Navino, and F. Cisarò. 2007. Effects of symbiotic preparations on constipated irritable bowel syndrome symptoms. Acta Biomed. 78:111-116. [PubMed] [Google Scholar]
- 18.Erra-Pujada, M., P. Debeire, F. Duchiron, and M. J. O'Donohue. 1999. The type II pullulanase of Thermococcus hydrothermalis: molecular characterization of the gene and expression of the catalytic domain. J. Bacteriol. 181:3284-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Favier, C. F., E. E. Vaughan, W. M. de Vos, and A. D. Akkermans. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hafer, A., S. Krämer, S. Duncker, M. Krüger, M. P. Manns, and S. C. Bischoff. 2007. Effect of oral lactulose on clinical and immunohistochemical parameters in patients with inflammatory bowel disease: a pilot study. BMC Gastroenterol. 7:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatada, Y., K. Igarashi, K. Ozaki, K. Ara, J. Hitomi, T. Kobayashi, S. Kawai, T. Watabe, and S. Ito. 1996. Amino acid sequence and molecular structure of an alkaline amylopullulanase from Bacillus that hydrolyzes alpha-1,4 and alpha-1,6 linkages in polysaccharides at different active sites. J. Biol. Chem. 271:24075-24083. [DOI] [PubMed] [Google Scholar]
- 22.Janecek, S., B. Svensson, and B. Henrissat. 1997. Domain evolution in the alpha-amylase family. J. Mol. Evol. 45:322-331. [DOI] [PubMed] [Google Scholar]
- 23.Ji, G. E., H. K. Han, S. W. Yun, and S. L. Rhim. 1992. Isolation of amylolytic Bifidobacterium sp. Int.-57 and characterisation of amylase. J. Microbiol. Biotechnol. 2:85-91. [Google Scholar]
- 24.Kang, S., C. Vieille, and J. G. Zeikus. 2005. Identification of Pyrococcus furiosus amylopullulanase catalytic residues. Appl. Microbiol. Biotechnol. 66:408-413. [DOI] [PubMed] [Google Scholar]
- 25.Kim, C. H., and Y. S. Kim. 1995. Substrate specificity and detailed characterization of a bifunctional amylase-pullulanase enzyme from Bacillus circulans F-2 having two different active sites on one polypeptide. Eur. J. Biochem. 227:687-693. [DOI] [PubMed] [Google Scholar]
- 26.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kok, R. G., A. de Waal, F. Schut, G. W. Welling, G. Weenk, and K. J. Hellingwerf. 1996. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl. Environ. Microbiol. 62:3668-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolida, S., and G. R. Gibson. 2007. Prebiotic capacity of inulin-type fructans. J. Nutr. 137:2503-2506. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Law, J., G. Buist, A. Haandrikman, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leathers, T. D. 2003. Biotechnological production and applications of pullulan. 2003. Appl. Microbiol. Biotechnol. 62:468-473. [DOI] [PubMed] [Google Scholar]
- 32.Leenhouts, K. J., J. Kok, and G. Venema. 1991. Lactococcal plasmid pWV01 as an integration vector for lactococci. Appl. Environ. Microbiol. 57:2562-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macfarlane, G. T., H. Steed, and S. Macfarlane. 2008. Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J. Appl. Microbiol. 104:305-344. [DOI] [PubMed] [Google Scholar]
- 34.Machovic, M., and S. Janecek. 2006. The evolution of putative starch-binding domains. FEBS Lett. 580:6349-6356. [DOI] [PubMed] [Google Scholar]
- 35.Marchesi, J., and F. Shanahan. 2007. The normal intestinal microbiota. Curr. Opin. Infect. Dis. 20:508-513. [DOI] [PubMed] [Google Scholar]
- 36.Mathupala, S. P., S. E. Lowe, S. M. Podkovyrov, and J. G. Zeikus. 1993. Sequencing of the amylopullulanase (apu) gene of Thermoanaerobacter ethanolicus 39E, and identification of the active site by site-directed mutagenesis. J. Biol. Chem. 268:16332-16344. [PubMed] [Google Scholar]
- 37.Mazé, A, M. O'Connell-Motherway, G. F. Fitzgerald, J. Deutscher, and D. van Sinderen. 2007. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73:545-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Riordan, K. 1998. Studies on antimicrobial activity and genetic diversity of Bifidobacterium species: molecular characterization of a 5.75 kb plasmid and a chromosomally encoded recA gene homologue from Bifidobacterium breve. Ph.D. thesis. National University of Ireland, Cork, Ireland.
- 39.Plant, A. R., R. M. Clemens, H. W. Morgan, and R. M. Daniel. 1987. Active-site- and substrate-specificity of Thermoanaerobium Tok6-B1 pullulanase. Biochem. J. 246:537-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A. C. Pittet, M. C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 101:2512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quigley, E. M. 2007. Bacterial flora in irritable bowel syndrome: role in pathophysiology, implications for management. J. Dig. Dis. 8:2-7. [DOI] [PubMed] [Google Scholar]
- 42.Rastall, R. A., G. R. Gibson, H. S. Gill, F. Guarner, T. R. Klaenhammer, B. Pot, G. Reid, I. R. Rowland, and M. E. Sanders. 2005. Modulation of the microbial ecology of the human colon by probiotics, prebiotics and synbiotics to enhance human health: an overview of enabling science and potential applications. FEMS Microbiol. Ecol. 52:145-152. [DOI] [PubMed] [Google Scholar]
- 43.Rhim, S. L., M. S. Park, and G. E. Ji. 2006. Expression and secretion of Bifidobacterium adolescentis amylase by Bifidobacterium longum. Biotechnol. Lett. 3:163-168. [DOI] [PubMed] [Google Scholar]
- 44.Russell, W. M., and T. R. Klaenhammer. 2001. Efficient system for directed integration into the Lactobacillus acidophilus and Lactobacillus gasseri chromosomes via homologous recombination. Appl. Environ. Microbiol. 67:4361-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 10:944-945. [DOI] [PubMed] [Google Scholar]
- 46.Ryan, S. M., G. F. Fitzgerald, and D. van Sinderen. 2006. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl. Environ. Microbiol. 72:5289-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 48.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sebaihia, M., B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeño-Tárraga, H. Wang, M. T. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell, and J. Parkhill. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779-786. [DOI] [PubMed] [Google Scholar]
- 50.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ton-That, H., L. A. Marraffini, and O. Schneewind. 2004. Protein sorting to the cell wall envelope of gram-positive bacteria. Biochim. Biophys. Acta 1694:269-278. [DOI] [PubMed] [Google Scholar]
- 52.Vaughan, E. E., M. C. de Vries, E. G. Zoetendal, K. Ben-Amor, D. L. Akkermans, and W. M. de Vos. 2002. The intestinal LABs. Antonie van Leeuwenhoek 82:341-352. [PubMed] [Google Scholar]
- 53.Ventura, M., R. Zink, G. F. Fitzgerald, and D. van Sinderen. 2005. Gene structure and transcriptional organization of the dnaK operon of Bifidobacterium breve UCC 2003 and application of the operon in bifidobacterial tracing. Appl. Environ. Microbiol. 71:487-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ventura, M., M. O'Connell-Motherway, S. Leahy, J. A. Moreno-Munoz, G. F. Fitzgerald, and D. van Sinderen. 2007. From bacterial genome to functionality; case bifidobacteria. Int. J. Food Microbiol. 120:2-12. [DOI] [PubMed] [Google Scholar]
- 55.Wang, X., P. L. Conway, I. L. Brown, and A. J. Evans. 1999. In vitro utilization of amylopectin and high-amylose maize (amylomaize) starch granules by human colonic bacteria. Appl. Environ. Microbiol. 65:4848-4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wells, J. M., P. W. Wilson, and R. W. Le Page. 1993. Improved cloning vectors and transformation procedure for Lactococcus lactis. J. Appl. Bacteriol. 74:629-636. [DOI] [PubMed] [Google Scholar]
- 57.Wronkowska, M., M. Soral-Smietana, and E. Biedrzycka. 2008. Utilization of resistant starch of native tapioca, corn and waxy corn starches and their retrograded preparations by Bifidobacterium. Int. J. Food Sci. Nutr. 59:80-87. [DOI] [PubMed] [Google Scholar]
- 58.Zoetendal, E. G., E. E. Vaughan, and W. M. de Vos. 2006. A microbial world within us. Mol. Microbiol. 59:1639-1650. [DOI] [PubMed] [Google Scholar]