Abstract
A PCR-denaturing gradient gel electrophoresis (DGGE) method was used to examine on-farm sources of Clostridium cluster I strains in four dairy farms over 2 years. Conventional microbiological analysis was used in parallel to monitor size of clostridial populations present in various components of the milk production chain (soil, forage, grass silage, maize silage, dry hay, and raw milk). PCR amplification with Clostridium cluster I-specific 16S rRNA gene primers followed by DGGE separation yielded a total of 47 operational taxonomic units (OTUs), which varied greatly with respect to frequency of occurrence. Some OTUs were found only in forage, and forage profiles differed according to farm location (southern or northern Québec). More clostridial contamination was found in maize silage than in grass silage. Milk represented a potential environment for certain OTUs. No OTU was milk specific, indicating that OTUs originated from other environments. Most (83%) of the OTUs detected in raw milk were also found in grass or maize silage. Milk DGGE profiles differed according to farm and sampling year and fit into two distinct categories. One milk profile category was characterized by the presence of a few dominant OTUs, the presence of which appeared to be more related to farm management than to feed contamination. OTUs were more varied in the second profile category. The identities of certain OTUs frequently found in milk were resolved by cloning and sequencing. Clostridium disporicum was identified as an important member of clostridial populations transmitted to milk. Clostridium tyrobutyricum was consistently found in milk and was widespread in the other farm environments examined.
Clostridia are ubiquitous in terrestrial environments. On the basis of 16S rRNA gene sequence analyses, 73 out of 152 validly described species fall within cluster I, often referred to as Clostridium sensu stricto (9, 39). Spores of clostridia belonging to cluster I are responsible for spoilage of cheeses with long ripening times, causing the so-called late blowing defect (17, 22, 38). Specifically, species able to convert lactic acid to butyric acid, carbon dioxide, and hydrogen at relatively low pH are detrimental (22, 38). Clostridium tyrobutyricum is considered the primary cause of late blowing. Other clostridia able to produce butyric acid have also been detected in milk and cheeses, especially Clostridium beijerinckii, Clostridium butyricum, and Clostridium sporogenes (21, 22, 29).
Silage is closely associated with the late blowing defect and is identified as the main source of milk contamination (19, 22, 29, 38). When silage fermentation conditions are not prone to rapid pH decrease and maintenance of uniformly anaerobic conditions, germination of clostridial spores and subsequent vegetative cell multiplication can occur (29, 33). Other farm environments may also contain clostridia. Based on cultivation or DNA similarity, cluster I species have been identified on forage surfaces (14, 20), as endophytic organisms of gramineous plants (25, 26), in grass and maize silage (29, 32), in the rumen (3, 39), in manure (12, 24, 29), and in milk and cheese (22, 29).
Applying cultivation-dependent methods to the study of clostridia in farm environments, contamination levels were assessed and hot spots were highlighted which favor clostridial growth and multiplication (18, 29, 33, 36, 38). Molecular methods have been applied to the examination of the late blowing spoilage process in cheese (7, 19, 21, 22). However, little is known about clostridial occurrence, population structure, and taxonomical composition in dairy farm environments. The present study aimed at examining the occurrence and dispersion of farm-related Clostridium species and their transmission pathways. Hence, the main objective of this study was to detect C. tyrobutyricum and other Clostridium cluster I species along the milk production chain (including soil, forage, silages, hay, and raw milk) under circumstances of silage feeding, using a cultivation-independent method.
MATERIALS AND METHODS
Sampling procedure.
Samples were collected from four dairy farms in Québec during two consecutive years. Six putative sources of Clostridium were sampled: soil, forage, dry hay, grass silage, maize silage, and raw milk. Two farms were located in a southern agricultural zone of Québec (Centre-du-Québec; farms A and B) and two in a northern zone (Abitibi-Témiscamingue, farms C and D; see the supplemental material for a description of farm characteristics). Samples were collected at different periods of the year. Soil and forage samples were collected at five constant locations per field in June, July, August, and September. Feed (dry hay, grass, and maize silage), and raw milk samples were collected in December, February, and March on a specific 13-day schedule. Feed samples were collected twice a day on days 1, 3, 9, and 11. Milk was sampled from the bulk tank in the morning of days 3, 5, 11, and 13. Silages and dry hay were sampled before being mixed together and served to the herd. Day 1 of the sampling schedule was set on a milk collection day, and raw milk samples were collected before the bulk tank was emptied. All samples were kept frozen at −20°C until analyzed (see below).
Determination of bacterial counts in soil and silage.
Clostridial spore numbers were estimated on reinforced clostridial agar medium (RCA; Oxoid, Basingstoke, England) containing 50 μg neutral red (Sigma, St. Louis, MO) and 200 μg d-cycloserine (Sigma) per ml. Soil (10 g), forage, dry hay, grass, and maize silages (20 g) were suspended in 90 ml (soil) or 180 ml (forage, hay, and silages) of sterile 0.2% (wt/vol) peptone water (Bacto Peptone; BD Diagnostic Systems, Sparks, MD) and homogenized in a stomacher (Stomacher 400 lab blender mixer; Seward, Worthing, United Kingdom), twice for 1 min at maximum speed. A fraction (10 ml) of the resulting suspensions was heated for 10 min in a water bath maintained at 70°C. Serial 10-fold dilutions were then prepared in sterile 0.2% peptone water and plated in duplicate on RCA medium with neutral red and d-cycloserine. Cultures were incubated for 7 days at 28°C in an anaerobic chamber (Thermo Electron Corporation, Waltham, MA). Colonies showing evidence of gold-yellow fluorescence under UV light and having negative catalase activity were considered Clostridium spp. Catalase activity was evaluated directly on RCA cultures placed in a conventional hood for at least 1 h before spraying of H2O2. Bubbles detected on colonies were indicative of catalase activity. Statistical analyses of plate counts from the various samples were performed on the log transformations, using JMP software (SAS Institute, Cary, NC). Results were compared using the Tukey-Kramer test with a 0.05 significance level. Counts in the range of 20 to 200 colonies per plate were used for statistical analysis. This corresponded to a detection limit of 2 log10.
Clostridium culture conditions and genomic DNA isolation.
Reference strains used in this study were C. beijerinckii ATCC 6015, Clostridium tyrobutyricum ATCC 25755, and C. sporogenes ATCC 11437. Clostridium spores were transferred to tryptic soy broth (Difco Laboratories, Cockeysville, MD), and the resulting cultures were incubated at 28°C for 72 h in an anaerobic chamber. For DNA extraction, 2 ml of culture was collected and centrifuged at 13,000 × g for 10 min. Genomic DNA was isolated from the pelleted cells with an Ultraclean soil DNA isolation kit (MoBio Laboratories, Carlsbad, CA), and the presence of DNA was confirmed on a 1% agarose gel containing ethidium bromide.
Genomic DNA extraction from farm samples.
Bacterial genomic DNA was extracted from the various farm samples with the Ultraclean soil DNA isolation kit. All samples were extracted individually, and the resulting DNA preparations were either pooled or analyzed separately as indicated below. For soil samples, 0.5 to 0.8 g of soil was added to the reaction tube depending on moisture content. For plant material, 20 g of fresh material (forage, grass, and maize silages) or 10 g of dry hay was suspended in 100 ml of sterile peptone water and homogenized (twice for 1 min each time at maximum speed) in a stomacher. From this mixture, 25 ml was taken and centrifuged (15,000 × g, 15 min), and DNA was extracted from the pellet. Raw milk samples (100 ml) were defatted by mixing with sterile 25% sodium citrate (6 ml), agitated (200 rpm, 5 min), and centrifuged (15,000 × g, 15 min). Supernatant and cream were removed, and pellets were used for DNA extraction. The concentration and quality of the extracted nucleic acids were determined by electrophoresis on a 1% agarose gel containing ethidium bromide. For soil, forage, dry hay, grass silage, and maize silage, genomic DNA extractions from a sample type were pooled for a given farm and collection period. Raw milk samples were analyzed individually. Thus, per farm and per year, the procedure yielded a maximum of four pooled soil DNA samples (each pooled soil DNA sample being assembled from all individual soil DNA samples from a particular farm and a particular collection period corresponding to June, July, August, or September; see above). Likewise, the maximum number of pooled DNA samples per farm and per year was four for forage and three for each type of feed (dry hay and silages). In the case of milk, a maximum of 24 DNA samples per farm and per year were obtained and analyzed individually.
Nested PCR amplification of Clostridium cluster I DNA.
The nested PCR strategy comprised a first amplification of most of the 16S rRNA gene (1.5 kb) using the universal eubacterial primers pA and pH′ (11). PCR amplifications were performed in a final volume of 50 μl, which included 5 μl of 10× ThermoPol PCR buffer (New England Biolabs, Ipswich, MA), a deoxynucleoside triphosphate mix (0.2 mM), primers (0.2 μM), 1 U of Taq polymerase (New England Biolabs, Ipswich, MA), and 1 μl of genomic DNA. Amplifications were performed in a Cetus DNA thermal cycler 480 (Perkin-Elmer, Waltham, MA). Template DNA was generally diluted 10-fold for forage, grass silage, maize silage, and dry hay samples, 100-fold for soil samples, and 1,000-fold for raw milk samples. Reaction mixtures were heated to 94°C for 5 min before the addition of the polymerase and cycled at 94°C for 1 min, 55°C for 1 min, and 72°C for 1.5 min. Cycles were repeated 35 times for raw milk samples and 30 times for the other samples. Finally, 5 μl of each PCR product was used for visualization on a 1% agarose gel containing ethidium bromide. Negative (without DNA) and positive (with DNA from reference strains) controls were included in every amplification.
A second round of PCR was then performed using the Clostridium-specific primers S-*-Chis-0150-a-S-23 and S-*-Cbot-0983-a-A-21, internal to the first primer set (35). Templates were 10- to 100-fold-diluted products from the first 16S rRNA gene amplification. Reaction mixtures and thermal cycles were as described above, with the appropriate annealing temperature for 28 cycles.
Separation of Clostridium amplicons by DGGE.
Clostridium cluster I-specific amplicons obtained by nested PCR amplification (see above) were diluted 10-fold and reamplified with primers pC and pD′, targeting the V3 region of the 16S rRNA gene and producing a 220-bp PCR product suitable for denaturing gradient gel electrophoresis (DGGE) analysis (11). A GC clamp (35) was incorporated in the 5′ region of the pC primer. The PCR mixture was as described above. A touchdown PCR protocol was used, in which the annealing temperature was decreased from 65°C to 55°C for 20 cycles, followed by 10 additional cycles at 55°C. After visualization of amplicons by gel electrophoresis in a 1% agarose gel, PCR products were submitted to DGGE analysis using the DCode universal mutation detection system (Bio-Rad Laboratories, Hercules, CA). Electrophoresis was performed in a 1-mm acrylamide gel (8% [wt/vol] acrylamide-bis-acrylamide, 37.5:1) containing a 35-to-70% denaturant gradient (100% denaturant corresponds to 7 M urea and 40% [vol/vol] deionized formamide) increasing in the direction of the electrophoretic run with a stacking gel on top. PCR products (25 μl) were loaded and migrated at 60 V for 16 h in Tris-acetate buffer (0.04 M Tris-acetate and 0.001 M EDTA, pH 8.0). Gels were stained for 15 min with SYBR gold (Invitrogen, Carlsbad, CA), visualized under UV illumination, and digitized using a GeneSnap apparatus (Syngene, Frederick, MD).
Gel normalization and analysis.
A standardization procedure was used to minimize migration differences between DGGE gels. This procedure relied on a migration ladder which was obtained by mixing amplicons of the V3 regions of three Clostridium reference strains (see above) amplified as described previously. To extend the range of the normalization ladder, V3 regions from four lactic acid bacteria frequently found in silage and milk samples were included in the ladder. These were Lactobacillus plantarum ATCC 4008, Leuconostoc mesenteroides subsp. mesenteroides ATCC 10877, Lactobacillus lactis ATCC 11454, and Pediococcus acidilactici UL5. PCR products suitable for DGGE were obtained from these lactic acid bacteria by using the primer pair pA-pH′ followed by pC-pD′.
The fingerprinting II Informatix software (Bio-Rad, Hercules, California) was used to generate a standardized database. A 5% band intensity threshold was set for the band selection process. Cluster analysis was performed on densitometric curves using the Pearson correlation coefficient and Ward distance calculations. A band-matching process, based on a 1.5% position tolerance, was used to obtain presence-absence matrixes, allowing the classification of individual bands according to their positions in gels and calculation of their frequency among farm samples.
Cloning and sequencing of the 16S rRNA gene from dominant Clostridium operational taxonomic units (OTUs).
Selected Clostridium cluster I-specific amplicons that generated prominent and frequently revealed DGGE bands in milk, were cloned for sequence analysis. Amplicons (820 bp) obtained with Clostridium cluster I-specific primers (see above) were cloned by ligation to the pGEM-T Easy vector system (Promega, Madison, WI) and transformed into competent Escherichia coli cells (JM109) according to the manufacturer's instructions. Plasmid purification was carried out with a QIAprep spin miniprep kit (Qiagen, Mississauga, Ontario, Canada). Recombinant clones were then examined by DGGE to match an individual clone with one of the migration positions already present in the database.
A selection of clones representing the various DGGE banding positions was sequenced. Sequencing reactions were performed with a BigDye Terminator cycle sequencing kit 3.1 with a genetic analyzer (3130XL; Applied Biosystems, Foster City, CA) by the Plate-forme d'Analyses Biomoléculaires (Université Laval, Québec, Canada). All clones were sequenced in both directions. Sequences were aligned using ClustalW2 (6), available from EMBL-EBI (Hinxton, United Kingdom). To exclude chimeric clones, sequences were screened using Chimera Check of the Ribosomal Database Project (8) before being assigned to taxonomic groups. Sequence identity searches were performed with the FASTA algorithm (30) against the European Molecular Biology Laboratory nucleotide database (EMBL-EBI, Hinxton, United Kingdom) and confirmed by BLASTn 2.2.16 (2) against the GenBank database at the National Center for Biotechnology Information (Bethesda, MD).
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the GenBank database under accession no. EU478466 to EU478488.
RESULTS
Culture-dependent detection of clostridia in dairy farms.
For soil samples, clostridial plate counts varied from the detection limit (2 log10) to a maximum of 5.2 log10 CFU g−1, with a median of 4.0 log10. Clostridial numbers stayed below the detection limit for forage and dry hay samples. Wide differences between clostridial numbers in grass and those in maize silage were noted. In grass silage, clostridial numbers ranged from below the detection limit to a maximum of 6.89 log10 CFU g−1. In this case, however, the median remained under the detection limit. Less variability in clostridial numbers was observed for maize silage, which ranged from below the detection limit to a maximum of 4.69 log10 CFU g−1. For maize silage clostridial counts, the median was well above the detection limit, at 4.45 log10 CFU g−1. In fact, 63% of grass silage samples, but only 8% of maize silage samples, were under the detection limit. The clostridial counts in milk consistently remained below the detection limit.
Efficiency of the PCR protocol and DGGE normalization.
The nested PCR protocol allowed amplification of DNA from reference clostridial strains (C. beijerinckii ATCC 6015, C. sporogenes ATCC 11437, and C. tyrobutyricum ATCC 25755) but not from nonclostridial reference strains (L. plantarum ATCC 4008, L. mesenteroides subsp. mesenteroides ATCC 10877, L. lactis ATCC 11454, and P. acidilactici UL5). Upon DNA amplification with the universal primers pC and pD′ (see Materials and Methods), each reference strain produced a single DGGE band, and the combination of these bands constituted a normalization ladder.
Detection of putative Clostridium OTUs in farm samples.
Using the nested PCR protocol, putative Clostridium cluster 1-specific amplicons were obtained for 164 farm and milk samples (72.6% of the total). The amplification-negative samples were randomly distributed among the various sample types. Putative Clostridium amplicons were subjected to DGGE analysis, and the resulting profiles were normalized using the normalization ladder (see Materials and Methods).
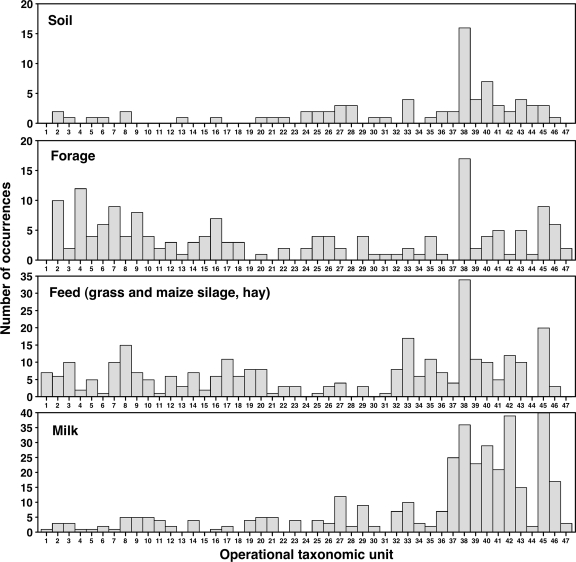
The melting profiles revealed various migration positions for the PCR products obtained from the different farm samples. A presence-absence matrix generated using a band-matching process was used to group bands in classes, according to their melting behavior in gels. A specific band class corresponded to an OTU, and a database that contained all observed OTUs, numbered from 1 to 47, was created. OTUs were variously distributed among farm samples (Fig. 1). Some of the OTUs exhibited a melting behavior identical to that of a Clostridium reference strain.
FIG. 1.
Histograms showing the number of occurrences of the 47 OTUs distinguished in DGGE profiles obtained from the complete set of PCR-positive dairy farm samples. Number of PCR-positive samples for each series: soil samples, 24; forage samples, 21; feed samples, 53 (including 22 grass silage, 14 maize silage, and 17 dry hay samples); raw milk samples, 66.
The OTUs varied greatly with respect to frequency of occurrence. Two OTUs were particularly frequent and well distributed in every type of sample. These were OTU 38 and OTU 45, which were detected in 63 and 44%, respectively, of all farm samples analyzed. By contrast, some other OTUs were rarely detected.
Distribution of OTUs in soil, forage, and feed samples.
Of the 47 OTUs included in the database, 43 were found in the field samples, including the soil and/or the forage samples. Specifically, 30 OTUs were found in soil and 41 in forage samples. Of these, 26 were detected in both materials, whereas 15 were found only in forage samples (Fig. 1). Thus, forage may be a privileged environment for some clostridial communities.
DGGE profiles from dry hay included 33 out of the 47 OTUs observed in the present study. Patterns from silage material showed 34 OTUs in grass and 24 OTUs in maize silage. Some differences were noted between grass silage profiles according to farm localization, as the dominant OTU in silages from southern farms (A and B), OTU 38, was mostly absent from silages from northern farms. Instead, these were dominated by OTU 33, which was rarely detected in southern grass silages (data not shown).
Clostridial OTUs in raw milk.
It appears that milk offered a potential environment for certain OTUs but discriminated against others that were better represented in other materials (Fig. 1). Of the 47 OTUs distinguished in the various samples from the four farms, 41 (87%) were detected in raw milk samples (Fig. 1). No OTU was milk specific, implying that a putative origin for all of the milk OTUs could be inferred by current DGGE profile analysis. Of the OTUs revealed in raw milk, 17% were not detected in grass or maize silage. This suggests that at least in these particular cases, milk contamination arose from a source other than silage.
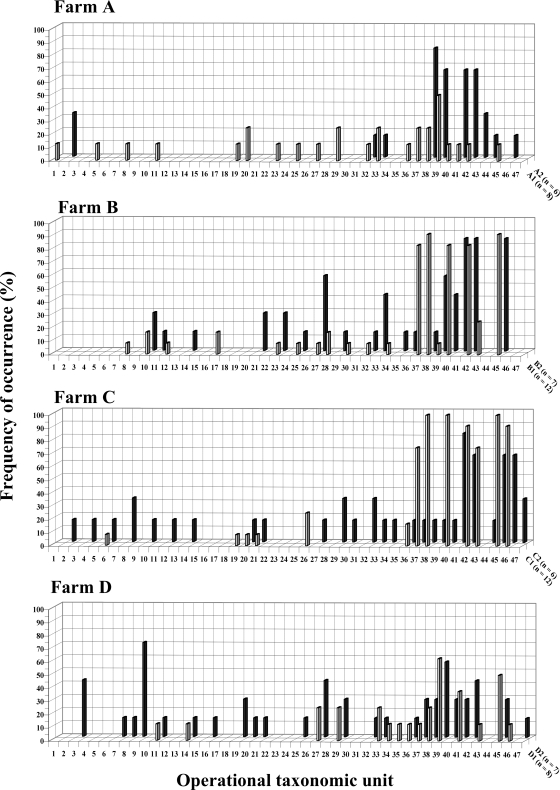
Analysis of raw milk DGGE profiles on a per-farm basis led to the classification of the profiles into two categories (Fig. 2). For the first year of sampling, farms B and C could be distinguished from farms A and D with respect to dominant OTUs. In particular, OTUs 37, 38, 40, 42, and 45 were detected in more than 80% of all raw milk samples from the two former farms. Other OTUs were less frequently detected in milk from those two farms (Fig. 2). In contrast, OTUs 42 and 45 were not dominant in milk samples from farms A and D, and OTU 40 was mostly absent. The latter two farms presented different DGGE banding patterns but shared the same dominant band, OTU 39. This second profile category was characterized by diversified patterns with a more even distribution (Fig. 2) than that of farms B and C.
FIG. 2.
Farm-specific three-dimensional histograms showing frequency of occurrence (percent PCR-positive samples [z axis]) of each of the 47 OTUs (x axis) detected by PCR-DGGE in milk samples during the 2 years of sampling (y axis). The year number designation (1 or 2) is preceded by the farm designation (A, B, C, or D) and followed by the number of samples (n) in the corresponding category.
Although first-year samples showed that OTUs 37, 38, 40, 42, and 45 were dominant in raw milk from farms B and C but not from farms A and D, no difference was noted with respect to prevalence of these OTUs in field or feed samples from the four farms (data not shown). Thus, it appeared that contamination of raw milk by those OTUs in farms B and C was related to farm management rather than to feed contamination.
For a given farm, milk DGGE profiles differed between years (Fig. 2). While profiles from farms B and D remained quite stable during both years, a shift was observed in farms A and C. For farm A, the number of detected OTUs decreased from 20 in year 1 to 10 in year 2, among which 4 were more frequently detected. In contrast, farm C OTU composition shifted from a specific profile containing few dominant OTUs in the first year to a highly diversified and well-distributed profile in the second year.
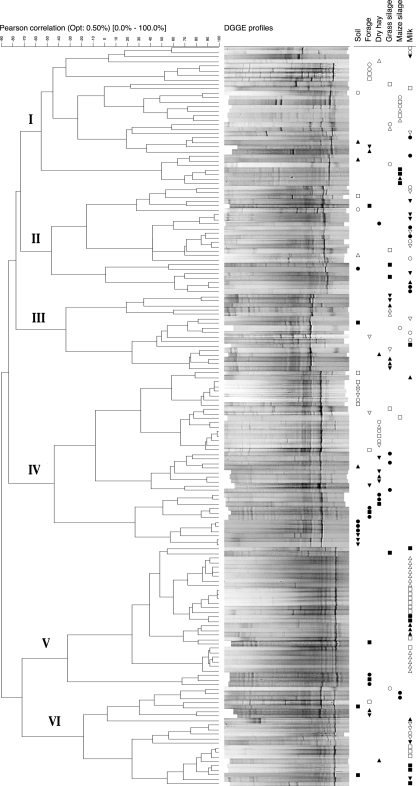
Cluster analysis of DGGE profiles.
The normalized DGGE profiles for all samples analyzed were represented as densitometric curves, which were then subjected to cluster analysis using Pearson's coefficient and Ward's algorithm. The DGGE profiles were classified into six groups (Fig. 3) based on a cutoff value of −70 on the similarity scale. Whereas profile group I was varied in its composition, groups II, III, IV, V, and VI were each characterized by the presence of one or two dominant OTUs. Profile group II was constituted by a majority of milk samples collected on farms A and D (Fig. 3), and its most intense bands corresponded to OTUs 38 and 39. Grass silage samples contributed 53% of the profile group III members, where the strongest densitometric peak corresponded to OTU 33. Soil and dry hay samples represented 70% of all samples comprising profile group IV. In this group, major bands corresponded to OTUs 37 and 38. Finally, groups V and VI mainly included DGGE profiles from raw milk samples, which represented 74% of all samples found in these clusters. The most intense band in profile group V and VI samples corresponded to OTU 45. In total, 59% of all milk OTUs belonged to profile groups V and VI. Most other milk samples clustered generally in groups I, II, and III, along with different field and feed samples.
FIG. 3.
Cluster analysis of DGGE patterns obtained from the various dairy environments (soil, forage, grass silage, maize silage, dry hay, and raw milk), carried out using Pearson's coefficient and Ward's algorithm. The dendrogram illustrating the results of the analysis is presented on the left, with defined profile group numbers on the extreme left. Corresponding profiles are shown adjacent to their localization in the dendrogram. The origin of samples is indicated by a symbol: (○) farm A, year 1; (•) farm A, year 2; (□) farm B, year 1; (▪) farm B, year 2; (▵) farm C, year 1; (▴) farm C, year 2; (▿) farm D, year 1; (▾) farm D, year 2. Symbols are arranged according to sample type (top right).
An overall examination of the clustering results suggests that the various farm environments examined here had defining effects on clostridial communities. In the case of raw milk, which provides a preferential environment for OTUs related to profile groups V and VI (Fig. 3), clostridial community definition may result from milk physicochemical properties, or from hygienic measures restricting access to the bulk tank.
Clostridium species identification and association with OTUs.
Some of the OTUs coincided with DGGE bands yielded by Clostridium reference strains included in the normalization ladder (see above for the composition of the ladder). Thus, on the basis of DGGE migration patterns, OTUs 32, 33, and 39 corresponded to C. beijerinckii ATCC 6015, C. tyrobutyricum ATCC 25755, and C. sporogenes ATCC 11437, respectively.
In addition to this observation of coincident bands, cloning and DNA sequencing were also used to identify some of the most frequent OTUs observed in raw milk. Clones of Clostridium-specific amplification products were obtained from certain samples, reprofiled by DGGE to establish a clone-OTU match, and sequenced over their 0.82-kb length. DNA from bands sharing the same DGGE melting profile was cloned independently from different samples. Sequence similarity searches against databases allowed the identification of 11 OTUs (Table 1). BLAST searches against GenBank gave high similarity levels (>98%) for all clones except one (95%).
TABLE 1.
Sequence similarities of Clostridium clones corresponding to OTUs detected in raw milk
| OTUa | Most closely related organism(s) | Similarity (%) |
|---|---|---|
| 32 | C. tyrobutyricum | 99 |
| 33 | C. tyrobutyricum | 99 |
| 35 | C. tyrobutyricum | 99 |
| 37 | “C. favososporum,” Clostridium sp. strain 915-1 | 98 |
| 37 | C. tyrobutyricum | 98 |
| 38 | C. sporogenes, C. botulinum | 99 |
| 38 | C. tyrobutyricum | 99 |
| 38 | C. magnum | 98 |
| 39 | “C. favososporum,” Clostridium sp. strain 915-1 | 99 |
| 40 | “C. autoethanogenum,” “C. ragsdalei” | 95 |
| 42 | C. disporicum | 98 |
| 45 | C. disporicum | 99 |
| 46 | C. disporicum | 99 |
| 47 | C. disporicum | 98 |
16S rRNA genes from milk samples were amplified by using Clostridium-specific primers (see Materials and Methods) and cloned as 820-bp inserts. Correspondence between clones and OTUs was established by DGGE analysis, and sequences from selected clones were used for comparative analysis against GenBank. Entries for the same OTU appear on separate lines when they correspond to both different clones and different species.
Clone sequence analysis suggests that distinct OTUs share homology to DNA from the same species. For example, clones corresponding to OTUs 42, 45, 46, and 47 showed more than 98% similarity with C. disporicum. However, these four OTUs differed with respect to both distribution in the various sample types and farms and frequency of occurrence. Alignment of the 170-bp sequences represented in the DGGE bands showed that only one or two nucleotides differed among these four OTUs, this difference being sufficient to change the melting behavior during DGGE. Similarly, we identified six OTUs for which the closest relative in GenBank was C. tyrobutyricum, with 99% similarity.
Two criteria were used to determine correspondence between OTU and clostridial species, namely, coincidence in migration pattern between OTUs and certain components of the normalization ladder (see above) and DNA sequence analysis (Table 1). Some OTUs were not completely resolved following this analysis. These unresolved OTUs were 32 (C. tyrobutyricum and C. beijerinckii), 37 (C. tyrobutyricum and “Clostridium favososporum”), 38 (C. tyrobutyricum, C. sporogenes, and C. magnum) and 39 (“C. favososporum” and C. sporogenes). As mentioned above, OTU 38 was the most frequent in dairy farms. DNA sequence analysis shows that, in fact, three different Clostridium species comigrate at this position, two of which, C. tyrobutyricum and C. sporogenes, are associated with late blowing of cheese. Hence, it appears that three clostridial species contribute to the high prevalence of this particular OTU in farm samples.
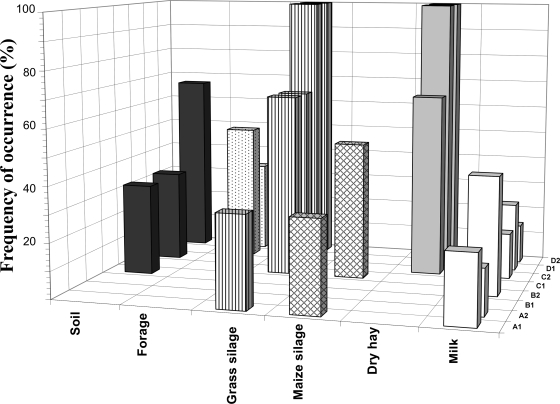
Identity of OTU 33 was confirmed by comigration with the pure culture C. tyrobutyricum ATCC 25755 and by DNA cloning and sequencing (see above). This prompted an examination of the on-farm distribution of this C. tyrobutyricum-specific OTU, which appeared to be widespread in the various environments examined and frequently found in the bulk tank (Fig. 4).
FIG. 4.
Three-dimensional histogram illustrating the frequency of occurrence (percent PCR-positive samples) of OTU 33 with respect to sample type for the four farms (A, B, C, and D) and sampling years (1 and 2). OTU 33 was identified as C. tyrobutyricum by sequencing analysis (Table 1) and observation of comigration with the DGGE amplicon from C. tyrobutyricum ATCC 25755.
DISCUSSION
Previous studies examined populations, subgroups, or species of clostridia in landfills using temporal temperature gradient gel electrophoresis (35), in plant tissues using terminal restriction fragment length polymorphism analysis (26), in cheese using DGGE (7, 22), and in human fecal samples using DGGE combined with clone library analysis (31). In the present work, the occurrence of Clostridium cluster I in the dairy ecosystem was investigated by PCR-DGGE. To our knowledge, this is the first culture-independent study of clostridial presence in the dairy production chain, encompassing various elements of the milk production chain from the field to the bulk tank.
Results of Clostridium cluster I-specific PCR analyses suggest that clostridia are nearly ubiquitous members of the dairy ecosystem. They were detected in ca. 75% of the samples analyzed, independently of the environment considered. While Clostridium DNA was detected by PCR in every sample type, enumeration results showed that high population densities occurred in soil and silage but not in forage, hay, or milk. Results showed wide differences between population numbers in grass silage and those in maize silage. Counts were, on average, 2 orders of magnitude higher in maize silage. This result is in accordance with previous findings that identified corn silage as an important source of butyric spores, especially following aerobic deterioration (37).
Every environment sampled yielded a variety of clostridial OTUs. In addition to those OTUs that were detected, others might have been present, but in such low numbers that they were not prominent in the DGGE profiles (4, 15, 16). This may be the case, in particular, with soil samples that might harbor small populations of dormant clostridia.
Several authors have reported comigration of amplicons from different species in DGGE gels (5, 13, 22). This may correspond to closely related species harboring related rRNA gene sequences that remain poorly separated (5), or to relatively different sequences that nevertheless display the same melting behavior (5, 10, 31). In particular, members of Clostridium cluster I exhibit relatively high levels of intracluster similarity (>92%) despite having markedly different phenotypes (9, 39). In the present study, some clostridial OTUs defined by DGGE correspond to more than one species.
Conversely, different OTUs exhibited high levels of similarity with sequences of the same species (e.g., four OTUs corresponded to C. disporicum and four OTUs to C. tyrobutyricum). Previous studies yielded similar results (34), and such multiple bands were attributed to intraspecies 16S rRNA gene heterogeneity (5, 7, 10, 22, 28), to differences among strains from the same species (31), or to PCR artifacts or heteroduplex formation (28). Some Clostridium species are also known to possess multiple rrn operons, which may generate multiple OTUs (1). In principle, a single base difference between two DNA fragments may lead to distinct DGGE migration behaviors (10). The multiple banding patterns observed here for some of the clostridial species may be attributed to one or several of these factors. However, sequence analysis of DGGE bands ruled out the possibility of extensive PCR artifacts altering the conclusions of the present work.
The various farm environments examined have defining effects on clostridial communities. For instance, forage appeared to be a privileged environment for certain clostridia. Indeed, various clostridial OTUs were detected in forage samples but not in soil. It was recently demonstrated that some clostridia occur as N-fixing endophytes in gramineous plant tissues, where their growth might be supported by oxygen consumption by associated bacteria (25).
Variation according to geographical area was observed in some sample types other than milk. Clostridial communities from the forage samples were placed in different profile groups according to geographical (northern or southern) farm localization. Differences between clostridial populations from grass silages were also noted. These differences might be explained by variations in plant species (which included gramineous and legume species), soil physicochemical properties (27), and climate.
Milk offered a potential environment for certain OTUs but discriminated against others that were well represented in other materials. Comparison of OTU occurrence in raw milk suggested that two categories of DGGE profiles existed. Milk from a particular farm either yielded DGGE profiles belonging to the same category over the two consecutive sampling years or shifted from one profile category to another. The first profile category, encountered in farms B and C for the first year of sampling, was characterized by strong dominating bands and few minor bands (Fig. 2). The same dominant OTUs were found in both farms, and these presumably represent numerically prominent clostridia in the bulk tank. Their lower frequency on other farms suggests that their massive presence may be due to constant management factors or animal behavior. Patterns were repeated in morning and night samples, between sampling periods and between different farms. Such putative OTU occurrence-determining factors appear to be independent of the geographical location and farm characteristics, which varied greatly on both farms where the profiles with dominant OTUs were observed. In contrast, the second profile category was defined by a relatively even frequency of diversified OTUs. This OTU occurrence pattern partly reflects the clostridial composition found in other farm environments, with an attenuation effect presumably resulting from milking processes or milk properties, which restrict OTU access to or survival in milk. Variability in DGGE profiles belonging to this second category between morning and night samples, periods, and farms suggests that in this case, contamination is the result of sporadic introductions.
Shifts in DGGE profile category were observed from one year to the next in farms A and C. Such changes again suggest that factors, perhaps related to agricultural practice or animal behavior, were determinant in shaping clostridial populations in milk bulk tanks. For example, a few highly contaminated cows can significantly affect bulk tank clostridial numbers (38). With regard to this, it is noteworthy that OTUs 42 and 45, which are prominent in samples of the DGGE profile category with dominant OTUs, share 98% similarity with C. disporicum. DNA highly homologous to C. disporicum DNA was found in swine manure, notably in the biofilm fraction of aerated manure (23).
The observations presented here help delineate a farm clostridial reservoir, from which bacteria may be transferred to milk across a sanitation barrier. The components of this reservoir, including soil, growing plants, hay, and silage, contribute variously to the final composition of clostridial milk populations. A putative source for all of the detected milk OTUs was inferred from the DGGE profile analysis. This suggests that no OTU was milk or animal specific and that clostridial milk contamination depends on contamination from outside sources. Using classical microbiology methods, other authors have reached similar conclusions (29, 37).
No correlation between the prevalence of particular clostridial OTUs in nonmilk samples and that in milk samples was observed. This reflects the effect of a sanitation barrier and indicates that certain attributes are required of the clostridia for their successful transfer to and survival in the bulk tank. C. tyrobutyricum, which has been implicated as the main agent of late blowing of cheese, was detected in all farm environments examined, but its presence and frequency of occurrence varied on a per-farm basis (Fig. 4). This species was consistently found in milk, and further studies on the clostridial transmission capacity of the milk production chain and associated environments will help design source-directed measures for contamination control.
Supplementary Material
Acknowledgments
We thank Alan J. McCarthy and Robert Lockhart (University of Liverpool) for their help in elaboration of the protocols and Gisèle LaPointe (Université Laval) for helpful suggestions throughout the course of this work. We are grateful to Patrick Laplante and Sandra Martel for technical assistance.
This work was supported by FQRNT (project 2003-NO-938980), Novalait, and Agriculture and Agri-Food Canada.
Footnotes
Published ahead of print on 29 August 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Polz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Microbiol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Attwood, G., D. Li, D. Pacheco, and M. Tavendale. 2006. Production of indolic compounds by rumen bacteria isolated from grazing ruminants. J. Appl. Microbiol. 100:1261-1271. [DOI] [PubMed] [Google Scholar]
- 4.Bent, S. J., J. D. Pierson, and L. J. Forney. 2007. Measuring species richness based on microbial community fingerprints: the emperor has no clothes. Appl. Environ. Microbiol. 73:2399-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodelier, P. L. E., M. Meima-Franke, G. Zwart, and H. J. Laanbroek. 2005. New DGGE strategies for the analyses of methanotrophic microbial communities using different combinations of existing 16S rRNA-based primers. FEMS Microbiol. Ecol. 52:163-174. [DOI] [PubMed] [Google Scholar]
- 6.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocolin, L., N. Innocente, M. Biasutti, and G. Comi. 2004. The late blowing in cheese: a new molecular approach based on PCR and DGGE to study the microbial ecology of the alteration process. Int. J. Food Microbiol. 90:83-91. [DOI] [PubMed] [Google Scholar]
- 8.Cole, J., B. Chai, T. Marsh, R. Farris, Q. Wang, S. Kulam, S. Chandra, D. McGarrell, T. Schmidt, G. Garrity, and J. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. E. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 4:812-826. [DOI] [PubMed] [Google Scholar]
- 10.Crosby, L. D., and C. S. Criddle. 2003. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. BioTechniques 34:1-9. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elsayed, S., and K. Zhang. 2005. Bacteremia caused by Clostridium intestinale. J. Clin. Microbiol. 43:2018-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ercolini, D. 2004. PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J. Microbiol. Methods 56:297-314. [DOI] [PubMed] [Google Scholar]
- 14.Flythe, M. D., and J. B. Russell. 2004. The effect of pH and a bacteriocin (bovicin HC5) on Clostridium sporogenes MD1, a bacterium that has the ability to degrade amino acids in ensiled plant materials. FEMS Microbiol. Ecol. 47:215-222. [DOI] [PubMed] [Google Scholar]
- 15.Forney, L. J., X. Zhou, and C. J. Brown. 2004. Molecular microbial ecology: land of the one-eyed king. Curr. Opin. Microbiol. 7:210-220. [DOI] [PubMed] [Google Scholar]
- 16.Godon, J.-J. 2003. Actualité des méthodes d'analyse sans a priori pour l'étude des émergences. INRA, Laboratoire de Biotechnologie de l'Environnement, Narbonne, France.
- 17.Herman, L. M., J. H. De Block, and G. M. Waes. 1995. A direct PCR detection method for Clostridium tyrobutyricum spores in up to 100 milliliters of raw milk. Appl. Environ. Microbiol. 61:4141-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jonsson, A. 1990. Enumeration and confirmation of Clostridium tyrobutyricum in silages using neutral red, D-cycloserine, and lactate dehydrogenase activity. J. Dairy Sci. 73:719-725. [DOI] [PubMed] [Google Scholar]
- 19.Klijn, N., F. F. Nieuwenhof, J. D. Hoolwerf, C. B. van der Waals, and A. H. Weerkamp. 1995. Identification of Clostridium tyrobutyricum as the causative agent of late blowing in cheese by species-specific PCR amplification. Appl. Environ. Microbiol. 61:2919-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lango, Z., and H. Heinonentanski. 1995. Occurrence of Clostridium tyrobutyricum in cattle slurry and fresh forage grasses. Bioresour. Technol. 53:189-191. [Google Scholar]
- 21.Le Bourhis, A. G., J. Doré, J.-P. Carlier, J.-F. Chamba, M.-R. Popoff, and J.-L. Tholozan. 2007. Contribution of C. beijerinckii and C. sporogenes in association with C. tyrobutyricum to the butyric fermentation in Emmental type cheese. Int. J. Food Microbiol. 113:154-163. [DOI] [PubMed] [Google Scholar]
- 22.Le Bourhis, A. G., K. Saunier, J. Dore, J. P. Carlier, J. F. Chamba, M. R. Popoff, and J. L. Tholozan. 2005. Development and validation of PCR primers to assess the diversity of Clostridium spp. in cheese by temporal temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 71:29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leung, K., and E. Topp. 2001. Bacterial community dynamics in liquid swine manure during storage: molecular analysis using DGGE/PCR of 16S rDNA. FEMS Microbiol. Ecol. 38:169-177. [Google Scholar]
- 24.McGarvey, J. A., W. G. Miller, S. Sanchez, and L. Stanker. 2004. Identification of bacterial populations in dairy wastewaters by use of 16S rRNA gene sequences and other genetic markers. Appl. Environ. Microbiol. 70:4267-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minamisawa, K., K. Nishioka, T. Miyaki, B. Ye, T. Miyamoto, M. You, A. Saito, M. Saito, W. L. Barraquio, N. Teaumroong, T. Sein, and T. Sato. 2004. Anaerobic nitrogen-fixing consortia consisting of clostridia isolated from gramineous plants. Appl. Environ. Microbiol. 70:3096-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyamoto, T., M. Kawahara, and K. Minamisawa. 2004. Novel endophytic nitrogen-fixing clostridia from the grass Miscanthus sinensis as revealed by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 70:6580-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nunan, N., T. J. Daniell, B. K. Singh, A. Papert, J. W. McNicol, and J. I. Prosser. 2005. Links between plant and rhizoplane bacterial communities in grassland soils, characterized using molecular techniques. Appl. Environ. Microbiol. 71:6784-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogier, J. C., O. Son, A. Gruss, P. Tailliez, and A. Delacroix-Buchet. 2002. Identification of the bacterial microflora in dairy products by temporal temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 68:3691-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pahlow, G., R. E. Muck, F. Driehuis, and S. J. W. H. O. Elferink. 2003. Microbiology of ensiling, p. 31-93. In D. R. Buxton, R. E. Muck, and J. H. Harrison (ed.), Silage science and technology. American Society of Agronomy/Crop Science Society of America/Soil Science Society of America, Madison, WI.
- 30.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen, J., B. Zhang, G. Wei, X. Pang, H. Wei, M. Li, Y. Zhang, W. Jia, and L. Zhao. 2006. Molecular profiling of the Clostridium leptum subgroup in human fecal microflora by PCR-denaturing gradient gel electrophoresis and clone library analysis. Appl. Environ. Microbiol. 72:5232-5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sparo, M. D., and R. A. Mallo. 2001. Evaluacion de la flora bacteriana en un ensilado natural de maiz (evaluation of the bacterial flora in natural corn silage). Rev. Argent. Microbiol. 33:75-80. [PubMed] [Google Scholar]
- 33.te Giffel, M. C., A. Wagendorp, A. Herrewegh, and F. Driehuis. 2002. Bacterial spores in silage and raw milk. Antonie van Leeuwenhoek 81:625-630. [DOI] [PubMed] [Google Scholar]
- 34.Tourova, T. P. 2003. Copy number of ribosomal operons in prokaryotes and its effect on phylogenetic analyses. Microbiology 72:389-402. [PubMed] [Google Scholar]
- 35.Van Dyke, M. I., and A. J. McCarthy. 2002. Molecular biological detection and characterization of Clostridium populations in municipal landfill sites. Appl. Environ. Microbiol. 68:2049-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vissers, M. 2007. Modeling to control spores in raw milk. Ph.D. thesis. Wageningen University, Wageningen, The Netherlands.
- 37.Vissers, M. M. M., F. Driehuis, M. C. T. Giffel, P. D. Jong, and J. M. G. Lankveld. 2007. Concentrations of butyric acid bacteria spores in silage and relationships with aerobic deterioration. J. Dairy Sci. 90:928-936. [DOI] [PubMed] [Google Scholar]
- 38.Vissers, M. M. M., F. Driehuis, M. C. Te Giffel, P. De Jong, and J. M. Lankveld. 2006. Improving farm management by modeling the contamination of farm tank milk with butyric acid bacteria. J. Dairy Sci. 89:850-858. [DOI] [PubMed] [Google Scholar]
- 39.Wiegel, J., R. Tanner, and F. Rainey. 2006. An introduction to the family Clostridiaceae, p. 654-678. In M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 4. Springer, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.