Abstract
Cyanase catalyzes the decomposition of cyanate into CO2 and ammonium, with carbamate as an unstable intermediate. The cyanase of Pseudomonas pseudoalcaligenes CECT5344 was negatively regulated by ammonium and positively regulated by cyanate, cyanide, and some cyanometallic complexes. Cyanase activity was not detected in cell extracts from cells grown with ammonium, even in the presence of cyanate. Nevertheless, a low level of cyanase activity was detected in nitrogen-starved cells. The cyn gene cluster of P. pseudoalcaligenes CECT5344 was cloned and analyzed. The cynA, cynB, and cynD genes encode an ABC-type transporter, the cynS gene codes for the cyanase, and the cynF gene encodes a novel σ54-dependent transcriptional regulator which is not present in other bacterial cyn gene clusters. The CynS protein was expressed in Escherichia coli and purified by following a simple and rapid protocol. The P. pseudoalcaligenes cyanase showed an optimal pH of 8.5°C and a temperature of 65°C. An insertion mutation was generated in the cynS gene. The resulting mutant was unable to use cyanate as the sole nitrogen source but showed the same resistance to cyanate as the wild-type strain. These results, in conjunction with the induction pattern of the enzymatic activity, suggest that the enzyme has an assimilatory function. Although the induction of cyanase activity in cyanide-degrading cells suggests that some cyanate may be generated from cyanide, the cynS mutant was not affected in its ability to degrade cyanide, which unambiguously indicates that cyanate is not a central metabolite in cyanide assimilation.
Cyanate is a toxic compound produced by industry, but it is also generated from some metabolites, such as urea and carbamoylphosphate (10). Cyanase (EC 4.2.1.104) is an enzyme that catalyzes the decomposition of cyanate into CO2 and ammonium. The nucleophilic reactant that attacks and breaks down the cyanate is bicarbonate, with carbamate as an unstable intermediate (3). From this reaction, the importance of bicarbonate in the process and the role of carbonic anhydrase in recycling carbon dioxide into bicarbonate can be deduced (13). Probably for this reason, the cyanase (cynS) and carbonic anhydrase genes are often clustered together (9).
At least three physiological roles have been attributed to cyanase activity, i.e., nitrogen assimilation, cyanate detoxification, and metabolism regulation. Since the enzyme catalyzes the direct formation of ammonium from cyanate, cyanase activity allows some bacteria to utilize cyanate as a nitrogen source. All heterotrophic bacteria able to assimilate cyanate have cyanase activity (40, 18). This enzyme has also been found in cyanobacteria (11, 27) and plants (1).
The role of cyanase in detoxification is based on the toxicity of cyanate at relatively low concentrations (10, 18). This toxicity is mainly due to the reactivity of isocyanate, which is in equilibrium with cyanate and carbamoylates some nucleophilic groups of proteins (10, 31). Carbamoylation of enzymes like carbamoylphosphate synthetase is a classic example of this process, but it has been also described for hormones and structural proteins, in which it causes both functional and structural changes (16, 30). Protein carbamoylation by cyanate, especially in the eye and kidney, causes severe health problems in mammals (17, 26, 31). In addition, cyanate may chelate metal centers in some enzymes, such as carbonic anhydrase, superoxide dismutase, and carboxypeptidase A (10). In the case of nitrate reductase activity, cyanate has been shown to be a competitive inhibitor of nitrate, and due to its oxidative character, cyanate also reactivates the reductively inactivated form of the enzyme (7, 8). Finally, a regulatory role for cyanate in the context of nitrogen metabolism has been proposed (39). This function is based on the fact that the cyanate concentration in ammonium-grown cells is relatively high due to the spontaneous decomposition of carbamoylphosphate. Therefore, the cyanate concentration may reflect the nitrogen status of the cell.
Cyanate and its derivatives have been widely used as herbicides as well as precursors in the synthesis of polymers (14, 16). It is worth noting that the greatest disaster in the world due to a chemical-industry accident was caused in 1984 in Bhopal (India) by the escape of methyl-isocyanate. Another way that cyanate is released into the environment is spontaneous cyanide photooxidation and, alternatively, the oxidative treatment of cyanide-containing wastes (28). Consequently, cyanate and cyanide are frequently cocontaminants. Moreover, cyanate has been proposed to be an intermediate in cyanide assimilation (12, 24). However, although the incorporation of oxygen in the enzymatic conversion of cyanide has been demonstrated (43), the putative oxygenase catalyzing the monooxygenation of cyanide has never been purified, nor has the corresponding gene been cloned. Cyanase is not essential for the degradation of cyanide in Pseudomonas fluorescens, since an uncharacterized mutant lacking cyanase activity is able to degrade cyanide (19).
The main aim of this work was to investigate the role of cyanase in cyanide assimilation by Pseudomonas pseudoalcaligenes CECT5344. Cyanate metabolism was characterized at both the genetic and biochemical levels, with the conclusion that the cyanase of this strain has an assimilatory role but is not essential for cyanide assimilation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. P. pseudoalcaligenes strains were grown either in Luria-Bertani (LB) medium or in the defined mineral salts medium M9 (24) without ammonium and citrate and supplemented with 50 mM acetate as the carbon source and an adjusted pH of 9.5. The appropriate nitrogen source was added from sterilized stocks at the indicated concentrations. Escherichia coli strains were grown in LB medium. For growth on solid media, 1.5% bacteriological agar was added. Cells were incubated at 30°C (P. pseudoalcaligenes) or 37°C (E. coli) on a rotary shaker at 230 rpm. The media for antibiotic-resistant strains were supplemented with ampicillin (100 μg/ml), kanamycin (25 μg/ml), nalidixic acid (10 μg/ml) or gentamicin (Gm; 20 μg/ml), as appropriate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristic(s)a | Reference or source |
|---|---|---|
| P. pseudoalcaligenes strains | ||
| CECT5344 | Wild type; uses cyanide as N source | 24 |
| CECT5344N | Spontaneous mutant resistant to nalidixic acid | This work |
| CECT5344N lacking the cyn gene | Gm-directed mutant in the cynS gene | This work |
| E. coli strains | ||
| DH5α | Lac[minus]; host for most of the plasmids | 32 |
| S17-1 | Tra+; host for the mobilizable mob plasmids | 35 |
| Plasmids | ||
| pGEM-T | Vector (Apr) used for cloning fragments amplified by PCR | Promega |
| pBluescript SK(+) | Multipurpose cloning vector (Apr) | Stratagene |
| pK18mobδE | Suicide vector in Pseudomonas spp. (Kmr) | 33 |
| pMS255 | Vector containing a Gm resistance cassette | 4 |
| pVIC1 | pK18mobδE derivative containing the 1.3-kb fragment of the cynS gene | This work |
| pVIC2 | pVC1 derivative with the cynS gene interrupted by the Gm cassette | This work |
| pMH1 | pBluescript SK(+) harboring the SalI fragment (1.5 kb) that contains the entire cynS gene | This work |
Apr, ampicillin resistant; Kmr, kanamycin resistant.
Analytical determinations.
Bacterial growth was monitored by following the optical density of the culture at 600 nm (OD600). The nitrate, nitrite, and ammonium concentrations were determined as previously described (24). The concentration of free cyanide was determined colorimetrically (24). Determination of the cyanate concentration was based on its chemical conversion into ammonium as follows: 0.1 ml of 6 M HCl was added to 0.9 ml of the sample, and the mixture was boiled for 1 min at 100°C. After the mixture cooled, the cyanate concentration was measured as the difference in the ammonium concentration before and after treatment. The protein concentration was determined by using a modified version of the Lowry method (34).
Cyanase activity assay.
Cyanase (EC 4.2.1.104) was assayed by the method described by Anderson (3). One milliliter of the reaction mixture included 50 mM Tris-HCl buffer (pH 8.5), 3 mM NaHCO3, and the appropriate volume of cell extract. The cell extract was obtained by disrupting the reaction in the cells by two passages through a French pressure cell at 120 MPa and removing the cell debris by centrifugation at 20,000 × g for 20 min. The addition of 2 mM KCNO (potassium cyanate) started the reaction, and the reaction mixture was incubated at 65°C for 5 to 10 min. Finally, the concentration of ammonium formed from cyanate was determined as previously described (24). One unit of activity is defined as the amount of enzyme producing one micromole of ammonium per minute under assay conditions.
Nucleic acid manipulations and sequence analysis.
DNA manipulation was performed according to the methods of Sambrook et al. (32). A 100-bp fragment of the cynS gene was amplified by PCR (Expand high fidelity PCR; Roche; Mastercycler personal; Eppendorf), using the degenerate primers Cyn1F 5′-GATTCCAACTGACCCG(A-T)(C-T)GAT(G-C)TATCGCTTC-3′ and Cyn2R 5′-CGCTC(A-G)(A-C)ATGATGCCATCGCCAAATTT(C-T)TC-3′, with P. pseudoalcaligenes CECT5344 genomic DNA as a template. To design the primers, the CynS sequences from E. coli (P58704) and Pseudomonas aeruginosa (ZP_00975105) were aligned, and the C-terminal conserved domain (see Fig. 4) was used to obtain a DNA consensus sequence in the nucleotide 86 to 96 and 105 to 115 regions (E. coli nomenclature) for the forward and reverse primers, respectively. The PCR program included an initial step of denaturation at 94°C for 2 min, 30 cycles of denaturation at 94°C for 30 s each, annealing at 50°C for 1 min, and elongation at 72°C for 1 min. The PCR product was digoxigenin labeled and used as a probe for hybridization with SalI-digested genomic DNA of P. pseudoalcaligenes CECT5344. A positive 1.5-kb fragment was cloned into plasmid pBluescript II KS and transformed into E. coli DH5α, using standard protocols (32).
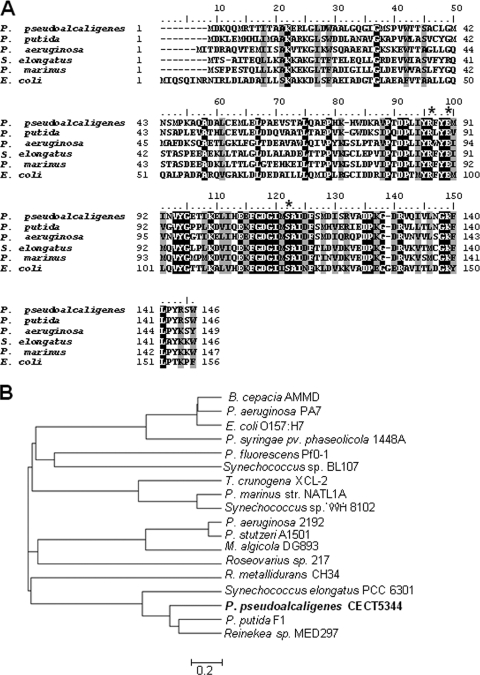
FIG. 4.
(A) Multiple amino acid sequence alignment of cyanases from several bacteria, P. pseudoalcaligenes CECT5344 (EF 451798), P. putida F1 (YP_001268549), P. aeruginosa 2192 (ZP_00975588), S. elongatus PCC 6301 (YP_172699), P. marinus strain NATL1A (YP_001013899), and E. coli O157:H7 (P58704). Identical residues are in black, and similar residues are in gray. The R, E, and S residues of the catalytic triad are marked by asterisks. (B) Phylogenetic tree of bacterial cyanases. In addition to the bacterial strains described for panel A, the tree includes Burkholderia cepacia AMMD (YP_777825), P. aeruginosa PA7 (NP-250742), Pseudomonas syringae pv. phaseolicola 1448A (YP_275568), P. fluorescens Pf0-1 (YP_349098), Synechococcus sp. BL107 (ZP_01469110), Thiomicrospira crunogena (YP_390311), Synechococcus sp. WH 8102 (NP_898579), Pseudomonas stutzeri (YP_001174036), Marinobacter algicola (ZP_01892318), Roseovarius sp. 217 (ZP_01036876), Ralstonia metallidurans (YP_587992), and Reinekea sp. (ZP_01113316). The tree was generated with MEGA 4.1 software.
To obtain more information about the P. pseudoalcaligenes cyn gene cluster, genomic DNA was digested with SacI and ligated into pBluescript previously digested with SacI. PCR was carried out with this ligation mixture as a template and the primers CynLF (5′-TCGAGCGAGCGGTTCACCAGAAAGTCCACGA-3′) and T3MJ (5′-GCGCAATTAACCCTCACTAAAGGGAACA-3′). The PCR program had an initial step of denaturation at 98°C for 3 min, 30 cycles of denaturation at 98°C for 30 s each, annealing at 65°C for 1 min, and elongation at 72°C for 4 min. A positive 1.7-kb fragment was isolated, and further analyses were performed. The genomic DNA was digested with the restriction enzymes ApaI and SmaI and ligated into pBluescript previously digested with ApaI/SmaI. PCR was carried out with this ligation mixture as a template and the primers T7-long (5′-ACGACTCACTATAGGGCGAATTGG-3′) and CynLF6 (5′-ACTCGGCCAATAGCGTCATGCAGCGT-3′). The PCR program used was the same as that described above. A positive 3.8-kb fragment was isolated.
DNA sequences were analyzed with the software programs DNA Strider version 1.1 and SeqEd v. 1.03 from the Genetics Computer Group at the University of Wisconsin (5). Database searches and peptide sequence alignments were performed with BLAST (2) and CLUSTAL W (41) tools, respectively.
Construction of a cyanase-deficient mutant.
In order to generate a mutant lacking the cynS gene, a spontaneous nalidixic acid-resistant strain of P. pseudoalcaligenes was previously obtained. To inactivate cynS by insertion, primers Cyn3 (5′-AAAAGGTACCGTAACCACCTCGTGGACTTTCTG-3′) and Cyn4 (5′-AAAAAAGCTTGTTGAGGTAGGCAGTGACCG-3′) (the KpnI and HindIII restriction sites created to facilitate cloning procedures are underlined) were designed to target the regions flanking cynS. A 1.3-kb PCR product containing the cynS gene was obtained and then cloned into the KpnI/HindIII sites in the kanamycin-resistant plasmid pK18mobδE, resulting in the plasmid pVIC1. To generate a cynS::Gm mutant, the aacC1 gentamicin resistance cassette isolated from the EcoRI-digested vector pMS255 (4) was cloned into the EcoRI site of the central region of cynS in pVIC1, resulting in the plasmid pVIC2. This plasmid was used for conjugational matings with the nalidixic acid-resistant P. pseudoalcaligenes strain CECT5344 to obtain a cyanase-deficient strain (lacking the cynS gene). The authenticity of the insertion was confirmed by PCR with the primers Cyn3 and Cyn4.
Purification of cyanase.
E. coli cells carrying the plasmid pMH1 (Table 1) were cultured in LB medium, harvested, resuspended in Tris-HCl buffer (pH 8), and disrupted by two passages through a French pressure cell at 120 MPa. Crude extracts were prepared by centrifugation at 20,000 × g for 20 min to remove cell debris. The resulting cell extract was initially heated to 70°C for 15 min. After the mixture was cooled to 4°C, the precipitated proteins were removed by centrifugation at 20,000 × g for 15 min, and the resulting supernatant was subjected to ammonium sulfate fractionation. The supernatant was brought to 40% saturation with ammonium sulfate by the stepwise addition of the salt. After being gently stirred for 30 min, the suspension was centrifuged at 20,000 × g for 20 min. The supernatant fraction was recovered and brought to 55% ammonium sulfate saturation, stirred, and centrifuged as above. The resulting pellet was resuspended in a minimal volume of Tris-HCl 50 mM (pH 8.5) buffer, and the pellet and buffer were loaded into a PD-10 molecular exclusion column (Pharmacia Biotech) in order to remove the ammonium salts. The fractions with cyanase activity were pooled, concentrated by ultrafiltration (Ultrafree-0.5; Millipore), and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (20) to test their purity.
Nucleotide sequence accession number.
The P. pseudoalcaligenes DNA sequence of the region discussed in this paper has been annotated and deposited in the EMBL/DDBJ/GenBank databases under accession number EF451798.
RESULTS
Growth of P. pseudoalcaligenes CECT5344 with cyanate and regulation of cyanase activity.
P. pseudoalcaligenes CECT5344 was able to use cyanate as the sole nitrogen source with a generation time of 5 h, which doubled times observed with ammonium (2.5 h) and nitrate (2.6 h) as nitrogen sources (data not shown). However, the maximal cellular growth (OD600) was very similar with these three nitrogen sources (data not shown). Cyanase activity was present in cell extracts from cyanate-grown cells, but it was undetectable in extracts from cells grown with ammonium, nitrate, arginine, or ornithine as N sources, as well as in those grown in LB media (Table 2). The presence of azide, to which this bacterium is resistant (24), did not induce cyanase activity (Table 2). In contrast, urea, cyanide, and some cyanometallic complexes induced the activity at a higher level than that observed in cyanate-grown cells (Table 2). The enzyme was also induced, although at a low level, in nitrogen-free media (Table 2). Subcellular fractionation of the cells revealed that the protein has a cytoplasmic location (data not show).
TABLE 2.
Cyanase activity in P. pseudoalcaligenes CECT5344 cells grown with different nitrogen sourcesa
| Nitrogen source(s) | Cyanase activity (U/g of protein) |
|---|---|
| Cyanate | 562 ± 53 |
| Ammonium | 0 |
| Ammonium + azideb | 0 |
| Ammonium + cyanate | 0 |
| Nitrate | 0 |
| Nitrate + azideb | 0 |
| Nitrate + cyanate | 495 ± 30 |
| NaCN | 960 ± 64 |
| [Cu(CN)4]2− | 10.000 ± 140 |
| Urea | 1.813 ± 110 |
| l-Arginine | 0 |
| l-Ornithine | 0 |
| −Nc | 41 ± 20 |
| LB | 0 |
| LB + NaCN | 0 |
The cells were cultured with the indicated nitrogen sources (10 mM, except cyanide, which was 2 mM) up to the late exponential growth phase. Cyanase activity from the different cell extracts was measured as indicated in Materials and Methods. The data presented are the averages of the results of five independent experiments.
Cells were cultured in the presence of 2 mM sodium azide.
The minus sign indicates that cells were cultured under nitrogen limitation conditions (2 mM ammonium).
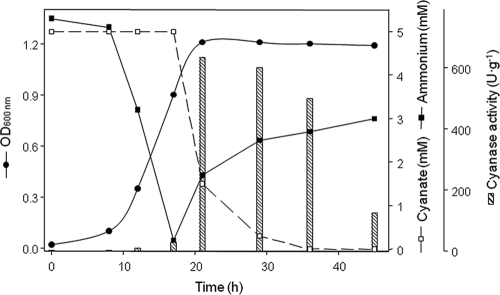
To further study the regulation of cyanate metabolism in strain CECT5344, cyanate uptake and cyanase activity were measured in media containing cyanate and some additional N sources. When ammonium and cyanate were supplied together as nitrogen sources, the cyanate was consumed only after the ammonium was assimilated (Fig. 1), which coincided with the induction of cyanase activity, which was not detected until this time, and with the production of ammonium, probably from the consumed cyanate (Fig. 1). However, the addition of ammonium to cells growing with cyanate affected neither the cyanate consumption nor the cell growth rate (data not shown). In contrast, when the cells were grown in mineral media containing cyanate and nitrate or nitrite, cyanate was the preferred nitrogen source and nitrate or nitrite were taken up only after the cyanate was completely consumed (data not shown).
FIG. 1.
Induction of cyanase activity in media containing both ammonium and cyanate simultaneously. Cells were cultured in 1 liter of M9 media containing ammonium and cyanate at a 5-mM final concentration as N sources. At the indicated times, the amount of cell growth (OD600 nm) was determined and 50 ml of the cultures was harvested by centrifugation and used to determine the level of cyanase activity (dashed bars) as indicated in Materials and Methods. Cyanate and ammonium concentrations in the supernatants were determined. The data correspond to a single experiment, and two other independent experiments gave similar results.
On the basis of these data, it can be concluded that ammonium exerts a negative effect on cyanase induction, whereas full induction of the enzyme requires the presence of cyanate or cyanide. To determine the minimal amount of cyanate necessary for the induction of cyanase activity, cells were grown with nitrate as the nitrogen source to avoid the repressive effect of ammonium and were then transferred to fresh mineral media with increasing cyanate concentrations. As shown in Fig. 2, the minimal cyanate concentration to fully induce the cyanase activity was 100 μM. Intermediate concentrations between 100 μM and 5 mM cyanate gave essentially the same level of induction of cyanase activity (data not shown). As expected, the activity was not detectable in cells grown with nitrate alone, whereas basal activity was detected in cells kept in nitrogen-depleted media.
FIG. 2.
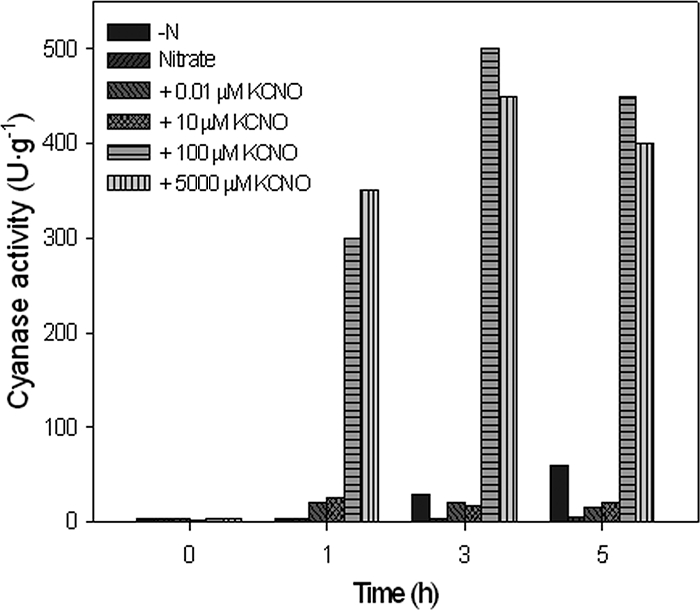
Effect of cyanate concentration on the induction of cyanase activity. The cells were pregrown with 5 mM nitrate as the sole nitrogen source and were collected by centrifugation at the mid-exponential growth phase. After being washed twice in nitrogen-free media, the cells were resuspended in fresh media up to an OD600 of 0.35. The culture was separated into six flasks that were treated with increasing amounts of cyanate. One flask, kept as a control, had no cyanate added (-N), and 5 mM nitrate was added to another flask (Nitrate). At the indicated times, 50-ml aliquots from each culture were collected, and the cyanase activities in the corresponding cell extracts were measured. The experiment was repeated three times with similar results.
Isolation and characterization of the cyn gene cluster of P. pseudoalcaligenes CECT5344 and generation of a cyanase-defective mutant strain.
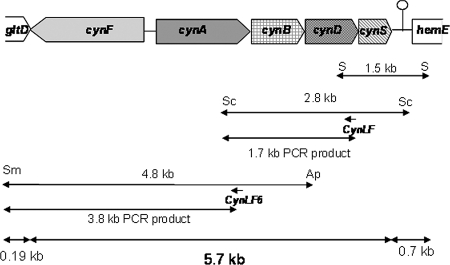
To clone the cynS gene from P. pseudoalcaligenes, the degenerated primers Cyn1F and Cyn2R were designed from conserved domains of the cyanases from E. coli and P. aeruginosa as indicated in the Materials and Methods section. They were used to amplify a DNA fragment of approximately 100 bp, which was used to further isolate a 1.5-kb SalI fragment containing the whole cynS gene (Fig. 3). DNA sequencing of this fragment revealed that the cynS gene was flanked by two putative open reading frames. The 3′ end of the upstream open reading frame (302 bp) overlapped 8 nucleotides of the start codon of cynS. The product of this putative gene showed similarity with CynD, an ATP-dependent component of ABC-type transporters for several oxyanions, such as nitrate, sulfonate, and bicarbonate. A noncoding region of 116 nucleotides that contains a putative transcription termination sequence is located downstream from the cynS gene (Fig. 3). This putative transcription terminator is located 42-bp upstream from the start codon of the open reading frame downstream from cynS, which codes for a protein 96% identical to the HemE uroporfirinogen decarboxylase (EC 4.1.1.37) from Pseudomonas mendocina (Pmen_0552). The CynS amino acid sequence comprises 146 residues, which present high similarity with cyanases from Pseudomonas putida F1, Synechococcus elongatus PCC6301 (59% and 43% identities, respectively), and other bacteria (Fig. 4). P. pseudoalcaligenes CECT5344 CynS shared 42% identity with the cyanase from E. coli, and the residues proposed to constitute the catalytic triad of the enzyme are conserved (Fig. 4). The 1.7-kb PCR fragment overlapped 303 bp of the 3′ end of the cynD gene included in the 1.5-kb SalI fragment and comprises 791 bp of the 5′ end of the cynD gene, the whole cynB gene, and 116 bp of the 3′ end of the cynA gene (Fig. 3). The cynD gene product has 302 amino acids and, as mentioned, shares homology with CynD proteins related to nitrate, sulfonate, and bicarbonate transport. The P. pseudoalcaligenes CynD product shows 63% identity with P. aeruginosa 2192 CynD (ZP_00975104) and 51% identity with Synechocystis strain CynD (NP_442736). The cynB gene codes for a putative membrane protein (278-amino-acid residues) that shows 63% identity with the transmembrane component of a putative ABC-type nitrate, sulfonate, or bicarbonate transporter of P. aeruginosa 2192 (ZP_00975103) and 58% identity with the permease component of a cyanate transporter of Prochlorococcus marinus (NP_892490). The 3.8-kb PCR fragment comprises the whole cynA gene, a 268-bp promoter region, the whole cynF gene, and 191 bp of the 3′ end of the gltD gene, which codes for the small subunit of the glutamate synthase (Fig. 3). The cynA gene codes for the periplasmic component of the ABC-type transporter (455-amino-acid residues) and shows more similarity with the periplasmic component of nitrate transporters (NrtA) than with cyanate transporters (CynA). Thus, P. pseudoalcaligenes CynA shared 60% identity with Rhodopseudomonas palustris NtrA (NP_947457) and 58% identity with Bradyrhizobium sp. NtrA (NP_772374). The cynF gene codes for a σ54-dependent transcriptional regulator of 648-amino-acid residues, which belongs to the FIS (factor for inversion stimulation) family of regulatory proteins. P. pseudoalcaligenes CynF shared 59% identity with R. palustris CynF (YP_485671) and 58% identity with Bradyrhizobium japonicum CynF (NP_772375).
FIG. 3.
The 5.7-kb cyn gene cluster of P. pseudoalcaligenes CECT5344 involved in cyanate assimilation. The 1.5-kb SalI (S) DNA fragment includes 0.7 kb of the hemE gene, a 143-bp noncoding region, the whole cynS gene, and 0.3 kb of the cynD gene. The 2.8-kb SacI (Sc) fragment comprises 0.4 kb of the hemE gene, the whole cynS, cynD, and cynB genes, and 116 bp of the cynA gene. The 4.8-kb ApaI (Ap)/SmaI (Sm) fragment includes 231 bp of the cynB gene, the whole cynB, cynA, and cynF genes, and 191 bp of the gltD gene. Between the cynA and cynF genes, a 268-bp promoter region is found. The 1.7-kb and 3.8-kb PCR fragments mentioned in the text are also shown. The position of a putative transcription terminator downstream from the cynS gene is also indicated.
In order to check the function of the CynS protein as well as its possible participation in cyanide degradation, the cloned gene region was used to generate a cynS insertion mutant. As indicated in the Materials and Methods section, the double-recombination mutant was selected by its resistance to Gm and nalidixic acid and its sensitivity to kanamycin. This mutant strain was further analyzed by PCR by using the Cyn3 and Cyn4 primers to confirm the double-recombination event (data not shown). This CynS− mutant did not grow with cyanate as the sole nitrogen source but retained the ability to grow in mineral media with cyanide, ammonium, nitrate, or nitrite as the sole nitrogen source. From these data, it can be concluded that cyanase is involved in cyanate metabolism but is not directly involved in cyanide assimilation.
Since the CynS− mutant was unable to grow on cyanate and since ammonium may exert a negative regulatory effect on the expression of several genes, the possible role of cyanase in the detoxification of cyanate was checked in media containing nitrate as the nitrogen source. Surprisingly, in the presence of nitrate both the mutant and the wild-type strains were resistant to up to 50 mM cyanate (Fig. 5). Nevertheless, the lag period of growth in both strains increased proportionally with the cyanate concentration in the media (data not shown). In addition, the wild-type strain was able to grow with 100 mM cyanate as the sole nitrogen source (data not shown) but not with cyanate and nitrate (Fig. 5), which suggests that the simultaneous presence of nitrate and cyanate exerts an inhibitory effect on cell growth.
FIG. 5.
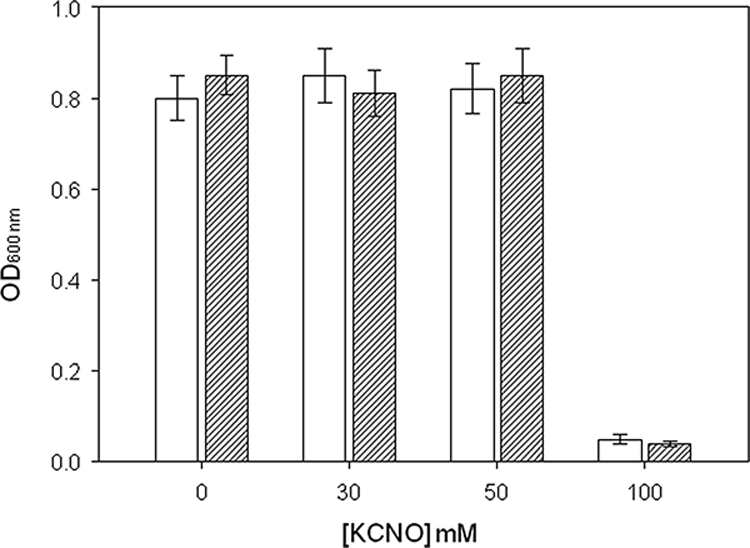
Tolerance of the wild type (white bars) and cynS− mutant (dashed bars) strains of P. pseudoalcaligenes CECT5344 to cyanate. Cells were cultured in mineral medium with 10 mM potassium nitrate as the nitrogen source supplemented with the indicated concentration of cyanate. The optical densities of the cultures at 600 nm (OD600 nm)were taken 48 h after inoculation. The experiments were run in triplicate.
Purification and biochemical characterization of the cyanase from P. pseudoalcaligenes CECT5344.
The cyanase of P. pseudoalcaligenes CECT5344 was expressed in E. coli DH5α transformed with the pMH1 plasmid (Table 1). The DH5α strain did not show cyanase activity in LB medium. The expressed enzyme was purified to apparent electrophoretic homogeneity following a simple and rapid protocol that takes advantage of the thermostability of the enzyme (Fig. 6). The purified protein showed a specific activity of 445 U·mg−1, which corresponds to a purification factor of almost 1,000 times the specific activity observed in the parental strain (Table 2).
FIG. 6.
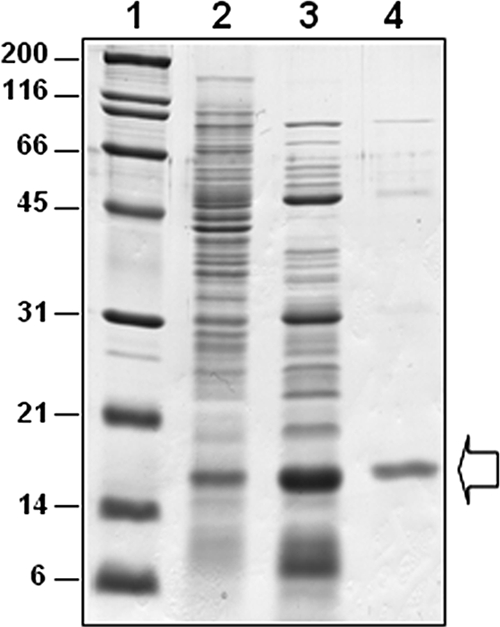
Purification of the cyanase from P. pseudoalcaligenes CECT5344 expressed in E. coli DH5α. Cyanase was heterologously expressed in E. coli and purified as indicated in Materials and Methods. The different lanes in the gel subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis correspond to molecular weight markers (lane 1), the cell extract (lane 2), the supernatant after centrifugation of a heated (70°C for 30 min) cell extract (lane 3), and 45% to 60% of the ammonium sulfate fraction of the heated extract (lane 4). The arrow indicates the location of the cyanase on the gel.
The pattern of regulation of the cyanase in P. pseudoalcaligenes is consistent with that of an enzyme of assimilatory type that is required for cyanate assimilation. However, the phenotype of the CynS− mutant strain does not support the possible involvement of cyanase in the assimilation of cyanide. The biochemical properties of the enzyme are shown in Table 3. The thermostability of the enzyme, as well as its inhibition by pHMB (p-hydroxymercuribenzoate) and azide Table 4, is especially remarkable. By contrast, the enzymatic activity was not inhibited by up to 10 mM concentrations of ammonium, cyanide, urea, nitrite, EDTA, dithioerythritol, or ferricyanide (data not shown). The presence of either cyanate or bicarbonate at a final concentration of 3 mM partially protected (about 25%) against thermal inactivation of the enzyme at 70°C (data not shown).
TABLE 3.
Biochemical characteristics of P. pseudoalcaligenes CynS partially purified from E. colia
| Kinetic parameter | Value |
|---|---|
| Km NaHCO3 (mM) | 0.67 |
| Km KCNO (mM) | 2.40 |
| Optimum temp (°C) | 65 |
| Optimum pH | 8.5 |
| Thermostabilityb | 30-70°C |
The cyanase activity was partially purified by heating the cell extract 30 min at 60oC. The apparent Km values were calculated by double-reciprocal plots.
The thermostability range represents temperatures at which the enzyme retains more than 25% of its original activity after 30 min. Higher temperatures resulted in the irreversible inactivation of the enzyme.
TABLE 4.
Inhibition of P. pseudoalcaligenes CynS partially purified from E. colia
| Inhibitor | % Inhibition |
|---|---|
| Nitrate | 22 |
| pHMB | 70 |
| Azide | 68 |
| NaCN | 0 |
| Thiocyanate | 5 |
Inhibitors were used at a final concentration of 10 mM. One hundred percent activity corresponds to 1,200 U·g−1.
DISCUSSION
In some cyanobacteria, the cyanase activity seems to be constitutively expressed (27), whereas in other strains, the transcription of the cyanase gene is negatively regulated by ammonium (11). In E. coli, the expression of the cynTSX operon is induced by exogenous cyanate through the positive regulator CynR, which belongs to the LysR family (37). Positive regulation by cyanate also takes place in the cyanide-degrading strain P. fluorescens NCIMB 11764, in which cyanase is present in cyanate-grown cells, even in the presence of ammonium (18). In P. pseudoalcaligenes CECT5344, a more-sophisticated regulatory control seems to exist, since cyanase was positively controlled by cyanate under nitrogen-limiting conditions and was negatively regulated by ammonium, even in the presence of cyanate (Table 2, Fig. 1). The negative effect exerted by ammonium on cyanate assimilation seems to take place at the level of gene expression, since the addition of ammonium to cells growing in cyanate had no effect on either cyanase activity or cyanate consumption (data not shown). Azide, a gratuitous inducer of the cyn operon in E. coli (21), does not seems to induce cyanase in P. pseudoalcaligenes (Table 2), but as azide also inhibited the enzymatic activity (Table 3), the possible induction of an inactivated enzyme cannot be completely discarded. Urea was a strong inducer of cyanase activity in P. pseudoalcaligenes (Table 2), probably because it slowly breaks down in cyanate. Nevertheless, the reason for the induction of cyanase activity in cells growing in cyanide is still an open question (Table 2). Nitrogen limitation conditions generated by cyanide, recently described for P. pseudoalcaligenes (25), are not sufficient to explain the high level of cyanase activity detected in cells growing in cyanide, which was similar to that observed in cells growing in cyanate (Table 2, Fig. 2). The altruistic induction of cyanase activity by cyanide also seems unlikely to take place, since azide, a cyanide and cyanate analog, inhibited the cyanase activity but did not induce the enzyme in P. pseudoalcaligenes CECT5344 (Tables 2 and 3). In addition, the cyanase activity was undetectable in LB medium supplemented with cyanide (Table 2). Under these conditions, P. pseudoalcaligenes is unable to assimilate cyanide (24), suggesting that the inducer should be a metabolite produced from cyanide in the assimilation pathway. This metabolite could be cyanate, since the induction of the cyanase activity takes place at very low concentrations of cyanate (Fig. 2). We were unable to detect a cyanide monooxygenase activity that would convert cyanide into cyanate, but other cyanate-producing processes could be involved. For example, cyanohydrins, which have been proposed as intermediates in cyanide metabolism in this bacterium (24), or their decomposition products could also be responsible for the induction of cyanase in cyanide-grown cells.
The cyn gene clusters of E. coli and P. aeruginosa share the presence of the carbonic anhydrase gene cynT (36). Both cyanate and bicarbonate are substrates of the cyanase enzyme, and the role of carbonic anhydrase activity is probably to supply bicarbonate to the cyanase reaction in cells growing in cyanate (9). In E. coli K12, the cynS gene is also clustered with the cynX gene, which encodes a hydrophobic protein that may be a cyanate transporter belonging to the MSF family (29). Here, we determined that the cyn cluster of P. pseudoalcaligenes CECT5344 shows a different gene organization (Fig. 3). In contrast to that described for E. coli K12 and P. aeruginosa PAO1, the cynS gene is not adjacent to cynT. The cyn gene organization in strain CECT5344 is similar to that found in S. elongatus PCC7942, where cynS is clustered with a putative ABC-type cyanate transporter closely related to a nitrate/nitrite transporter (6, 11). In S. elongatus, bicarbonate is supplied by the CO2-concentrating mechanism (6), but as far as we know, there is no CO2-concentrating mechanism in P. pseudoalcaligenes. Therefore, although the cynA, cynB, and cynD genes could encode a cyanate transporter, it could also be proposed that the the cynA, cynB, and cynD genes from CECT5344 code for a putative bicarbonate transporter, provided that the bicarbonate concentration is relatively high at the alkaline pH at which this bacteria thrives. On the other hand, the only gene in the cyn cluster of P. pseudoalcaligenes that codes for a putative regulatory protein (cynF) does not show identity with the cynR gene that codes for the transcriptional activator described for other bacterial cyn clusters.
The amino acid sequence of P. pseudoalcaligenes CynS comprises 146 residues, and its alignment with other cyanases reveals that the C-terminal region is highly conserved (Fig. 4A). The residues proposed by Walsh et al. (42) to constitute the catalytic triad of the enzyme (R96, E99, and S122 in E. coli nomenclature) are also conserved in the CynS protein from P. pseudoalcaligenes (Fig. 4A). Phylogenetic analysis revealed that P. pseudoalcaligenes CynS is closely related to several cyanases of the pseudomonad and cyanobacterium groups. However, this cyanase distribution seems to be independent of the bacterial group (Fig. 4A). Among all the cyanases that have so far been described, only the enzyme from E. coli has been purified to electrophoretic homogeneity (3, 38). The enzyme from P. pseudoalcaligenes CECT5344 was partially purified in order to design a cyanate biosensor (23). Taking advantage of its thermostability, in this study we purified the cyanase from strain CECT5344 by following a simple purification protocol after its heterologous expression in E. coli (Fig. 6). The purified enzyme has an approximate molecular mass of 16 kDa, which is in agreement with the size predicted from its sequence and is very similar to the monomeric molecular mass of other bacterial cyanases.
The cyanase activity was located in the cytoplasm, as described for E. coli (15). The specific activity of the enzyme in cell extracts from cells grown with cyanate (around 560 U·g−1; Table 2) was higher than that described for P. fluorescens NCIMB 11764 (168 U·g−1) (18). The cyanase from P. pseudoalcaligenes has some biochemical properties that are different from those of the cyanases described up to the present. Thus, the enzyme is insensitive to 10 mM thiocyanate (Table 3), a competitive inhibitor of some cyanases (44). The cyanase from P. pseudoalcaligenes showed an optimum temperature of 65°C and an optimum pH of 8.5 (Table 3), data that are far different from that for the cyanases characterized so far. Although the cyanase from E. coli was shown to be thermostable (3, 40), its optimum temperature in the assay was 37°C (3), whereas the cyanase from P. fluorescens NCIB 11664 was optimally assayed at 30°C (18). On the other hand, the optimum pH of the enzyme from E. coli is close to neutral (7 to 7.4; references 40 and 3, respectively), which is near the pH employed to assay the enzyme from P. fluorescens NCIMB 11764 (7.5 [18]). The biochemical properties of the cyanase in cell extracts from P. pseudoalcaligenes were similar to those obtained with the enzyme heterologously expressed in E. coli. The inhibition of cyanase activity by pHMB suggests the involvement of essential sulfhydryl groups in the protein (Table 3). By contrast, there is evidence that free sulfhydryl groups are not required for catalytic activity in E. coli (22).
Cyanate is closely related to cyanide, since both chemicals are single-carbon, N-containing compounds that can be interconverted by a single redox reaction. Cyanate was proposed to be an intermediate in cyanide metabolism in P. fluorescens NCIMB 11764 (12). In a previous work, we reported for the first time the induction of the cyanase activity in P. pseudoalcaligenes CECT5344 cyanide-grown cells (24). Here, we clearly show that cyanate is not a key intermediate in the degradation of cyanide, since the cynS mutant is still able to use cyanide as the sole nitrogen source. However, cyanide could be a direct inducer of cyanase, or more likely, low amounts of cyanate may be formed during cyanide metabolism, thus explaining cyanase induction in the presence of cyanide.
Cyanate has been shown to be relatively toxic, even for bacteria able to use it as a N source. For example, the growth of P. fluorescens NCIMB 11764 was partially inhibited by cyanate at concentrations higher than 5 mM, and total inhibition was observed at a 20-mM concentration (18). By contrast, P. pseudoalcaligenes CECT5344 was able to use cyanate as the sole nitrogen source at concentrations up to 100 mM. The resistance did not depend on the cyanase activity, since the wild-type strain and the CynS− mutant were equally resistant to cyanate (Fig. 5), thus discarding a protective function of cyanase against cyanate, as suggested for E. coli (10). Therefore, this result, together with the regulatory pattern of the enzyme, suggests that the cyanase of P. pseudoalcaligenes CECT5344 has an assimilatory role.
Acknowledgments
This work was funded by the Ministerio de Ciencia y Tecnología (grants BMC2002-04126-C03-01, BMC2002-04126-C03-03, and BIO2005-077741), Junta de Andalucía (grant CVI-1728), and Junta de Extremadura (grants 2PR04A022 and PRI07A097). V.M.L.-A. was the recipient of a fellowship from the Ministerio de Educación y Ciencia, and M.D.R. holds a postdoctoral fellowship from the Junta de Andalucía (Spain).
We gratefully acknowledge the help of Maria Dolores Luque de Castro and B. Vallejo-Pecharromán for some of the analytical determinations. We also thank GEMASUR S. L. and Kinbauri España S. L. for their fruitful collaboration.
Footnotes
Published ahead of print on 15 August 2008.
REFERENCES
- 1.Aichi, M., I Nishida, and T. Omata. 1998. Molecular cloning and characterization of a cDNA encoding cyanase from Arabidopsis thaliana. Plant Cell. Physiol. Suppl. 39:S135. [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myenrs, and D. J. Lipman. 1990. Basic local alignment tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, P. M. 1980. Purification and properties of the inducible enzyme cyanase. Biochemistry 19:2882-2888. [DOI] [PubMed] [Google Scholar]
- 4.Becker, A., M. Schmidt, W. Jäger, and A. Pühler. 1995. New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene 162:37-39. [DOI] [PubMed] [Google Scholar]
- 5.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for VAX. Nucleic Acid Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Espie, G. S., F. Jalali, T. Tong, N. J. Zacal, and A. K.-C. So. 2007. Involvement of the cynABDS operon and the CO2-concentrating mechanism in the light-dependent transport and metabolism of cyanate by cyanobacteria. J. Bacteriol. 189:1013-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerrero, M. G., J. M. Vega, E. Leadbetter, and M. Losada. 1973. Preparation and characterization of a soluble nitrate reductase from Azotobacter chroococcum. Arch. Mikrobiol. 91:287-304. [DOI] [PubMed] [Google Scholar]
- 8.Guerrero, M. G., J. M. Vega, and M. Losada. 1981. The assimilatory nitrate-reducing system and its regulation. Annu. Rev. Plant Physiol. 32:169-204. [Google Scholar]
- 9.Guilloton, M., J. Korte, A. F. Lamblin, J. A. Fuchs, and P. M. Anderson. 1992. Carbonic anhydrase in Escherichia coli. J. Biol. Chem. 267:3731-3734. [PubMed] [Google Scholar]
- 10.Guilloton, M., and F. Karst. 1987. Isolation and characterization of E. coli mutants lacking inducible cyanase. J. Gen. Microbiol. 133:645-653. [DOI] [PubMed] [Google Scholar]
- 11.Harano, Y., I. Suzuki, S. Maeda, T. Kaneko., S. Tabata, and T. Omata. 1997. Identification and nitrogen regulation of the cyanase gene from the cyanobacteria Synechocystis sp. strain PCC 6803 and Synechococcus sp. strain PCC 7942. J. Bacteriol. 179:5744-5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris, R. E., and C. J. Knowles. 1983. The conversion of cyanide to ammonia by extracts of a strain of Pseudomonas fluorescens that utilizes cyanide as source of nitrogen for growth. FEMS Microbiol. Lett. 20:337-341. [Google Scholar]
- 13.Johnson, W. C., and P. M. Anderson. 1987. Bicarbonate is a recycling substrate for cyanase. J. Biol. Chem. 262:9021-9025. [PubMed] [Google Scholar]
- 14.Koshiishi, I., Y. Mamura, and T. Imanari. 1997. Cyanate causes depletion of ascorbate in organisms. Biochim. Biophys. Acta 1336:566-574. [DOI] [PubMed] [Google Scholar]
- 15.Kozliak, E. I., M. B. Guilloton, M. Gerami-Nejad, J. A. Fuchs, and P. M. Anderson. 1994. Expression of proteins encoded by the Escherichia coli cyn operon: carbon dioxide-enhanced degradation of carbonic anhydrase. J. Bacteriol. 176:5711-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraus, L. M., and A. P. Kraus. 1998. The search for the uremic toxin: the case for carbamoylation of amino acids and proteins. Wien. Klin. Wochenschr. 110:521-530. [PubMed] [Google Scholar]
- 17.Kraus, L. M., L. Gaber, C. R. Handorf, H. P. Marti, and A. P. Kraus, Jr. 2001. Carbamoylation of glomerular and tubular proteins in patients with kidney failure: a potential mechanism of ongoing renal damage. Swiss Med. Wkly. 131:139-145. [DOI] [PubMed] [Google Scholar]
- 18.Kunz, D. A., and O. Nagappan. 1989. Cyanase-mediated utilization of cyanate in Pseudomonas fluorescens NCIMB 11764. Appl. Environ. Microbiol. 55:256-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunz, D. A., C.-S. Wang, and J.-L. Che. 1994. Alternative routes of enzymatic cyanide metabolism in Pseudomonas fluorescens NCIMB 11764. Microbiology 140:1705-1712. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 21.Lamblin, A.-F. J., and J. A. Fuchs. 1994. Functional analysis of the Escherichia coli K-12 cyn operon transcriptional regulation. J. Bacteriol. 176:6613-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Little, R. M., and P. M. Anderson. 1987. Structural properties of cyanase. Denaturation, renaturation, and role of sulfhydryls and oligomeric structure in catalytic activity. J. Biol. Chem. 262:10120-10126. [PubMed] [Google Scholar]
- 23.Luque-Almagro, V. M., R. Blasco, J. M. Fernandez-Romero, and M. D. de Castro. 2003. Flow-injection spectrophotometric determination of cyanate in bioremediation processes by use of immobilised inducible cyanase. Anal. Bioanal. Chem. 377:1071-1078. [DOI] [PubMed] [Google Scholar]
- 24.Luque-Almagro, V. M., M. J. Huertas, M. Martinez-Luque, C. Moreno-Vivian, M. D. Roldan, L. J. Garcia-Gil, F. Castillo, and R. Blasco. 2005. Bacterial degradation of cyanide and its metal complexes under alkaline conditions. Appl. Environ. Microbiol. 71:940-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luque-Almagro, V. M., M. J. Huertas, M. D. Roldán, C. Moreno-Vivián, M. Martínez-luque, R. Blasco, and F. Castillo. 2007. The cyanotrophic bacterium Pseudomonas pseudoalcaligenes CECT5344 responds to cyanide by defence mechanisms against iron deprivation, oxidative damage and nitrogen stress. Environ. Microbiol. 9:1541-1549. [DOI] [PubMed] [Google Scholar]
- 26.Martin, S., and J. J. Harding. 1989. Site of carbamoylation of bovine γ-II-crystallin by potassium [14C]cyanate. Biochem. J. 262:909-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, A. G., and G. S. Espie. 1994. Photosynthetic metabolism of cyanate by the cyanobacterium Synechococcus UTEX 625. Arch. Microbiol. 162:151-157. [Google Scholar]
- 28.Nowakowska, M., M. Sterzel, and K. Szczubialka. 2006. Photosensitized oxidation of cyanide in aqueous solutions of photoactive modified hydroxyethylcellulose. J. Polym. Environ. 14:59-64. [Google Scholar]
- 29.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pieniazek, A., and K. Gwoździński. 2003. Carbamylation of proteins leads to alteration in the membrane structure of erythrocytes. Cell. Mol. Biol. Lett. 8:127-131. [PubMed] [Google Scholar]
- 31.Qian, M., J. W. Eaton, and S. P. Wolff. 1997. Cyanate-mediated inhibition of neutrophil myeloperoxidase activity. Biochem. J. 326:159-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 33.Schäfer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 34.Shakir, F. K., D. Audilet, A. J. Drake, and K. M. Shakir. 1994. A rapid protein determination by modification of the Lowry procedure. Anal. Biochem. 216:232-233. [DOI] [PubMed] [Google Scholar]
- 35.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 36.Sung, Y.-C., and J. A. Fuchs. 1988. Characterization of the cyn operon in Escherichia coli K12. J. Biol. Chem. 263:14769-14775. [PubMed] [Google Scholar]
- 37.Sung, Y.-C., and J. A. Fuchs. 1992. The E. coli K12 cyn operon is positively regulated by a member of the lysR family. J. Bacteriol. 174:3645-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sung, Y.-C., P. M. Anderson, and J. A. Fuchs. 1987. Characterization of high-level expression and sequencing of the Escherichia coli K-12 cynS gene encoding cyanase. J. Bacteriol. 169:5224-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki, I., T. Sugiyama, and T. Omata. 1996. Regulation by cyanate of the genes involved in carbon and nitrogen assimilation in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 178:2688-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taussig, A. 1960. The synthesis of the induced enzyme “cyanase” in E. coli. Biochim. Biophys. Acta 44:510-519. [DOI] [PubMed] [Google Scholar]
- 41.Thompson, J., D. Higgins, and T. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walsh, M. A., Z. Otwinowski, A. Perrakis, P. M. Anderson, and A. Joachimiak. 2000. Structure of cyanase reveals that a novel dimeric and decameric arrangement of subunits is required for formation of the enzyme active site. Structure 8:505-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, C.-S, D. A. Kunz, and B. J. Venables. 1996. Incorporation of molecular oxygen and water during enzymatic oxidation of cyanide by Pseudomonas fluorescens NCIMB 11764. Appl. Environ. Microbiol. 62:2195-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood, A. P., D. P. Kelly, I. R. McDonald, S. L. Jordan, T. D. Morgan, S. Khan, J. C. Murrell, and E. Borodina. 1998. A novel pink-pigmented facultative methylotroph, Methylobacterium thiocyanatum sp. nov., capable of growth on thiocyanate or cyanate as sole nitrogen sources. Arch. Microbiol. 169:148-158. [DOI] [PubMed] [Google Scholar]