Abstract
Escherichia coli strains isolated from fecal samples were screened to examine changes in phenotypic and genotypic characteristics including antimicrobial susceptibility, clonal type, and carriage of resistance determinants. The goal of this 197-day study was to investigate the influence of administration of chlortetracycline alone (T) or in combination with sulfamethazine (TS) on the development of resistance, dissemination of defined strain types, and prevalence of resistance determinants in feedlot cattle. Inherent tetracycline resistance was detected in cattle with no prior antimicrobial exposure. Antimicrobial administration was not found to be essential for the maintenance of inherently ampicillin-resistant and tetracycline-resistant (Tetr) E. coli in control animals; however, higher Tetr E. coli shedding was observed in animals subjected to the two treatments. At day 0, high tetracycline (26.7%), lower sulfamethoxazole-tetracycline (19.2%), and several other resistances were detected, which by the finishing phase (day 197) were restricted to ampicillin-tetracycline (47.5%), tetracycline (31.7%), and ampicillin-tetracycline-sulfamethoxazole (20.8%) from both treated and untreated cattle. Among the determinants, blaTEM1, tet(A), and sul2 were prevalent at days 0 and 197. Further, E. coli from day 0 showed diverse antibiogram profiles and strain types, which by the finishing phase were limited to up to three, irrespective of the treatment. Some genetically identical strains expressed different phenotypes and harbored diverse determinants, indicating that mobile genetic elements contribute to resistance dissemination. This was supported by an increased linked inheritance of ampicillin and tetracycline resistance genes and prevalence of specific strains at day 197. Animals in the cohort shed increasingly similar genotypes by the finishing phase due to animal-to-animal strain transmission. Thus, characterizing inherent resistance and propagation of cohort-specific strains is crucial for determining antimicrobial resistance in cattle.
In recent years, the development of antimicrobial resistance (AR) in intensive livestock production has been increasingly reported (17). Consequently, the role of antimicrobial administration and the extent to which it affects the development of resistance in animals are receiving much attention. Antimicrobial usage for livestock can be for therapeutic, prophylactic, metaphylactic, or growth promotion purposes (26). Reportedly, 90% of the antimicrobials used in animal agriculture are for growth promotion and prophylaxis (26) and this widespread use is suggested to be an important contributor to the emergence, selection, and dissemination of antimicrobial-resistant bacteria, as indicated by several recent studies (9, 13, 28, 38). Antimicrobial-resistant bacteria, including Escherichia coli, are frequently isolated from the commensal gut flora of food animals (1, 2), and although the resistance they carry may not be a problem per se, the transfer of resistance elements to zoonotic pathogens inhabiting the gut has serious implications for animal and human health. Subtherapeutic use of antimicrobials is known to select for resistance (9, 14), but evidence suggests that development of resistance is far more complex and that prevalence of resistant bacterial populations cannot be simply related to antimicrobial usage (29, 30) since antimicrobial withdrawal does not always reduce this resistance (5, 16). Further, the relationship between subtherapeutic usage and maintenance of resistance within the gut bacterial population has not been well characterized.
Commensal E. coli is often used as an indicator organism to assess the extent and type of resistance in the gastrointestinal tract since it plays a dynamic role in the ecology of multidrug-resistant bacteria and has been shown to be a reservoir of resistance (8, 42). Studies have demonstrated that young cattle have a higher prevalence of resistant fecal E. coli than older stock held on the same farm (12, 27), while carriage of ampicillin-resistant (Ampr) E. coli declines with calf age (22). Elucidating how young animals are affected by continuous subtherapeutic antimicrobial administration and defining the dynamics of acquisition of resistance in E. coli are essential for establishing the mechanism(s) of resistance transmission in feedlot cattle. Serotyping and resistance profiling have provided useful information relating to E. coli populations in cattle (20); however, these techniques have their limitations in establishing the movement of strain types. We used a pulsed-field gel electrophoresis (PFGE) fingerprinting method to discriminate between E. coli strains to understand the persistence of strain types and linked the derived genotypes to antibiogram profiles (ABGs; susceptibility testing) for assessing the movement of transferable resistance elements between strains. We chose to monitor Ampr and tetracycline (TE)-resistant (Tetr) commensal E. coli due to increasing resistance to these antimicrobials in humans.
Using healthy beef cattle maintained for 197 days in a feedlot setting, we investigated the influence of administration of dietary chlortetracycline alone (T) or in combination with sulfamethazine (TS) on resistance selection and distribution of resistance determinants. Further, we examined if resistance was disseminated by specific E. coli strain types between animals in the cohort. The concentrations of the antimicrobial agents used were similar to those of growth promoters used in the cattle industry. The specific objectives were addressed by monitoring (i) the shedding of Ampr and Tetr E. coli over time and assessing the effects of time and individual antimicrobial treatments on the two E. coli populations, (ii) the prevalence and stability of E. coli genetic and phenotypic diversity over time between treatments, and (iii) the distribution and emergence of common TE (tet), ampicillin (amp), and sulfonamide (sul) determinants in untreated versus treated beef cattle.
MATERIALS AND METHODS
Animals, diets, and treatments.
The 240 crossbred steer calves (6 to 8 months old, weighing 150 ± 20 kg) used in this study, which had no previous history of antimicrobial administration, were from Deseret Ranches (Raymond, Alberta, Canada). Animals were transported over 2 consecutive days by truck and upon arrival were housed in 24 pens at the Lethbridge Research Center experimental feedlot. Calves were arbitrarily assigned to one of the following treatments: (i) no antimicrobial agent (n = 50; control), (ii) 350 mg head−1 day−1 chlortetracycline (Aureomycin-100G; Alpharma Inc., Bridgewater, NJ) (n = 40; T), (iii) 350 mg head−1 day−1 each chlortetracycline and sulfamethazine (Aureo S-700G; Alpharma Inc.) (n = 50; TS). Due to pen limitations, the T treatment could be replicated only four times and thus for uniformity, samples from 40 animals each from the control and treatment groups were processed. The antimicrobial agents were selected based on the commonality of their use in the Canadian feedlot industry and were fed at the concentrations recommended by the manufacturers. The treatments were arranged in a semirandomized complete block design, with each block consisting of a separate pen containing 10 steers. All antimicrobials supplemented the diet as top dressing which was mixed and spread manually over the feed surface during morning feeding, and animals were fed once daily. The pen setting was similar to any large industrial feedlot such that all animals could feed at the same time in their respective feed bunks. Water troughs were shared by animals in adjacent pens, and the treatments were arranged such that only animals that received the same treatment could drink from the same water troughs, preventing cross contamination.
For first 80 days, the steers were fed a typical forage-based background diet (70% barley silage, 25% barley grain, and 5% dry matter supplemented with vitamins and minerals) (Fig. 1). Thereafter, animals were shifted over a 21-day transition period from the background diet to a grain-based finishing diet (84% barley grain, 10% barley silage, and 5% supplements). The finishing diet was administered for an additional 124 days. The antimicrobials were administered continually for 197 days starting on day 0 (after fecal collection) and were withdrawn 28 days prior to slaughter. All animals were cared for according to the guidelines set out by the Canadian Council on Animal Care (7).
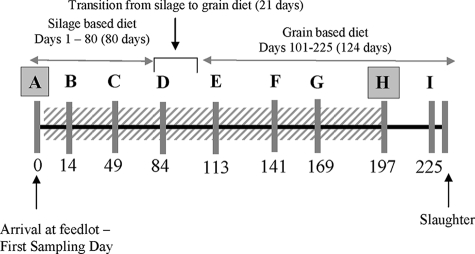
FIG. 1.
Schematic representation of the time line of the experiment, outlining the arrival of animals, diets, and sampling days. The letters A to I represent sampling times (gray bars). The subtherapeutic antimicrobials were administered via top dressing (diagonal hatching) continuously for 197 days. Isolates from periods A (background diet) and H (finishing diet) were analyzed via susceptibility testing and PFGE and for resistance determinants. The numbers below the time periods represent the numbers of days into the trial. The drawing is not to scale.
Fecal sampling.
Fecal samples were collected from each animal nine times in time periods A, B, C (background diet), D (diet transition period), E, F, G, H (finishing diet), and I (off antimicrobials). For fecal sampling, cattle were restrained in a constraint device, and a deep fecal sample was obtained from the rectum via a fecal grab. All fecal samples were collected in sterile tubes, placed on ice, and transported within 1 h to the laboratory for plating. Fresh gloves were used for fecal collection from each animal.
Isolation, identification, and enumeration of E. coli.
Each fecal sample (10 g wet weight) was aseptically placed in a sterile stomacher bag (Fisher Scientific, Ottawa, Ontario, Canada) containing 90 ml of 1× phosphate-buffered saline and mixed for 2 min (230 rpm, room temperature) with a Stomacher (Seward Ltd., Worthing, West Sussex, United Kingdom). The resulting slurries were 10-fold serially diluted, and 100 μl of the appropriate dilution was plated onto the following standard media for E. coli isolation in duplicate: MacConkey agar containing no antibiotics (Difco, Sparks, MD) for total E. coli isolation, MacConkey agar containing ampicillin (32 μg ml−1) for Ampr E. coli, and MacConkey agar containing TE hydrochloride (16 μg ml−1) for isolation of Tetr E. coli. These antimicrobial concentrations are recommended breakpoints for E. coli according to the CLSI (11). The plates were incubated at 39°C for 24 h, and individual colonies were enumerated to calculate CFU counts by taking into account the number of observed colonies and the dilution factor. The numbers of CFU per gram (wet weight) of total, Ampr, and Tetr E. coli populations were calculated. These counts were used for statistical analysis in determining the effects of antimicrobial treatments and time on total, Ampr, and Tetr E. coli strain shedding, as well as to assess treatment-time interactions. Two isolates per animal from nonselective (MacConkey agar containing no antibiotics) plates were arbitrarily selected and confirmed to be E. coli with API 20E (bioMérieux Inc., Durham, NC). These positive isolates were streaked onto Trypticase soy agar (Difco) and grown at 39°C for 24 h. Each isolate was stored in 20% glycerol at −80°C until processed.
During standardization, we found that clonal types from individual animals were generally stable and hence one isolate per animal was used for characterization, which included PFGE analysis, susceptibility profiling, and characterization of resistance determinants. Thus, 80 isolates from the growing (period A, n = 40) and finishing (period H, n = 40) periods were analyzed for each group (control, T, and TS).
Susceptibility testing.
The antimicrobial susceptibilities (phenotypes) of E. coli isolates from periods A and H to a panel of 12 antimicrobials was determined with a disk diffusion assay following CLSI standards (11). For this purpose, Mueller-Hinton II agar (Difco) was used and cells were harvested from the surface of the medium with a cotton swab after 24 h growth at 37°C. Cells were suspended in sterile saline (0.85% NaCl), cell density was adjusted to a 0.5 McFarland turbidity standard, and the diluted cells were plated. Following incubation, zone sizes were measured to two decimal points and used for quantitative analysis. Isolates resistant to two or more antimicrobials were defined as dual resistant or multiresistant, respectively. E. coli ATCC 25922 (American Type Culture Collection, Manassas, VA) was included in each assay as a control strain. Antimicrobial agents were tested with BD BBL Sensi-Disc antimicrobial susceptibility test discs (Becton Dickinson & Co., Sparks, MD) with the breakpoints (micrograms per milliliter) indicated as follows: amoxicillin-clavulanic acid (20 and 10, respectively), ampicillin (AMP; 10), amikacin (30), ceftriaxone (30), cefoxitin (30), ciprofloxacin (5), gentamicin (10), kanamycin (30), nalidixic acid (30), streptomycin (STR; 10), tetracycline (TE; 30), and trimethoprim-sulfamethoxazole (SXT; 1.25 and 23.75, respectively). The ABG of each isolate was established based on the resistance(s) observed.
Genotyping.
PFGE was conducted to assess the genetic diversity of E. coli and to analyze strain transmission in the control and treatment groups during periods A and H. Isolates were subtyped on the basis of PFGE patterns (PPs) of XbaI-cleaved chromosomal DNA in accordance with the standard protocol established by the Centers for Disease Control and Prevention (PulseNet; Centers for Disease Control and Prevention, Atlanta, GA). DNA fragments were resolved by electrophoresis on 1% SeaKem Gold agarose gels (Cambrex BioScience, Rockland, ME) with a contour-clamped homogeneous electric field DRII PFGE apparatus (Bio-Rad Laboratories, Mississauga, Ontario, Canada), with 0.5× Tris-borate-EDTA (BioBasics Inc., Edmonton, Alberta, Canada) as the running buffer, for 17 h at 14°C with a linearly ramped switching time from 2.2 to 55 s and a voltage of 6.0 V cm−1. Gels were stained with 0.5 mg liter−1 ethidium bromide (Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada), destained in distilled water for 2 h, and photographed with an AlphaImager gel doc (version 4.1.0; Alpha Innotech FC, San Leandro, CA). XbaI-digested Salmonella enterica serovar Branderup H9812 was used as a marker and included in the first, middle, and last lanes of each gel to account for run-to-run variability. Comparison of digested profiles to identify restriction endonuclease digestion pattern clusters (REPCs) was performed with BioNumerics software, version 4.0 (Applied Maths, Austin, TX). Fingerprints were clustered by using the Dice coefficient evaluated by the unweighted-pair group method. A tolerance and optimization of 1% was allowed to account for gel differences. Each isolate was assigned a PP identification number. Isolates that were >90% related were considered highly related and were grouped as a cluster group. Patterns that did not fall into any particular REPC were also assigned a pattern identification number, and if they were detected only once during the trial they were considered unique.
Characterization of resistance determinants.
Based on the ABGs E. coli isolates from periods A and H were characterized for genes responsible for ampicillin resistance (Ampr), TE resistance (Tetr), dual resistance (Ampr Tetr), and sulfonamide resistance (Sulr). Individual isolates that showed resistance were screened for related resistance determinants (amp, tet, or sul). This was done by using multiplex PCR for three β-lactamase-encoding genes (blaOXA1, blaPSE1, and blaTEM1), 14 TE resistance-encoding alleles [tet(A), tet(B), tet(C), tet(D), tet(E), tet(G), tet(K), tet(L), tet(M), tet(O), tet(S), tetA(P), tet(Q), and tet(X)], and 3 sul alleles (sul1, sul2, and sul3). Details of primers, annealing temperatures, and amplicon sizes are provided in Table 1. Plasmids with known, sequenced genes were used as positive controls (Table 1).
TABLE 1.
PCR primers and control strains used for identification of Ampr, Tetr, and Sulr determinants
| Plasmid | Resistance gene | PCR primer sequence (5′ → 3′) | Amplicon size (bp) | Accession no. | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|---|
| SU01 | pse1 | CGC TTC CCG TTA ACA AGT AC CTG GTT CAT TTC AGA TAG CG | 419 | M69058 | 60 | 18 |
| SU05 | oxa1 | AGC AGC GCC AGT GCA TCA ATT CGA CCC CAA GTT TCC | 708 | AJ009819 | 60 | 18 |
| SU07 | tem1 | TTG GGT GCA CGA GTG GGT TAA TTG TTG CCG GGA AGC | 503 | AF126482.1 | 60 | 18 |
| pSL18 | tet(A) | GCT ACA TCC TGC TTG CCT TC CAT AGA TCG CCG TGA AGA GG | 210 | X61367 | 59.5 | 31 |
| pRT11 | tet(B) | TTG GTT AGG GGC AAG TTT TG GTA ATG GGC CAA TAA CAC CG | 659 | J01830 | 59.5 | 31 |
| pBR322 | tet(C) | CTT GAG AGC CTT CAA CCC AG ATG GTC GTC ATC TAC CTG CC | 418 | J01749 | 59.5 | 31 |
| pSL106 | tet(D) | AAA CCA TTA CGG CAT TCT GC GAC CGG ATA CAC CAT CCA TC | 787 | L06798 | 59.5 | 31 |
| pSL1504 | tet(E) | AAA CCA CAT CCT CCA TAC GC AAA TAG GCC ACA ACC GTC AG | 278 | L06940 | 59.5 | 31 |
| pJA8122 | tet(G) | GCT CGG TGG TAT CTC TGC TC AGC AAC AGA ATC GGG AAC AC | 468 | S52437 | 59.5 | 31 |
| PAT102 | tet(K) | TCG ATA GGA ACA GCA GTA CAG CAG ATC CTA CTC CTT | 169 | S67449 | 59.5 | 31 |
| pVB.A15 | tet(L) | TCG TTA GCG TGC TGT CAT TC GTA TCC CAC CAA TGT AGC CG | 267 | U17153 | 59.5 | 31 |
| pJ13 | tet(M) | GTG GAC AAA GGT ACA ACG AG CGG TAA AGT TCG TCA CAC AC | 406 | X90939 | 59.5 | 31 |
| pUOA1 | tet(O) | AAC TTA GGC ATT CTG GCT CAC TCC CAC TGT TCC ATA TCG TCA | 515 | Y07780 | 59.5 | 31 |
| pAT451 | tet(S) | CAT AGA CAA GCC GTT GAC C ATG TTT TTG GAA CGC CAG AG | 667 | C92946 | 59.5 | 31 |
| pJIR39 | tetA(P) | CTT GGA TTG CGG AAG AAG AG ATA TGC CCA TTT AAC CAC GC | 676 | L20800 | 59.5 | 31 |
| pNFD13-2 | tet(Q) | TTA TAC TTC CTC CGG CAT CG ATC GGT TCG AGA ATG TCC AC | 904 | X58717 | 59.5 | 31 |
| pBS5 | tet(X) | CAA TAA TTG GTG GTG GAC CC TTC TTA CCT TGG ACA TCC CG | 468 | M37699 | 59.5 | 31 |
| amr130 | sul1 | CGG CGT GGG CTA CCT GAA CG GCC GAT CGC GTG AAG TTC CG | 433 | AF071413 | 69 | 25 |
| amr130 | sul2 | CGG CTC AAG GCA GAT GGC ATT GCG TTT GAT ACC GCG ACC CGT | 285 | M36657 | 69 | 25 |
| rlo44 | sul3 | GAG CAA GAT TTT TGG AAT CG CTA ACC TAG GGC TTT GGA TAT | 790 | AJ459418 | 53 | 19 |
To obtain a PCR template, individual E. coli isolates grown on MacConkey agar were resuspended in 25 μl of UltraPure DNase- and RNase-free water (Invitrogen, Burlington, Ontario, Canada) and lysed by boiling for 2 min. The suspension was centrifuged (12,000 × g for 2 min), and 1 μl supernatant was used as the template. The PCR mixture (25 μl) contained 1× reaction buffer and 1 U Platinum Taq polymerase (Invitrogen). The primers and deoxynucleoside triphosphate concentrations used were as specified in the original protocols cited below. The amplification conditions used for amp were as described previously (18). For amplification of tet genes, the conditions outlined by Ng et al. (31) were used. The PCR cycling conditions used for sul1 and sul2 were as described previously (25), while the amplification conditions used for sul3 were those described by Hammerum et al. (19).
To ensure the absence of PCR inhibitors in lysate, a positive control that amplified a 200-bp fragment of the 16S rRNA gene was included in each PCR run with the primer sets and conditions described previously (25). All PCRs were performed on a PTC 100 thermocycler (MJ Research Inc., Watertown, MA), and each run included a negative control (no template DNA) and an appropriate positive control (control plasmid for the tet, amp, or sul determinant). All PCR products (18 μl) were resolved on 1.5% agarose gels containing ethidium bromide by standard procedures. A 100-bp DNA ladder (MBI Fermentas, Burlington, Ontario, Canada) was used to determine product sizes.
Statistical analysis.
E. coli CFU counts were averaged from replicate plates. The counts were log transformed, and the mean and variance were calculated for each pen before performing the weighted analyses, with the inverse of the variance for each pen being used as the weight. For assessing treatment effect, counts from four time periods (A, D, E, and H) were analyzed for the total, Ampr, and Tetr E. coli populations. For assessing time effects, the numbers of CFU were analyzed by a weighted analysis of variance by the MIXED procedure (36) with treatment, time, and their interaction in the model as fixed effects and pen nested in treatment as the random effect. Time was treated as a repeated-measures effect to account for potential correlations among the various times. Various types of variance-covariance structures were fitted, and the one with the lowest Akaike information criterion value was used for the final analysis. Linear and quadratic orthogonal polynomials were used to check the time effect for trends. The UNIVARIATE procedure was used to check the residuals for normality and for potential outliers. When an outlier was detected, it was removed before the final analysis was performed. P values were used to estimate significant differences between various treatments over time, and P values of ≤0.05 were considered to indicate significant differences.
Cluster analysis was performed on PFGE clonal types by jackknife comparison between REPCs within a treatment. The resulting PPs were characterized by using the criteria described by Tenover et al. (40).
RESULTS AND DISCUSSION
Characterization of Ampr and Tetr E. coli strain.
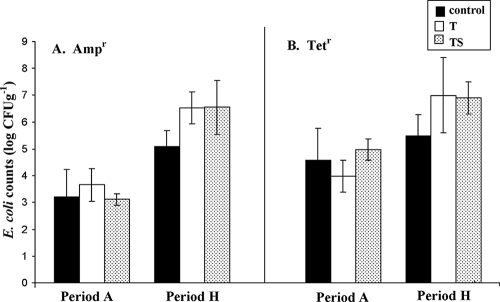
Ampr and Tetr E. coli strains were detected throughout this study. Both Ampr and Tetr E. coli strain numbers were higher in period H than in period A (Fig. 2), in contrast to a previous study, in which a decline in Ampr E. coli counts with animal age was observed (22). Tetr E. coli counts were higher than Ampr E. coli counts for the control and treatment groups in periods A and H (Fig. 2). Overall, higher numbers of Tetr and Ampr E. coli bacteria were shed in period H when either T or TS was administered compared to the control. However, continued detection of Ampr and Tetr E. coli in time period H in control animals suggests that antimicrobial administration may not be the only factor contributing to continued inherent resistance past day 0 and other aspects such as a change in animal diet from forage to grain could help in sustaining resistance, as also suggested previously (41). A drop in pH in the rumen and the gastrointestinal tract occurs when cattle are shifted from a forage-based background diet to a grain-based finishing diet. Increasing evidence corroborates that a low pH supports AR E. coli proliferation and implicates diet as playing a crucial role in controlling bacterial populations from animals with no prior antimicrobial exposure (4). Our observations are corroborated by previous studies in which Ampr and Tetr E. coli strains were frequently detected in poultry, swine, and cattle despite the restricted use of antimicrobials (18, 24).
FIG. 2.
Ampr (A) and Tetr (B) E. coli counts (log CFU g−1 [wet weight]) in periods A and H with no antibiotic treatment (control), 350 mg head−1 day−1 chlortetracycline (T), and 350 mg head−1 day−1 each chlortetracycline and sulfamethazine (TS).
As in any commercial feedlot, antimicrobial administration was terminated 28 days prior to animal slaughter. In order to assess the effect of antimicrobial withdrawal, comparison of counts in period H versus those in period I indicated a marginal (<0.5 log CFU g−1) decline in Ampr and Tetr E. coli strains in the T and TS groups (P > 0.5) (data not shown). This indicates that Ampr and Tetr E. coli can persist for at least 28 days in the gastrointestinal tract and could be disseminated during animal slaughter. These observations are in agreement with other studies in which withdrawal of antimicrobials did not necessarily result in an immediate decrease in the prevalence of resistant bacteria (10, 16, 32, 34).
Periods A, D, E, and H were used to assess the effects of antimicrobial treatment and time on E. coli counts. Least-square differences revealed significant treatment effects on Tetr E. coli (P < 0.001), and a significant effect of time on all three (total, Ampr, and Tetr) E. coli populations (P < 0.001) was observed. Analysis revealed a significant treatment-time interaction for Tetr (P < 0.001) but not for Ampr E. coli (P = 0.9532). Orthogonal contrasts revealed significant linear relationships with time for Tetr E. coli in the control (P < 0.001) and TS (P < 0.001) groups and a significant quadratic relationship for the T group (P < 0.001). Although the highest Tetr E. coli increase per week from period A period to H was observed in the TS group (0.14 log CFU g−1 week−1), followed by the control group (0.11 log CFU g−1 week−1), the highest counts (absolute numbers) of Tetr E. coli bacteria shed (CFU per gram) were observed in the T group and followed the trend T > TS > control. We found limited information in the literature on the effect of TE administration on Tetr E. coli shedding over time.
Susceptibility profiles.
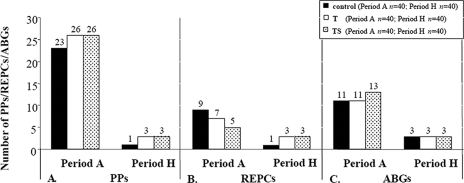
All of the isolates examined in this study carried determinants of resistance to at least one of the antimicrobials tested, as also observed previously in domesticated animals (38, 39). The antibiogram diversity, irrespective of the treatments, decreased as the trial progressed and animals adapted to a finishing diet (period H) (Fig. 3C). Within the control population in period A, isolates displayed 11 ABGs, with SXT TE being a common phenotype found in 22.5% of the isolates (Table 2). By period H, only three ABGs were recorded (Fig. 3C), with AMP TE detected in 50% of the isolates (Table 2). Isolates from the T treatment group in period A also displayed 11 ABGs, which decreased to 3 by period H (Fig. 3C). For this treatment, however, higher frequencies of detection of TE and SXT TE (30 and 27.5% of the isolates) in period A and AMP TE and TE (37.5% of the isolates for both phenotypes) in period H were observed (Table 2). In the TS treatment group, period A isolates displayed 13 ABGs, which also decreased to 3 by period H (Fig. 3C); the most common phenotype in period A was TE (30% of the isolates), which by period H had changed to AMP TE (55% of the isolates) (Table 2). In the literature, we did not find any evidence of a decrease in phenotypic diversity with animal age; however, a higher prevalence of resistant fecal E. coli in younger than older animals held within the same farm has been previously reported (3, 20, 21, 27, 30). Khachatryan et al. (27) have suggested that the intestinal physiology of young animals favors certain niche-specific clones which are retained in older animals. Environmental sources have also been implicated as playing a role in the inoculation of particular strain types, with active competition in the bovine gut leading to the expansion of these niche-specific strains (28). We know for certain that the animals in this study were not exposed to any antimicrobials prior to arrival at our feedlot and that the 6- to 8-month-old animals had diverse phenotypes on day 0. Therefore, animal-to-animal transmission of selected phenotypes and strains (discussed later) seems to better fit predominate detection of TE and AMP TE by period H in the control and treatment groups. A fitness advantage of strains carrying these prevalent phenotypes, however, cannot be ruled out.
FIG. 3.
Numbers of PPs (A) REPCs (B), and ABGs (C) in tested isolates from periods A and H within the control group and the T and TS treatment groups.
TABLE 2.
Frequencies of detection of particular antibiogram patterns during periods Aa and Ha in control group and T and TS treatment groups
| Group (no. of animals) and ABGb | Frequency | % of isolates within:
|
ABGb | Frequency | % of isolates within:
|
||
|---|---|---|---|---|---|---|---|
| Treatment group (n = 40) | Period Ac (n = 120) | Treatment group (n = 40) | Period Hc (n = 120) | ||||
| Control (40) | |||||||
| SXT TE | 9 | 22.5 | 19.2 | AMP TE | 20 | 50 | 47.5 |
| TE | 8 | 20 | 26.7 | TE | 16 | 40 | 31.7 |
| AMP SXT TE | 6 | 15 | 7.5 | AMP TE SXT | 4 | 10 | 20.8 |
| AMC AMP CIP TE | 5 | 12.5 | 7.5 | ||||
| AMP | 4 | 10 | 5 | ||||
| FOX SXT TE | 3 | 7.5 | 5 | ||||
| AMC CIP TE | 1 | 2.5 | 1.7 | ||||
| AMP CIP SXT | 1 | 2.5 | 3.4 | ||||
| AMP CF FOX | 1 | 2.5 | 2.5 | ||||
| AMP SXT | 1 | 2.5 | 1.7 | ||||
| AMP TE | 1 | 2.5 | 3.4 | ||||
| T (40) | |||||||
| TE | 12 | 30 | AMP TE | 15 | 37.5 | ||
| SXT TE | 11 | 27.5 | TE | 15 | 37.5 | ||
| AMC TE | 3 | 7.5 | 3.4 | AMP TE SXT | 10 | 25 | |
| AMP CIP SXT | 3 | 7.5 | |||||
| FOX SXT TE | 3 | 7.5 | |||||
| CIP TE | 3 | 7.5 | 8.3 | ||||
| AMC CIP TE | 1 | 2.5 | |||||
| AMP CF | 1 | 2.5 | 1.7 | ||||
| AMP CF FOX | 1 | 2.5 | |||||
| AMP TE | 1 | 2.5 | |||||
| CF CIP TE | 1 | 2.5 | 0.8 | ||||
| TS (40) | |||||||
| TE | 12 | 30 | AMP TE | 22 | 55 | ||
| CIP TE | 7 | 17.5 | AMP TE SXT | 11 | 27.5 | ||
| AMC AMP CIP TE | 4 | 10 | TE | 7 | 17.5 | ||
| AMP SXT TE | 3 | 7.5 | |||||
| SXT TE | 3 | 7.5 | |||||
| AMC AMP | 2 | 5 | 1.7 | ||||
| AMP | 2 | 5 | |||||
| AMP TE | 2 | 5 | |||||
| AMP CF FOX | 1 | 2.5 | |||||
| AMC TE | 1 | 2.5 | |||||
| AMP CF | 1 | 2.5 | |||||
| AMP CF TE | 1 | 2.5 | 0.8 | ||||
| AMP SXT | 1 | 2.5 | |||||
n = 120.
Abbreviations: ABG, antibiogram profile; AMC, amoxicillin-clavulanic acid; AMP, ampicillin; AMK, amikacin; CF, ceftriaxone; FOX, cefoxitin; CIP, ciprofloxacin; GEN, gentamicin; KAN, kanamycin; NAL, nalidixic acid; STR, streptomycin; TE, tetracycline; SXT, trimethoprim-sulfamethoxazole.
Takes into account all three treatments.
Previous studies of cattle indicate a higher prevalence of certain phenotypes, which varies from study to study but is found to be consistent within a trial. Certain core phenotypes were found to be common in this study, irrespective of the treatment, such as SXT TE (19.2%) and TE (26.7%) in period A and TE (31.7%) and AMP TE (47.5%) in period H (Table 2). These commonly detected phenotypes were different from those reported in previous investigations. For example, STR TE, STR SXT, and SXT TE were found in 60.6 to 77.3% of the isolates and STR SXT TE was found in 60.4% of the isolates from calves (23). However, STR SXT TE, another common phenotype recovered from cattle (28), was not detected in our study in any time period. We found a significant increase in the proportion of isolates with the phenotype AMP TE (P = 0.026) over time but no significant difference in the other phenotypes examined, including TE (P = 0.315). Data from period H showed significant acquisition of ampicillin resistance, which was always linked to either TE or SXT resistance, irrespective of the treatment (Table 2). We also observed sustained TE resistance in the control, T, and TS groups even though resistance to other antimicrobials was not detected by period H. Persistent detection of the TE resistance phenotype in control animals also supports our data on Tetr E. coli counts in periods A and H.
Temporal distribution of strains.
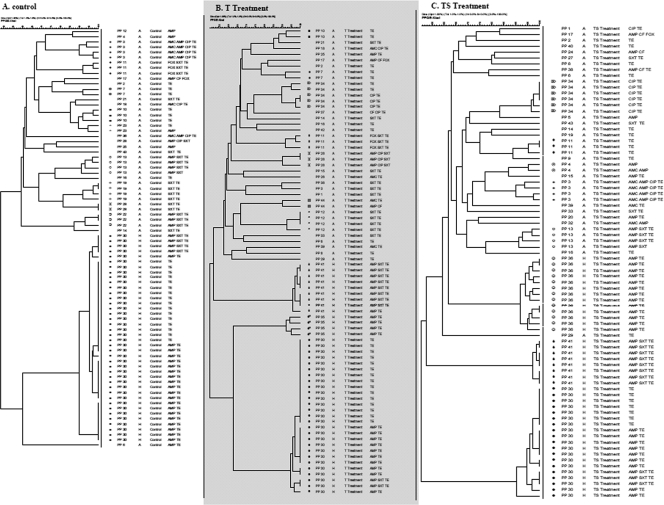
In order to better understand strain prevalence, isolates were genotyped and correlated to ABGs. Given a similarity coefficient of 90% (Dice index), PFGE data revealed 44 distinct PPs among the 240 isolates examined (Fig. 4). When comparing PFGE data in period A, some PPs (3 and 11) were detected in all of the treatment groups whereas others were specific to a particular treatment group. Further, some isolates within the control (n = 13), T (n = 20), and TS (n = 21) treatment groups did not cluster into any REPC. Unique isolates (n = 8) were also detected (Fig. 4).
FIG. 4.
Dendrograms showing the relationship between isolates shed in period A (day 0; n = 40) and those shed in period H (day 197; n = 40) by the control (A), T treatment (B), and TS treatment (C) groups. PPs, time periods, ABGs, and REPCs (>90% banding similarity) in the indicated time periods are shown. REPCs are separated by different symbols among the three treatment groups.
Within the control group, 23 different PPs clustered into nine distinct REPCs in period A and, interestingly, by period H were limited to PP 30 and formed a single REPC (Fig. 3A and B and 4A). Close examination of PP 30 revealed that this genotype was shared by two distinct phenotypes: TE and AMP TE. Based on the strain and resistance gene analysis, we found that this particular strain had acquired the additional ampicillin resistance over time (as discussed later). For the T treatment group, 26 different PPs in period A clustered into seven REPCs, which by period H were reduced to 3 predominant PPs (30, 35, 41) and REPCs (Fig. 3A and B and 4B). Similarly, isolates from period A in the TS treatment group showed 26 different PPs, which clustered into five REPCs and by period H were limited to three PPs (30, 36, 41) and REPCs (Fig. 3A and B and 4C).
The reduction in the numbers of PPs and REPCs correlates well with the limited number of ABGs by period H, indicating overall lower strain diversity by the finishing phase in cattle in a feedlot setting. This decrease in genetic diversity was not a treatment effect since it was also observed in animals in the control group. Instead, it appears to be due to strain selection and propagation of selected strain types via animal-to-animal transmission. In order to validate this strain transmission hypothesis, the PFGE types of individual animals were tracked over time and E. coli isolates from animals in the same pens were observed to cluster into related REPCs, indicative of sufficient movement of E. coli between animals. We found that animals within a pen, among different pens, and also within this cohort started shedding E. coli strains with similar genotypic characteristics over time. Given these results, environmental sources are speculated to play a role in strain transmission among cattle. The animals in the control and treatment groups were housed spatially separate from each other, and no contact was possible between animals on different treatments and even among some replicates of the same treatment. Some nose-to-nose and body contact between animals in adjacent pens and within the same pen could be responsible for possible strain transmissions, but this limited contact cannot explain the presence of the same strain type in spatially separate pens. The animal-to-animal transmission is further validated by the diverse PPs of animals in period A which had little to no chance of strain transmission at day 0. It has also been established by other studies that the administration of antimicrobials selects for resistant bacteria which are subsequently transferred through contaminated food or water and that the feedlot environment is a crucial source of new strains (23). Similarly, in our study we found that antimicrobial administration to beef cattle selects for resistant E. coli and we speculate the feedlot environment plays a critical role in resistance dissemination. The exact route of strain transmission was not characterized in this study, but fecal material is considered a possible source. Our results are in contrast to those of a previous study in which animals in interspersed pens maintained distinct E. coli populations (28).
We recognize that using a single isolate per animal may have its limitations. Therefore, upon observing similar clonal types in animals on different treatments, we PFGE typed an additional isolate from each animal in the T and TS groups in period H. We found that these supplementary isolates grouped in the already described phenotypes and PPs. In the past, single isolates have been used to assess the temporal diversity of antimicrobial-resistant E. coli without compromising the trends over time (6, 23). In addition, the repeated measure offered by four replicates in each treatment group gave us a statistically significant sample size, confirming the trends, and showed that an increase in sample size did not result in an increased genotype number, as also previously observed (23).
Distribution of resistance genes in E. coli.
Isolates from periods A and H displaying ampicillin resistance (Ampr, n = 19), TE resistance (Tetr, n = 116), dual resistance (Ampr Tetr; n = 105), or SXT resistance (Sulr, n = 69) upon susceptibility testing were characterized for resistance alleles in order to understand the distribution of genes and their acquisition over time. In many cases, we found that genotypically similar strains did not necessarily carry the same resistance determinants.
β-Lactamase genes.
Ampr alone was observed exclusively in 19 of the period A isolates and was never observed among isolates from period H (Table 3). Upon testing for β-lactamase genes, we found that blaTEM1 was present in 14 of these isolates while blaPSE1 was detected in the remaining 5 (Table 3). blaTEM1 has been previously detected as a predominant allele in beef cattle (18).
TABLE 3.
PCR detection of Ampr and Tetr determinants in isolates from periods A and Ha
| Phenotype tested and allele(s) | No. of isolates
|
|||||
|---|---|---|---|---|---|---|
| Ampr (n = 19)
|
Tetr (n = 116)
|
Ampr Tetr (n = 105)
|
||||
| Period A (n = 19) | Period H (n = 0) | Period A (n = 78) | Period H (n = 38) | Period A (n = 23) | Period H (n = 82) | |
| Ampr | ||||||
| blaPSE1 | 5 | 0 | NAd | NA | 4 | 27 |
| blaOXA1 | 0 | 0 | NA | NA | 0 | 0 |
| blaTEM1 | 14 | 0 | NA | NA | 19 | 55 |
| Tetr | ||||||
| tet(A)b | NA | NA | 35 | 14 | 7 | 34 |
| tet(B)b | NA | NA | 15 | 7 | 4 | 13 |
| tet(C)b | NA | NA | 8 | 5 | 3 | 9 |
| tet(A) tet(B)c | NA | NA | 9 | 5 | 4 | 11 |
| tet(B) tet(C)c | NA | NA | 5 | 5 | 2 | 4 |
| tet(A) tet(C)c | NA | NA | 6 | 2 | 3 | 11 |
Isolates that displayed Ampr, Tetr, or Ampr Tetr dual resistance upon susceptibility testing were selected for resistance allele characterization.
Single resistance.
Dual resistance.
NA, not applicable.
TE resistance-encoding genes.
Alleles tet(K), tet(L), tet(M), tet(O), tet(P), tet(Q), tet(S), tet(X), and tetA(P) were never detected, and some of these have only been be found in gram-positive bacteria. tet(A) was the predominant allele detected in period A (44.9%) and period H (36.8%); tet(B) was also prevalent but at lower frequencies in periods A (19.2%) and H (18.4%) (Table 3). Taking into consideration both periods A and H, the tet(A) and tet(B) resistance alleles accounted for 42.2 and 31.9% of the TE resistance among isolates. Overall, the frequency of detection of the tet alleles was tet(A) > tet(B) > tet(C), being detected in 40.7, 17.6, and 11.3% of the total isolates, respectively. Similar to our results, Guerra et al. (18) found tet(A) and tet(B) to be the predominant alleles detected in 66 and 42% of the isolates examined from cattle, swine, and poultry. Like the ABGs, the prevalence of individual determinants responsible for specific resistances varies from study to study. For instance, tet(B) was the predominant determinant detected in 93% of the isolates and tet(A) was detected to a lesser extent (7%) in dairy cattle (37).
Several Tetr isolates (n = 32) from periods A and H contained more than one tet allele (dual tet resistance) (Table 3). tet(A) tet(B) was detected in periods A (11.5%) and H (13.2%); combined tet(B) tet(C) and tet(A) tet(C) were also observed.
An increase in the number of dual-resistant Ampr Tetr isolates from period A (21.9%) to period H (78.1%) and a marked reduction in the number of isolates carrying a single resistance, either Ampr or Tetr alone, in period H was clearly observed and not restricted to a few individual animals or pens. These results are corroborated by ABGs, which showed an increased prevalence of isolates carrying AMP TE. Of the 105 isolates which tested positive for combined Ampr Tetr (Table 3), 23 belonged to period A and 82 belonged to period H. tet(A) was detected frequently in periods A (30.4%) and H (41.5%), while blaTEM1 conferred ampicillin resistance in periods A (82.6%) and H (67.1%). blaOXA1 and blaPSE1 were never detected within these dual-resistant isolates. These results indicate an increase in combined tet(A) and blaTEM1 acquisition by period H. As noted via genotyping previously, predominant detection of PP 30 in period H with phenotype AMP TE or TE suggested that this strain type either acquired or lost Ampr over time to display this phenotype. Resistance gene distribution supports the acquisition of Ampr within PP 30 (Fig. 4; Table 3). Other investigators have reported a similar genetic linkage of antimicrobial drug resistance genes (39). As observed for Tetr single resistance, Ampr Tetr isolates also showed an increase in dual TE resistance genes, viz., tet(A) tet(B), tet(B) tet(C), and tet(A) tet(C), in periods A to H (Table 3).
Sulfonamide genes.
Of the isolates examined from periods A and H, 69 exhibited Sulr (Table 4). sul2 was detected at a higher frequency (47.8%) than sul1 (17.4%) (Table 4). Similar higher sul2 than sul1 detection has been reported earlier in humans and swine (19) and attributed to a fitness advantage in clinical human E. coli (15). sul3 was never detected in this study but has been previously reported in E. coli from pigs (18, 19, 33, 35). Dual sul1 sul2 genes were also observed in isolates from both periods A (31.9%) and H (40%) (Table 4).
TABLE 4.
PCR detection of Sulr determinants in isolates from periods A and H that displayed Sulr upon susceptibility testing
| Allele(s) | No. of resistant isolates (total n = 69)
|
|
|---|---|---|
| Period A (n = 44) | Period H (n = 25) | |
| sul1a | 7 | 5 |
| sul2a | 23 | 10 |
| sul3a | 0 | 0 |
| sul1 sul2b | 14 | 10 |
Single resistance.
Dual resistance.
In conclusion, results from the present study indicate that fecal E. coli populations in beef cattle in a feedlot setting are temporally variable and that antimicrobial selection pressure is not essential for the maintenance of inherent Ampr and Tetr E. coli. Nevertheless, usage of chlortetracycline alone or in combination with sulfamethazine results in higher Tetr E. coli shedding. Inherent resistance in other animals of the cohort affects resistance dissemination and strain acquisition. By the finishing phase, an increased combined ampicillin and TE resistance was observed and cattle in the cohort shed E. coli with resistance limited to the AMP SXT TE, AMP TE, and TE phenotypes, irrespective of treatment. Genotyping results confirmed strain movement from animal to animal in the cohort to be responsible for this limited diversity and supports the hypothesis that strain transmission among animals plays a crucial role in transmitting AR. The transmission of selected antimicrobial-resistant E. coli genotypes indicates that feedlot farms are important reservoirs of resistance. Detection of inherent and acquired resistance in E. coli from beef cattle in this study proves the importance of monitoring as an essential component of AR management strategies.
Acknowledgments
The technical assistance of Lorna Selinger, Lisa Yang, and the barn staff is gratefully acknowledged. We thank B. Guerra and R. Helmuth, Federal Institute for Risk Assessment (BFR), Germany; P. Boerlin, University of Guelph, Canada; D. Taylor, University of Alberta, Edmonton, Canada; J. I. Rood, Monash University, Australia; V. Burdett, Duke University, Durham, NC; and A. Salyers, University of Illinois, for providing some of the control strains. We acknowledge the assistance of V. Boras and her team at the Lethbridge Regional Hospital in susceptibility testing.
This research was supported by a GAPS grant from Agriculture and Agri-Food Canada (AAFC) and an A-base grant (AAFC) to R.S.
Footnotes
Published ahead of print on 22 August 2008.
Contribution 38707054 from Agriculture and Agri-Food Research Centre, Lethbridge, Alberta, Canada.
REFERENCES
- 1.Berge, A. C., E. R. Atwill, and W. M. Sischo. 2003. Assessing antibiotic resistance in fecal Escherichia coli in young calves using cluster analysis techniques. Prev. Vet. Med. 61:91-102. [DOI] [PubMed] [Google Scholar]
- 2.Berge, A. C., E. R. Atwill, and W. M. Sischo. 2005. Animal and farm influences on the dynamics of antibiotic resistance in faecal Escherichia coli in young dairy calves. Prev. Vet. Med. 69:25-38. [DOI] [PubMed] [Google Scholar]
- 3.Berge, A. C., D. A. Moore, and W. M. Sischo. 2006. Field trial evaluating the influence of prophylactic and therapeutic antimicrobial administration on antimicrobial resistance of fecal Escherichia coli in dairy calves. Appl. Environ. Microbiol. 72:3872-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bettelheim, K. A., A. Kuzevski, R. A. Gilbert, D. O. Krause, and C. S. McSweeney. 2005. The diversity of Escherichia coli serotypes and biotypes in cattle faeces. J. Appl. Microbiol. 98:699-709. [DOI] [PubMed] [Google Scholar]
- 5.Borgen, K., G. S. Simonsen, A. Sundsfjord, Y. Wasteson, Ø. Olsvik, and H. Kruse. 2000. Continuing high prevalence of VanA-type vancomycin-resistant enterococci on Norwegian poultry farms three years after avoparcin was banned. J. Appl. Microbiol. 89:478-485. [DOI] [PubMed] [Google Scholar]
- 6.Brun, E., G. Holstad, H. Kruse, and J. Jarp. 2002. Within-sample and between-sample variation of antimicrobial resistance in fecal Escherichia coli isolates from pigs. Microb. Drug Resist. 8:385-391. [DOI] [PubMed] [Google Scholar]
- 7.Canadian Council on Animal Care. 2003. Guide to the care and use of experimental animals, vol. 1, 2nd ed. Canadian Council on Animal Care, Ottawa, Ontario, Canada. http://www.ccac.ca./en/CCAC_Programs/Guidelines_Policies/GUIDES/ENGLISH/toc_v1.htm (accessed on 10 January 2008).
- 8.Carson, A. C., B. L. Shear, M. R. Ellersieck, and A. Asfaw. 2001. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 67:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catry, B., H. Laevens, L. A. Devriese, G. Opsomer, and A. De Kruif. 2003. Antimicrobial resistance in livestock. J. Vet. Pharmacol. Ther. 26:81-93. [DOI] [PubMed] [Google Scholar]
- 10.Chaslus-Dancla, E., G. Gerbaud, M. Lagorce, J. P. Lafont, and P. Courvalin. 1987. Persistence of an antibiotic resistance plasmid in intestinal Escherichia coli of chickens in the absence of selective pressure. Antimicrob. Agents Chemother. 31:784-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial disk susceptibility tests—ninth edition: approved standard M02-A9. Clinical and Laboratory Standards Institute, Wayne, PA.
- 12.DeFrancesco, K. A., R. N. Cobbold, D. H. Rice, T. E. Besser, and D. D. Hancock. 2004. Antimicrobial resistance of commensal Escherichia coli from dairy cattle associated with recent multi-resistant salmonellosis outbreaks. Vet. Microbiol. 98:55-61. [DOI] [PubMed] [Google Scholar]
- 13.Donaldson, S. C., B. A. Straley, N. V. Hegde, A. A. Sawant, C. DebRoy, and B. M. Jayarao. 2006. Molecular epidemiology of ceftiofur-resistant Escherichia coli isolates from dairy calves. Appl. Environ. Microbiol. 72:3940-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunlop, R. H., S. A. McEwen, A. H. Meek, R. C. Clarke, W. D. Black, and R. M. Friendship. 1998. Associations among antimicrobial drug treatments and antimicrobial resistance of fecal Escherichia coli of swine on 34 farrow-to-finish farms in Ontario, Canada. Prev. Vet. Med. 34:283-305. [DOI] [PubMed] [Google Scholar]
- 15.Enne, V. I., P. M. Bennett, D. M. Livermore, and L. M. C. Hall. 2004. Enhancement of host fitness by the sul2-encoding plasmid p9123 in the absence of selective pressure. J. Antimicrob. Chemother. 53:958-963. [DOI] [PubMed] [Google Scholar]
- 16.Enne, V. I., D. M. Livermore, P. Stephens, and L. M. C. Hall. 2001. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357:1325-1328. [DOI] [PubMed] [Google Scholar]
- 17.Gilchrist, M. J., C. Greko, D. B. Wallinga, G. W. Beran, D. G. Riley, and P. S. Thorne. 2007. The potential role of concentrated animal feeding operations in infectious disease epidemics and antibiotic resistance. Environ. Health Perspect. 115:313-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerra, B., E. Junker, A. Schroeter, B. Malorny, S. Lehman, and R. Helmuth. 2003. Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine, and poultry. J. Antimicrob. Chemother. 52:489-492. [DOI] [PubMed] [Google Scholar]
- 19.Hammerum, A. M., D. Sandvang, S. R. Andersen, A. M. Seyfarth, L. J. Porsbo, N. Frimodt-Møller, and O. E. Heuer. 2006. Detection of sul1, sul2 and sul3 in sulphonamide resistant Escherichia coli isolates obtained from healthy humans, pork and pigs in Denmark. Int. J. Food Microbiol. 106:235-237. [DOI] [PubMed] [Google Scholar]
- 20.Hinton, M., A. H. Linton, and A. J. Hedges. 1985. The ecology of Escherichia coli in calves reared as dairy-cow replacements. J. Appl. Bacteriol. 58:131-138. [DOI] [PubMed] [Google Scholar]
- 21.Howe, K., and A. H. Linton. 1976. A longitudinal study of Escherichia coli in cows and calves with special reference to the distribution of O-antigen types and antibiotic resistance. J. Appl. Bacteriol. 40:331-340. [DOI] [PubMed] [Google Scholar]
- 22.Hoyle, D. V., H. I. Knight, D. J. Shaw, K. Hillman, M. C. Pearce, J. C. Low, G. J. Gunn, and M. E. J. Woolhouse. 2004. Acquisition and epidemiology of antimicrobial-resistant Escherichia coli in a cohort of newborn calves. J. Antimicrob. Chemother. 53:867-871. [DOI] [PubMed] [Google Scholar]
- 23.Hoyle, D. V., C. M. Yates, M. E. Chase-Topping, E. J. Turner, S. E. Davies, J. C. Low, G. J. Gunn, M. E. Woolhouse, and S. G. Amyes. 2005. Molecular epidemiology of antimicrobial-resistant commensal Escherichia coli strains in a cohort of newborn calves. Appl. Environ. Microbiol. 71:6680-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoyle, D. V., H. C. Davison, H. I. Knight, C. M. Yates, O. Dobay, G. J. Gunn, S. G. B. Amyes, and M. E. J. Woolhouse. 2006. Molecular characterisation of bovine faecal Escherichia coli shows persistence of defined ampicillin resistant strains and the presence of class 1 integrons on an organic beef farm. Vet. Microbiol. 115:250-257. [DOI] [PubMed] [Google Scholar]
- 25.Kerrn, M. B., T. Klemmensen, N. Frimodt-Møller, and F. Espersen. 2002. Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance. J. Antimicrob. Chemother. 50:513-516. [DOI] [PubMed] [Google Scholar]
- 26.Khachatourians, G. G. 1998. Agricultural use of antibiotics and the evolution and transfer of antibiotic-resistant bacteria. CMAJ 159:1129-1136. [PMC free article] [PubMed] [Google Scholar]
- 27.Khachatryan, A. R., D. D. Hancock, T. E. Besser, and D. R. Call. 2004. Role of calf-adapted Escherichia coli in maintenance of antimicrobial drug resistance in dairy calves. Appl. Environ. Microbiol. 70:752-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khachatryan, A. R., T. E. Besser, D. D. Hancock, and D. R. Call. 2006. Use of a nonmedicated dietary supplement correlates with increased prevalence of streptomycin-sulfa-tetracycline-resistant Escherichia coli on a dairy farm. Appl. Environ. Microbiol. 72:4583-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, C. Y., B. E. Langlois, and K. A. Dawson. 1993. Detection of tetracycline resistance determinants in pig isolates from three herds with different histories of antimicrobial agent exposure. Appl. Environ. Microbiol. 59:1467-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathew, A. G., M. A. Beckmann, and A. M. Saxton. 2001. A comparison of antibiotic resistance in bacteria isolated from swine herds in which antibiotics were used or excluded. J. Swine Health Prod. 9:125-129. [Google Scholar]
- 31.Ng, L.-K., I. Martin, M. Alfa, and M. Mulvey. 2001. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15:209-215. [DOI] [PubMed] [Google Scholar]
- 32.O'Brien, T. F. 2002. Emergence, spread, and environmental effect of antimicrobial resistance: how use of an antimicrobial anywhere can increase resistance to any antimicrobial anywhere else. Clin. Infect. Dis. 34(Suppl. 3):S78-S84. [DOI] [PubMed] [Google Scholar]
- 33.Perreten, V., and P. Boerlin. 2003. A new sulphonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 47:1169-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rollins, L. D., S. A. Gaines, D. W. Pocurull, H. D. Mercer, and L. T. Frobish. 1976. Persistence of transferable drug resistance in the lactose-fermenting enteric flora of swine following antimicrobial feeding. Can. J. Comp. Med. 40:175-183. [PMC free article] [PubMed] [Google Scholar]
- 35.Sáenz, Y., L. Briñas, E. Domínguez, J. Ruiz, M. Zarazaga, J. Vila, and C. Torres. 2004. Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob. Agents Chemother. 48:3996-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.SAS Institute Inc. 2005. SAS online doc 9.1.3. SAS Institute Inc., Cary, NC.
- 37.Sawant, A. A., N. V. Hegde, B. A. Straley, S. C. Donaldson, B. C. Love, S. J. Knabel, and B. M. Jayarao. 2007. Antimicrobial-resistant enteric bacteria from dairy cattle. Appl. Environ. Microbiol. 73:156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sayah, R. S., J. B. Kaneene, Y. Johnson, and R. Miller. 2005. Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal fecal samples, human septage, and surface water. Appl. Environ. Microbiol. 71:1394-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, J. L., D. J. V. Drum, Y. Dai, J. M. Kim, S. Sanchez, J. J. Maurer, C. L. Hofacre, and M. D. Lee. 2007. Impact of antimicrobial usage on antimicrobial resistance in commensal Escherichia coli strains colonizing broiler chickens. Appl. Environ. Microbiol. 73:1404-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Baale, M. J., J. M. Sargeant, D. P. Gnad, B. M. DeBey, K. F. Lechtenberg, and T. G. Nagaraja. 2004. Effect of forage or grain diets with or without monensin on ruminal persistence and fecal Escherichia coli O157:H7 in cattle. Appl. Environ. Microbiol. 70:5336-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Donkersgoed, J., K. Manninen, A. Potter, S. McEwen, V. Bohaychuk, S. Klashinsky, A. Deckert, and R. Irwin. 2003. Antimicrobial susceptibility of hazard analysis critical control point Escherichia coli isolates from federally inspected beef processing plants in Alberta, Saskatchewan, and Ontario. Can. Vet. J. 44:723-728. [PMC free article] [PubMed] [Google Scholar]