Abstract
Polaromonas sp. strain JS666 can grow on cis-1,2-dichloroethene (cDCE) as a sole carbon and energy source and may be useful for bioremediation of chlorinated solvent-contaminated sites. Analysis of the genome sequence of JS666 (5.9 Mb) shows a bacterium well adapted to pollution that carries many genes likely to be involved in hydrocarbon and xenobiotic catabolism and metal resistance. Clusters of genes coding for haloalkane, haloalkanoate, n-alkane, alicyclic acid, cyclic alcohol, and aromatic catabolism were analyzed in detail, and growth on acetate, catechol, chloroacetate, cyclohexane carboxylate, cyclohexanol, ferulate, heptane, 3-hydroxybenzoate, hydroxyquinol, gentisate, octane, protocatechuate, and salicylate was confirmed experimentally. Strain JS666 also harbors diverse putative mobile genetic elements, including retrons, inteins, a miniature inverted-repeat transposable element, insertion sequence transposases from 14 families, eight genomic islands, a Mu family bacteriophage, and two large (338- and 360-kb) plasmids. Both plasmids are likely to be self-transferable and carry genes for alkane, alcohol, aromatic, and haloacid metabolism. Overall, the JS666 genome sequence provides insights into the evolution of pollutant-degrading bacteria and provides a toolbox of catabolic genes with utility for biotechnology.
There are few cultured representatives of the genus Polaromonas (Betaproteobacteria, family Comamonadaceae), and only four well-established Polaromonas species exist. They are the type strain Polaromonas vacuolata (21), from Antarctic seawater; the subsurface isolate P. naphthalenivorans (24); the tap water isolate P. aquatica; and the hydrogen-oxidizing chemolithotroph P. hydrogenivorans (37). Polaromonas isolates tend to be moderately psychrotrophic, oligotrophic, and slow growing, traits that likely impede their isolation and characterization by standard methods. We are just beginning to understand the environmental roles and ecological niches of polaromonads, but recent studies suggest that they are widespread (27) and influential in pollutant degradation (1, 10, 11, 24, 27).
Polaromonas strains can biodegrade important groundwater contaminants, including petroleum hydrocarbons (6, 24, 38) and chlorinated ethenes (6, 10, 38). Petroleum hydrocarbons include aliphatic, aromatic, and polycyclic compounds, all of which are naturally occurring but problematic due to their toxicity and persistence (3). Chlorinated ethenes are essentially xenobiotic and are synthesized for the manufacture of plastics (vinyl chloride) and for use as solvents (tetrachloroethene, trichloroethene, and cis-dichloroethene [cDCE]). Cleanup of sites contaminated with chlorinated ethenes and/or petroleum hydrocarbons by using physical or chemical methods is difficult and expensive, so there has been intense interest in finding bacteria capable of degrading these pollutants for use in bioremediation.
Polaromonas sp. strain JS666 was isolated from a granular activated-carbon filter treating chloroethene-contaminated groundwater and is capable of growth-coupled cDCE oxidation and cometabolic degradation of diverse chlorinated aliphatic pollutants (10). As a result, strain JS666 is potentially useful for bioaugmentation of chlorinated-ethene-contaminated sites because growth of JS666 on cDCE in the subsurface would essentially convert the pollutant into a catalyst (i.e., new cells), leading to theoretically increasing degradation rates over time. Transfer of the biodegradative activity of JS666 to in situ conditions requires a better understanding of the physiology and biochemistry of the organism, especially with respect to the enzymes involved in organochlorine degradation, the ability to degrade or resist other pollutants, the range of nutrient sources used, and the types of environmental conditions preferred.
The metabolic and phylogenetic novelty of Polaromonas sp. strain JS666 and its potential for use in bioremediation provided the impetus for complete genome sequencing. Here, we describe a bioinformatic and phenotypic analysis that reveals diverse metabolic capabilities of JS666 that may be useful for bioremediation and biocatalysis. We also describe the roles of mobile genetic elements in shaping the genome and metabolic abilities of the bacterium.
MATERIALS AND METHODS
Construction, isolation, and sequencing of small-insert and large-insert DNA libraries.
Whole-genome shotgun sequencing and finishing was carried out at the U.S. Department of Energy Joint Genome Institute as described on the Joint Genome Institute website (http://www.jgi.doe.gov/sequencing/protocols/index.html). Briefly, genomic DNA was randomly sheared with a hydroshear device (Genemachines, San Carlos, CA), and the fragments were blunt-end repaired using T4 polymerase and Klenow fragment. Fragments were size selected by agarose gel electrophoresis, purified from the gel (Qiaquick; Qiagen Corporation, Valencia, CA), and ligated into pUC18 (∼3-kb inserts), pMCL200 (∼7-kb inserts), or pCC1Fos (Epicentre, Madison, WI) (∼35-kb inserts).
Ligation reactions were used to transform Escherichia coli DH10B, and the resulting colonies containing plasmids were picked into 384-well plates containing LB and glycerol. Template DNA for sequencing was produced by amplifying plasmid subclones using rolling-circle amplification (Templiphi; GE Healthsciences, Piscataway, NJ) as described previously (14) or Sprintprep (Agencourt Biosciences, Beverly, MA) magnetic-bead DNA purification. Subclone inserts were sequenced from both ends using universal primers and Energy Transfer (GE Healthsciences, Piscataway, NJ) or Big Dye (ABI, Foster City, CA) terminator chemistry.
Sequence assembly and gap closure.
A total of 78,720 sequencing reads from the three libraries (3 kb, 33,792; 8 kb, 39,936; 40 kb, 4,992) were attempted and assembled with PHRAP (18) to produce a draft assembly containing 63 contigs. An additional 970 finishing reads were sequenced to resolve misassemblies, close gaps, and polish low-quality regions. The finished genome consists of one chromosome (5.2 Mb) and two plasmids (360 kb and 338 kb) containing no gaps and an error rate of less than 1 in 50,000 bases. Dupfinisher (C. S. Han and P. Chain, presented at the International Conference on Bioinformatics and Computational Biology, June 26-29, 2006) and transposon bombing (Epicentre Biotechnologies, Madison, WI) were used to resolve misassemblies.
Sequence analysis and automated annotation.
Gene modeling was performed by using the Critica (4), Glimmer (12), and Generation (http://compbio.ornl.gov/public/tools) modeling packages. The results were combined, a BLASTP search of the translations versus GenBank's nonredundant (NR) database was conducted, and the best NR match was used to select the preferred gene model. If there was not a BLAST match, the longest gene model was retained. Overlapping gene models were manually examined, and genes with BLAST matches were retained. The revised gene-protein set was searched against the KEGG GENES, Pfam, PROSITE, PRINTS, PRoDom, and COGs databases, in addition to the BLASTP versus NR databases. Genes were functionally categorized using the KEGG and COGs hierarchies. Manual revisions were made to the machine annotation as appropriate. Chromosome and plasmid maps were generated using the program CGview (40). The IslandPath interface (20) was used to search for genomic islands based on dinucleotide bias and unusual G+C content (default settings, ±3.48% G+C), presence of an integrase gene, and proximity to a tRNA gene. Inteins were identified by the presence of a homing endonuclease domain and flanking conserved regions (A, B, F, and G) in the N- and C-terminal splicing domains; these analyses were performed by Francine Perler of New England Biolabs (34).
Growth experiments.
Either minimal salts medium (MSM) (10) or R2A medium (Oxoid) was used for culture growth. Fed-batch growth substrate tests were done in MSM, using test compounds (≥97% purity) as sole sources of carbon and energy. Polaromonas sp. strain JS666 cells grown to mid-exponential phase on succinate, cDCE, or ethanol were inoculated (optical density at 600 nm [OD600] = 0.01 to 0.07) into 40 ml of MSM in 160-ml serum bottles with air headspace. Each compound was tested over a range of starting concentrations from 0.2 to 10 mM, with the concentration of volatile compounds estimated as if they were entirely dissolved in the aqueous phase. Bottles were sealed with Teflon-coated butyl rubber septa and aluminum crimp caps (Wheaton) and incubated at 20 to 22°C with shaking at 190 to 250 rpm. At least two independent growth experiments were conducted for each substrate, including a control with no carbon source and uninoculated controls for compounds yielding colored breakdown products (catechol, protocatechuate, ferulate, and gentisate). Chemolithotrophic thiosulfate and carbon monoxide oxidation cultures were also prepared as described above with the following exceptions: sodium thiosulfate (0 to 40 mM) or CO (10% of the headspace) was added as the energy source, and sodium bicarbonate (10 mM) was added as a carbon source. Duplicate sets of growth experiments were conducted for both lithotrophic and mixotrophic growth; in the latter case, ethanol (5 to 10 mM) was provided as a limited source of organic carbon.
Anaerobic nitrate reduction cultures were prepared as described above, except 160 ml of MSM was used, leaving no headspace in the serum bottles, and the medium was purged with nitrogen gas to create anaerobic conditions. Ethanol (5 mM) was provided as a carbon source, and samples were removed while the bottles were being purged with nitrogen gas.
Auxanography plates were prepared with 1.5% Noble agar in MSM (made with high-performance liquid chromatography grade water). Exponentially growing succinate cultures were diluted in MSM to an OD600 of 0.1, and 100 μl was spread on each plate. Solid-phase test compounds (5 mg) were deposited on the edges of the plates to allow a concentration gradient to form in the agar, and the starting locations of the compounds were marked. The plates were sealed with Parafilm, inverted, and incubated at 20 to 22°C for 2 to 8 weeks. All compounds were tested with duplicate plates in two independent experiments, as were the no-carbon and no-inoculum controls.
PFGE.
Agarose plugs were prepared and pulsed-field gel electrophoresis (PFGE) was performed as described previously (28), except that succinate-grown JS666 cells were suspended in PFGE grade agarose at an OD600 of 3.
Analytical methods.
Growth was indirectly estimated in liquid cultures by using the OD600. Following auxanography, cells were scraped from the plates and then resuspended in MSM and washed, and total protein was analyzed by a previously described UV protein assay (10). Heptane, hexane, octane, 1,2-dichloroethane, and cDCE were analyzed in headspace samples (100 μl) by gas chromatography under isothermal conditions with flame ionization detection and a J&W Scientific GSQ (30-m by 0.53-mm inside diameter) column. Nitrate and nitrite concentrations in anaerobic cultures were measured on a Dionex DX600 ion chromatograph with a Dionex IonPac AS18 hydroxide-selective anion-exchange column.
Nucleotide sequence accession numbers.
The finished, annotated sequence was deposited in GenBank under accession numbers CP000316 to CP000318 and is also available at http://genome.jgi-psf.org/finished_microbes/poljs/poljs.info.html.
RESULTS
Major features of the genome.
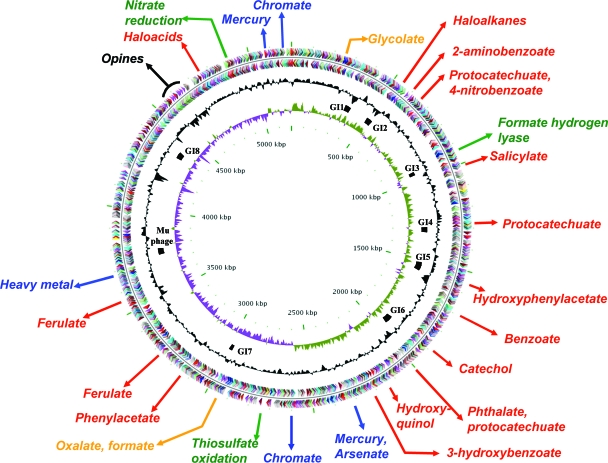
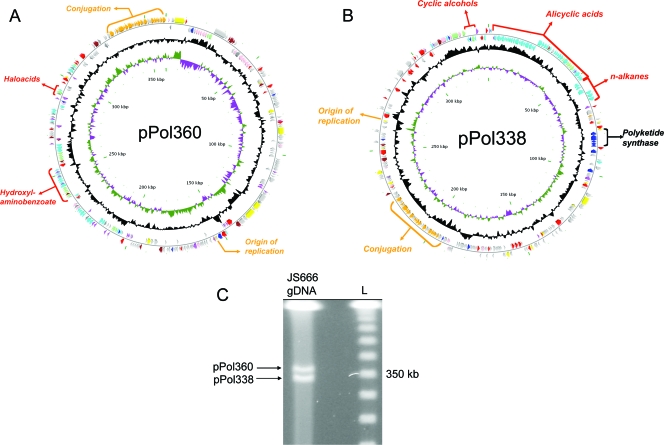
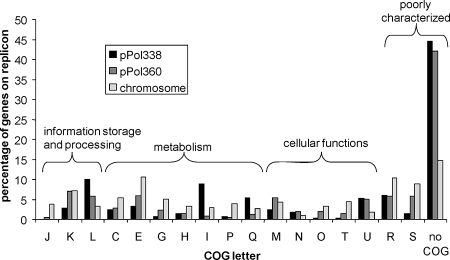
The Polaromonas sp. strain JS666 genome (5.9 Mb) (Table 1) consists of a circular chromosome (5.2 Mb) (Fig. 1) and two circular plasmids, pPol360 (360 kb) (Fig. 2A) and pPol338 (338 kb) (Fig. 2B) with a total of 5,569 predicted protein-encoding genes (Table 1). Only a single rRNA operon is present (Bpro_R0044 to Bpro_R0040). Analysis of clusters of orthologous groups (COGs) across the three replicons (Fig. 3) revealed several trends. Translation-related COGs (J) were restricted to the chromosome, while hypothetical proteins (no COG) were concentrated on the plasmids (42 to 45% of all plasmid open reading frames). The proportions of COGs involved in inorganic-ion transport and metabolism (P), signal transduction (T), and most metabolic functions (C, E, G, H, and P) were higher on the chromosome, while the proportions of DNA-related functions (L) and trafficking/secretion (U) COGs were higher on the plasmids. Plasmid pPol338 was enriched in COGs involved in lipid (I) and secondary (Q) metabolism.
TABLE 1.
Features of the Polaromonas sp. strain JS666 genomea
| Feature | Value |
|---|---|
| Total size (bases) | 5,898,676 |
| Chromosome | 5,200,264 |
| Plasmid 1 (pPol360) | 360,405 |
| Plasmid 2 (pPol338) | 338,007 |
| Gene density (no. of genes per kb) | 0.944 |
| Avg gene length (bases) | 931 |
| G+C% | 61.97 |
| No. of protein-coding genes | 5,569 |
| No. of genes with function prediction | 4,099 |
| No. of conserved hypothetical genes | 1,388 |
| No. of hypothetical genes (ORFans) | 82 |
| No. of pseudogenes | 116 |
| Coding region (%) | 99.2 |
| rRNA | 1 operon, arranged 16S-tRNAIle-tRNAAla-23S-5S |
| tRNA | 44 |
| tmRNA | 1 |
| Putative mobile genetic elements (no.) | |
| Plasmids | 2 |
| Transposases | 55 |
| Phage | 1 |
| Genomic islands | 8 |
| Retrons | 6 |
| Inteins | 2 |
| Xenobiotic/hydrocarbon metabolism | |
| Total monooxygenases | 16 |
| Total no. of P450s | 4 |
| Total no. of dioxygenases | 43 |
| Total no. of glutathione S-transferases | 21 |
| Total no. of hydrolases | 111 |
| Total no. of hydratases | 27 |
| Total no. of dehalogenases | 9 |
Derived, in part, from the DOE-JGI IMG server (http://img.jgi.doe.gov).
FIG. 1.
The chromosome of Polaromonas sp. strain JS666. Approximate locations of important gene clusters are indicated outside of the first circle, which depicts coding regions on the plus strand colored by functional category: translation, ribosomal structure, and biogenesis (yellow); transcription (magenta); DNA replication, recombination, and repair (red); cellular processes (sky blue); signal transduction (lavender); energy production and conversion (green); carbon and carbohydrate metabolism (blue); amino acid transport and metabolism (orange); nucleotide transport and metabolism (light red); coenzyme metabolism (pink); lipid metabolism (cyan); secondary-metabolite biosynthesis, transport, and catabolism (dark green); general function prediction only (brown); function unknown (dark gray); and no COG (light gray). Moving toward the center, the second circle depicts coding regions on the minus strand (with the same color scheme as the plus strand). The third circle shows G+C content (deviation from average), and the fourth circle G+C skew in purple and olive. Genomic islands (GI1 to GI8) and the Mu phage are indicated by black bars between the third and fourth circles. The scale (in kbp) is indicated on the innermost circle.
FIG. 2.
(A) The 360-kb plasmid pPol360 of Polaromonas sp. strain JS666. (B) The 338-kb plasmid pPol338 of Polaromonas sp. strain JS666. Approximate locations of important metabolic gene clusters, plasmid replication, and conjugation genes are indicated outside of the first circle, which depicts coding regions on the plus strand colored by functional category: plasmid transfer (orange); plasmid replication and maintenance (yellow); transport (light green); DNA replication, recombination, and repair (red); metabolism (cyan); biosynthesis (dark blue); regulators (purple); cell envelope and membrane biogenesis (pink); general function prediction (brown); conserved hypothetical (dark gray); and no COG (light gray). Moving toward the center, the second circle depicts coding regions on the minus strand (with the same color scheme as the plus strand). The third circle shows G+C content (deviation from average), and the fourth circle G+C skew in purple and olive. The scale (in kbp) is indicated on the innermost circle. The base pair numbering for both plasmids begins near the putative origins of replication (i.e., repB [Bpro_5513] in pPol338 and trfA [Bpro_5045] in pPol360). (C) PFGE of genomic DNA (gDNA) from Polaromonas sp. strain JS666 depicting the presence of two extrachromosomal genetic elements. L, 50-kb lambda ladder.
FIG. 3.
Functional distribution of COGs among replicons in the JS666 genome sequence. COG designations: J, translation and ribosomal structure/biogenesis; K, transcription; L, DNA replication, recombination, and repair; C, energy production/conversion; E, amino acid transport/metabolism; G, carbohydrate transport/metabolism; H, coenzyme metabolism; I, lipid metabolism; P, inorganic-ion transport/metabolism; Q, secondary metabolism; M, cell envelope biogenesis outer membrane; N, cell motility and secretion; O, posttranslational modification and protein turnover; T, signal transduction mechanisms; U, intracellular trafficking and secretion; R, general function prediction only; and S, function unknown.
Plasmids.
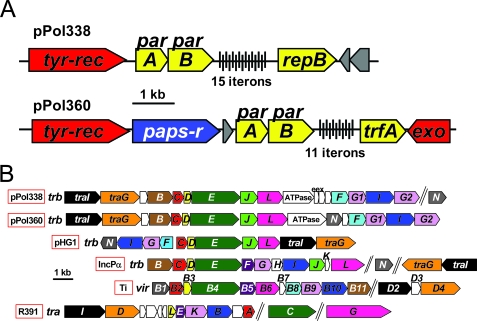
Due to the importance of plasmids in disseminating hydrocarbon and xenobiotic metabolism genes, we examined pPol338 and pPol360 in detail. The plasmids have similar sizes (360 kb and 338 kb), conjugative-transfer functions, and stabilization systems but differ in the putative replication regions (Fig. 2; see Table S1D and E in the supplemental material). The presence and sizes of pPol338 and pPol360 were corroborated by contour-clamped homogeneous electric field-PFGE (Fig. 2C). However, the bands observed on PFGE corresponded to linear forms of the two plasmids. This phenomenon could be explained by spontaneous production of linear DNA from circular DNA during plug preparation or electrophoresis (5). The putative replication region of pPol338 (Fig. 4A) encodes an initiator, RepB (Bpro_5513), similar to that of Burkolderia minor chromosomes (26 to 36% amino acid identity) and the Paracoccus plasmid pTAV3 (23% amino acid identity) and is associated with 15 direct repeats of a 19-bp iteron (GAAGGTATCCGTTTCTGGG) and a parA homolog. The replication region of pPol360 contains an IncPα-like initiator (Bpro_5045; 28% amino acid identity to trfA of RP4/RK2) associated with 11 direct repeats of a 22-bp iteron (MRNCGTGTTNGWCCGCTAATAG), parA and parB genes, and an exonuclease.
FIG. 4.
(A) Comparison of the putative plasmid replication regions of pPol338 and pPol360. (B) Alignment of the putative plasmid conjugation gene regions of pPl338 and pPol360 with the conjugation gene regions from the hydrogen catabolic plasmid HG1, IncP-α and Ti plasmids, and the integrative conjugative element R391.
Very similar conjugative-transfer genes (trb, tra, and pil) are found on both pPol360 and pPol338 (Fig. 4B), suggesting that both plasmids are independently conjugative. The level of inferred amino acid identity ranges from low (Trb and Tra, 22 to 41%) to high (Pil, 64 to 93%). The trb and tra genes in the JS666 plasmids are arranged similarly to transfer genes of IncPα and Ti plasmids, but the closest database matches (25 to 30% amino acid identity) are to gene products from the hydrogen catabolic plasmid pHG1 (Trb), integrative conjugative elements (R391, SXT; Tra), and IncH plasmids (R27 and R478; Trh). The pil genes of the JS666 plasmids (pilLNOPQRST1MT2V) are similar in arrangement to pilus biosynthesis genes of Pseudomonas aeruginosa elements (pKLC102 and PAPI-1) and the IncI1 plasmid R64.
Both pPol338 and pPol360 carry multiple putative stabilization systems, including Burkholderia-like parAB gene pairs (Bpro_5043-Bpro_5044 and Bpro_5514-Bpro_5515) (15), solitary parB genes (Bpro_5053 and Bpro_5341), resolvases (Bpro_5040, Bpro_5516, Bpro_5529, and Bpro_5548), and DNA methylases (Bpro_5027 and Bpro_5389). pPol338 also carries a stbB antitoxin homolog (Bpro_5318) and a higAB-like killer-antidote gene pair (Bpro_5380-Bpro_5381). Other putative “backbone” genes present on both pPol338 and pPol360 include primase (Bpro_5218 and Bpro_5493), helicase (Bpro_5077 and Bpro_5490), exonuclease VIII (Bpro_5046 and Bpro_5358), and ribonucleotide reductase (Bpro_5207 and Bpro_5497) genes.
Other mobile genetic elements.
Diverse transposases are present in the JS666 genome, consisting of 55 genes of 40 different types (see Table S1A in the supplemental material). They include representatives of 14 insertion sequence (IS) families, a Mu-like transposase, and four Tn3-like transposases. Associations between transposases and genes of interest for biodegradation and biocatalysis were noted in many cases (see Table S2 in the supplemental material and below). There are eight putative genomic islands (see Table S1B in the supplemental material) and a 39-kb Mu family phage (Fig. 1; see Table S1C in the supplemental material). Several of the genomic islands contain genes of interest to biodegradation and biocatalysis, including aromatic-compound utilization (GI1), haloacid dehalogenase (GI2), and cytochrome P450 (GI5). Two types of putative retrons (type II introns) are present in the JS666 genome. The first type is present in four chromosomal copies (Bpro_949, Bpro_4601, Bpro_4617, and Bpro_4899) and contains a reverse transcriptase of 564 to 569 amino acids (aa) with a C-terminal endonuclease domain, while the second type has two copies on pPol338 (Bpro_5394 and Bpro_5396) associated with an IS605 family transposase (Bpro_5398) and a reverse transcriptase of 456 to 461 aa. JS666 carries two inteins, one (351 aa) in the DnaB helicase (Bpro_3033) and the other (380 aa) in a ribonucleoside reductase (Bpro_4588). Both retrons and inteins have potential uses in biotechnology, for example, as a source of “homing” endonucleases (39). Three copies of the 132- to 169-bp miniature inverted-repeat transposable element (MITE) “Nezha” are present in the JS666 genome (50). This MITE is related to cyanobacterial MITEs and is not found in other bacterial genomes, suggesting a recent horizontal gene transfer event from cyanobacteria to JS666 (50).
Central metabolism and C1 and C2 metabolism.
The JS666 genome contains complete glycolytic and Entner-Doudoroff pathways and a complete tricarboxylic acid cycle. The pentose phosphate pathway appears to be missing 6-phosphogluconolactonase. Glyoxalate pathway genes encoding isocitrate lyase (Bpro_2101) and malate synthase (Bpro_4517) are present, as expected for an organism isolated on a C2 compound (cDCE). Oxalate and glycolate catabolic genes were also noted (see Table S2 in the supplemental material). Two ribulose bisphosphate carboxylase homologs (Bpro_0032 and Bpro_0093) are in the genome; however, a phosphoribulokinase homolog was not found, suggesting that JS666 does not fix carbon via the Calvin cycle. Alternative pathways for carbon fixation, such as the C4-dicarboxylic acid cycle, the 3-hydroxypropionate pathway, the reverse tricarboxylic acid cycle, and the acetyl-coenzyme A (CoA) pathway, are either missing or incomplete, suggesting that JS666 is not capable of autotrophic CO2 fixation.
The presence of a monofunctional (CoxMSL-like) carbon monoxide dehydrogenase (Bpro_0576 to Bpro_0578) and the lack of a complete CO2 fixation pathway suggest that JS666 uses carbon monoxide (CO) as an energy source in the presence of an organic carbon source (i.e., mixotrophy). However, experiments with 10% CO in the headspace and 0 to 10 mM ethanol as a carbon source failed to show growth of JS666 on CO as a carbon and/or energy source. Genes encoding a putative tungsten-containing formate dehydrogenase are present, suggesting growth on formate as a carbon and/or energy source (see Table S2 in the supplemental material) (this was not tested experimentally).
Alternative electron donors and acceptors.
The presence of a gene cluster similar to the E. coli K-12 nar operon (narGHJI) (Bpro_4592 to Bpro_4599) (see Table S2 and Fig. S1 in the supplemental material) raises the possibility that JS666 uses nitrate as an alternative electron acceptor for growth. Incubating JS666 under anaerobic conditions with 5 mM ethanol and 5 to 35 mM sodium nitrate resulted in nitrite production (0.5 to 0.8 mM) but failed to show the use of nitrate as an electron acceptor for growth.
The coincidence of a putative hydrogenase gene cluster related to E. coli membrane-bound respiratory hydrogenases 3 and 4 (Bpro_0916 to Bpro_0921) (see Table S2 in the supplemental material) and several possible formate dehydrogenase genes (Bpro_0937 to Bpro_0942) suggests that JS666 possesses a fermentative formate hydrogenlyase system that converts formate into molecular hydrogen and carbon dioxide. However, this possibility was not tested experimentally.
The presence of potential sulfur oxidation genes (Bpro_2625 to Bpro_2631), encoding proteins with 25 to 50% identity to those from mixotrophic sulfur-oxidizing Paracoccus pantotrophus (16) could indicate the ability to grow chemolithotrophically by oxidizing thiosulfate, sulfite, hydrogen sulfide, and/or elemental sulfur in the presence of an organic carbon source (see Table S2 and Fig. S2 in the supplemental material). However, growth tests with thiosulfate (0 to 40 mM) as an electron donor and bicarbonate (10 mM) or ethanol (0 to 5 mM) as a carbon source were negative (Table 2).
TABLE 2.
Growth substrate range of Polaromonas sp. strain JS666
| Substrate | Growtha | Initial concn (mM)b | Incubation time (days)c |
|---|---|---|---|
| Acetate | 6.5d | 10 | 5 |
| Benzoate | − | 2 | 30 |
| Benzoate | + | Auxanography | 30 |
| Carbon monoxide | − | 10% headspacee | 17 |
| Carbon monoxide (+10 mM ethanol) | − | 10% headspace | 17 |
| Catechol | − | 2 | 30 |
| Catechol | + | Auxanography | 30 |
| Chloroacetaldehyde | − | 0.2-10 | 50 |
| Chloroacetate | 2.2 | 6 | 35 |
| Cyclohexane | − | 0.2-10 | 19 |
| Cyclohexaneacetic acid | − | 0.2-10 | 33 |
| Cyclohexanecarboxylate | 4.6 | 6 | 7 |
| Cyclohexanol | 2.7 | 1.2 | 8 |
| 1,2-Dichloroethane | − | 0.2-10 | 14 |
| cis-1,2-Dichloroethene | 4.6 | 1.3 | 15 |
| Dalapon (2,2-dichloropropionate) | − | 0.2-10 | 57 |
| Ethane | − | 0.2-10 | 30 |
| Ethanol (aerobic) | 5.3 | 10 | 17 |
| Ethanol (anaerobic + NO3−) | − | 5-35 (NO3−) | 41 |
| Ethylcyclohexane | − | 0.2-10 | 54 |
| Ferulate | 1.8 | 2 | 15 |
| Formate/H2 | Not tested | Not tested | Not tested |
| Gentisate | 4.1 | 2 | 15 |
| Glycolate | Not tested | Not tested | Not tested |
| Heptane | 2.1 | 0.9 | 13 |
| Hexane | − | 0.2-10 | 6 |
| 3-Hydroxybenzoate | 4.7 | 2 | 17 |
| 4-Hydroxybenzoate | − | 2 | 30 |
| 2-Hydroxylaminobenzoate | Not tested | Not tested | Not tested |
| Hydroxyquinol | +/− | Auxanography | 30 |
| Naphthalene | − | 2 | 30 |
| 2-Nitrobenzoate | − | 2 | 30 |
| 4-Nitrobenzoate | − | 2 | 30 |
| Octane | 0.6 | 0.2 | 7 |
| Opines | Not tested | Not tested | Not tested |
| Oxalate | Not tested | Not tested | Not tested |
| Phenylacetate | Not tested | Not tested | Not tested |
| o-Phthalate | − | 2 | 30 |
| Propane | − | 0.2-10 | 34 |
| Protocatechuate | 4.9 | 2 | 15 |
| Salicylate | 2.1 | 0.6 | 5 |
| Succinate | 4.8 | 10 | 6 |
| Thiosulfate | − | 10-40 | 29 |
| Thiosulfate (+5 mM ethanol) | − | 10-40 | 29 |
The numbers indicate the highest number of observed exponential doublings of OD600 in liquid medium. +, positive for growth on auxanography plates (see Materials and Methods for details on auxanography); −, negative for growth; +/−, growth was inconclusive.
The concentration range tested is reported in cases where growth was negative. When growth was confirmed, the concentration showing the highest number of exponential doublings is reported.
In cases where growth was negative, the time when the experiment was abandoned is reported. In cases where growth was positive, the time when a maximum OD600 was observed is reported.
The starting OD600 was 0.002 in this experiment.
Volume of gas added equaled 10% of the headspace in the serum bottle.
Metal resistance.
Resistance to the toxicity of heavy metals is an important trait for microbes intended for application at contaminated sites. We identified 20 putative metal resistance genes at five locations in the JS666 chromosome. Bioinformatic analysis suggests that JS666 is likely to be resistant to mercury, arsenate, chromate, and other heavy metals (Fig. 1; see Table S2 in the supplemental material). Genes for resistance to mercury (merRTPCADE; Bpro_2232 to Bpro_2226) and arsenic (arsHCRC; Bpro_2220 to Bpro_2217) are clustered on the chromosome. The metal resistance region was associated with transposases of the IS110 family (Bpro_2223) and the IS4 family (Bpro_2203). A second copy of merRTP (Bpro_4798 to Bpro_4796) is found at a second chromosomal location, flanked by ISL3 and IS91 family transposases (Bpro_4792 and Bpro_4805). The association of resistance genes with transposable elements is a common feature of metal-resistant bacteria and likely indicates relatively recent acquisition of these genes by JS666.
Metabolism of haloalkanes and haloacids.
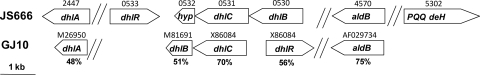
Microbial hydrolytic dehalogenase enzymes are important in the metabolism of diverse pollutants, such as chlorinated alkanes and aromatics (23). JS666 possesses putative haloalkane biodegradation genes, the products of which are 48 to 75% identical to those in the dichloroethane-degrading Xanthobacter autotrophicus strain GJ10 (Fig. 1, 2, and 5; see Table S2 in the supplemental material) (22, 46). They include homologs of haloalkane dehalogenase DhlA (Bpro_0521, Bpro_0547, and Bpro_2447), haloalkanoate dehalogenase DhlB (Bpro_0530 and Bpro_5186), the regulator DhlR (Bpro_0533 and Bpro_5183), and the uptake protein DhlC (Bpro_0531 and Bpro_5185). The presence of an apparently complete set of dichloroethane catabolic genes in JS666 is surprising, since previous tests (10) showed that cDCE-grown cells attacked dichloroethane but did not grow on the substrate. Further attempts to test the growth of JS666 on dichloroethane in this study were also negative (Table 2). Nearly identical dhlRBC clusters are located on the chromosome (Bpro_0530 to Bpro_0533) and on pPol360 (Bpro_5183 to Bpro_5186) within a duplicated 9.9-kb region that shares 97% nucleotide identity. The duplication could have been mediated by nearby transposases (Bpro_518 and Bpro_5194-Bpro_5195). Duplication of haloalkanoate dehalogenase genes may have been significant for the evolution of cDCE biodegradation in JS666.
FIG. 5.
Organization of haloalkane and haloacid degradation genes of JS666 and X. autotrophicus strain GJ10. dhlA, haloalkane dehalogenase; dhlR, sigma-54-dependent transcriptional regulator; hyp, hypothetical protein; dhlC, putative uptake protein; dhlB, haloalkane dehalogenase; aldB, chloroacetaldehyde dehydrogenase; PQQ deH, pyrroloquinoline quinone alcohol dehydrogenase. Bpro gene numbers are shown above the JS666 genes, GenBank nucleotide accession numbers are shown above the GJ10 genes, and percentage amino acid identities to JS666 homologs are shown under the GJ10 genes.
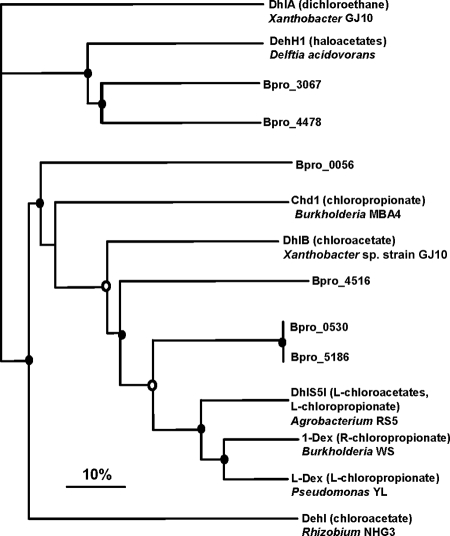
A phylogenetic tree of putative JS666 haloacid dehalogenase genes (Fig. 6) suggests that two groups of dehalogenases are present. Bpro_0530, Bpro_5183, Bpro_0056, and Bpro_4516 group with enzymes that are active on both haloacetates and halopropionates (EC 3.8.1.2), while Bpro_3067 and Bpro_4478 are more related to a dehalogenase that displays activity only with haloacetates (EC 3.8.1.3). Growth experiments revealed that JS666 grew slowly on chloroacetate but did not grow on dalapon (2,2-dichloropropionic acid) (Table 2).
FIG. 6.
Phylogenetic tree depicting the relationship between known dehalogenases and dehalogenase homologs in the JS666 genome. An alignment of 360 amino acids (including gaps) was made with ClustalX and converted into a neighbor-joining tree, which was visualized with TreeView using the haloalkane dehalogenase DhlA as the outgroup. The filled circles at nodes indicate bootstrap values of >95%, while the open circles at nodes indicate bootstrap values of 75% to 95%. Bootstrap values lower than 75% are not shown. The bar represents 10% sequence difference.
Metabolism of n-alkanes, alicyclic acids, and cyclic alcohols.
A large region of pPol338 (77 kb; Bpro_5563 to Bpro_5571 and Bpro_5257 to Bpro_5308) is apparently devoted to metabolism of alkanes, cycloalkanes, and cyclic alcohols (Fig. 2; see Table S2 in the supplemental material). This catabolic gene region has an atypically high G+C content (Fig. 2B) and is associated with several transposase genes (Bpro_5309, Bpro_5569, Bpro_5570, Bpro_5572, and Bpro_5545), suggesting relatively recent acquisition.
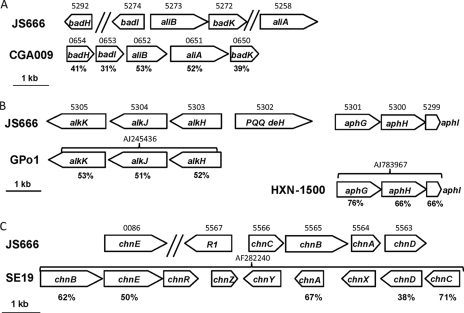
The n-alkane pathway is most likely initiated by the Bpro_5301 product, a probable cytochrome P450 with 76% amino acid identity to a Mycobacterium hydroxylase that attacks C5 to C10 alkanes (43). Downstream gene products (Bpro_5303 to Bpro_5305) are 51 to 53% similar to aldehyde dehydrogenase AlkH, alcohol dehydrogenase AlkJ, and acyl-CoA synthetase AlkK in Pseudomonas putida GPo1 (Fig. 7A). A second alcohol dehydrogenase gene of the pyrroloquinoline quinone type (Bpro_5034) is also present, suggesting the ability to metabolize both primary and secondary alcohols, as observed in Pseudomonas butanovora (48). JS666 did not use ethane, propane, or hexane for growth, but growth on heptane and octane was observed (Table 2).
FIG. 7.
(A) Organization of alicyclic acid biodegradation genes of JS666 and R. palustris CGA009. badH, 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase; badI, 2-ketocyclohexanecarboxyl-CoA hydrolase; aliB, cyclohexanecarboxyl-CoA dehydrogenase; badK, enoyl-CoA hydratase; aliA, cyclohexanecarboxylate-CoA ligase. JS666 gene numbers (Bpro) are shown above the JS666 genes, CGA009 gene numbers (RPA) are shown above the genes, and percentage amino acid identities to JS666 homologs are shown under the CGA009 genes. (B) Organization of n-alkane biodegradation genes of JS666, P. putida GPo1, and Mycobacterium sp. strain HXN-1500. alkK, acyl-CoA synthetase; alkJ, alcohol dehydrogenase; alkH, aldehyde dehydrogenase; PQQ deH, pyrroloquinoline quinone alcohol dehydrogenase; aphG, cytochrome P450 alkane hydroxylase; aphH, ferredoxin reductase; aphI, ferredoxin. JS666 gene numbers (Bpro) are shown above the JS666 genes, GenBank nucleotide accession numbers are shown above the GPo1 and HXN-1500 genes, and percentage amino acid identities to JS666 homologs are shown under the GPo1 and HXN-1500 genes. (C) Organization of cyclic alcohol biodegradation genes of JS666 and Acinetobacter sp. strain SE19. R1, sigma-54-dependent regulator; chnC, 6-hexanolactone hydrolase; chnB, cyclohexanone monooxygenase; chnA, cyclohexanol dehydrogenase; chnD, 6-hydroxyhexanoic acid dehydrogenase; chnE, 6-oxo-hexanoate dehydrogenase; chnR, araC-like regulator; chnZ, hypothetical protein; chnY, pilin invertase; chnX, hypothetical protein. Bpro gene numbers are shown above the JS666 genes, GenBank nucleotide accession numbers are shown above the SE19 genes, and percentage amino acid identities to JS666 homologs are shown under the SE19 genes.
Alicyclic acids are produced as microbial metabolites of petroleum hydrocarbons (31). Putative alicyclic acid biodegradation genes similar to cyclohexane carboxylate biodegradation genes in Rhodopseudomonas palustris strain CGA009 are encoded immediately adjacent to the n-alkane degradation genes in JS666 (Fig. 2 and 7B; see Table S2 in the supplemental material), suggesting that JS666 might attack cycloalkyl-n- alkanes in addition to n-alkanes. JS666 grew well on cyclohexane carboxylate, but not cyclohexane acetate or ethylcyclohexane (Table 2).
The oxidation of cyclohexanol to adipic acid is an industrially important reaction for nylon synthesis and is catalyzed by a Baeyer-Villiger monooxygenase capable of performing regio- and stereoselective oxygenations (45). Plasmid pPol338 encodes a Baeyer-Villiger monooxygenase (Bpro_5565) as part of a putative cyclohexanol biodegradation gene cluster (Fig. 2 and 7C; see Table S2 in the supplemental material) similar to that of Acinetobacter sp. strain SE19 (9). Growth experiments confirmed that strain JS666 uses cyclohexanol as a carbon and energy source (Table 2).
Aromatic metabolism.
Diverse natural and xenobiotic aromatic compounds are oxidized via the β-ketoadipate and gentisate pathways (19), and the corresponding genes are found in many bacterial genomes. Phenylpropenoid compounds, such as ferulate, are used by plants to make lignin, which is the second most abundant polymer on Earth (after cellulose). In JS666, multiple clusters of central β-ketoadipate pathway genes are present (Bpro_626 to Bpro_628, Bpro_1228 to Bpro_1331, Bpro_1855 to Bpro_1857, Bpro_1901 to Bpro_1906, and Bpro_5171 to Bpro_5173), in addition to gene clusters encoding related peripheral catabolic pathways, including ferulate (Bpro_3111 to Bpro_3118 and Bpro_3385 to Bpro_3387), benzoate (Bpro_1617 to Bpro_1633) (see Fig. S3 in the supplemental material), 4-nitrobenzoate (Bpro_682 and Bpro_5125), phthalate (Bpro_1908 to Bpro_1912), and hydroxyquinol (Bpro_2063-Bpro_2064) (Fig. 1; see Table S2 in the supplemental material).
Ferulate.
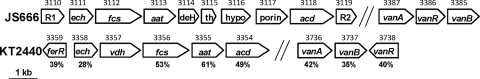
Strain JS666 possesses putative ferulate catabolic genes similar to those described in Pseudomonas sp. strains HR199 (32) and KT2440 (Fig. 1 and 8), but the ferulate operon of JS666 lacks an obvious vanillate dehydrogenase (vdh) gene and has four additional genes between aat and acd. The JS666 ferulate operon is flanked by two regulatory genes, one of which resembles FerR of KT2440. While a ferulate pathway that proceeds through vanillin has been proposed for strain HR199 (32) and also seems plausible for KT2440 (25), the JS666 ferulate pathway more likely proceeds via vanillyl-CoA due to the lack of vdh and the presence of a thioesterase gene (see Fig.S4 in the supplemental material). Growth experiments confirmed that JS666 could grow on ferulate (Table 2).
FIG. 8.
Organization of ferulate and vanillate biodegradation genes of JS666 and P. putida KT2440. R1, GntR-type regulator; ech, enoyl-CoA hydratase; fcs, feruloyl-CoA synthetase; aat, β-ketothiolase; deH, dehydrogenase; th, thioesterase; hypo, hypothetical protein; acd, acyl-CoA dehydrogenase; R2 and ferR, marR-type regulators; vanA, vanillate demethylase oxygenase subunit; vanB, vanillate demethylase reductase subunit; vanR, gntR-like regulator; vdh, vanillate dehydrogenase. JS666 gene numbers (Bpro) are shown above the JS666 genes, KT2440 gene numbers (PP) are shown above the KT2440 genes, and percentage amino acid identities to JS666 homologs are shown under the KT2440 genes.
Protocatechuate.
Genes encoding the protocatechuate branch of the β-ketoadipate pathway are dispersed throughout the JS666 chromosome. This is similar to the pattern observed in the genomes of Ralstonia metallidurans strain CH34 (Fig. 1 and 9) and P. putida KT2440 (25) and unlike the tightly clustered genes seen in Acinetobacter baylyi strain ADP1 (26, 47) (Fig. 9) or Agrobacterium tumefaciens A348 (33). The pcaBCD genes in JS666 appear to be embedded in an aromatic degradation gene cluster consisting of two dioxygenases, a hydrolase, a racemase, and an esterase. One copy of pcaIJ is associated with the thiolase gene pcaF, while another is linked to a second pcaC homolog and putative phthalate degradation genes (see Table S1 in the supplemental material and below). Growth experiments in liquid media confirmed that JS666 uses protocatechuate as a growth substrate (Table 2).
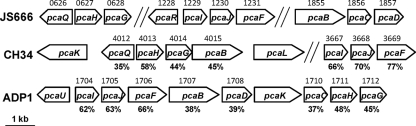
FIG. 9.
Organization of protocatechuate biodegradation genes of JS666, Cupriavidus metallidurans CH34, and A. baylyi ADP1. pcaQ, LysR family regulator; pcaH, protocatechuate 3,4-dioxygenase beta subunit; pcaG, protocatechuate 3,4-dioxygenase alpha subunit; pcaR, pobR family regulator; pcaI, β-ketoadipate:succinyl-CoA transferase alpha subunit; pcaJ, β-ketoadipate:succinyl-CoA transferase beta subunit; pcaF, β-ketoadipyl-CoA thiolase; pcaB, β-carboxy-cis,cis-muconate lactonizing enzyme; pcaC, γ-carboxymuconolactone decarboxylase; pcaD, β-ketoadipate enol-lactone hydrolase. JS666 gene numbers (Bpro) are indicated above the JS666 genes, CH34 gene numbers (Rmet) are shown above the CH34 genes, ADP1 gene numbers (ACIAD) are shown above the ADP1 genes, and percentage amino acid identities to JS666 homologs are shown under the CH34 and ADP1 genes.
Catechol.
Two possible catechol 1,2-dioxygenases are present in the JS666 genome: Bpro_1822, with 34% identity to catA of A. baylyi strain ADP1, and Bpro_0357, with 41% amino acid identity to tcpC of Ralstonia eutropha JMP134. Either of these dioxygenases could potentially allow growth on catechol when combined with the activities of pcaDIJF (see above), a cis,cis-muconate lactonizing enzyme (catB), and a muconolactone isomerase (catC). Bpro_4404 is a likely catB candidate, but no catC homolog was identified. Interestingly, despite the apparent lack of a catC homolog and the failure to observe growth on catechol in liquid growth media (Table 2), growth was observed approximately 30 to 50 mm from the catechol addition point on auxanography plates (see Fig. S5 in the supplemental material). This was confirmed by protein analysis, in which catechol-fed auxanography plates contained 0.95 to 1.20 mg protein per plate while control plates (with no exogenous carbon source added) contained 0.44 to 0.70 mg protein per plate.
Benzoate.
A cluster of 16 genes encodes the putative aerobic benzoate biodegradation pathway in strain JS666 (Fig. 1; see Table S1 in the supplemental material). The cluster bears similarity to the aerobic benzoate degradation genes in Azoarcus evansii strain KB 740 (see Fig. S3 in the supplemental material), as well as the anaerobic genes of R. palustris strain CGA009. Growth on benzoate was not observed in liquid media (Table 2) but was observed via auxanography (photo not shown). Total protein on benzoate-fed auxanography plates ranged from 0.92 to 0.96 mg per plate, while total protein on control plates (with no exogenous carbon source added) ranged from 0.44 to 0.70 mg per plate.
Phthalate.
A putative phthalate biodegradation gene cluster in strain JS666 consists of genes encoding a phthalate 4,5-dioxygenase (Bpro_1909 and Bpro_1910), 4,5-dihydro-4,5-dihydroxyphthalate dehydrogenase (Bpro_1908), and 4,5-dihydroxyphlatate decarboxylase (Bpro_1912). The encoded enzymes show 45 to 79% identity to those encoded on the P. putida PHT plasmid (30). The expected product of 4,5-dihydroxyphlatate decarboxylase is protocatechuate, which is expected to be further metabolized in strain JS666 (see Fig. S4 in the supplemental material). However, growth on o-phthalate was not observed (Table 2). The “phthalate” genes of JS666 could be involved in utilization of iso (meta)-phthalate or tere (para)-phthalate isomers, rather than ortho-phthalate or, alternatively, they may encode phthalate ester degradation, which requires the ester form for transport or induction.
Hydroxyquinol.
Bpro_2064 and Bpro_2063 encode a putative hydroxyquinol 1,2-dioxygenase and 3-hydroxy-cis,cis-muconate reductase, respectively (Fig. 1; see also Table S1 in the supplemental material). These encoded enzymes are expected to convert hydroxyquinol into β-ketoadipate, which is expected to be further metabolized (see Fig. S4 in the supplemental material). The possibility that JS666 uses hydroxyquinol as a growth substrate was not tested in liquid media. Auxanography experiments revealed evidence of growth on hydroxyquinol (see Fig. S6 in the supplemental material). Total protein on hydroxyquinol-fed plates ranged from 0.51 to 0.53 mg per plate, while total protein on control plates (with no exogenous carbon source added) ranged from 0.44 to 0.70 mg per plate. Therefore, growth of JS666 on hydroxyquinol remains unresolved.
Another major pathway for aromatic metabolism is the gentisate pathway, which is used when ring cleavage occurs between carboxyl- and hydroxyl-substituted carbon atoms (19). Gentisate biodegradation genes were noted in the genome (Bpro_981-Bpro_982, Bpro_991, and Bpro_2125-Bpro_2126), and growth on gentisate was confirmed (Table 2). Aromatic compounds that appear to be degraded via the gentisate pathway in strain JS666 include salicylate (Bpro_980 and Bpro_983 to Bpro_985) and 3-hydroxybenzoate (Bpro_2128 and Bpro_3591).
Salicylate and 3-hydroxybenzoate.
The chromosomally located salicylate pathway in strain JS666 (Fig. 1) bears similarity to the pathway described in naphthalene-degrading Ralstonia sp. strain U2 (51) (Fig. 10). Growth experiments in liquid media confirmed that JS666 used salicylate as a growth substrate (Table 2). The putative JS666 salicylate 5-hydroxylase subunits (Bpro_0983 and Bpro_0984) are 79% (large subunit) and 59% (small subunit) identical to the U2 salicylate 5-hydroxylase. One major difference between the two gene clusters is that a naphthalene dioxygenase is cotranscribed with the salicylate degradation genes in strain U2, while in strain JS666, there is no evidence for a naphthalene dioxygenase, but a 2-component oxygenase similar to vanillate demethylase is encoded immediately downstream (Fig. 10). One plausible hypothesis (not tested experimentally) is that the JS666 oxygenase enables oxidative demethylation of o-anisic acid (2-methoxyphenol) to yield salicylate. The observation of growth on gentisate, along with the discovery of additional putative salicylate/3-hydroxybenzoate hydroxylases in the JS666 genome (Bpro_2128 and Bpro_3591), raised the possibility that JS666 could grow on 3-hydroxybenzoate (see Fig. S4 in the supplemental material). This was confirmed experimentally (Table 2).
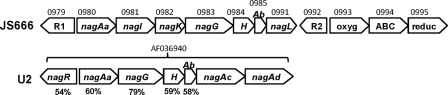
FIG. 10.
Organization of salicylate/gentisate biodegradation genes in JS666 and Ralstonia sp. strain U2. R1, LysR family regulator; nagAa, ferredoxin reductase; nagI, gentisate 1,2-dioxygenase; nagK, fumarylpyruvate hydrolase; nagG, salicylate 5-hydroxylase large oxygenase component; H, salicylate 5-hydroxylase small oxygenase component (nagH); Ab, ferredoxin nagAb; nagL, maleylpyruvate isomerase; R2, TetR regulator; oxyg, oxygenase; ABC, ABC transporter; reduc, reductase; nagR, LysR family regulator; nagAc, napththalene dioxygenase large subunit; nagAd, napththalene dioxygenase small subunit. JS666 gene numbers (Bpro) are shown above the JS666 genes, GenBank nucleotide accession numbers are shown above the U2 genes, and percentage amino acid identities to JS666 homologs are shown under the U2 genes.
Other predicted aromatic biodegradation genes present include those for assimilation of 2-aminobenzoate (Bpro_587 to Bpro_593) (see Table S2 in the supplemental material), phenylacetate (Bpro_2983 to Bpro_3000) (see Table S2 and Fig. S4 in the supplemental material), and 2-hydroxylaminobenzoate (Bpro_5124 to Bpro_5140) (see Table S2 and Fig. S4 in the supplemental material). Most of the aromatic catabolic genes of JS666 are chromosomal and not linked to obvious mobile genetic elements. Exceptions are the protocatechuate genes pcaRIJF (associated with transposases Bpro_1224-Bpro_1225), pcaC (two homologs on pPol360), and the hydroxylaminobenzoate gene cluster (on pPol360, flanked by transposases Bpro_5122 and Bpro_5143).
Opine metabolism.
The biosynthesis and catabolism of opines (unusual amino acid derivatives) is central to the interaction of the pathogen Agrobacterium with host plants, and opine catabolism is also found in other plant-associated bacteria. A large cluster of genes likely to encode the uptake and catabolism of opines (octopine, nopaline, and mannopine) is found on the JS666 chromosome (Fig. 1; see also Table S2 in the supplemental material). The opine genes Bpro_4331 to Bpro_4362 were located in a putative genomic island (GI8) (see Table S1B in the supplemental material) that also contains genes for amino acid metabolism and transport. The data suggest that JS666 could occupy a plant-associated niche, which would have relevance for potential applications in combined microbial/plant bioremediation.
DISCUSSION
The JS666 genome, the first to be sequenced from a Polaromonas species, has revealed unexpected metabolic capabilities (e.g., cyclic alkane degradation) and allowed the prediction of likely environmental niches (e.g., the rhizosphere) and tolerances (e.g., heavy metals), which permits more intelligent future application of the microbe as an agent of bioremediation or biocatalysis. The importance of plasmids in disseminating unusual metabolic capabilities is highlighted by the pPol338 and pPol360 plasmids of JS666, which appear to be self-transmissible elements encoding pathways for alkane, cycloalkane, cyclic alcohol, haloalkanoate, and amino-aromatic metabolism. Based on this genome analysis and the phenotypic differences between strain JS666, the type strain P. vacuolota and the naphthalene-degrading P. naphthalenivorans, we propose that strain JS666 be named Polaromonas chloroethenica.
Catabolic pathways were assigned only in cases where whole clusters of catabolic genes were present (not single genes) and where good BLAST matches for the majority of such genes were obtained. In several cases, growth substrates predicted by genomic analyses were confirmed experimentally (Table 2). An auxanography approach confirmed that JS666 uses catechol and benzoate as growth substrates, even though liquid growth experiments were negative. Despite extensive washing and dilutions, we observed apparent pinpoint colonies on control plates (i.e., JS666 plated on MSM-Noble agar with no exogenous carbon source). This suggests that JS666 grows on trace constituents in the solid media. However, differential growth on plates amended with a substrate in comparison to the no-carbon control plates was confirmed by protein analysis.
Our inability to observe growth on other predicted substrates could indicate that the actual substrates are substituted variants or that the conditions used for testing were not optimal (e.g., toxic substrates that support growth only in a narrow concentration range). We have evidence for this in the case of salicylate, which initially tested negative at 2 mM but was positive at 0.6 mM. In cases where we were unable to demonstrate growth on the predicted compounds experimentally, likely catabolic pathways were still presented to provide a basis for future work, which could, for example, aim to test close structural analogs of the compounds we have experimentally tested (e.g., chlorobenzoate instead of benzoate) or substrates that were unavailable due to cost/legal restrictions (e.g., phenylacetate). In addition, identification of putative catabolic genes and pathways is important to provide targets for heterologous expression and protein purification because such strategies do not depend on whole pathways being functional, only on the activities of single or few gene products.
The replication and transfer genes of the JS666 plasmids are not closely related to those of experimentally studied plasmids, but some similarities are apparent. The presence of an IncPα-like replication determinant (trfA) linked to haloacid dehalogenase genes in pPol360 highlights the major role played by IncP plasmids in bacterial adaptation to pollutants (7, 13, 42). The similarity of pPol338 replication genes to those of Burkholderia small chromosomes indicates that gene transfer between Polaromonas and Burkholderia is likely, which is of interest due to the importance of members of the genus Burkholderia as plant and human pathogens and agents of bioremediation (8). The presence of extensive and well-conserved plasmid transfer and mobilization functions on both pPol338 and pPol360 is notable, suggesting both elements are independently capable of conjugative transfer. Establishing the host range and transfer properties of the JS666 plasmids is significant for bioremediation due to their likely roles in moving catabolic genes among bacteria in situ. The biology of the JS666 plasmids is also of interest for the development of new cloning vectors for betaproteobacteria.
Transposable elements play a major role in biodegradation pathway evolution and are often found associated with catabolic plasmids (29, 41, 49). While we did not attempt to define the inverted repeats associated with ISs and transposons, abundant and diverse transposable elements are clearly present in the JS666 genome, especially on the two plasmids. The duplication of dehalogenase genes on pPol360 and the chromosome suggests a recent transposase-mediated event, which may have been involved in the evolution of halogenated hydrocarbon metabolism (e.g., cDCE). A large region of pPol360 (76 kb) containing the β-ketoadipate, hydroxylaminobenzoate, and haloalkanoate metabolic genes is flanked by identical IS3 family transposases (Bpro_5122 and Bpro_5204) and could represent a compound (type I) catabolic transposon. Transposase genes are found throughout the alkane/cycloalkane/cyclic-alcohol gene region of pPol338, suggesting that ISs have also contributed to the evolution of aliphatic hydrocarbon catabolic abilities in JS666.
Based on our genome analysis, we propose that the acquisition of plasmids pPol338 and pPol360 played a role in the evolution of the cDCE biodegradation pathway in JS666. These large plasmids have persisted in JS666 despite an extended enrichment and isolation process (10), providing circumstantial evidence that they contribute to growth on cDCE. The presence of an alkane monooxygenase gene on pPol338 is consistent with the prior observation that cDCE-grown JS666 cells produce epoxyethane from ethene (10). A plausible cDCE assimilation pathway could arise from the intersection of oxygenase and dehydrogenase genes from alkane pathways (pPol338) and hydrolytic dehalogenase genes from haloalkanoate pathways (pPol360 and chromosomal). We have observed that the cDCE-degrading phenotype is unstable in media lacking cDCE (T. E. Mattes and N. V. Coleman, unpublished data), which would be consistent with plasmid- or transposon-borne catabolic genes, but this observation could alternatively reflect poor regulatory control over a newly evolved metabolic pathway. Another possibility is that gene amplification processes (36) contribute to the unstable phenotype.
The JS666 genome encodes many enzymes that are potentially useful for biocatalysis, including the cytochrome P450 monooxygenase (Bpro_5299 to Bpro_5301) (44), the ferulate/vanillate system (Bpro_3111 to Bpro_3118 and Bpro_3385 to Bpro_3387) (17, 35), and the cyclohexanone monooxygenase-like gene (Bpro_5565) (2). Metabolic engineering of the alkane, ferulate, and cyclohexanol biodegradation pathways of JS666 could theoretically be used to produce the fragrances limonene and vanillin and the nylon monomer adipic acid, respectively. Other JS666 genes of strong biotechnological interest not discussed in detail here include a polyketide synthase region on pPol338, which is similar to cyanobacterial toxin gene clusters; several P450 oxygenases (Bpro_1581, Bpro_4920, and Bpro_2807); a halogenase (Bpro_3311); an amidase (Bpro_571); and a nitrilase (Bpro_3376).
Supplementary Material
Acknowledgments
This work was supported by the Engineering Research Centers Program of the National Science Foundation under NSF award number EEC-0310689, the Biotechnology By-Products Consortium, start-up funds for T.E.M, and funding from a University of Sydney Sesqui postdoctoral fellowship for N.V.C.
We thank Andy Holmes and Erich Lanka for helpful discussions.
Footnotes
Published ahead of print on 22 August 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alfreider, A., C. Vogt, and W. Babel. 2002. Microbial diversity in an in situ reactor system treating monochlorobenzene contaminated groundwater as revealed by 16S ribosomal DNA analysis. Syst. Appl. Microbiol. 25:232-240. [DOI] [PubMed] [Google Scholar]
- 2.Alphand, V., G. Carrea, R. Wohlgemuth, R. Furstoss, and J. M. Woodley. 2003. Towards large-scale synthetic applications of Baeyer-Villiger monooxygenases. Trends Biotechnol. 21:318. [DOI] [PubMed] [Google Scholar]
- 3.Atlas, R. M. 1981. Microbial degradation of petroleum hydrocarbons: an environmental perspective. Microbiol. Mol. Biol. Rev. 45:180-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badger, J. H., and G. J. Olsen. 1999. CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16:512-524. [DOI] [PubMed] [Google Scholar]
- 5.Barton, B. M., G. P. Harding, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235-240. [DOI] [PubMed] [Google Scholar]
- 6.Bucher, J. R., G. Cooper, J. K. Haseman, C. W. Jameson, M. Longnecker, F. Kamel, R. Maronpot, H. B. Matthews, R. Melnick, R. Newbold, R. W. Tennant, C. Thompson, and M. Waalkes. 2005. Report on carcinogens, 11th ed. U.S. Department of Health and Human Services, Washington, DC.
- 7.Burlage, R. S., L. A. Bemis, A. C. Layton, G. S. Sayler, and F. Larimer. 1990. Comparative genetic organization of incompatibility group P degradative plasmids. J. Bacteriol. 172:6818-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chain, P. S. G., V. J. Denef, K. T. Konstantinidis, L. M. Vergez, L. Agullo, V. L. Reyes, L. Hauser, M. Cordova, L. Gomez, M. Gonzalez, M. Land, V. Lao, F. Larimer, J. J. LiPuma, E. Mahenthiralingam, S. A. Malfatti, C. J. Marx, J. J. Parnell, A. Ramette, P. Richardson, M. Seeger, D. Smith, T. Spilker, W. J. Sul, T. V. Tsoi, L. E. Ulrich, I. B. Zhulin, and J. M. Tiedje. 2006. Inaugural article: Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. USA 103:15280-15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, Q., S. M. Thomas, K. Kostichka, J. R. Valentine, and V. Nagarajan. 2000. Genetic analysis of a gene cluster for cyclohexanol oxidation in Acinetobacter sp. strain SE19 by in vitro transposition. J. Bacteriol. 182:4744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman, N. V., T. E. Mattes, J. M. Gossett, and J. C. Spain. 2002. Biodegradation of cis-dichloroethene as the sole carbon source by a beta-proteobacterium. Appl. Environ. Microbiol. 68:2726-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connon, S. A., A. Tovanabootr, M. Dolan, K. Vergin, S. J. Giovannoni, and L. Semprini. 2005. Bacterial community composition determined by culture-independent and -dependent methods during propane-stimulated bioremediation in trichloroethene-contaminated groundwater. Environ. Microbiol. 7:165-178. [DOI] [PubMed] [Google Scholar]
- 12.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dennis, J. J. 2005. The evolution of IncP catabolic plasmids. Curr. Opin. Biotechnol. 16:291. [DOI] [PubMed] [Google Scholar]
- 14.Detter, J. C., J. M. Jett, S. M. Lucas, E. Dalin, A. R. Arellano, M. Wang, J. R. Nelson, J. Chapman, Y. Lou, D. Rokhsar, T. L. Hawkins, and P. M. Richardson. 2002. Isothermal strand-displacement amplification applications for high-throughput genomics. Genomics 80:691-698. [DOI] [PubMed] [Google Scholar]
- 15.Dubarry, N., F. Pasta, and D. Lane. 2006. ParABS systems of the four replicons of Burkholderia cenocepacia: new chromosome centromeres confer partition specificity. J. Bacteriol. 188:1489-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedrich, C. G., D. Rother, F. Bardischewsky, A. Quentmeier, and J. Fischer. 2001. Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. Environ. Microbiol. 67:2873-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasson, M. J., Y. Kitamura, W. R. McLauchlan, A. Narbad, A. J. Parr, E. L. H. Parsons, J. Payne, M. J. C. Rhodes, and N. J. Walton. 1998. Metabolism of ferulic acid to vanillin. J. Biol. Chem. 273:4163-4170. [DOI] [PubMed] [Google Scholar]
- 18.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 19.Harwood, C. S., and R. E. Parales. 1996. The beta-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 20.Hsiao, W., I. Wan, S. J. Jones, and F. S. L. Brinkman. 2003. IslandPath: aiding detection of genomic islands in prokaryotes. Bioinformatics 19:418-420. [DOI] [PubMed] [Google Scholar]
- 21.Irgens, R. L., J. J. Gosink, and J. T. Staley. 1996. Polaromonas vacuolata gen. nov., sp. nov., a psychrophilic, marine, gas vacuolate bacterium from Antarctica. Int. J. Syst. Bacteriol. 46:822-826. [DOI] [PubMed] [Google Scholar]
- 22.Janssen, D. B., F. Pries, J. van der Ploeg, B. Kazemier, P. Terpstra, and B. Witholt. 1989. Cloning of 1,2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10 and expression and sequencing of the dhlA gene. J. Bacteriol. 171:6791-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssen, D. B., F. Pries, and J. R. van der Ploeg. 1994. Genetics and biochemistry of dehalogenating enzymes. Annu. Rev. Microbiol. 48:163-191. [DOI] [PubMed] [Google Scholar]
- 24.Jeon, C. O., W. Park, P. Padmanabhan, C. DeRito, J. R. Snape, and E. L. Madsen. 2003. Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment. Proc. Natl. Acad. Sci. USA 100:13591-13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jimenez, J. I., B. Minambres, J. L. Garcia, and E. Diaz. 2002. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 4:824-841. [DOI] [PubMed] [Google Scholar]
- 26.Kowalchuk, G. A., G. B. Hartnett, A. Benson, J. E. Houghton, K.-L. Ngai, and L. N. Ornston. 1994. Contrasting patterns of evolutionary divergence within the Acinetobacter calcoaceticus pca operon. Gene 146:23-30. [DOI] [PubMed] [Google Scholar]
- 27.Loy, A., W. Beisker, and H. Meier. 2005. Diversity of bacteria growing in natural mineral water after bottling. Appl. Environ. Microbiol. 71:3624-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattes, T. E., N. V. Coleman, J. M. Gossett, and J. C. Spain. 2005. Physiological and molecular genetic analyses of vinyl chloride and ethene biodegradation in Nocardioides sp. strain JS614. Arch. Microbiol. 183:95-106. [DOI] [PubMed] [Google Scholar]
- 29.Nojiri, H., M. Shintani, and T. Omori. 2004. Divergence of mobile genetic elements involved in the distribution of xenobiotic-catabolic capacity. Appl. Microbiol. Biotechnol. 64:154. [DOI] [PubMed] [Google Scholar]
- 30.Nomura, Y., M. Nakagawa, N. Ogawa, S. Harashima, and Y. Oshima. 1992. Genes in PHT plasmid encoding the initial degradation pathway of phthalate in Pseudomonas putida. J. Ferment. Bioeng. 74:333. [Google Scholar]
- 31.Ougham, H. J., and P. W. Trudgill. 1982. Metabolism of cyclohexaneacetic acid and cyclohexanebutyric acid by Arthrobacter sp. strain CA1. J. Bacteriol. 150:1172-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Overhage, J., H. Priefert, and A. Steinbüchel. 1999. Biochemical and genetic analyses of ferulic acid catabolism in Pseudomonas sp. strain HR199. Appl. Environ. Microbiol. 65:4837-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parke, D. 1997. Acquisition, reorganization, and merger of genes: novel management of the beta-ketoadipate pathway in Agrobacterium tumefaciens. FEMS Microbiol. Lett. 146:3-12. [Google Scholar]
- 34.Perler, F. B. 2000. InBase, the intein database. Nucleic Acids Res. 28:344-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Priefert, H., J. Rabenhorst, and A. Steinbüchel. 1997. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J. Bacteriol. 179:2595-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reams, A. B., and E. L. Neidle. 2004. Selection for gene clustering by tandem duplication. Annu. Rev. Microbiol. 58:119-142. [DOI] [PubMed] [Google Scholar]
- 37.Sizova, M., and N. Panikov. 2007. Polaromonas hydrogenivorans sp. nov., a psychrotolerant hydrogen-oxidizing bacterium from Alaskan soil. Int. J. Syst. Evol. Microbiol. 57:616-619. [DOI] [PubMed] [Google Scholar]
- 38.Squillace, P. J., M. J. Moran, W. W. Lapham, C. V. Price, R. M. Clawges, and J. S. Zogorski. 1999. Volatile organic compounds in untreated ambient groundwater of the United States, 1985-1995. Environ. Sci. Technol. 33:4176-4187. [Google Scholar]
- 39.Stoddard, B. L. 2006. Homing endonuclease structure and function. Q. Rev. Biophys. 38:49. [DOI] [PubMed] [Google Scholar]
- 40.Stothard, P., and D. S. Wishart. 2005. Circular genome visualization and exploration using CGView. Bioinformatics 21:537-539. [DOI] [PubMed] [Google Scholar]
- 41.Top, E. M., and D. Springael. 2003. The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 14:262. [DOI] [PubMed] [Google Scholar]
- 42.Trefault, N., R. De la Iglesia, A. M. Molina, M. Manzano, T. Ledger, D. Perez-Pantoja, M. A. Sanchez, M. Stuardo, and B. Gonzalez. 2004. Genetic organization of the catabolic plasmid pJP4 from Ralstonia eutropha JMP134 (pJP4) reveals mechanisms of adaptation to chloroaromatic pollutants and evolution of specialized chloroaromatic degradation pathways. Environ. Microbiol. 6:655-668. [DOI] [PubMed] [Google Scholar]
- 43.van Beilen, J. B., E. G. Funhoff, A. van Loon, A. Just, L. Kaysser, M. Bouza, R. Holtackers, M. Rothlisberger, Z. Li, and B. Witholt. 2006. Cytochrome P450 alkane hydroxylases of the CYP153 family are common in alkane-degrading eubacteria lacking integral membrane alkane hydroxylases. Appl. Environ. Microbiol. 72:59-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Beilen, J. B., R. Holtackers, D. Luscher, U. Bauer, B. Witholt, and W. A. Duetz. 2005. Biocatalytic production of perillyl alcohol from limonene by using a novel Mycobacterium sp. cytochrome P450 alkane hydroxylase expressed in Pseudomonas putida. Appl. Environ. Microbiol. 71:1737-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Beilen, J. B., F. Mourlane, M. A. Seeger, J. Kovac, Z. Li, T. H. M. Smits, U. Fritsche, and B. Witholt. 2003. Cloning of Baeyer-Villiger monooxygenases from Comamonas, Xanthobacter and Rhodococcus using polymerase chain reaction with highly degenerate primers. Environ. Microbiol. 5:174-182. [DOI] [PubMed] [Google Scholar]
- 46.van der Ploeg, J., and D. B. Janssen. 1995. Sequence analysis of the upstream region of dhlB, the gene encoding haloalkanoic acid dehalogenase of Xanthobacter autotrophicus GJ10. Biodegradation 6:257-263. [DOI] [PubMed] [Google Scholar]
- 47.Vaneechoutte, M., D. M. Young, L. N. Ornston, T. De Baere, A. Nemec, T. Van Der Reijden, E. Carr, I. Tjernberg, and L. Dijkshoorn. 2006. Naturally transformable Acinetobacter sp. strain ADP1 belongs to the newly described species Acinetobacter baylyi. Appl. Environ. Microbiol. 72:932-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vangnai, A. S., D. J. Arp, and L. A. Sayavedra-Soto. 2002. Two distinct alcohol dehydrogenases participate in butane metabolism by Pseudomonas butanovora. J. Bacteriol. 184:1916-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wyndham, R. C., E. C. Alisa, H. N. Cindy, and C. P. Michelle. 1994. Catabolic transposons. Biodegradation 5:323. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, F., T. Tran, and Y. Xu. 2008. Nezha, a novel active miniature inverted-repeat transposable element in cyanobacteria. Biochem. Biophys. Res. Commun. 365:790-794. [DOI] [PubMed] [Google Scholar]
- 51.Zhou, N.-Y., S. L. Fuenmayor, and P. A. Williams. 2001. nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J. Bacteriol. 183:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.