Abstract
The putative source of hide contamination for 236 cattle in Scotland followed from the farm through to slaughter was determined using phage and verocytotoxin type data. The majority of cattle (84%) were found to have subtypes of Escherichia coli O157 on their hide that had not been found previously in any animal from the farm of origin, strongly suggesting that contamination occurred once animals had left the farm of origin. Using logistic regression analysis, several variables and factors were found to be strongly associated (P < 0.01) with cross-contamination of cattle hides at the univariate level; commercial transport to slaughter, transport with other animals, use of a crush, line automation, and increasing slaughterhouse throughput were all risk factors, while feeding hay in lairage, processing an animal earlier in a slaughter cohort, and cleaning the landing area poststunning were protective. In the multivariable model, with the slaughterhouse and the farm group included as random effects, factors associated with the cross-contamination of cattle hides were identified. Transport to the slaughterhouse by a commercial hauler had a borderline-significant association with increased odds of an animal having a cross-contaminated hide (odds ratio [OR] [95% confidence interval {CI}] = 5.7 [0.99, 33.0]; P = 0.05). At the slaughterhouse, providing hay to cattle waiting in lairage (OR [95% CI] = 0.04 [<0.01, 1.04]; P = 0.05) and cleaning the landing area (OR [95% CI] = 0.03 [<0.01, 1.15,]; P = 0.06) also had a borderline-significant association with decreased odds of an animal having a cross-contaminated hide. Although the prevalence of carcass contamination remains very low, targeted intervention at the preslaughter stage may have the potential to reduce further the risk to public health.
Escherichia coli O157 is an important food-borne pathogen, in part because of its low infectious dose in humans, the severity of disease outcomes in affected people, and its widespread prevalence in cattle. In Scotland, E. coli O157 has had a relatively high profile over the past two decades due to several outbreaks of the disease. However, the majority of cases of human illness are sporadic rather than outbreak related. The incidence of sporadic E. coli O157 cases in Scotland is higher than those in England, Wales, and Northern Ireland. Between 1995 and 2000, there were between 4 and 10 cases of E. coli O157 illness per 100,000 population per year in Scotland, while in England and Wales there were between 1.27 and 2.08 cases per 100,000 population per year in the same time period (32). Cattle are asymptomatic carriers of E. coli O157, shedding the bacteria in their feces (6, 17). Earlier research has indicated that contact with livestock or their environment is the strongest risk factor for sporadic E. coli O157 cases in Scotland (18, 22-23, 37). Recently cattle hides were specifically identified as a potential food safety concern, since E. coli on the hide can be transferred to the carcass during the skinning process (3, 26). A study using marker bacteria inoculated on a small number of cattle hides entering the slaughterhouse demonstrated that the bacteria become widely disseminated throughout the slaughterhouse environment (8). In previous work, 256 cattle were followed from the farm to the slaughterhouse, and it was found that more than half of the animals (55%) had contaminated hides postexsanguination (24). The current analysis examines the origins of that hide contamination using E. coli O157 subtype data to determine where and how animal hides became contaminated. In terms of considering control strategies, it is important to know whether contamination arises from an animal's own feces or from those of other animals that it may come in contact with. Strains of E. coli O157 can persist in farm, feedlot, transport vehicle, or slaughterhouse environments (21, 33), and consequently, contamination may occur at different points. In terms of prevention, it is especially important to examine the origins of cross-contamination to determine where intervention strategies may be most effectively applied, whether at the slaughterhouse, farm, or individual animal level.
Previous studies have found identical pulsed-field gel electrophoresis (PFGE) patterns for isolates taken from slaughterhouse environments, transport trailers, other cattle's colons, cattle hides, and carcasses, highlighting the significance and extent of cross-contamination taking place in the preslaughter environment (1, 2, 7, 40). This analysis uses a multilevel hierarchical model to examine the factors associated with Scottish animals having a hide contaminated from external sources (cross-contamination) at slaughter, that is, sources other than the farm of origin, the animal itself, or other animals from the herd of origin.
MATERIALS AND METHODS
Sample collection.
Thirty-four farms participated in the study on a voluntary basis; farms that were visited as part of a prevalence study (16) (February 2002 to February 2004) and that sent cattle directly to slaughterhouses in Scotland were invited to enroll their animals. The 12 slaughterhouses participating in the study were those that received animals from these farms. A total of 256 cattle were followed from farm to slaughterhouse. Farm and slaughterhouse visits were conducted over a period of 22 months, April 2002 to February 2004.
Farm visits.
The majority of visits were made either the day before or the day of animal departure for slaughter. Several farms were visited on more than one occasion—three farms were visited twice, and one farm was visited three times. A questionnaire was completed at the farm, recording details on animal descriptors, such as breed and age, and preslaughter management practices, such as clipping of the belly hair. Belly clipping is a widely used intervention to improve the visible hygiene of animals presented for slaughter in the United Kingdom. In addition, farm characteristics (such as housing and feed) and the mode of transport to the slaughterhouse were recorded. Fecal samples were obtained per rectum from all of the cattle going to slaughter and identified for each animal by means of the ear tags. The fecal samples were transported to the laboratory in a cooler.
Slaughterhouse visits.
Visits were made on the day that the study cattle were scheduled for slaughter. A questionnaire was completed to document preslaughter conditions for the study animals. This included slaughterhouse characteristics, such as the average daily number of animals slaughtered, lairage conditions, and slaughter hall features, and questions on the preslaughter management of animals, such as whether they were held in lairage and for how long, if feed was provided, and the position on the processing line (killing order). The slaughterhouse killing capacity was calculated, using the questionnaire information, as the number of animals slaughtered on the day of the visit divided by the average daily number of animals slaughtered. This was used as a measure of the slaughterhouse activity on the day of the visit; slaughterhouses with a value of 1.0 were operating at routine capacity, whereas slaughterhouses with values below or above 1.0 were operating below or above capacity, respectively. In the slaughterhouse, animals entered the slaughter queue in farm groups, and the E. coli O157 hide contamination status of each carcass (before hide removal) immediately preceding and immediately following each group was assessed. Thus, contamination information was available for the hides of every animal/carcass followed from the farm and for the carcass directly before and directly after it on the line, allowing assessment of clustering of contamination on the processing line. For this purpose, an additional 84 carcasses were sampled on the processing line at the slaughterhouse only (i.e., there was no farm information for these carcasses).
Four different samples were obtained from study carcasses on the processing line after killing: hide, rectal content (after evisceration), and each side of the carcass (also after evisceration and after carcass splitting). Hide samples were obtained from the brisket area of each carcass immediately after exsanguination. This area was chosen because it is an area that has been identified previously as one of the most heavily contaminated (25, 31) and it is one of the initial opening cut sites for removal of the hide. The area along the breastbone between the front legs (along the incision line for hide removal) was sampled using a moist swab. Samples were placed in 80 ml of sterile buffered peptone water and transported to the laboratory in a cooler with ice packs. The method used for recovery of E. coli O157 from the hide samples was validated using sterile buffered peptone water spiked with E. coli O157 strain 6252 (39).
Isolation, enumeration, and laboratory analysis.
The methods of Chapman et al. (5) were used to isolate E. coli O157 from fecal samples, with the modification of using non-antibiotic-enriched buffered peptone water during enrichment (38).
Upon arrival at the laboratory, hide samples were refrigerated overnight at 4°C. The following day, samples were enriched for 6 h at 37°C using buffered peptone water. The samples were then frozen at −80°C for a minimum of 6 months (39). Upon thawing, immunomagnetic separation was used to identify E. coli O157 strains as described by Ternent et al. (39). A positive sample was defined as one testing positive for verocytotoxin-producing Escherichia coli O157. The Scottish E. coli O157 Reference Laboratory (Edinburgh) confirmed the identification of positive colonies and performed phage typing and PFGE on the samples.
Typing of the verocytotoxin genes (VT1 and VT2 genes) in positive E. coli O157 isolates was accomplished using PCR (29). Phage typing was carried out following standard procedures described by Khakhria et al. (20). For the majority of the samples, a single colony was isolated and tested. However, to obtain an indication of how representative a single colony would be of the sample tested, every 20th sample was duplicate tested. Two aliquots were plated on separate plates, and from each a single colony was isolated and tested in the manner described above, the phage type and verocytotoxin type determined, and the results compared.
Definition of the outcome variable.
The dichotomous outcome variable concerned the presence or absence of E. coli O157 cross-contamination on an individual animal's hide. The farms that participated in this study were also participants in at least one of two previously conducted prevalence studies (16, 28). The first study was conducted from March 1998 to May 2000, whereas the second study (during which the farms were recruited for the subsequent slaughter component) was conducted during the same time period as the slaughterhouse study, from February 2002 to February 2004, as part of the same research project. Information regarding the phage and verocytotoxin type (VT type) of E. coli O157 from samples taken in these two prevalence studies was available. Putative cross-contamination was defined as an instance in which the phage/VT type of E. coli O157 isolated from an animal's hide was not the same as that of any sample that had been recorded (either in the current or previous studies) on the farm from which the animal was sent to slaughter. The outcome was determined to be negative if the hide sample result was negative for E. coli O157, if it matched the subtype of an E. coli O157 sample from the animal's own fecal sample or the fecal or hide sample of an animal in its farm group, or if it matched the subtype of an E. coli O157 sample previously recorded on the farm of origin. The farm group, in this context, is defined as a group of cattle from the same farm, sent to slaughter at the same time. A conservative definition of the outcome variable was used; since it was not possible to determine whether or not an animal's hide had become contaminated by an extrinsic animal (not from its own farm of origin) but with an E. coli O157 subtype identical to one previously found on the farm of origin, this occurrence of cross-contamination would not be considered. Potentially, therefore, the true prevalence of E. coli O157 hide cross-contamination was underestimated.
Statistical analysis.
A generalized linear mixed model was fitted to the data using multivariable logistic regression and the statistical software Stata 9.1 (36). Records of individual cattle in which there were missing values were removed, leaving 236 cattle in the reduced data set, from 32 farms, going to 11 slaughterhouses. Data for many variables were collected during the course of the slaughterhouse study; however, only variables that were considered relevant for the identification of risk factors for hide cross-contamination once animals left the farm were examined. Predictor variables included those at the level of individual animals, farms, and slaughterhouses. Categorical variables with more than two levels were coded as dummy variables. Univariate analyses were conducted by regressing each predictor variable individually on the outcome variable using logistic regression. Those predictor variables with a P value of <0.25 were offered to a multivariable model. The “gllamm” function of Stata 9.1 was used, with adaptive quadrature specified (30). The data set included groups of animals sent from 32 farms to 11 slaughterhouses; as a consequence, both “group” and “slaughterhouse” were modeled as random effects. Variables with P values of >0.05 were removed sequentially from the model, starting with the least-significant variable. Inclusion or exclusion of variables from the model was determined by an examination of both the Akaike's information criterion of the full and reduced models and the likelihood ratio test. To avoid overfitting the data, interactions between the variables in the main effects model were not examined due to the small number of observations (236 individual cattle) and the inclusion of two random effects. Intraclass correlation coefficients (ICCs) were calculated for both random effects. The ICC is a measure of the variability among groups within the random effects and so provides an estimate of the correlation between observations within the same group (in this case, within the same farm group or within the same slaughterhouse).
RESULTS
Animal characteristics and management factors.
The E. coli O157 subtype data relating to the hide samples are shown in Table 1, and the risk factors examined are shown in Table 2. The number of carcasses processed and the prevalence of E. coli O157 hide contamination at each slaughterhouse in the study are presented in Table 3. A breakdown of the potential risk factors by hide cross-contamination status and the univariate analysis results are presented in Tables 4 and 5.
TABLE 1.
Phage and VT types of E. coli O157 in the 236 hide samples taken from Scottish cattle at slaughter, 2002 to 2004
| Hide sample phage type | Hide sample VT type | Frequency [no. (%)] | No. of farms (fecal sample)a |
|---|---|---|---|
| —b | — | 106 (44.9) | 32 (all) |
| 21/28 | VT2 | 57 (24.2) | 10 |
| 8 | VT1VT2 | 21 (8.9) | 1 |
| 4 | VT2 | 14 (5.9) | 0 |
| 2 | VT1VT2 | 11 (4.7) | 0 |
| 2 | VT2 | 9 (3.8) | 3 |
| 32 | VT2 | 7 (3.0) | 0 |
| 8 | VT1 | 6 (2.5) | 0 |
| 34 | VT2 | 2 (0.9) | 4 |
| RDNC | VT1VT2 | 2 (0.9) | 0 |
| Untypeable | 1 (0.4) | 0 | |
| Total | 236 (100) |
Number of farms for which the subtype was detected in fecal samples, either in the slaughterhouse study or one of the two previous prevalence studies (total, 32 farms).
—, no E. coli O157 was found.
TABLE 2.
Potential risk factors considered in study examining E. coli O157 hide cross-contamination of 236 cattle at slaughter in Scotland, 2002 to 2004
| Variable (abbreviated name) | Description |
|---|---|
| Transport | Transport from the farm to the slaughterhouse (dichotomous): by farmer or by commercial hauler |
| Crush | Use of a crush (restraining crate) by the slaughterhouse to restrain individual animals when reading ear tags (dichotomous): crush not used or crush used |
| Feed in lairage | Provision of hay in the slaughterhouse lairage to animals that are not slaughtered immediately upon arrival (dichotomous): feed not provided or hay provided |
| Mechanics | Mechanics of the processing line (dichotomous): automated line or manually operated line |
| Travel with | Travel with animals from other farms on the journey to the slaughterhouse (dichotomous): did not travel with animals from other farms or did travel with animals from other farms |
| Land area | Slaughterhouse practice regarding the landing area (area of the floor where carcasses fall immediately after stunning, dichotomous): landing area not cleaned after each carcass or landing area cleaned after each carcass using a squeegee |
| Lairage | Animals held in lairage after arriving at the slaughterhouse and prior to slaughter (dichotomous): animals not held in lairage or animals held in lairage |
| Clipped | Clipping of the belly hide of animals prior to transport to the slaughterhouse, to make animals appear more visually clean (dichotomous): animals not clipped or animals clipped |
| Line no. | Position on the processing line (killing order, continuous) |
| Throughput | Average no. of animals slaughtered per day in the slaughterhouse (continuous) |
| Agea | Age (mo) of each study animal (continuous) |
| Capacity | Slaughterhouse capacity (no. of animals slaughtered on day of researchers' visit/avg daily no. slaughtered at that slaughterhouse, dichotomous): capacity of <1.0 or capacity of >1.0 |
| Time | Time (h) from when animals left the farm to when hide sample was taken at the slaughterhouse (continuous) |
At the time of this study, slaughter of cattle older than 30 months for human consumption was prohibited under the transmissible spongiform encephalopathy regulations in force in the United Kingdom.
TABLE 3.
Number of hide samples positive for E. coli O157 contamination per slaughterhouse in the reduced data set of 236 carcasses in Scotland, 2002 to 2004
| Slaughterhouse no. | Carcasses' hides processed
|
||
|---|---|---|---|
| No. E. coli O157 positive | Total no. sampled | % E. coli O157 positive | |
| 1 | 32 | 34 | 94 |
| 2 | 14 | 15 | 93 |
| 3 | 0 | 14 | 0 |
| 4 | 15 | 22 | 68 |
| 5 | 1 | 3 | 33 |
| 6 | 10 | 19 | 53 |
| 7 | 24 | 37 | 65 |
| 8 | 0 | 18 | 0 |
| 9 | 2 | 3 | 67 |
| 10 | 1 | 5 | 20 |
| 11 | 31 | 66 | 47 |
| Total | 130 | 236 | 55 |
TABLE 4.
Univariate analysis of categorical variables considered in analysis of risk factors for hide cross-contamination of 236 cattle with E. coli O157 at slaughter in Scotland, 2002 to 2004
| Variable | Category | No. of cattle | Hide cross-contamination
|
P value | |
|---|---|---|---|---|---|
| No. (%) negative | No. (%) positive | ||||
| Transport | Farmer | 87 | 60 (69) | 27 (31) | |
| Hauler | 149 | 67 (45) | 82 (55) | <0.001 | |
| Crush | No | 178 | 107 (60) | 71 (40) | |
| Yes | 58 | 20 (34) | 38 (66) | 0.001 | |
| Feed | None | 214 | 106 (50) | 108 (50) | |
| Hay | 22 | 21 (95) | 1 (5) | 0.003 | |
| Mechanics | Automated | 198 | 93 (47) | 105 (53) | |
| Manual | 38 | 34 (89) | 4 (11) | <0.001 | |
| Travel with | No | 87 | 60 (69) | 27 (31) | |
| Yes | 149 | 67 (45) | 82 (55) | <0.001 | |
| Land area | Nothing | 200 | 92 (46) | 108 (54) | |
| Cleaned | 36 | 35 (97) | 1 (3) | <0.001 | |
| Lairage | Not held in lairage | 17 | 7 (41) | 10 (59) | |
| Held in lairage | 219 | 120 (55) | 99 (45) | 0.255 | |
| Clipped | No | 101 | 49 (49) | 52 (51) | |
| Yes | 135 | 78 (58) | 57 (42) | 0.260 | |
| Capacity | No | 121 | 69 (57) | 52 (43) | |
| Yes | 115 | 58 (50) | 57 (50) | 0.216 | |
TABLE 5.
Univariate analysis of continuous variables considered in analysis of risk factors for hide cross-contamination of 236 cattle with E. coli O157 at slaughter in Scotland, 2002 to 2004
| Variable | Range | Median | SD | Direction of effect | P value |
|---|---|---|---|---|---|
| Line no. | 1-536 | 73 | 106 | Positive | <0.001 |
| Throughput (animals/day) | 25-400 | 200 | 107 | Positive | <0.001 |
| Age (mo) | 12-30 | 23 | 4.4 | 0.519 | |
| Time (h) | <1-48.9 | 4.1 | 10.4 | 0.478 |
Putative source of hide contamination.
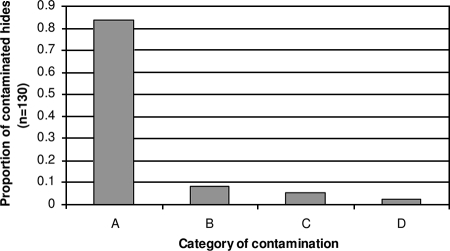
Five percent of the farm fecal samples, 6% of the gut samples, and <1% of the carcass samples obtained from the study animals were positive for E. coli O157. Of the hide samples, 130 of the 236 cattle (55%) had contaminated hides. Using phage types and VT types to classify the contaminated hides, only 5% of study animals with contaminated hides appeared to be “self-contaminated” (i.e., had the same subtype on the hide as in fecal or rectal content samples) (Fig. 1). Nine percent of contaminated hides were contaminated with subtypes that had been found on the farm of origin in either of the two previous prevalence studies, while 2% of the contaminated hides had subtypes matching the subtypes in a fecal sample or hide sample of at least one animal in the same farm group. Eighty-four percent of the study animals that were found to have E. coli O157 on their hides were contaminated with subtypes not previously recorded from any animal from the farm of origin.
FIG. 1.
Putative origin of Escherichia coli O157 hide contamination of 236 Scottish beef cattle at slaughter, as determined by phage/VT type. A, cross-contaminated; B, contaminated with subtype previously found on farm of origin; C, self-contaminated; D, contaminated by animals in the same farm group.
Significant univariable associations.
Several variables and factors were found to be strongly associated (P < 0.01) with cross-contamination of cattle hides at the univariate level: commercial transport to slaughter, transport with other animals, use of a crush, line automation, and increasing slaughterhouse throughput were all risk factors, while feeding hay in lairage, processing an animal earlier in a slaughter cohort, and cleaning the landing area poststunning were protective.
Multivariable logistic regression.
Transport to the slaughterhouse by a commercial hauler, as opposed to transport by the farmer, was associated with a borderline-significant increase in the odds of a hide being cross-contaminated (odds ratio [OR] [95% confidence interval {CI} = 5.7 [0.99, 33.0]; P = 0.05). In the slaughterhouse, providing hay to animals in lairage (OR [95% CI] = 0.04 [<0.01, 1.04]; P = 0.05) and cleaning the landing area from the stunning crate between each animal (OR [95% CI] = 0.03 [<0.01, 1.15]; P = 0.06) were associated with borderline-significant decreases in the odds of hide cross-contamination (Table 6). ICCs for farm group and slaughterhouse were calculated to be 0.56 and 0.20, respectively.
TABLE 6.
Final mixed-model logistic regression analysis of hide cross-contamination data set
| Factor | Coefficient | SE | OR (95% CI) | P value |
|---|---|---|---|---|
| Transport | ||||
| Farmer | 1.0 | |||
| Hauler | 1.7 | 0.9 | 5.7 (0.99, 33.0) | 0.05 |
| Feed in lairage | ||||
| No feed | 1.0 | |||
| Hay | −3.2 | 1.7 | 0.04 (2.0e−03, 1.04) | 0.05 |
| Landing area practice | ||||
| Not wiped between carcasses | 1.0 | |||
| Wiped between carcasses | −3.4 | 1.8 | 0.03 (9.1e−04, 1.15) | 0.06 |
Duplicate testing results.
For 14 hide samples, two colonies were isolated and tested. Five were positive for E. coli O157, were confirmed positive in both samples, and had matching phage types and VT types. All nine negative samples were negative for both samples.
DISCUSSION
The observation that the great majority of animals with contaminated hides were contaminated with subtypes never previously recorded on the farm of origin suggests that cross-contamination was a significant phenomenon in this study population. This was also the conclusion of a study examining the effects of transportation and the lairage environment on the prevalence and diversity of E. coli O157:H7 cattle hide contamination (1). Hide contamination increased from 50.3 to 94.4% from sampling pretransport at the feedlot to sampling poststunning and -exsanguination at the processing plant. The number of highly contaminated hides also increased, and only 29% of the isolates collected at the processing plant matched those collected pretransport (indicating cross-contamination had occurred). Cattle hide contamination by E. coli O157 is a potential threat to human health via the food chain, since contaminated hides have been shown to be a likely route of carcass contamination (3, 14, 26). Therefore, it is important to determine when and how the hides become contaminated, as this indicates the level at which interventions may be most effectively applied (e.g., individual animal, farm, and slaughterhouse). If the major source of contamination was shown to be from the farm of origin (i.e., hides contaminated with E. coli O157 from the animal's own fecal matter or that of a herdmate), this would suggest that the most effective interventions should be directed toward reducing fecal shedding on particular “problem” farms, that is, farms that don't necessarily have high prevalences or concentrations of E. coli O157 but ones on which some of the animals are shedding for a prolonged period of time (i.e., many consecutive months). However, the finding that the majority of contaminated hides were contaminated with subtypes not found on the animals' farms of origin suggests that such a focused approach may not be effective. In these circumstances, a more global farm-based strategy, such as universal vaccination or therapy, or intervention further along the processing chain, such as adjusting transport conditions and/or intervention in the slaughterhouse, would be required. Childs et al. (7) found genotypic matches between colonic isolates from cattle and those from environmental samples, suggesting that animals that are shedding are doing so in a way that widely distributes bacteria throughout the preharvest environment. It was also found that E. coli O157:H7 isolates from hide samples of cattle genetically matched isolates found in environmental samples. There are many points in the beef processing chain where hides may become contaminated: on the farm, during transport, during unloading at the slaughterhouse, in lairage, and on the lairage-to-slaughter route (34). Since only 6% of animals were found to be shedding on the farm and less than 5% of animals had E. coli O157 in their terminal rectum at the slaughterhouse in this study, a combination of individual animal/herd-level prevention of shedding and sanitization of environmental reservoirs may prove useful.
A number of categorical and continuous variables were strongly associated with cross-contamination at the univariable level. All significant variables were plausible potential risk or protective factors, although the apparent protective effect of a manual line was interesting and somewhat unexpected. However, this variable, as well as the position in line, slaughterhouse throughput, feeding of hay, traveling with others, and use of a crush, dropped out of the subsequent multivariable model, suggesting the apparent associations are explained by the remaining variables.
A nearly sixfold increase in the odds of an animal having a cross-contaminated hide was observed when a commercial hauler took animals to the slaughterhouse as opposed to their being transported by the farmer. The importance of transport with respect to hide contamination has been previously discussed (1, 7, 10, 12, 24, 40). In the current study, transport of groups from more than one farm occurred only with commercial haulage (24), and this may have contributed to the significance of this factor, providing the opportunity for direct hide-to-hide or hide-to-trailer-to-hide cross-contamination, transferring foreign subtypes from one hide to another. Tutenel et al. (40) hypothesized that a major factor mediating the spread of hide contamination is direct contact between animals after they leave the farm. A study examining cattle behavior in housing found that there was an average of 13 instances per hour of an animal grooming another (direct contact), although it is not known whether this behavior is similar among animals in transport (27). Another possible contributor to indirect cross-contamination is inadequate cleaning of vehicles between shipments of cattle from different farms. Since E. coli O157 can remain viable and infectious for up to 2 months in fecal samples (41), thorough cleaning and disinfecting of trailers after each journey to the slaughterhouse may reduce the prevalence of cross-contaminated hides. The potential for transport trailers to be sources of E. coli O157 contamination has been demonstrated using molecular epidemiological techniques. Using PFGE, Childs et al. (7) found that E. coli O157:H7 isolates obtained from transport trailer side walls matched those from cattle hides within a slaughterhouse. This study also found that isolates could be genetically matched between transport trailers, areas of the slaughterhouse, and the colons of some animals. Another study found at least one positive E. coli O157:H7 sample from each of the seven trailers in the study (1).
Providing hay in lairage was negatively associated with hide cross-contamination. The point estimate of the odds ratio for this factor was 0.04, and the significance was borderline (P = 0.05). This result was similar to that reported in a previous publication (24) regarding general hide contamination. Although other studies have found that diet has a sparing effect on the prevalence of shedding of E. coli (9) and E. coli O157 (15, 35), the length of time that most study animals spent in lairage in the current study (mean, 3 h; range, 0 to 45.2 h) would not be sufficient for any feeding regimen to have a known biological effect for the majority of cattle. A potential confounding factor is that in the United Kingdom, only animals spending more than 12 h in lairage must be provided with feed. It is possible that it is this “resting” period that may be responsible for the decrease in hide contamination, or perhaps this factor is simply a proxy for some other factor not measured in the study.
Another factor that was found to be protective was cleaning of the landing area from the stunning crate after each animal had been stunned. This factor was also only borderline significant (P = 0.06), with an associated odds ratio of 0.03. Cleaning the landing area, in the slaughterhouses included in the study, involved mopping the area with a squeegee after each animal had been removed following stunning. Our results suggest that this practice may be beneficial in reducing the risk of cross-contamination. However, in some slaughterhouse configurations this would not be easily implemented since animals fall onto a tubular steel raised cradle. Elder et al. (11) detected the presence of E. coli O157 on cattle hides following stunning, suggesting the landing area as a plausible reservoir of contamination. Small et al. (34) and Collis et al. (8) also suggested the landing area is a key site for hide cross-contamination. The results presented here support these hypotheses. The landing area cleaning methods employed by the slaughterhouses in this study did not involve any antimicrobial measures; it is plausible that simply wiping down the landing area and removing gross fecal contamination was sufficient to reduce cross-contamination.
The ICCs associated with the random effects provide an indication of the degree of variability between the different farm groups and between the different slaughterhouses with respect to the outcome. The ICC of 0.56 for farm group indicates that 56% of the variability in the occurrence of cross-contaminated hides was due to differences between farm groups; that is, different farm groups had widely varying prevalences of cross-contaminated hides. This wide variation may be explained by the differences in transport methods used to take groups to the slaughterhouse or may be due to some factor or factors not measured. The ICC for slaughterhouse was 0.20, and consequently, there does not appear to be a high level of variation in the occurrence of E. coli O157 cross-contaminated cattle hides between the 11 slaughterhouses in this data set.
There are several limitations of this study that should be explored. While only one colony was isolated from each sample for laboratory analysis in this study, animals can be colonized by more than one strain of E. coli O157 (26), although this appears to be a rare occurrence. Therefore, if more isolates had been typed per sample, it is possible that a different estimate of cross-contamination prevalence would have been found. However, although it is only a small number, the five positive hide samples that were duplicate tested had identical phage and verocytotoxin types. Although there is evidence that particular subtypes can persist on a farm over long periods of time (21, 33), new strains of E. coli O157 can also appear on farms (33). Thus, the use of historical data on subtypes on the farm may not necessarily reflect the current contamination profile of the farm, although cattle derived from it should be representative of the farm. Although the first prevalence study was completed 2 years prior to the commencement of the slaughterhouse study, the second prevalence study was conducted contemporaneously (farms for the slaughterhouse study were recruited from participants of the second prevalence study). However, the most surprising finding was that the majority of E. coli O157 isolates discovered on the hide did not match those found in the fecal or gut samples of the animal or those from others in its farm group; the historical data were included as a convenient additional source of information. Other studies have shown that most farms appear to have E. coli O157 strains with unique patterns (using PFGE); however, it has also been reported that several farms can harbor strains with identical genetic patterns, although this again does not appear to be a common occurrence (33). In our approach, this would cause the apparent prevalence of hide cross-contamination to be lower than the true prevalence. It is likely that the prevalence of cross-contaminated hides in this study was underestimated, since phage and VT typing were used to type the isolates sampled. Isolates may have the same phage type/VT type but different PFGE patterns. In addition, no environmental samples were taken (e.g., from the transport trailers or the slaughterhouse environment) to compare with the hide samples. It is therefore not known whether there were any dominant or persistent strains in the environment that would match the observed E. coli O157 subtype patterns of the hide samples tested. E. coli O157 contamination of cattle lairages has been demonstrated to persist from one day to the next, despite routine cleaning (34). Tutenel et al. (40) reported that even after 3 days of zero slaughter activity, E. coli O157 could still be isolated from two aprons and the stunning crate before the start of slaughtering.
Although several farm groups had low numbers of cattle, this represents all the animals comprising a slaughter group from that farm on that day (median, 5 animals per farm group; range, 1 to 19). This is typical of the system that operates in Scotland, with numerous relatively small family-run operations. This is in contrast to the large-scale feedlots typical of beef production in other parts of the world.
It is possible, since only one area of the hide was sampled (the brisket), that if other carcass sites were tested, more risk factors might be revealed. However, the brisket has been found to be one of the most contaminated sites on the hides of cattle (31), and so the analysis described here likely represents one of the most important sites of hide contamination. Further, the brisket represents an area for the opening cut site for skinning the animal and subsequently for splitting the sternum to access the thorax for purposes of evisceration. Contamination may be transferred from this site to others on the carcass, and roll-back of the hide, which may occur during the removal process, could also transfer bacteria from the brisket to other parts of the carcass (31).
Conclusions.
In this study, cross-contamination appeared to be the predominant mechanism for hide contamination of cattle with Escherichia coli O157. This suggests that it is not sufficient for individual farmers to eliminate E. coli O157 from their herd but rather that there should be a coordination of intervention strategies aimed at reducing the prevalence of hide contamination and/or preventing contact between animals from different sources after they leave the farm of origin. Further research is required to better understand how the provision of hay in lairage and the wiping down of the landing area have protective effects against E. coli O157 hide cross-contamination of cattle.
Acknowledgments
This work was supported by the Wellcome Trust-funded International Partnership Research Awards in Veterinary Epidemiology (IPRAVE) study entitled “Epidemiology and evolution of Enterobacteriaceae infections in humans and domestic animals.” Alison Mather's time on this project was supported by the Department for Environment, Food and Rural Affairs (DEFRA) Veterinary Research Training Initiative (VTRI) and by a Postgraduate Scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC).
We are grateful to the other members of the IPRAVE consortium for their assistance and advice. We acknowledge the advice of Lesley Allison of the Scottish E. coli Reference Laboratory and Mary Locking of Health Protection Scotland.
Footnotes
Published ahead of print on 22 August 2008.
REFERENCES
- 1.Arthur, T. M., J. M. Bosilevac, D. M. Brichta-Harhay, M. N. Guerini, N. Kalchayanand, S. D. Shackelford, T. L. Wheeler, and M. Koohmaraie. 2007. Transportation and lairage environment effects on prevalence, numbers, and diversity of Escherichia coli O157:H7 on hides and carcasses of beef cattle at processing. J. Food Prot. 70:280-286. [DOI] [PubMed] [Google Scholar]
- 2.Avery, S. M., E. Liebana, M. L. Hutchison, and S. Buncic. 2004. Pulsed field gel electrophoresis of related Escherichia coli O157 isolates associated with beef cattle and comparison with unrelated isolates from animals, meats and humans. Int. J. Food Microbiol. 92:161-169. [DOI] [PubMed] [Google Scholar]
- 3.Bell, R. G. 1997. Distribution and sources of microbial contamination on beef carcasses. J. Appl. Microbiol. 82:292-300. [DOI] [PubMed] [Google Scholar]
- 4.Reference deleted.
- 5.Chapman, P. A., D. J. Wright, and C. A. Siddons. 1994. A comparison of immunomagnetic separation and direct culture for the isolation of verocytotoxin-producing Escherichia coli O157 from bovine feces. J. Med. Microbiol. 40:424-427. [DOI] [PubMed] [Google Scholar]
- 6.Chapman, P. A., C. A. Siddons, A. T. Cerdan Malo, and M. A. Harkin. 1997. A 1-year study of Escherichia coli O157 in cattle, sheep, pigs and poultry. Epidemiol. Infect. 119:245-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Childs, K. D., C. A. Simpson, W. Warren-Serna, G. Bellenger, B. Centrella, R. A. Bowling, J. Ruby, J. Stefanek, D. J. Vote, T. Choat, J. A. Scanga, J. N. Sofos, G. C. Smith, and K. E. Belk. 2006. Molecular characterization of Escherichia coli O157:H7 hide contamination routes: feedlot to harvest. J. Food Prot. 69:1240-1247. [DOI] [PubMed] [Google Scholar]
- 8.Collis, V. J., C.-A. Reid, M. L. Hutchison, M. H. Davies, K. P. A. Wheeler, A. Small, and S. Buncic. 2004. Spread of marker bacteria from the hides of cattle in a simulated livestock market and at an abattoir. J. Food Prot. 67:2397-2402. [DOI] [PubMed] [Google Scholar]
- 9.Diez-Gonzalez, F., T. R. Callaway, M. G. Kizoulis, and J. B. Russell. 1998. Grain feeding and the dissemination of acid-resistant Escherichia coli from cattle. Science 281:1666-1668. [DOI] [PubMed] [Google Scholar]
- 10.Duffy, G., S. B. O'Brien, E. Carney, J. J. Sheridan, D. A. McDowell, and I. S. Blair. 2005. Characterisation of E. coli O157 isolates from bovine hide and beef trimming in Irish abattoirs by pulsed field gel electrophoresis. J. Microbiol. Methods 60:375-382. [DOI] [PubMed] [Google Scholar]
- 11.Elder, R. O., J. E. Keen, G. R. Siragusa, G. A. Barkocy-Gallagher, M. Koohmaraie, and W. W. Laegreid. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. USA 97:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fegan, N., G. Higgs, P. Vanderlinde, and P. Desmarchelier. 2005. An investigation of Escherichia coli O157 contamination of cattle during slaughter at an abattoir. J. Food Prot. 68:451-457. [DOI] [PubMed] [Google Scholar]
- 13.Foster, G., G. F. Hopkins, G. J. Gunn, H. E. Ternent, F. Thomson-Carter, H. I. Knight, D. J. L. Graham, V. Edge, and B. A. Synge. 2003. A comparison of two pre-enrichment media prior to immunomagnetic separation for the isolation of E. coli O157 from bovine faeces. J. Appl. Microbiol. 95:155-159. [DOI] [PubMed] [Google Scholar]
- 14.Gill, C. O., J. C. McGinnis, and J. Bryant. 1998. Microbial contamination of meat during the skinning of beef carcase hindquarters at three slaughtering plants. Int. J. Food Microbiol. 42:175-184. [DOI] [PubMed] [Google Scholar]
- 15.Gregory, N. G., L. H. Jacobson, T. A. Nagle, R. W. Muirhead, and G. J. Leroux. 2000. Effect of preslaughter feeding system on weight loss, gut bacteria, and the physico-chemical properties of digesta in cattle. N.Z. J. Agric. Res. 43:351-361. [Google Scholar]
- 16.Gunn, G. J., I. J. McKendrick, H. E. Ternent, F. Thomson-Carter, G. Foster, and B. A. Synge. 2007. An investigation of factors associated with the prevalence of verocytotoxin producing Escherichia coli O157 shedding in Scottish beef cattle. Vet. J. 174:554-564. [DOI] [PubMed] [Google Scholar]
- 17.Hancock, D. D., D. H. Rice, L. A. Thomas, D. A. Dargatz, and T. E. Besser. 1997. Epidemiology of Escherichia coli O157 in feedlot cattle. J. Food Prot. 60:462-465. [DOI] [PubMed] [Google Scholar]
- 18.Innocent, G. T., D. J. Mellor, S. A. McEwen, W. J. Reilly, J. Smallwood, M. E. Locking, D. J. Shaw, P. Michel, D. J. Taylor, W. B. Steele, G. J. Gunn, H. E. Ternent, M. E. Woolhouse, and S. W. J. Reid. 2005. Spatial and temporal epidemiology of sporadic human cases of Escherichia coli O157 in Scotland, 1996-1999. Epidemiol. Infect. 133:1033-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reference deleted.
- 20.Khakhria, R., D. Duck, and H. Lior. 1990. Extended phage-typing scheme for Escherichia coli O157:H7. Epidemiol. Infect. 105:511-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeJeune, J. T., T. E. Besser, D. H. Rice, J. L. Berg, R. P. Stilborn, and D. D. Hancock. 2004. Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: predominance and persistence of specific clonal types despite massive cattle population turnover. Appl. Environ. Microbiol. 70:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Locking, M., L. Allison, L. Rae, K. Pollack, and M. Hanson. 2006. VTEC infections and livestock-related exposures in Scotland, 2004. Euro. Surveill. 11:E060223.4. [DOI] [PubMed] [Google Scholar]
- 23.Locking, M. E., S. J. O'Brien, W. J. Reilly, E. M. Wright, D. M. Campbell, J. E. Coia, L. M. Browning, and C. N. Ramsay. 2001. Risk factors for sporadic cases of Escherichia coli O157 infection: the importance of contact with animal excreta. Epidemiol. Infect. 127:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mather, A. E., G. T. Innocent, S. A. McEwen, W. J. Reilly, D. J. Taylor, W. B. Steele, G. J. Gunn, H. E. Ternent, S. W. J. Reid, and D. J. Mellor. 2007. Risk factors for hide contamination of Scottish cattle at slaughter with Escherichia coli O157. Prev. Vet. Med. 80:257-270. [DOI] [PubMed] [Google Scholar]
- 25.McEvoy, J. M., A. M. Doherty, M. Finnerty, J. J. Sheridan, L. McGuire, I. S. Blair, D. A. McDowell, and D. Harrington. 2000. The relationship between hide cleanliness and bacterial numbers on beef carcasses at a commercial abattoir. Lett. Appl. Microbiol. 30:390-395. [DOI] [PubMed] [Google Scholar]
- 26.McEvoy, J. M., A. M. Doherty, J. J. Sheridan, F. M. Thomson-Carter, P. Garvey, L. McGuire, I. S. Blair, and D. A. McDowell. 2003. The prevalence and spread of Escherichia coli O157:H7 at a commercial beef abattoir. J. Appl. Microbiol. 95:256-266. [DOI] [PubMed] [Google Scholar]
- 27.McGee, P., L. Scott, J. J. Sheridan, B. Earley, and N. Leonard. 2004. Horizontal transmission of Escherichia coli O157:H7 during cattle housing. J. Food Prot. 67:2651-2656. [DOI] [PubMed] [Google Scholar]
- 28.Pearce, M. C., D. Fenlon, J. C. Low, A. W. Smith, H. I. Knight, J. Evans, G. Foster, B. A. Synge, and G. J. Gunn. 2004. Distribution of Escherichia coli O157 in bovine fecal pats and its impact on estimates of the prevalence of fecal shedding. Appl. Environ. Microbiol. 70:5737-5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollard, D., W. Johnson, H. Lior, S. Tyler, and K. Rozee. 1990. Rapid and specific detection of verotoxin genes in Escherichia coli by the polymerase chain-reaction. J. Clin. Microbiol. 28:540-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabe-Hesketh, S., A. Skrondal, and A. Pickles. 2006. Generalized linear latent and mixed models (GLLAMM) for Stata. StataCorp, College Station, TX.
- 31.Reid, C.-A., A. Small, S. M. Avery, and S. Buncic. 2002. Presence of food-borne pathogens on cattle hides. Food Control 13:411-415. [Google Scholar]
- 32.Reilly, W. J. 2001. Task force on E. coli O157 final report, p. 169. Scottish Centre for Infection and Environmental Health, Glasgow, United Kingdom. http://www.ecoli-uk.com/Download/ecolitaskfinreport.pdf.
- 33.Rice, D. H., K. M. McMenamin, L. C. Pritchett, D. D. Hancock, and T. E. Besser. 1999. Genetic subtyping of Escherichia coli O157 isolates from 41 Pacific Northwest USA cattle farms. Epidemiol. Infect. 122:479-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Small, A., C.-A. Reid, S. M. Avery, N. Karabasil, C. Crowley, and S. Buncic. 2002. Potential for the spread of Escherichia coli O157, Salmonella, and Campylobacter in the lairage environment at abattoirs. J. Food Prot. 65:931-936. [DOI] [PubMed] [Google Scholar]
- 35.Stanton, T. L., and D. Schutz. 2000. Effect of switching from high grain to hay five days prior to slaughter on finishing cattle performance. Colorado State University Research Report. Colorado State University, Fort Collins, CO.
- 36.StataCorp. 2006. STATA v. 9.1 statistics/data analysis. StataCorp, College Station, TX.
- 37.Strachan, N. J., G. M. Dunn, and I. D. Ogden. 2002. Quantitative risk assessment of human infection from Escherichia coli O157 associated with recreational use of animal pasture. Int. J. Food Microbiol. 75:39-51. [DOI] [PubMed] [Google Scholar]
- 38.Synge, B. A., H. E. Ternent, G. F. Hopkins, D. J. L. Graham, H. I. Knight, G. Foster, V. L. Edge, and G. J. Gunn. 1998. A comparison of buffered peptone water with and without antibiotics for the isolation of E. coli O157 from bovine faeces using immunomagnetic separation, p. 171. In G. Duffy, P. Garvey, J. Coia, Y. Wasterson, and D. A. McDowell (ed.), Concerted action CT98-3935: verocytotoxigenic E. coli in Europe. 1. Methods for verocytotoxigenic E. coli. Teagasc, National Food Centre, Dunsinea, Castleknock, Dublin, Ireland.
- 39.Ternent, H. E., G. T. Innocent, L. M. Filshie, D. J. Taylor, W. B. Steele, S. A. McEwen, W. J. Reilly, G. J. Gunn, S. W. J. Reid, and D. J. Mellor. 2004. Frozen storage of Escherichia coli O157 in buffered peptone water and its detection on bovine carcasses. J. Food Prot. 67:40-45. [DOI] [PubMed] [Google Scholar]
- 40.Tutenel, A. V., D. Pierard, J. Van Hoof, and L. De Zutter. 2003. Molecular characterization of Escherichia coli O157 contamination routes in a cattle slaughterhouse. J. Food Prot. 66:1564-1569. [DOI] [PubMed] [Google Scholar]
- 41.Wang, G., T. Zhao, and M. P. Doyle. 1996. Fate of enterohemorrhagic Escherichia coli O157:H7 in bovine feces. Appl. Environ. Microbiol. 62:2567-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]