Abstract
Studies in eukaryotes and prokaryotes have revealed that gene expression is not only controlled through altering the rate of transcription but also through varying rates of translation and mRNA decay. Indeed, the expression level of a protein is strongly affected by the steady state level of its mRNA. RNA decay can, along with transcription, play an important role in regulating gene expression by fine-tuning the steady state level of a given transcript and affecting its subsequent decoding during translation. Alterations in mRNA stability can in turn have dramatic effects on cell physiology and as a consequence the fitness and survival of the organism. Recent evidence suggests that mRNA decay can be regulated in response to environmental cues in order to enable the organism to adapt to its changing surroundings. Bacteria have evolved unique post transcriptional control mechanisms to enact such adaptive responses through: 1) general mRNA decay, 2) differential mRNA degradation using small non-coding RNAs (sRNAs), and 3) selective mRNA degradation using the tmRNA quality control system. Here, we review our current understanding of these molecular mechanisms, gleaned primarily from studies of the model gram negative organism E. coli, that regulate the stability and degradation of normal and defective transcripts.
Keywords: RNA quality control, SmpB, tmRNA, trans-translation, RNase R, RNA decay
1. Introduction
As with other biomolecules, the great variety of RNA species produced by the bacterial cell require quality control mechanisms to ensure proper folding and function. In addition, the role of mRNA as a template for protein synthesis adds greater significance to mRNA quality control. The translation of a faulty transcript without adequate quality assurance measures might lead to the accumulation of aberrant protein products that could be detrimental to the cell. While much data have been generated recently on the mechanisms of eukaryotic mRNA quality control, especially on that of nonsense mediated decay (NMD, reviewed elsewhere in this issue), the related topics in prokaryotes are comparatively less explored. Generally, bacterial mRNAs are not post-transcriptionally spliced, nor do they exhibit the 5’-cap structures of their eukaryotic counterparts. As such, post-transcriptional quality control processes of prokaryotic mRNA are distinct from the corresponding processes in eukaryotes. This review focuses on issues related to post-transcriptional processing, targeting, and degradation of bacterial mRNAs including those facilitated by small regulatory RNAs, with special emphasis on tmRNA and trans-translation.
2. Bacterial mRNA decay
A comparison of the stability of bacterial and eukaryotic mRNAs reveals that bacterial mRNAs have a comparatively brief existence. The half-lives of most bacterial mRNAs range from 40 seconds to 60 minutes, whereas the half-lives of some eukaryotic mRNAs can be as long as several days [1]. Variation in the stability of transcripts has an important role in the control of protein expression within the cell, as long-lived transcripts are generally subject to more rounds of translation than those with a shorter half-life. Several factors play a role in controlling the lifespan of specific mRNAs by regulating their propensity to be degraded. Although the detailed mechanism of prokaryotic mRNA decay remains to be fully elucidated, both 5’-and 3’-end dependent degradation pathways have been described [2]. The rate of mRNA turnover, the nucleases involved, and the directionality of degradation seem all to depend on the bacterial species, the rate of translation of the mRNA, as well as sequence and structural elements present within the transcript [3–6].
2.1. 5’-terminus dependant endonucleases
One general pathway for prokaryotic mRNA decay, using E. coli as a model system, has been well characterized. It begins with an endonucleolytic cleavage at one or more defined sites within the transcript in a 5’-end dependent manner. The 5’-terminus of the mRNA appears to be of particular importance in initiating the decay process. Yet, E. coli lacks any processive 5’-3’-exoribonucleases [7] – although a 5’-3’-exoribonuclease activity has been reported in Bacillus subtilis [4]. The most commonly cited experiments that demonstrate the importance of the 5’-end in E. coli mRNA decay are those concerning the ompA transcript. The ompA transcript has a long half-life (for an E. coli mRNA) of 15–20 minutes [8] that is dependent on its 133-nt 5’-untranslated region (5’-UTR). Fusion of the ompA 5’-UTR sequence to the 5’-end of a heterologous mRNA lengthens the half-life of the mRNA [9, 10]. Further investigation of this effect has revealed that a stem-loop structure at the 5’-end of the mRNA, a high degree of ribosomal occupancy near the initiation codon, and the presence of ribosomes translating the protein coding region are all factors that affect the rate of endonucleolytic cleavage of the mRNA, slowing its rate of decay [8]. Interestingly, addition of a region of unpaired, single stranded nucleotides prior to the stem-loop structure reverses the mRNA stabilization effect of the ompA 5′-UTR [11]. Based on these and related studies a model has been proposed whereby 5’-end dependant decay of primary transcripts in E. coli is initiated by an endoribonuclease that requires a single stranded 5’-landing site to gain access to internal cleavage sites.
Of the known E. coli endoribonucleases, RNase E is thought to be the principle nuclease responsible for this cleavage [12]. In the absence of RNase E activity there is stabilization of a wide range of mRNAs [13–15], which supports the idea that RNase E is the main endoribonuclease in this process. However, other endonucleases also participate in initiating mRNA decay. The endonucleases that have been implicated in these alternate pathways are perhaps more specialized and include RNase III, which cleaves double stranded RNAs [16], RNase G, which has similar specificity to RNase E but is responsible for far fewer cleavages in vivo [17, 18], and RNase Z, which might have a more prominent role in organisms that lack RNase E function [19]. Although RNase E can act alone, it is part of a multiprotein complex called the degradosome and is thought to function in cooperation with these factors (see [20] and references therein). In addition to RNase E, the degradosome also contains polynucleotide phosphorylase (PNPase), an ATP-dependent helicase (RhlB) and enolase [20].
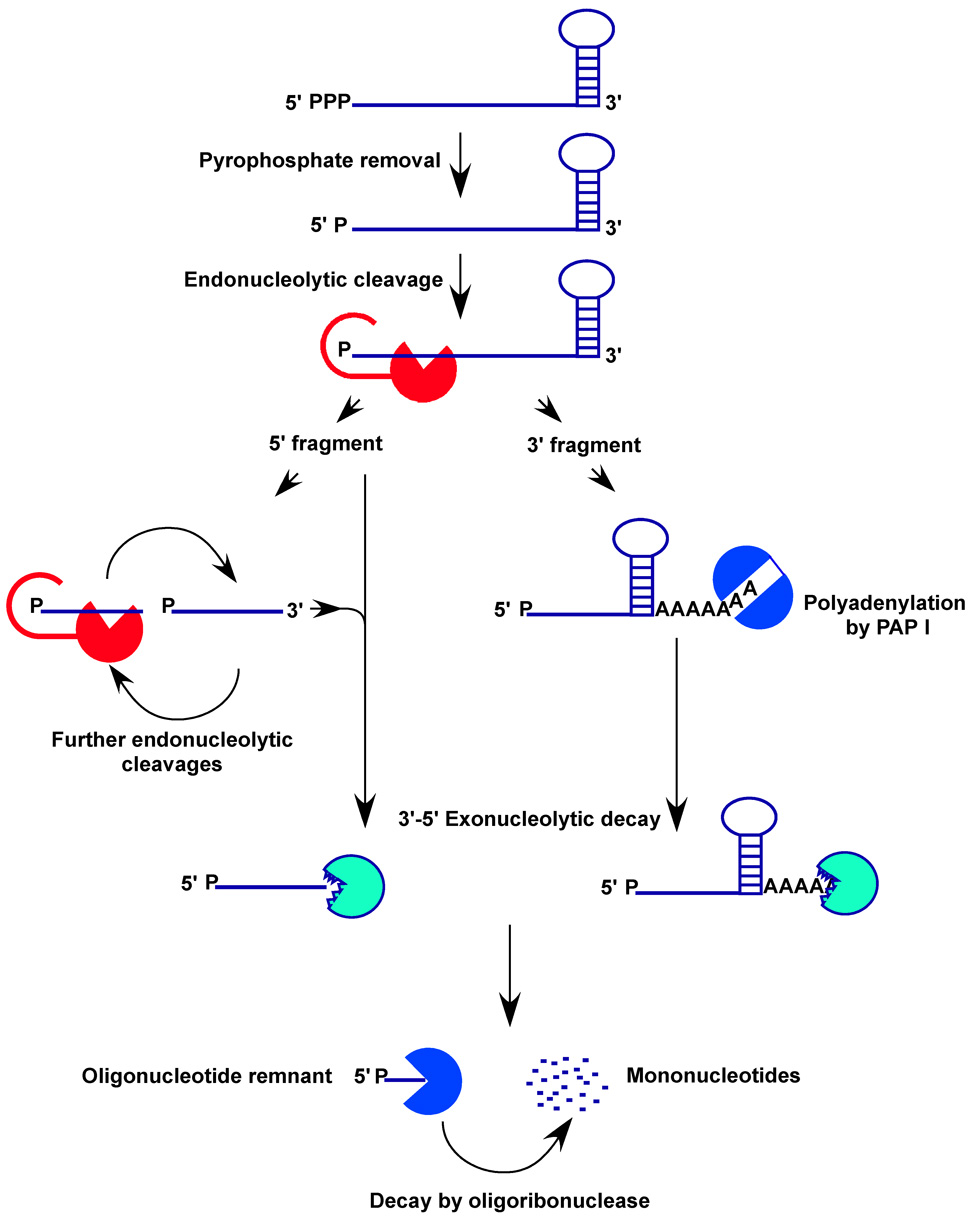
From a mechanistic perspective, RNase E requires 5’ single-stranded RNA of at least four nucleotides in length for efficient binding and degradation. RNase E also prefers 5’-monophosphorylated substrates for full activity [21], as natural substrates with terminal 5’-triphosphates are poorly cleaved [22]. The slow decay rate of 5’-triphosphorylated substrates led to the question of how mRNA decay is initiated by RNase E if its preferred substrates are monophosphorylated mRNA transcripts. Recent work has shown that there may be a rate limiting conversion prior to RNase E endonucleolytic cleavage, whereby the triphosphorylated 5’-terminus is converted to a monophosphorylated form [23]. It was recently demonstrated that the RppH protein, a pyrophosphohydrolase enzyme, catalyzes the 5’-pyrophosphate removal reaction [24]. In light of these findings, mRNA decay is initiated by a 5’-pyrophosphate removal step allowing an initial, internal endonucleolytic cleavage by RNase E – provided the 5’-terminus is available and not sequestered in a hairpin structure. This first internal cleavage then presents the substrates for a series of endo- and exo-nucleolytic reactions, rapidly reducing the mRNA to mononucleotides (Figure 1). The preferential decay of monophosphorylated substrates explains the observation that complete mRNA decay in E. coli occurs very rapidly after an initial 5’-end dependant endonucleolytic cleavage event. Rapid decay occurs due to the monophosphorylated 5’-end of the 3’-cleavage fragments, making these RNAs ideal substrates for RNase E. Furthermore, RNase E and the 3′-5′ exonucleases act cooperatively to rapidly degrade the cleaved transcript to mononucleotides. Furthermore, the cleavage dependent loss of the 5’-UTR and ribosome-binding site inhibits translation initiation and removes the shielding effect of translating ribosomes on the mRNA.
Figure 1. Schematic representation of a major pathway for mRNA decay in E. coli.
A typical primary transcript possesses a single stranded, triphosphorylated 5’-terminus and a 3’-end with a stem-loop structure. Decay is initiated by the RppH-dependent pyrophosphate removal step at the 5’-terminus. Internal, endonucleolytic cleavages are performed by RNase E, which requires the monophosphorylated 5’-end for catalytic activity. The monophosphorylated 5’-fragment is then subject to further endonucleolytic cleavages or 3’-5’ exonucleolytic decay by exoribonucleases such as RNase II, RNase R, and PNPase. Fragments that contain a 3’ stem-loop structure are polyadenylated by poly (A) polymerase (PAP I), allowing 3’-5’ exonucleolytic decay to be initiated by PNPase or RNase R. The final oligoribonucleotide product of this process is degraded to individual nucleotides by oligoribonuclease.
2.2. 3’-terminus and 3’-to-5’ exonucleases
mRNA decay in E. coli comprises a number of parallel pathways utilizing endoribonucleases and exoribonucleases, both on their own and in combination. Of the exoribonucleases present in E. coli, two enzymes, RNase II and PNPase, are thought to perform the bulk of 3′-to-5′ exoribonucleolytic decay. Both enzymes degrade the mRNA from the 3’-terminus, one nucleotide at a time, resulting in individual mononucleotides and a 5’-end oligonucleotide remnant of 2 to 5 nucleotides. The short oligonucleotide products of RNase II and PNPase action are converted to mono-nucleotides through the activity of oligoribonuclease (Fig. 1 and [25]). There are differing reports of the share of exonucleolytic decay performed by RNase II and PNPase. Recent studies suggest that although both RNases play prominent roles in mRNA degradation, PNPase may be a more significant contributor to the decay process [26, 27]. The ability to deal with RNA secondary structures is a known difference between RNase II and PNPase. RNase II can efficiently degrade single-stranded RNA but is incapable of unwinding secondary structures. PNPase is known to favor single stranded substrates and pause at stem-loop structures. Both enzymes are unable to bind substrates with fewer than 6–10 unpaired bases at the 3’-end [28]. PNPase, however, has been shown to have some ability to degrade secondary structural elements [29]. PNPase degradation of structured mRNA elements is thought to be aided via polyadenylation by poly (A) polymerase I (PAP I). PNPase might also require assistance from the RhlB helicase for particularly stable structures.
Polyadenylation of the 3’-end of a bacterial RNA, in contrast to eukaryotic mRNAs, has been shown to have a role in facilitating decay. Recent evidence suggests that endonucleolytic cleavage by RNase E provides the signals required for polyadenylation of the resultant fragments. This might be facilitated by direct targeting of PAP I to these fragments through its interaction with the RNase E degradosome [30]. It is suggested that addition of a single stranded 3’- poly-A tail to an mRNA promotes binding of PNPase. Upon binding to the poly-A tail, PNPase may then degrade the RNA until it reaches the structured region, whereupon it stalls. Stalled PNPase can then resume through the structured region if there is a temporary unwinding of the stem structure, possibly facilitated by the action of the RhlB helicase. Recent studies suggest that polyadenylation plays a much more pivotal role in bacterial RNA quality control than previously anticipated. The significance and extent of polyadenylation was highlighted by the finding that >90 % of transcripts in E. coli are modified by PAP I at some stage of their life cycle [31].
2.3. Role of stem-loop structures in regulating mRNA decay rates
The presence of a stem-loop in an mRNA can perform a regulatory function that influences the expression of particular genes by lengthening the half-lives of their transcripts. Bacterial mRNAs frequently possess a stem loop structure at their 3’-end. The slow rate at which 3’-5’ exoribonucleases can degrade such structures provides some degree of protection from decay. Indeed, the absence of a 3’-end structure, which can be brought about by an endonucleolytic cleavage, results in very rapid RNA decay [1]. Differential expression of ORFs present on polycistronic transcripts is also achieved via the inability of bacterial exoribonucleases to degrade highly structured Repeated Extragenic Palindromic (REP) elements. The presence of REP elements in a polycistronic message can protect upstream sequences from decay, lengthening their lifetimes relative to other downstream sequences [1].
3’-5’ exonucleolytic decay proceeds up to a stem loop, leaving shorter decay intermediates with a stem loop at the 3’-end. The degradation of these small intermediates is a critical step in the decay pathway. Recent work suggests that the decay of these REP stabilizers involves not just PAP I and PNPase, but also RhlB and the degradosome [32]. In particular, RhlB and RNase E, both components of the degradosome, have been shown to be necessary for REP-stabilizer degradation. One interesting question is how the bacterial mRNA decay machinery differentiates between primary transcripts and endonucleolytic cleavage products. A stem-loop at the 3’-end of a primary transcript protects it from exonucleolytic decay, yet the 3’-end of a 5’-fragment generated by endonucleolytic cleavage is more rapidly degraded. This degradation not only occurs through single stranded regions but also through structured regions, via the combined actions of RhlB, poly (A) polymerase and PNPase. In addition, studies have established that poly (A) polymerase activity in degradation of stem-loop regions is initiated by an RNase E-mediated cleavage event [33].
Recently an additional 3’-5’ exonuclease, RNase R, has been implicated in the decay of REP-stabilizers. RNase R is a hydrolytic 3’-5’ exonuclease with considerable sequence homology to RNase II. Yet unlike RNase II it has the ability to degrade through double stranded regions of RNA, including REP-stabilizers [34]. As with RNase II and PNPase, RNase R requires a single stranded 3’-end to bind and initiate decay. However, upon binding and initiation of exonucleolytic decay, RNase R proceeds much more efficiently through secondary structural elements within the RNA. Although RNase R is widely conserved in eubacteria and may conceivably play a critical role in RNA decay, little is known about its mode of action. Intriguingly, RNase R is induced under stress conditions in E. coli [35] and is associated with the degradosome under cold shock conditions in Pseudomonas syringae [36], indicating a role for the enzyme in mRNA decay under stress conditions. Consistent with this conclusion, Purusharth and colleagues recently reported that RNase R is essential for growth of Pseudomonas syringae at low temperature, and that under these conditions rnr mutants accumulate aberrant 5S and 16S rRNA variants exhibiting 3’-end processing defects [37]. These results suggest that RNase R may have a specialized role in 3’-5’ exonucleolytic decay that neither RNase II nor PNPase can fulfill.
3. Small non-coding RNA regulators in mRNA decay
Since the discovery of the phenomenon of RNA silencing in eukaryotes, small non-protein coding RNA (sRNA) regulators have become the subject of an explosively growing field of investigation. In comparison to eukaryotic small RNAs, our knowledge of similar types of regulators in prokaryotes is more limited. Nevertheless, sRNA regulators of mRNA stability and expression are present in prokaryotes and participate in a variety of key cellular functions including: adaptation to environmental and nutritional stresses, quorum sensing, virulence factor production, and plasmid maintenance [38–40].
A large number of sRNAs act by directly base pairing, albeit imperfectly, with short sequences in the 5’-UTR of target mRNAs. The known outcomes of such pairing include both repression of translation, coupled with rapid degradation of the target mRNA, as well as activation of translation, coupled with stabilization of the target mRNA. In most known cases of repression, sRNAs prevent initiation of translation by binding in the vicinity of the ribosome-binding site of the target mRNAs and blocking ribosomal access and translation initiation (Fig. 2). For instance, the well-characterized E. coli sRNAs OxyS, MicA, and MicC have been shown to directly interfere with 30S ribosome binding of target mRNAs [41, 42]. RyhB, the sRNA regulator of iron metabolism, which targets transcripts encoding various iron-binding proteins, is thought to function in this manner [40]. RyhB is induced under iron-depleted conditions, binds in the vicinity of the ribosome-binding site of its target mRNA and facilitates its selective degradation [43, 44]. It is likely that other known E. coli sRNAs, such as DsrA, MicF, SgrS, and Spot42, use a similar mechanism to affect the translation and decay of their target transcripts [45–47].
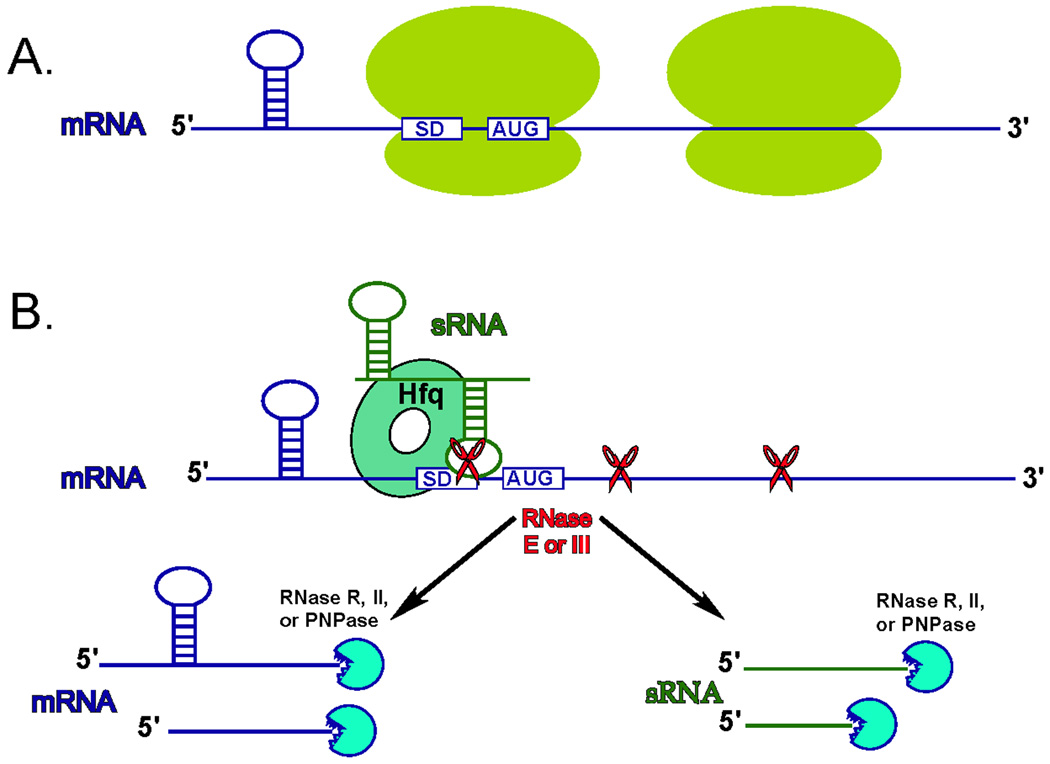
Figure 2. sRNA-mediated translation repression and mRNA decay.
A. In the absence of sRNA, translating ribosomes protect mRNA from endonucleolytic degradation by blocking endonuclease sensitive sites. B. sRNA regulator is expressed and bound by Hfq, or another chaperone protein, that stabilizes it and facilitates its binding to the vicinity of the Shine-Delgarno (SD) element and initiation codon of the target mRNA. sRNA binding leads to cleavage of both the mRNA and sRNA by RNase III and/or RNase E. The blocking of translation might also exposes cryptic RNase E- sensitive sites within the mRNA. Initial endonucleolytic cleavage generates 5’-monophospate ends that stimulate further cleavage by RNase E, followed by 3’ to 5’ exonucleolytic digestion of both mRNA and sRNA remnants.
Curiously, the stability and target binding activities of RyhB, and many other sRNAs, are in large part dependent on the presence of a ubiquitous and highly abundant small RNA chaperone Hfq [40, 42, 48–54]. Hfq belongs to a family of Sm-like proteins, which in eukaryotes take part in mRNA splicing. Hexameric Hfq binds with high affinity to A/U rich segments of sRNAs to promote efficient pairing with target mRNAs [49–51, 53–57]. Formation of the sRNA-mRNA complex not only inhibits translation of the target transcript, but also facilitates the degradation of both the target mRNA and sRNA [57, 58]. The degradation process is initiated by the endonucleolytic activity of either RNase III or RNase E. While RNase III can directly recognize the sRNA-mRNA duplex, it is not clear how RNase E is targeted to this complex (Fig. 2B). One possible mechanism is that the sRNA-mediated translation block unmasks RNase E sensitive sites that are usually protected by the translating ribosomes [59]. Another possible mechanism might involve changes in the mRNA secondary structure, induced by sRNA pairing, that make the target mRNA more susceptible to recognition and cleavage by RNase E. Several recent experiments suggest that RNase E is recruited to the sRNA regulatory complex via direct interaction with sRNA chaperone Hfq [48, 57, 60, 61]. Therefore, bacterial small regulatory RNAs perform an important mRNA quality control function by selectively removing transcripts whose encoded protein products are no longer needed in the cell.
4. tmRNA and translation quality control
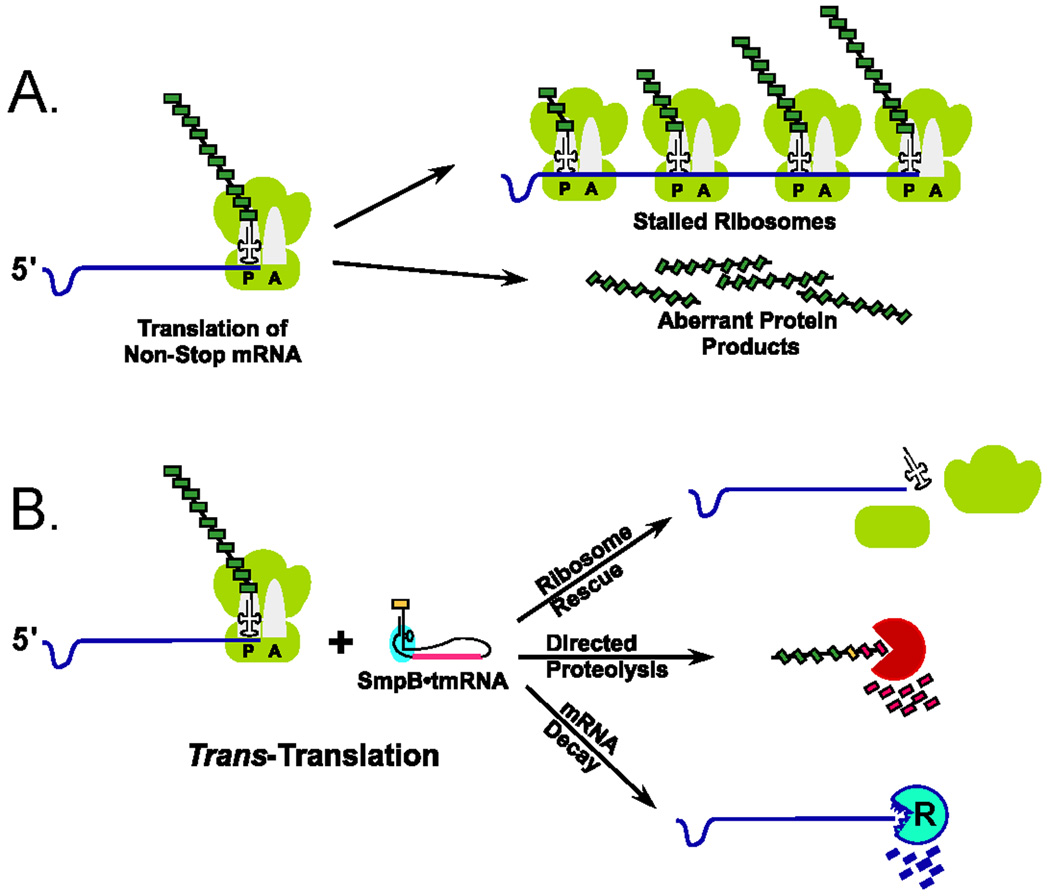
A small bacterial RNA that has received a great deal of attention is transfer messenger RNA (tmRNA). tmRNA along with its requisite protein partner Small protein B (SmpB) orchestrates an elegant translational control process termed trans-translation [62–71] Gene mutation, DNA damage, mRNA damage, and translational errors may all lead to ribosomes reaching the 3’-end of an mRNA without encountering an in-frame termination codon. This event could have two potentially hazardous consequences for the bacterium (Fig. 3A). First, since an in-frame stop codon is required to recruit the translation termination apparatus, mRNAs lacking in-frame stop codons lead to ribosome stalling and significant loss of translational efficiency. Secondly, aberrant protein products translated from incomplete mRNAs may be harmful to cells. The SmpB•tmRNA quality control system solves both problems by recognizing and rescuing stalled ribosomes, and directing the addition of a C-terminal proteolysis tag to incomplete protein products [64, 66, 71–79] (Fig. 3B). In addition, the SmpB•tmRNA system targets the aberrant mRNA for degradation, thus preventing future ribosome stalling events [62, 67, 69].
Figure 3. The consequences of translation of a non-stop mRNA, and how they are alleviated by trans-translation.
A. Translation of non-stop mRNAs leads to ribosome stalling, and as mRNAs are translated by poly-ribosomes, a single stalling event can sequester many ribosomes. Furthermore, stalling could lead to the release of aberrant protein products that may be harmful to the bacterium. B. The SmpB•tmRNA-mediated trans-translation process rescues stalled ribosomes and directs the degradation of the associated incomplete protein products. The SmpB•tmRNA system also facilitates selective decay of the causative non-stop mRNA, preventing future cycles of futile translation and ribosome stalling events.
4.1. tmRNA and SmpB
tmRNA is a unique bi-functional RNA molecule that exhibits features and activities similar to both tRNA and mRNA. The 5’-and 3’-ends fold to form a tRNA-like domain with sequence and structural similarity to tRNAAla [80–84]. The tRNA-like domain of tmRNA possesses an amino acid acceptor stem, a TΦC arm and a D-loop, but lacks an anticodon arm. Like tRNAAla, tmRNA can be charged with alanine through the action of alanyl-tRNA synthetase (Ala-RS) [84, 85]. tmRNA exhibits additional secondary structural elements in the form of four RNA pseudoknots (pk1-pk4). The specific functions of the tmRNA pseudoknots remain unclear, however several studies suggest that pk1 is important for tmRNA structure and possibly function [85–90]. Between pseudoknots 1 and 2 in the tmRNA sequence lies the mRNA-like open reading frame (ORF), which codes for a proteolytic degradation tag (ANDENYALAA in E. coli) followed by tandom UAA termination codons.
The essential protein partner of tmRNA, SmpB, is a small basic RNA binding protein that binds tmRNA with high affinity and specificity [73, 91]. SmpB is required for stable interactions of tmRNA with stalled ribosomes [73, 92]. The structure of SmpB protein has been solved by both NMR and X-ray crystallography [75, 93–95]. It has an antiparallel β-barrell type structure with an embedded oligonucleotide binding (OB) fold [75, 93–95]. The tmRNA binding residues of SmpB appear to be clustered on a unique surface of the protein [75, 91, 93]. Alterations that affect this surface of the protein abolish tmRNA binding acitivity both in vivo and in vitro [91]. SmpB also possesses a C-terminal tail that is unstructured in solution. However, the C-terminal tail is functionally indespensible for trans-translation and might gain structure upon ribosome binding [96, 97]. The C-terminal tail of SmpB seems to be involved in facilitating the tRNA-like activity of tmRNA, perhaps acting as an anti-codon arm mimic [62, 97]. SmpB variants that lack the C-terminal tail, or harboring specific mutations within this region, do not support trans-translation – despite being fully capable of binding tmRNA and delivering it to stalled ribosomes [97].
4.2. The mechanism of trans-translation
Trans-translation begins with recognition of stalled ribosomes by SmpB and tmRNA. The tRNA-like domain of tmRNA is charged with alanine by Ala-RS. The GTP bound form of elongation factor Tu (EF-Tu-GTP) recognizes alanylated-tmRNA, and a quaternary complex of SmpB•tmRNA•EF-Tu(GTP) binds in the A-site of stalled ribosomes. GTP is hydrolyzed, EF-Tu(GDP) is released, and the tRNA-like domain of tmRNA is accommodated into the ribosomal A-site. The growing polypeptide is then transferred onto the tRNA-like domain of tmRNA. Next, the stalled ribosome is promoted to disengage from the defecive mRNA and engage the mRNA-like sequence of tmRNA as a surrogate template. The tmRNA encoded ORF is translated normally until the ribosome reaches a termination codon at the end of the tmRNA reading frame. This allows for rapid translation termination, protein release, and ribosome recycling. The nascent polypeptide, now carrying an 11 amino acid C-terminal degradation tag, is efficiently recognized and degraded by cellular proteases. Several cellular proteases, including ClpXP, ClpAP, Lon, FtsH, and Tsp have been shown to be involved in degradation of tmRNA tagged proteins [63, 66, 98–100]. The SmpB•tmRNA system also facilitates the selective degradation of the aberrant mRNA by RNase R [67, 69, 101, 102] (see below).
4.3. SmpB•tmRNA substrates
What differentiates a stalled ribosome, making it a substrate for SmpB•tmRNA binding, remains an open question. A variety of situations which delay the progress of protein synthesis have been shown to elicit tagging by the SmpB•tmRNA system. Tagging can occur at the end of a non-stop mRNA, but also at internal positions on the mRNA [66, 103–105]. However, it is likely that in cases of internal tagging, a co-translational mRNA cleavage event leads to the generation of a non-stop mRNA [101, 106–109]. As such, one possible mechanism for the differentiation between normal and stalled ribosomes is the presence of mRNA sequence within or 3’-to the ribosomal A-site. Tagging has also been reported at positions corresponding to weak termination codons, presumably due to slow rates of release factor binding to the A-site, as compared to the rate of cognate tRNA A-site entry [105, 109–111]. Indeed, depletion of the cognate release factor has been shown to enhance the rate of tagging at a specific termination codon [112, 113]. In addition, tRNA scarcity can lead to SmpB•tmRNA-mediated tagging [67, 69, 104, 108]. This observation is highlighted by the discovery that overexpression of an mRNA containing tandem rare arginine codons leads to tagging, an effect that is rescued by driving increased expression of the cognate tRNA. Tagging has also been observed in response to programmed ribosome stalling events such as translation of the secM arrest sequence [114, 115]. Our current understanding is that various stress conditions could lead to unproductively stalled ribosomes. Such stalled ribosomes are not immediately a substrate for the SmpB•tmRNA mediated trans-translation process. They become a substrate only after an as yet unknown endonuclease cleaves the causitive mRNA within the A-site of the ribosme, freeing the A-site decodong center for efficient SmpB•tmRNA binding to commence trans-translation [62, 69, 107, 113].
4.4. Decay of defective mRNAs
In addition to the ribosome rescue and directed proteolysis activities in trans-translation, recent work has shown that tmRNA promotes the decay of mRNA transcripts that lead to ribosome stalling. Yamamoto et al. [102] were the first to report a role of tmRNA in mRNA decay. They utilized a non-stop crp mRNA reporter gene that was shown to engage the trans-translation machinery. They found that this non-stop transcript was rapidly degraded in cells containing tmRNA, while in cells lacking tmRNA it accumulated to higher steady state levels. This finding supported a role for tmRNA in facilitating the degradation of non-stop mRNAs.
Work in our laboratory has confirmed the role of trans-translation in promoting the degradation of faulty mRNAs that cause ribosome stalling [67, 69]. Interestingly, we have also found that specific sequence elements within tmRNA are required to facilitate the decay of non-stop transcripts engaged in trans-translation. Mehta et al. [67] demonstrated that certain nucleotides within the last three codons of the tmRNA encoded reading frame are required for efficient and selective decay of defective mRNAs. tmRNA variants carrying substitutions of these nucleotides that alter neither the secondary structure of the mRNA-like domain nor the amino acid sequence of the proteolysis tag were found to be defective in facilitating mRNA decay. Importantly, these mRNA decay-defective tmRNA variants showed similar expression levels and stability to wild type tmRNA. In addition, these tmRNA variants were fully competent in supporting the ribosome rescue and reporter protein tagging functions of tmRNA.
Furthermore, we have found that tmRNA-mediated mRNA decay requires the processive 3’-5’ exoribonuclease RNase R. RNase R has previously been shown to associate with a ribonucleoprotein complex consisting of ribosomal protein S1, phosphoribosyl pyrophosphate synthase, tmRNA and SmpB [116]. To test the putative involvement of processive 3’-5’ exoribonucleases, we made use of in vivo mRNA decay assays involving both non-stop and rare codon containing reporter mRNAs. We showed that RNase R, tmRNA and SmpB were all required for rapid decay of both non-stop and rare codon containing mRNAs [69]. In contrast, the steady state level of a related normal mRNA reporter (with a stop codon and no rare codons) was not effected by the presence or absence of tmRNA or RNase R [69]. These results suggested that a functional trans-translation system and RNase R are both required for the selective disposal of aberrant mRNAs that promote ribosome stalling. Three possible mechanisms were proposed for tmRNA-mediated decay of defective mRNAs. The first is that prior to disengagement of the mRNA from the rescued ribosome, interactions of certain nucleotides in the stop codon proximal segment of the tmRNA ORF with ribosomal elements promote binding, rearrangement or loading of RNase R onto the defective mRNA. The second possible mechanism involves the tmRNA ORF region interacting with other elements of tmRNA or other tmRNA-associated factors that then facilitate decay of the faulty mRNA by RNase R. The third explanation is that this region of the tmRNA reading frame interacts directly with RNase R to bring it to the 3’-terminus of the faulty mRNA and thus promote exonucleolytic decay of the transcript.
Several additional trans-translation related roles of RNase R have been reported. Cairrao et al. [117] have indicated that RNase R is required for the processing of the pre-tmRNA transcript to generate functional tmRNA under cold shock conditions. Furthermore, Hong et al. [118] analyzed degradation of a two piece Caulobacter crescentus tmRNA and have suggested that RNase R is required for the cell cycle dependent degradation of tmRNA. SmpB is thought to protect tmRNA from degradation by RNase R. As the levels of SmpB protein decrease at specific stages of Caulobacter crescentus cell cycle, RNase R gains access to the SmpB-protected 3’-ends of this two piece tmRNA to enact its degradation [118]. Therefore, the interplay between SmpB and RNase R is thought to regulate the cell cycle dependent stability and activity of Caulobacter crescentus tmRNA.
4.5. Physiological significance of the SmpB•tmRNA system
The genes encoding SmpB protein and tmRNA are represented in all sequenced bacterial genomes. The ssrA gene (encoding tmRNA) is not essential under ideal growth conditions in E. coli. ssrA deficient strains, however, exhibit slower growth at high temperature, slow recovery from carbon starvation, and reduced motility [84, 119]. Similarly, ssrA is not required for normal growth of Bacillus subtilis, however smpB and ssrA deletions lead to reduced growth rate at both low and high temperatures [120]. The SmpB•tmRNA translational quality control system is important for survival and pathogenesis of some bacterial species. Genome wide mutagenesis results suggest that the ssrA and smpB genes are essential in both Mycoplasma genitalium and Mycoplasma pneumoniae [121]. The ssrA gene is also thought to be essential for survival in Neisseria gonorrhoeae [122]. Although non-essential, smpB and ssrA are important for virulence in Salmonella enterica [123, 124], and Yersinia pseudotuberculosis [124]. Interestingly, in Yersinia pseudotuberculosis, loss of SmpB and tmRNA leads to severe defects in the expression and delivery of virulence effector proteins [124]. It is likely that this effect contributes to the loss of virulence observed in these mutants. Hence, the SmpB•tmRNA system is a prokaryotic-specific quality control mechanism that is important for survival and virulence of some pathogenic bacteria.
SmpB•tmRNA mediated tagging is also thought to serve as a mechanism for translational control of gene expression under normal growth conditions. A number of regulatory proteins have been shown to be tagged by the SmpB•tmRNA system, for example YbeL, GalE, RbsK, and the LacI repressor are all substrates of the SmpB•tmRNA system [105, 125]. In the case of LacI, it is thought that tagging of the repressor serves as a negative feedback mechanism to maintain optimal repressor concentrations. Consistent with this hypothesis, ssrA− cells exhibit a delay in induction of the lac operon [105, 125]. More recently, tmRNA has been implicated in the maintenance of cellular concentration of the E. coli stress sigma factor RpoS [126]. RpoS is upregulated in response to various cellular stresses. The stationary phase upregulation of RpoS is substantially reduced in the absence of tmRNA, and the effect appears to be at the level of RpoS translation [126].
4.6. Comparison of prokaryotic and eukaryotic translation surveillance mechanisms
Unproductive ribosome stalling is a very basic cellular event with a variety of causes and consequences. However, prokaryotic and eukaryotic cells deal with the phenomenon in distinct ways. Bacterial trans-translation, in comparison to eukaryotic mechanisms, is a comparitivly robust response to ribosome stalling, as non-stop mRNAs, rare codon containing mRNAs, as well as mRNAs with specific stall sequences are all targets for the SmpB•tmRNA system [66, 103, 104, 115]. In contrast, eukaryotic cells exhibit at least two distinct mechanisms for preventing ribosome stalling. The eukaryotic non-stop mRNA decay (NSD) pathway targets transcripts lacking in-frame termination codons [127–132], while the no-go decay (NGD) pathway deals with mRNAs containing internal stall sites (i.e., mRNAs with secondary structural elements such as stem loops and pseudoknots) [133]. NSD involves exosome mediated 3′-5′ mRNA decay, mediated by the C-terminal domain of Ski7p [127, 131] – although a 5′-3′ non-stop mRNA decay activity has been reported [128]. No-go decay is initiated by an mRNA cleavage event performed by an as yet unidentified nuclease and involves two proteins, Dom34p and Hbs1p, which exhibit similarity to the eukaryotic release factor eRF-1 [133]. Translation inhibition as well as nascent protein destabilization are thought to limit the accumulation of protein products translated from non-stop mRNAs [128–130, 132], but the fate of nascent polypeptides is unclear in no-go decay. It has been convincingly demonstrated that non-stop mRNAs lead to translational repression, as non-stop mRNA reporters accumulate in polysome fractions [128]. However, the mechanism of ribosome rescue and restoration of translational efficiency is unclear. Similarly, secondary structure elements present in mRNAs that elicit no-go decay must result in sequestration of ribosomes and it is, as yet, unclear how this unproductive stalling is alleviated.
Trans-translation is an elegant, simple and comprehensive mechanism for dealing with unproductive ribosome stalling. It can efficiently deal with different types of stalling events, including non-stop mRNAs, multiple rare codons, and engineered stall sequences. In doing this, the SmpB•tmRNA system promotes ribosome rescue, directs the degradation of aberrant protein products, and facilitates the decay of the causative non-stop mRNA (Fig. 3B). Considering the versatility and efficiency of this system, it seems surprising that such a complete mechanism for dealing with a host of universal cellular problems has been lost in eukaryotes. One possible explanation is that eukaryotes have evolved more efficient mechanisms for the individual activities performed by the SmpB•tmRNA system. Another possibility is that a trans-translation-like mechanism would interfere with productive ribosome stalling events. For instance, SmpB•tmRNA might interfere with translational control processes such as programmed ribosomal frameshifting or delayed translation to facilitate proper co-translational folding of nascent proteins. More details about the way eukaryotic cells deal with unproductive ribosome stalling events may help shed light on this issue.
5. Conclusions and Perspectives
The unique properties of bacterial mRNAs necessitate distinct prokaryotic mRNA decay and quality control mechanisms. We have discussed progress in the complex field of bacterial mRNA decay, and have shown that controlling the rate of transcript degradation represents a mechanism for control of gene expression. Bacterial mRNA decay involves 5’- and 3’-end dependent mechanisms facilitated by several exo- and endo-ribonucleases. Sequence and structural elements within the transcript regulate both its rate of decay and the primary nucleases involved. We also discussed the role of small non-coding RNAs in post-transcriptional gene regulation and described models for how these regulatory RNAs contribute to mRNA quality control and stability. Finally, we discussed the SmpB•tmRNA-mediated mechanism for translation quality control and selective mRNA decay. These mechanisms contribute both to providing additional routes for post-transcriptional control of gene expression and relieving cellular stresses caused by aberrant transcripts.
Although there has been rapid progress in our understanding of the role of ribonucleases in post-transcriptional control of mRNA stability and decay, clearly a tremendous amount is yet to be learned. It is also abundantly clear that a thorough understanding of the post-transcriptional regulation of gene expression will require studies not only in E. coli but also in other related and distant bacterial species. The recent discovery of 5’-to-3’ exonucleases in Bacillus subtilis, the RNase activity and the substrate range of the toxin–antitoxin systems, and the ribozyme-like activity of certain riboswitches highlight only a few examples of other novel mechanism that remain to be elucidated.
Acknowledgements
We thank members of the Karzai lab for helpful discussions and suggestions. We are grateful to Dr. Jorge L. Benach and members of The Center for Infectious Diseases for their continued support. We apologize to those colleagues whose work could not be cited due to space and citation number constraints. This work was supported in part by Grants (to AWK) from The National Institute of Health (GM65319, and AI055621), and The Pew Scholars Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Belasco JG, Brawerman G. Control of messenger RNA stability. San Diego: Academic Press; 1993. [Google Scholar]
- 2.Curtiss R, Neidhardt FC. Escherichia coli and Salmonella : cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. [Google Scholar]
- 3.Kushner SR. mRNA decay in prokaryotes and eukaryotes: different approaches to a similar problem. IUBMB Life. 2004;56:585–594. doi: 10.1080/15216540400022441. [DOI] [PubMed] [Google Scholar]
- 4.Mathy N, Benard L, Pellegrini O, Daou R, Wen T, Condon C. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- 5.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deutscher MP. Ribonuclease multiplicity, diversity, and complexity. J Biol Chem. 1993;268:13011–13014. [PubMed] [Google Scholar]
- 8.Arnold TE, Yu J, Belasco JG. mRNA stabilization by the ompA 5′ untranslated region: two protective elements hinder distinct pathways for mRNA degradation. RNA. 1998;4:319–330. [PMC free article] [PubMed] [Google Scholar]
- 9.Belasco JG, Nilsson G, von Gabain A, Cohen SN. The stability of E. coli gene transcripts is dependent on determinants localized to specific mRNA segments. Cell. 1986;46:245–251. doi: 10.1016/0092-8674(86)90741-5. [DOI] [PubMed] [Google Scholar]
- 10.Hansen MJ, Chen LH, Fejzo ML, Belasco JG. The ompA 5′ untranslated region impedes a major pathway for mRNA degradation in Escherichia coli. Mol Microbiol. 1994;12:707–716. doi: 10.1111/j.1365-2958.1994.tb01058.x. [DOI] [PubMed] [Google Scholar]
- 11.Emory SA, Bouvet P, Belasco JG. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992;6:135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- 12.Kushner SR. mRNA decay in Escherichia coli comes of age. J Bacteriol. 2002;184:4658–4665. doi: 10.1128/JB.184.17.4658-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babitzke P, Kushner SR. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc Natl Acad Sci U S A. 1991;88:1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mudd EA, Krisch HM, Higgins CF. RNase E, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: evidence that rne and ams are the same genetic locus. Mol Microbiol. 1990;4:2127–2135. doi: 10.1111/j.1365-2958.1990.tb00574.x. [DOI] [PubMed] [Google Scholar]
- 15.Taraseviciene L, Miczak A, Apirion D. The gene specifying RNase E (rne) and a gene affecting mRNA stability (ams) are the same gene. Mol Microbiol. 1991;5:851–855. doi: 10.1111/j.1365-2958.1991.tb00758.x. [DOI] [PubMed] [Google Scholar]
- 16.Schmeissner U, McKenney K, Rosenberg M, Court D. Removal of a terminator structure by RNA processing regulates int gene expression. J Mol Biol. 1984;176:39–53. doi: 10.1016/0022-2836(84)90381-4. [DOI] [PubMed] [Google Scholar]
- 17.Deana A, Belasco JG. The function of RNase G in Escherichia coli is constrained by its amino and carboxyl termini. Mol Microbiol. 2004;51:1205–1217. doi: 10.1046/j.1365-2958.2003.03905.x. [DOI] [PubMed] [Google Scholar]
- 18.Ow MC, Perwez T, Kushner SR. RNase G of Escherichia coli exhibits only limited functional overlap with its essential homologue, RNase E. Mol Microbiol. 2003;49:607–622. doi: 10.1046/j.1365-2958.2003.03587.x. [DOI] [PubMed] [Google Scholar]
- 19.Perwez T, Kushner SR. RNase Z in Escherichia coli plays a significant role in mRNA decay. Mol Microbiol. 2006;60:723–737. doi: 10.1111/j.1365-2958.2006.05124.x. [DOI] [PubMed] [Google Scholar]
- 20.Carpousis AJ. The RNA Degradosome of Escherichia coli: An mRNA-Degrading Machine Assembled on RNase E. Annu Rev Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 21.Jiang X, Belasco JG. Catalytic activation of multimeric RNase E and RNase G by 5′-monophosphorylated RNA. Proc Natl Acad Sci U S A. 2004;101:9211–9216. doi: 10.1073/pnas.0401382101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackie GA. Ribonuclease E is a 5′end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- 23.Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 25.Cannistraro VJ, Kennell D. The 5′ ends of RNA oligonucleotides in Escherichia coli and mRNA degradation. Eur J Biochem. 1993;213:285–293. doi: 10.1111/j.1432-1033.1993.tb17761.x. [DOI] [PubMed] [Google Scholar]
- 26.Deutscher MP, Reuven NB. Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci U S A. 1991;88:3277–3280. doi: 10.1073/pnas.88.8.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohanty BK, Kushner SR. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol Microbiol. 2003;50:645–658. doi: 10.1046/j.1365-2958.2003.03724.x. [DOI] [PubMed] [Google Scholar]
- 28.Coburn GA, Mackie GA. Differential sensitivities of portions of the mRNA for ribosomal protein S20 to 3′-exonucleases dependent on oligoadenylation and RNA secondary structure. J Biol Chem. 1996;271:15776–15781. doi: 10.1074/jbc.271.26.15776. [DOI] [PubMed] [Google Scholar]
- 29.McLaren RS, Newbury SF, Dance GS, Causton HC, Higgins CF. mRNA degradation by processive 3′-5′ exoribonucleases in vitro and the implications for prokaryotic mRNA decay in vivo. J Mol Biol. 1991;221:81–95. [PubMed] [Google Scholar]
- 30.Mohanty BK, Kushner SR. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol Microbiol. 1999;34:1094–1108. doi: 10.1046/j.1365-2958.1999.01673.x. [DOI] [PubMed] [Google Scholar]
- 31.Mohanty BK, Kushner SR. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006;34:5695–5704. doi: 10.1093/nar/gkl684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khemici V, Carpousis AJ. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol Microbiol. 2004;51:777–790. doi: 10.1046/j.1365-2958.2003.03862.x. [DOI] [PubMed] [Google Scholar]
- 33.Joanny G, Le Derout J, Brechemier-Baey D, Labas V, Vinh J, Regnier P, Hajnsdorf E. Polyadenylation of a functional mRNA controls gene expression in Escherichia coli. Nucleic Acids Res. 2007;35:2494–2502. doi: 10.1093/nar/gkm120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng ZF, Deutscher MP. An important role for RNase R in mRNA decay. Mol Cell. 2005;17:313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 35.Chen C, Deutscher MP. Elevation of RNase R in response to multiple stress conditions. J Biol Chem. 2005;280:34393–34396. doi: 10.1074/jbc.C500333200. [DOI] [PubMed] [Google Scholar]
- 36.Purusharth RI, Klein F, Sulthana S, Jager S, Jagannadham MV, Evguenieva-Hackenberg E, Ray MK, Klug G. Exoribonuclease R interacts with endoribonuclease E and an RNA helicase in the psychrotrophic bacterium Pseudomonas syringae Lz4W. J Biol Chem. 2005;280:14572–14578. doi: 10.1074/jbc.M413507200. [DOI] [PubMed] [Google Scholar]
- 37.Purusharth RI, Madhuri B, Ray MK. Exoribonuclease R in Pseudomonas syringae is essential for growth at low temperature and plays a novel role in the 3′ end processing of 16 and 5 S ribosomal RNA. J Biol Chem. 2007;282:16267–16277. doi: 10.1074/jbc.M605588200. [DOI] [PubMed] [Google Scholar]
- 38.Majdalani N, Vanderpool CK, Gottesman S. Bacterial small RNA regulators. Crit Rev Biochem Mol Biol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- 39.Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, FitzGerald DJ. Small RNA regulators and the bacterial response to stress. Cold Spring Harb Symp Quant Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Argaman L, Altuvia S. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J Mol Biol. 2000;300:1101–1112. doi: 10.1006/jmbi.2000.3942. [DOI] [PubMed] [Google Scholar]
- 42.Udekwu KI, Darfeuille F, Vogel J, Reimegard J, Holmqvist E, Wagner EG. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vecerek B, Moll I, Blasi U. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO J. 2007;26:965–975. doi: 10.1038/sj.emboj.7601553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masse E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darfeuille F, Reigadas S, Hansen JB, Orum H, Di Primo C, Toulme JJ. Aptamers targeted to an RNA hairpin show improved specificity compared to that of complementary oligonucleotides. Biochemistry. 2006;45:12076–12082. doi: 10.1021/bi0606344. [DOI] [PubMed] [Google Scholar]
- 46.Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu Rev Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- 47.Storz G, Opdyke JA, Wassarman KM. Regulating bacterial transcription with small RNAs. Cold Spring Harb Symp Quant Biol. 2006;71:269–273. doi: 10.1101/sqb.2006.71.033. [DOI] [PubMed] [Google Scholar]
- 48.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J Mol Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 52.Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Blasi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–1314. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moll I, Leitsch D, Steinhauser T, Blasi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 55.Arluison V, Folichon M, Marco S, Derreumaux P, Pellegrini O, Seguin J, Hajnsdorf E, Regnier P. The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur J Biochem. 2004;271:1258–1265. doi: 10.1111/j.1432-1033.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 56.Vecerek B, Moll I, Afonyushkin T, Kaberdin V, Blasi U. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol Microbiol. 2003;50:897–909. doi: 10.1046/j.1365-2958.2003.03727.x. [DOI] [PubMed] [Google Scholar]
- 57.Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O'Croinin T, Carroll RK, Kelly A, Dorman CJ. Roles for DNA supercoiling and the Fis protein in modulating expression of virulence genes during intracellular growth of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;62:869–882. doi: 10.1111/j.1365-2958.2006.05416.x. [DOI] [PubMed] [Google Scholar]
- 59.Afonyushkin T, Vecerek B, Moll I, Blasi U, Kaberdin VR. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005;33:1678–1689. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morita T, Mochizuki Y, Aiba H. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc Natl Acad Sci U S A. 2006;103:4858–4863. doi: 10.1073/pnas.0509638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dulebohn D, Choy J, Sundermeier T, Okan N, Karzai AW. Trans-translation: the tmRNA-mediated surveillance mechanism for ribosome rescue, directed protein degradation, and nonstop mRNA decay. Biochemistry. 2007;46:4681–4693. doi: 10.1021/bi6026055. [DOI] [PubMed] [Google Scholar]
- 63.Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haebel PW, Gutmann S, Ban N. Dial tm for rescue: tmRNA engages ribosomes stalled on defective mRNAs. Curr Opin Struct Biol. 2004;14:58–65. doi: 10.1016/j.sbi.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 65.Karimi R, Pavlov MY, Buckingham RH, Ehrenberg M. Novel roles for classical factors at the interface between translation termination and initiation. Mol Cell. 1999;3:601–609. doi: 10.1016/s1097-2765(00)80353-6. [DOI] [PubMed] [Google Scholar]
- 66.Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 67.Mehta P, Richards J, Karzai AW. tmRNA determinants required for facilitating nonstop mRNA decay. RNA. 2006;12:2187–2198. doi: 10.1261/rna.247706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moore SD, Sauer RT. The tmRNA System for Translational Surveillance and Ribosome Rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 69.Richards J, Mehta P, Karzai AW. RNase R degrades non-stop mRnas selectively in an SmpB-tmRNA-dependent manner. Mol Microbiol. 2006;62:1700–1712. doi: 10.1111/j.1365-2958.2006.05472.x. [DOI] [PubMed] [Google Scholar]
- 70.Tu GF, Reid GE, Zhang JG, Moritz RL, Simpson RJ. C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J Biol Chem. 1995;270:9322–9326. doi: 10.1074/jbc.270.16.9322. [DOI] [PubMed] [Google Scholar]
- 71.Withey JH, Friedman DI. A salvage pathway for protein structures: tmRNA and trans-translation. Annu Rev Microbiol. 2003;57:101–123. doi: 10.1146/annurev.micro.57.030502.090945. [DOI] [PubMed] [Google Scholar]
- 72.Gillet R, Felden B. Emerging views on tmRNA-mediated protein tagging and ribosome rescue. Mol Microbiol. 2001;42:879–885. doi: 10.1046/j.1365-2958.2001.02701.x. [DOI] [PubMed] [Google Scholar]
- 73.Karzai AW, Susskind MM, Sauer RT. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karzai AW, Roche ED, Sauer RT. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat Struct Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 75.Gutmann S, Haebel PW, Metzinger L, Sutter M, Felden B, Ban N. Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature. 2003;424:699–703. doi: 10.1038/nature01831. [DOI] [PubMed] [Google Scholar]
- 76.Atkins JF, Gesteland RF. A case for trans translation. Nature. 1996;379:769–771. doi: 10.1038/379769a0. [DOI] [PubMed] [Google Scholar]
- 77.Grzymski EC. tmRNA to the rescue. Nat Struct Biol. 2003;10:321. doi: 10.1038/nsb0503-321. [DOI] [PubMed] [Google Scholar]
- 78.Muto A, Ushida C, Himeno H. A bacterial RNA that functions as both a tRNA and an mRNA. Trends Biochem Sci. 1998;23:25–29. doi: 10.1016/s0968-0004(97)01159-6. [DOI] [PubMed] [Google Scholar]
- 79.Withey JH, Friedman DI. The biological roles of trans-translation. Curr Opin Microbiol. 2002;5:154–159. doi: 10.1016/s1369-5274(02)00299-0. [DOI] [PubMed] [Google Scholar]
- 80.Himeno H, Sato M, Tadaki T, Fukushima M, Ushida C, Muto A. In vitro trans translation mediated by alanine-charged 10Sa RNA. J Mol Biol. 1997;268:803–808. doi: 10.1006/jmbi.1997.1011. [DOI] [PubMed] [Google Scholar]
- 81.Himeno H, Nameki N, Tadaki T, Sato M, Hanawa K, Fukushima M, Ishii M, Ushida C, Muto A. Escherichia coli tmRNA (10Sa RNA) in trans-translation. Nucleic Acids Symp Ser. 1997:185–186. [PubMed] [Google Scholar]
- 82.Komine Y, Kitabatake M, Inokuchi H. 10Sa RNA is associated with 70S ribosome particles in Escherichia coli. J Biochem (Tokyo) 1996;119:463–467. doi: 10.1093/oxfordjournals.jbchem.a021264. [DOI] [PubMed] [Google Scholar]
- 83.Muto A, Sato M, Tadaki T, Fukushima M, Ushida C, Himeno H. Structure and function of 10Sa RNA: trans-translation system. Biochimie. 1996;78:985–991. doi: 10.1016/s0300-9084(97)86721-1. [DOI] [PubMed] [Google Scholar]
- 84.Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc Natl Acad Sci U S A. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nameki N, Tadaki T, Muto A, Himeno H. Amino acid acceptor identity switch of Escherichia coli tmRNA from alanine to histidine in vitro. J Mol Biol. 1999;289:1–7. doi: 10.1006/jmbi.1999.2754. [DOI] [PubMed] [Google Scholar]
- 86.Nameki N, Chattopadhyay P, Himeno H, Muto A, Kawai G. An NMR and mutational analysis of an RNA pseudoknot of Escherichia coli tmRNA involved in trans-translation. Nucleic Acids Res. 1999;27:3667–3675. doi: 10.1093/nar/27.18.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nameki N, Felden B, Atkins JF, Gesteland RF, Himeno H, Muto A. Functional and structural analysis of a pseudoknot upstream of the tag-encoded sequence in E. coli tmRNA. J Mol Biol. 1999;286:733–744. doi: 10.1006/jmbi.1998.2487. [DOI] [PubMed] [Google Scholar]
- 88.Nameki N, Tadaki T, Himeno H, Muto A. Three of four pseudoknots in tmRNA are interchangeable and are substitutable with single-stranded RNAs. FEBS Lett. 2000;470:345–349. doi: 10.1016/s0014-5793(00)01349-1. [DOI] [PubMed] [Google Scholar]
- 89.Nonin-Lecomte S, Felden B, Dardel F. NMR structure of the Aquifex aeolicus tmRNA pseudoknot PK1: new insights into the recoding event of the ribosomal trans-translation. Nucleic Acids Res. 2006;34:1847–1853. doi: 10.1093/nar/gkl111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tanner DR, Dewey JD, Miller MR, Buskirk AR. Genetic analysis of the structure and function of transfer messenger RNA pseudoknot 1. J Biol Chem. 2006;281:10561–10566. doi: 10.1074/jbc.M600167200. [DOI] [PubMed] [Google Scholar]
- 91.Dulebohn DP, Cho HJ, Karzai AW. Role of conserved surface amino acids in binding of SmpB protein to SsrA RNA. J Biol Chem. 2006;281:28536–28545. doi: 10.1074/jbc.M605137200. [DOI] [PubMed] [Google Scholar]
- 92.Sundermeier TR, Karzai AW. Functional SmpB-ribosome interactions require tmRNA. J Biol Chem. 2007;282:34779–34786. doi: 10.1074/jbc.M707256200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bessho Y, Shibata R, Sekine S, Murayama K, Higashijima K, Hori-Takemoto C, Shirouzu M, Kuramitsu S, Yokoyama S. Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proc Natl Acad Sci U S A. 2007;104:8293–8298. doi: 10.1073/pnas.0700402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dong G, Nowakowski J, Hoffman DW. Structure of small protein B: the protein component of the tmRNA-SmpB system for ribosome rescue. EMBO J. 2002;21:1845–1854. doi: 10.1093/emboj/21.7.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Someya T, Nameki N, Hosoi H, Suzuki S, Hatanaka H, Fujii M, Terada T, Shirouzu M, Inoue Y, Shibata T, Kuramitsu S, Yokoyama S, Kawai G. Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus. FEBS Lett. 2003;535:94–100. doi: 10.1016/s0014-5793(02)03880-2. [DOI] [PubMed] [Google Scholar]
- 96.Jacob Y, Sharkady SM, Bhardwaj K, Sanda A, Williams KP. Function of the SmpB tail in transfer-messenger RNA translation revealed by a nucleus-encoded form. J Biol Chem. 2005;280:5503–5509. doi: 10.1074/jbc.M409277200. [DOI] [PubMed] [Google Scholar]
- 97.Sundermeier TR, Dulebohn DP, Cho HJ, Karzai AW. A previously uncharacterized role for small protein B (SmpB) in transfer messenger RNA-mediated trans-translation. Proc Natl Acad Sci U S A. 2005;102:2316–2321. doi: 10.1073/pnas.0409694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Farrell CM, Grossman AD, Sauer RT. Cytoplasmic degradation of ssrA-tagged proteins. Mol Microbiol. 2005;57:1750–1761. doi: 10.1111/j.1365-2958.2005.04798.x. [DOI] [PubMed] [Google Scholar]
- 99.Herman C, Prakash S, Lu CZ, Matouschek A, Gross CA. Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol Cell. 2003;11:659–669. doi: 10.1016/s1097-2765(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 100.Herman C, Thevenet D, Bouloc P, Walker GC, D'Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH) Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sunohara T, Jojima K, Yamamoto Y, Inada T, Aiba H. Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. RNA. 2004;10:378–386. doi: 10.1261/rna.5169404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yamamoto Y, Sunohara T, Jojima K, Inada T, Aiba H. SsrA-mediated trans-translation plays a role in mRNA quality control by facilitating degradation of truncated mRNAs. RNA. 2003;9:408–418. doi: 10.1261/rna.2174803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hayes CS, Bose B, Sauer RT. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J Biol Chem. 2002;277:33825–38832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- 104.Roche ED, Sauer RT. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Roche ED, Sauer RT. Identification of endogenous SsrA-tagged proteins reveals tagging at positions corresponding to stop codons. J Biol Chem. 2001;276:28509–28515. doi: 10.1074/jbc.M103864200. [DOI] [PubMed] [Google Scholar]
- 106.Ivanova N, Pavlov MY, Felden B, Ehrenberg M. Ribosome rescue by tmRNA requires truncated mRNAs. J Mol Biol. 2004;338:33–41. doi: 10.1016/j.jmb.2004.02.043. [DOI] [PubMed] [Google Scholar]
- 107.Hayes CS, Sauer RT. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol Cell. 2003;12:903–911. doi: 10.1016/s1097-2765(03)00385-x. [DOI] [PubMed] [Google Scholar]
- 108.Li X, Hirano R, Tagami H, Aiba H. Protein tagging at rare codons is caused by tmRNA action at the 3′ end of nonstop mRNA generated in response to ribosome stalling. RNA. 2006;12:248–255. doi: 10.1261/rna.2212606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sunohara T, Jojima K, Tagami H, Inada T, Aiba H. Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J Biol Chem. 2004;279:15368–15375. doi: 10.1074/jbc.M312805200. [DOI] [PubMed] [Google Scholar]
- 110.Fujihara A, Tomatsu H, Inagaki S, Tadaki T, Ushida C, Himeno H, Muto A. Detection of tmRNA-mediated trans-translation products in Bacillus subtilis. Genes Cells. 2002;7:343–350. doi: 10.1046/j.1365-2443.2002.00523.x. [DOI] [PubMed] [Google Scholar]
- 111.Sunohara T, Abo T, Inada T, Aiba H. The C-terminal amino acid sequence of nascent peptide is a major determinant of SsrA tagging at all three stop codons. RNA. 2002;8:1416–1427. doi: 10.1017/s1355838202020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Asano K, Kurita D, Takada K, Konno T, Muto A, Himeno H. Competition between trans-translation and termination or elongation of translation. Nucleic Acids Res. 2005;33:5544–5552. doi: 10.1093/nar/gki871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li X, Yokota T, Ito K, Nakamura Y, Aiba H. Reduced action of polypeptide release factors induces mRNA cleavage and tmRNA tagging at stop codons in Escherichia coli. Mol Microbiol. 2007;63:116–126. doi: 10.1111/j.1365-2958.2006.05498.x. [DOI] [PubMed] [Google Scholar]
- 114.Garza-Sanchez F, Janssen BD, Hayes CS. Prolyl-tRNA(Pro) in the A-site of SecM-arrested ribosomes inhibits the recruitment of transfer-messenger RNA. J Biol Chem. 2006;281:34258–34268. doi: 10.1074/jbc.M608052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hayes CS, Bose B, Sauer RT. Stop codons preceded by rare arginine codons are efficient determinants of SsrA tagging in Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:3440–3445. doi: 10.1073/pnas.052707199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Karzai AW, Sauer RT. Protein factors associated with the SsrA.SmpB tagging and ribosome rescue complex. Proc Natl Acad Sci U S A. 2001;98:3040–3044. doi: 10.1073/pnas.051628298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cairrao F, Cruz A, Mori H, Arraiano CM. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol Microbiol. 2003;50:1349–1360. doi: 10.1046/j.1365-2958.2003.03766.x. [DOI] [PubMed] [Google Scholar]
- 118.Hong SJ, Tran QA, Keiler KC. Cell cycle-regulated degradation of tmRNA is controlled by RNase R and SmpB. Mol Microbiol. 2005;57:565–575. doi: 10.1111/j.1365-2958.2005.04709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Oh BK, Apirion D. 10Sa RNA, a small stable RNA of Escherichia coli, is functional. Mol Gen Genet. 1991;229:52–56. doi: 10.1007/BF00264212. [DOI] [PubMed] [Google Scholar]
- 120.Shin JH, Price CW. The SsrA-SmpB ribosome rescue system is important for growth of Bacillus subtilis at low and high temperatures. J Bacteriol. 2007;189:3729–3737. doi: 10.1128/JB.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, Venter JC. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 122.Huang C, Wolfgang MC, Withey J, Koomey M, Friedman DI. Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J. 2000;19:1098–1107. doi: 10.1093/emboj/19.5.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Baumler AJ, Kusters JG, Stojiljkovic I, Heffron F. Salmonella typhimurium loci involved in survival within macrophages. Infect Immun. 1994;62:1623–1630. doi: 10.1128/iai.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Julio SM, Heithoff DM, Mahan MJ. ssrA (tmRNA) plays a role in Salmonella enterica serovar Typhimurium pathogenesis. J Bacteriol. 2000;182:1558–1563. doi: 10.1128/jb.182.6.1558-1563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Abo T, Inada T, Ogawa K, Aiba H. SsrA-mediated tagging and proteolysis of LacI and its role in the regulation of lac operon. EMBO J. 2000;19:3762–3769. doi: 10.1093/emboj/19.14.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ranquet C, Gottesman S. Translational Regulation of the Escherichia coli Stress Factor RpoS: a Role for SsrA and Lon. J Bacteriol. 2007;189:4872–4879. doi: 10.1128/JB.01838-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 128.Inada T, Aiba H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 2005;24:1584–1595. doi: 10.1038/sj.emboj.7600636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ito-Harashima S, Kuroha K, Tatematsu T, Inada T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007;21:519–524. doi: 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Meaux S, Van Hoof A. Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. RNA. 2006;12:1323–1337. doi: 10.1261/rna.46306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 132.Wilson M, Meaux S, van Hoof A. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics. 2007 doi: 10.1534/genetics.107.073205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]