Abstract
During the adaptive immune response, antigen challenge triggers a million-fold increase in mutation rates in the variable region antibody genes. The frequency of mutation is causally and directly linked to transcription, which provides ssDNA and drives supercoiling that stabilizes secondary structures containing unpaired, intrinsically mutable bases. Simulation analysis of transcription in VH5 reveals a dominant 65 nt secondary structure in the non-transcribed strand containing six sites of mutable ssDNA that have also been identified independently in human B cell lines and in primary mouse B cells. This dominant structure inter-converts briefly with less stable structures and is formed repeatedly during transcription, due to periodic pauses and backtracking. In effect, this creates a stable yet dynamic “mutability platform” consisting of ever-changing patterns of unpaired bases that are simultaneously exposed and therefore able to coordinate mutagenesis. Such a complex of secondary structures may be the source of ssDNA for enzyme-based diversification, which ultimately results in high affinity antibodies.
Keywords: Somatic hypermutation, Transcription-directed mutagenesis, Secondary structures, Supercoiling
1. Introduction
When B cells recognize a foreign antigen, a series of successive events are initiated that ensure the rapid evolution of antibodies with high affinity for their target antigen. The frequency of point mutations during somatic hypermutation (SHM) increases to an estimated 10−3 mutations per base per generation, which is a million-fold higher than background in the rest of the genome (Rajewsky, 1996). Research in this field has primarily focused on enzyme-catalyzed antibody diversification, i.e., mechanisms by which point mutations can occur at all base positions due to the activities and interactions of activation-induced cytidine deaminase (AID) and multiple mutagenic pathways involving base excision, mismatch repair and error-prone DNA polymerases (reviewed in Barreto et al., 2005; Honjo et al., 2005; Longerich et al., 2006; Odegard and Schatz, 2006; Neuberger and Rada, 2007; Teng and Papavasiliou, 2007).
A number of studies indicate that transcription plays a key role in SHM by exposing ssDNA and thus allowing access to enzyme-based mutation (Bransteitter et al., 2003; Chaudhuri et al., 2003; Dickerson et al., 2003; Lebecque and Gearhart, 1990; Pham et al., 2003; Ramiro et al., 2003; Ronai et al., 2007; Shen and Storb, 2004; Yang et al., 2006; Yoshikawa et al., 2002). Transcription is required for SHM, and the initiation site defines the 5′ boundary of the hypermutable region (Betz et al., 1994; Peters and Storb, 1996; Rada et al., 1998; Tumas-Brundage and Manser, 1997). The frequency of point mutations correlates with levels of transgene-specific pre-mRNA (Fukita et al., 1998), and increased transcription levels correlate directly with mutation rate (Bachl et al., 2001; Bachl and Olsson, 1999). Transcription drives supercoiling (Liu and Wang, 1987), which stabilizes DNA secondary structures (SSs) containing unpaired bases that are chemically unstable and intrinsically mutable (Lindahl, 1993; Singer and Kusmierek, 1982). Such SS elements have been implicated as targets for AID. For example, it has been hypothesized that negatively supercoiled DNA, exposed in the context of transcription, produces ssDNA targets for AID. Alternatively, it has been proposed that AID binds directly to short stretches of ssDNA exposed by RNA polymerase. Yet, although the consensus in the field is that the availability of ssDNA substrate is likely AID-independent and precedes enzyme-based diversification (Ramiro et al., 2003; Ronai et al., 2007), the transcriptionally driven structural elements recognized by AID have not been characterized.
The present study aims to characterize these elements by using the mfg computer program to simulate transcription in VH genes. This program has repeatedly demonstrated predictive value in analyzing mechanisms of mutagenesis in SSs formed during transcription in both prokaryotes (Reimers et al., 2004; Schmidt et al., 2006; Wright et al., 2003; Wright, 2004) and eukaryotes (Wright et al., 2002; Wright et al., 2004; Wright et al., 2006). The role of DNA sequence in mutagenesis is simulated by folding successive segments of ssDNA to form SSs in the non-transcribed strand. Assuming that each fold must be initiated with an unpaired base, mfg selects the most stable SS [determined by mfold (Markham and Zuker, 2005)] in which each base is unpaired, thus modeling the competition between alternate SSs for shared nts during transcription in vivo. The resulting simulation of SHM reveals a complicated mechanism that involves pausing and “backtracking” of the folding process, in which the dominant highest stability SS forms repeatedly, providing maximum exposure for coordinated mutagenesis among 26 critical mutable bases. In the present study and in other systems, the SSs formed are too large to exist in transcription bubbles (Krasilnikov et al., 1999; Rahmouni and Wells, 1992), and are assumed to form in supercoiled DNA in the wake of the transcription complex (see Discussion and Materials and Methods).
Mfg analysis of VH5 has resulted in a model of SHM and coordinated mutagenesis in which experimentally-determined mutations most frequently occur in five highly mutable sites, or “block” mutations located in stable, exposed ssDNA segments of a 65 nt SS. This model has received independent support from a recent study of the same gene sequence (Ronai et al., 2007), in which ssDNA segments isolated from splenocyte DNA of immunized mice were identified by chemical means (i.e., bisulfite conversion assay). Specifically, the unpaired mutable Sites in our model are identical to those within the ssDNA segments described by Ronai et al. Hence our theoretical studies, which are strongly supported by both chemical and mutation frequency data, predict that each V region DNA template has evolved to expose critical mutable bases in highly stable SSs.
2. Materials and Methods
2.1 Genes analyzed
The sequence and mutation frequencies of the following Ig genes were included in our analyses: VH5 (GenBank accession numbers X92278 and M99684) (Zheng et al., 2005) and VH4 (GenBank accession number L10088). In VH5 the mfg nt number 1 corresponds to nt 311 in the GenBank sequence, and in VH4, the mfg nt number 1 corresponds to number 85 in the GenBank sequence.
2.2 The computer programs, mfold and mfg
The computer program, mfg, was created to predict the intrinsic mutability of unpaired bases in SSs of the non-transcribed strand during simulated transcription (Wright et al., 2003). Mfg interfaces with the mfold program, which simulates the melting of a segment of single-stranded nucleic acids in solution (Markham and Zuker, 2005) and reports all possible SSs that can form from each folded segment, in descending order of their stability. Free energies are computed by summing Boltzmann factors over every possible folded state. For each base in a specified window size, mfg selects and reports the most stable SS(s) in which that base is unpaired (which may or may not be a proximal sequence), and also reports the percent of total folds in which it is unpaired during transcription. The Mutability Index (MI) of each unpaired base is the product of these two variables, −ΔG and percent unpaired. This program is available on the web with directions for its use: http://biology.dbs.umt.edu/wright/upload/mfg.html. In a sliding window analysis mfg folds successive segments of the non-coding strand. Thus, in effect, nts are continuously removed from the 5′ end and added to the 3′ end. This process would involve a running competition for shared nts between the successive, inter-converting SSs of different stabilities and pause times. The major difference between mfold and mfg is that mfold simply folds successive single-stranded segments, while mfg must temporarily halt the folding process in the forward direction when a stem is encountered. Mfg must then find the most stable SS in which each paired base of the stem is unpaired. In the meantime, transcription and the folding window of the non-coding strand are moving forward.
2.3 Assumptions underlying the mfg computer program
Extensive experimental and theoretical work has clarified the role of free energy parameters governing superhelical strand separation transitions (Benham, 1996; Breslauer et al., 1986; Dayn et al., 1992; Krasilnikov et al., 1999). Computational methods are available for predicting DNA sites where torsional stress destabilizes the duplex and results in strand separation, ssDNA and SS formation. Secondary structures at destabilized sites result from inverted complementary sequences that hydrogen bond to form stems. Sequence segments that form SSs are clearly selected during evolution, as they occur in DNA about 10,000 times more frequently than predicted by chance (Lilley, 1980). In the present work and in other systems (Dayn et al., 1992; Lilley, 1980; Wright et al., 2002; Wright et al., 2003), these SSs are probably located in negatively supercoiled DNA in the wake of the transcription complex. Transcriptionally-driven SS formation in vivo has been quantitatively measured and is highly localized (Krasilnikov et al., 1999; Rahmouni and Wells, 1992). Evidence from templating mutations implicate 40–50 nt SSs (Wright et al., 2003) and atomic force microscopy has shown formation of SSs as large as 53 nts (Shlyakhtenko et al., 1998).
Dayn et al. (1992) have demonstrated that increased promoter activation and transcription levels correlate with the size of SSs formed, presumably due to increased availability of ssDNA. Using mfg, mutation rates in prokaryotes have been correlated with rates of transcription determined by mRNA half-lives and concentrations (Reimers et al., 2004), and transcription levels have been found to correlate with promoter strength and supercoiling (Schmidt et al., 2006). Thus, when simulating transcription using mfg, the window size is assumed to be directly proportional to the level of transcription, i.e., the number of nucleotides in ssDNA that form SSs in the non-transcribed strand. Using this program, it has been possible to establish good correlations between MIs and experimentally-determined mutation frequencies. Secondary structures have long been implicated in SHM (Rogozin and Diaz, 2004), and mutation frequencies are also known to correlate with transcription frequency. The present study describes mechanisms by which these correlations can occur during SHM.
3. Results
3.1 Secondary structures and coordinated mutagenesis in VH5
A large mutation database is essential for meaningful analyses of the relationship between base exposure and mutability. Another essential characteristic for clarifying the mechanism of coordinated mutagenesis is the presence of several “block” mutations within a single SS. Both of these attributes are found in the data for hypervariable region VH5 (Zheng et al., 2005), which was cloned from isolated germinal centers and has an atypically high number of mutations (7,491).
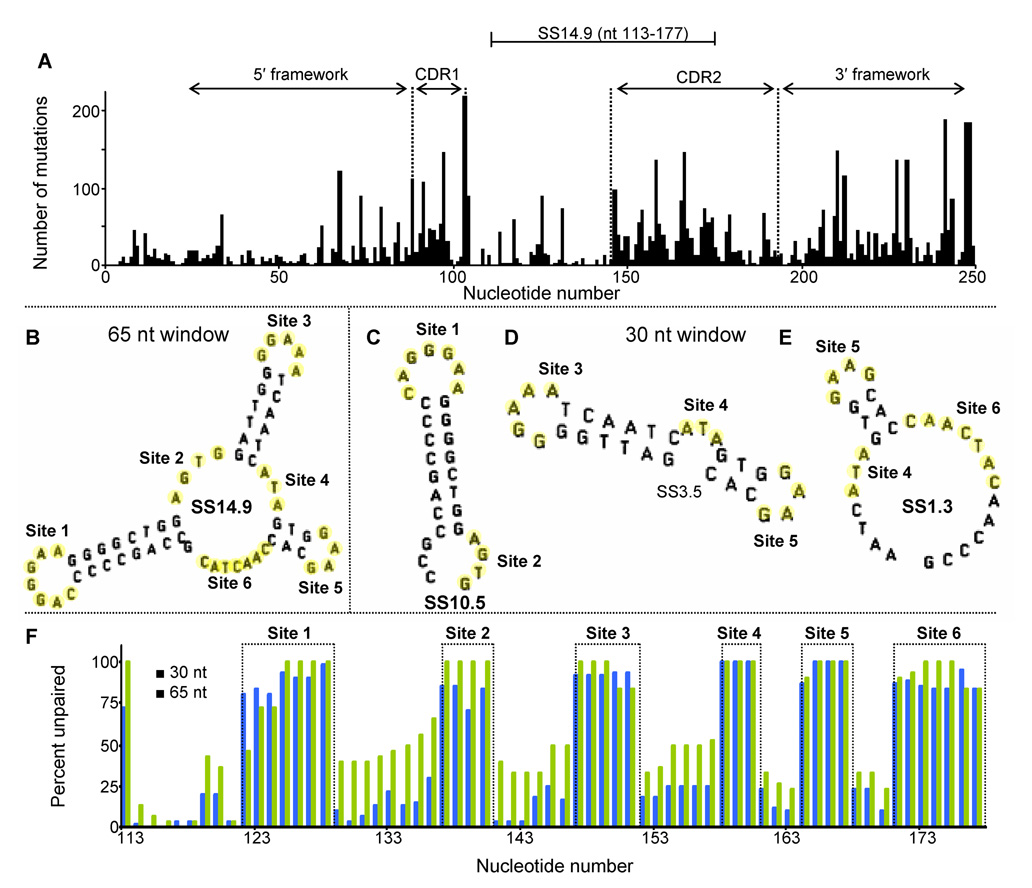
Mutation frequencies in the variable region of VH5 are especially high in complementarity-determining regions 1 (CDR1) and 2 (CDR2) and in the framework segment immediately 3′ to CDR2 (Fig. 1A). Simulations of transcription with a 65 nt window revealed a SS (nt 113–177) of particular interest, SS14.9 (Fig. 1B), referred to by its stability (−ΔG = 14.9 kcal / mol). The mutable sites (numbered 1–6, in red), composed of unpaired bases (yellow), are shown. These sites are also seen in the three relevant 30 nt structures (Fig. 1C–E), which presumably represent lower levels of transcription. The extent to which unpaired bases are exposed is remarkably similar and high at both levels of transcription (Fig. 1F). [Transcription levels are assumed to be directly proportional to the size of SSs (see Materials and Methods)].
Fig. 1. Base mutation frequencies and characteristics of secondary structures in VH5.
(A) Mutation frequencies in CDR1, CDR2, two framework regions, and SS14.9. (B) The 65 nt structure showing mutable sites (red) of unpaired bases (yellow). (C-to-E) The 30 nt SSs and mutable sites. (F) A comparison of the extent to which a base is unpaired during transcription in 65 nt (blue) and 30 nt (green) SSs in nts 113–177 from mfg output. See Materials and Methods.
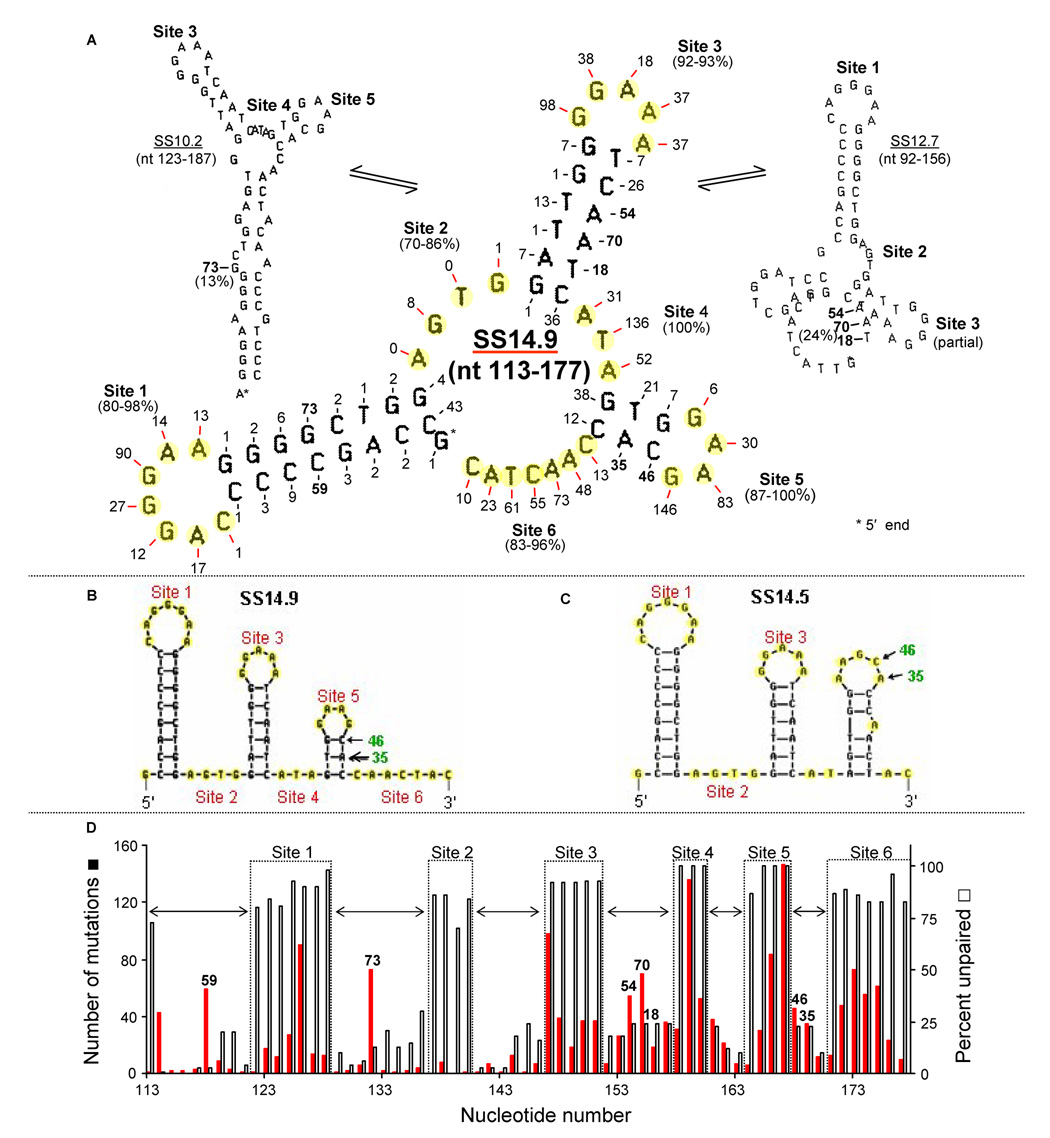
The central most stable VH5 65 nt structure (Fig. 2A) is SS14.9, which has six ssDNA Sites totaling 30 highly unpaired bases (% unpaired in parentheses under each Site). As paired bases are unlikely to mutate, mutations that are shown in stems actually occur in other structures in which those bases are unpaired. Two examples (Fig. 2A, smaller figures) are shown: If the highly mutable G between Sites 1 and 2 (73 mutations in green) is selected by mfg, the structure to the left (SS10.2) is shown. This base (G) is unpaired in only 13% of its folds. Similarly, if one of the As or the T in the stem between Sites 3 and 4 (54, 70, and 18 mutations in green) is selected, SS12.7 to the right is shown. This is the most stable SS in which those three bases are unpaired (in 24% of their folds) during transcription. In addition, mfold reveals two possible configurations for nt 113–177: SS14.9 (Fig. 2B, mfold’s depiction of SS14.9) and SS14.5 (Fig. 2C). The mutations (46 and 35 in green) in bases C and A in the stem of Site 5 are explained by the less stable configuration of SS14.5, in which these bases are unpaired (Fig. 2C). [When the transcribed strand is folded, the most stable and repetitive SS has a −ΔG of 13.2; thus the non-transcribed strand is energetically favored as the primary source of mutations.]
Fig. 2. Inter-conversion of SS14.9 with less stable SSs during transcription.
(A) The dominant SS14.9 flanked by two lower stability structures, SS10.2 (left) and SS12.7 (right). The number of mutations at each unpaired mutable base in SS14.9 is shown in red, and selected mutations in paired bases of stems are in green. SS10.2 is the most stable in which the G in the stem of Site 1 in SS14.9 (73 mutations, in green) and the three bases (AAT, green) in the stem of Site 3 (54, 70, and 18 mutations, respectively, in green), are unpaired. Also shown in green are mutations in the stem of Site 5 (46 and 35) that are reported (mfold) to be in a less stable conformation of nt 113–177. (B) The location of mutations 46 and 35 in SS14.9 depicted by mfold. (C) The less stable SS14.5 configuration of nt 113–177 depicted by mfold. (D) The relationship of mutation frequency and percent unpaired in SS14.9. Horizontal arrows indicate the location of stems, and high mutation numbers (green) in stem sequences indicate mutable bases that are located in SSs other than SS14.9.
SS14.9 is first formed just 3′ of CDR1 (Fig. 1A) and thereafter appears repeatedly by a “backtracking” mechanism (see below). Many unique, brief inter-conversions of SS14.9 occur to and from less stable SSs, such as SS10.2 and SS12.7, as shown in Figure 2A and in the computer output (Table 1). In general, there is a correlation between highly mutable and highly unpaired bases in SS14.9 during transcription (Fig. 2D). Site 2 is the only highly unpaired sequence that is not mutated, suggesting that it is a protected site (this may also be true of SS14.9 pre nt 107–113; see below). Horizontal arrows indicate the location of stems. The fourth column of Table 1 indicates the most stable fold in which each base of this sequence is unpaired. As seen by comparing the first and last columns, these folds may or may not be initiated at the location of that base in the sequence. That is, a pause in the folding process can occur, and some folds have backtracked in the 5′ direction.
Table 1.
Computer output of nt 107–177 derived from mfg
| nt number | Base | −ΔG a | Fold b | % unpaired | ||||
|---|---|---|---|---|---|---|---|---|
| 107 | G | 14.9 c | 107–171 | 56 | 58.3 | |||
| 108 | G | 14.9 c | 108–172 | 56 | ||||
| 109 | A | 14.9 c | 109–173 | 63 | ||||
| 110 | T | 14.9 c | 110–174 | 44 | ||||
| 111 | C | 14.9 c | 111–175 | 44 | ||||
| 112 | C | 14.9 c | 112–176 | 44 | ||||
| 113 | G | 14.9 c | 113–177 | 73 | ||||
| 114 | C | 5.2 | 50–114 | S-IB | 1 | |||
| 115 | C | 0.0 | 0 | 0 | ||||
| 116 | A | 0.0 | 0 | 0 | ||||
| 117 | G | 10.4 | 68–132 | 3 | ||||
| 118 | C | 10.4 | 69–133 | 3 | ||||
| 119 | C | 9.9 | 60–124 | 20 | ||||
| 120 | C | 9.9 | 61–125 | 20 | ||||
| 121 | C | 9.8 | 57–121 | 4 | ||||
| 122 | C | 14.9 | 107–171 | 80 | 81.7 | |||
| 123 | A | 14.9 | 108–172 | 84 | ||||
| 124 | G | 14.9 | 109–173 | 81 | ||||
| 125 | Site 1 | G | 14.9 | 110–174 | 93 | |||
| 126 | G | 14.9 | 111–175 | 90 | ||||
| 127 | A | 14.9 | 112–176 | 90 | ||||
| 128 | A | 14.9 | 113–177 | 98 | ||||
| 129 | G | 9.7 | 65–129 | S-IB | 10 | |||
| 130 | G | 9.3 | 66–130 | 4 | ||||
| 131 | G | 9.4 | 67–131 | 6 | ||||
| 132 | G | 10.2 | 123–187 | 13 | ||||
| 133 | C | 10.2 | 124–189 | 21 | ||||
| 134 | T | 12.2 | 70–134 | 13 | ||||
| 135 | G | 12.2 | 71–132 | 15 | ||||
| 136 | G | 11.8 | 115–179 | 30 | ||||
| 137 | A | 14.9 | 107–171 | 86 | 80.7 | |||
| 138 | Site 2 | G | 14.9 | 108–172 | 86 | |||
| 139 | T | 14.9 | 109–173 | 70 | ||||
| 140 | G | 14.9 | 110–174 | 84 | ||||
| 141 | G | 13.7 | 77–141 | S-IB | 3 | |||
| 142 | A | 13.7 | 78–142 | 3 | ||||
| 143 | T | 2.3 | 142–206 | 3 | ||||
| 144 | T | 14.2 | 80–144 | 18 | ||||
| 145 | G | 14.2 | 81–145 | 24 | ||||
| 146 | G | 14.2 | 82–146 | 16 | ||||
| 147 | G | 14.9 | 107–171 | 92 | 92 | |||
| 148 | G | 14.9 | 108–172 | 92 | ||||
| 149 | Site 3 | A | 14.9 | 109–173 | 92 | |||
| 150 | A | 14.9 | 110–174 | 93 | ||||
| 151 | A | 14.9 | 111–175 | 93 | ||||
| 152 | T | 11.6 | 88–152 | S-IB | 18 | |||
| 153 | C | 2.3 | 145–209 | 18 | ||||
| 154 | A | 12.7 | 92–156 | 24 | ||||
| 155 | A | 12.7 | 93–157 | 24 | ||||
| 156 | T | 12.7 | 94–158 | 24 | ||||
| 157 | C | 3.0 | 156–220 | 24 | ||||
| 158 | A | 14.9 | 107–171 | 100 | 100 | |||
| 159 | Site 4 | T | 14.9 | 108–172 | 100 | |||
| 160 | A | 14.9 | 109–173 | 100 | ||||
| 161 | G | 13.9 | 97–161 | S-IB | 23 | |||
| 162 | T | 13.9 | 98–162 | 12 | ||||
| 163 | G | 13.9 | 99–163 | 10 | ||||
| 164 | G | 14.9 | 107–171 | 87 | 95.7 | |||
| 165 | Site 5 | A | 14.9 | 108–172 | 100 | |||
| 166 | A | 14.9 | 109–173 | 100 | ||||
| 167 | G | 14.9 | 110–174 | 100 | ||||
| 168 | C | 13.6 | 104–168 | S-IB | 23 | |||
| 169 | A | 13.6 | 105–169 | 23 | ||||
| 170 | C | 2.1 | 170–234 | 10 | ||||
| 171 | C | 14.9 | 107–171 | 87 | ||||
| 172 | A | 14.9 | 108–172 | 89 | ||||
| 173 | A | 14.9 | 109–173 | 86 | ||||
| 174 | Site 6 | C | 14.9 | 110–174 | 83 | |||
| 175 | T | 14.9 | 111–175 | 83 | ||||
| 176 | A | 14.9 | 112–176 | 96 | ||||
| 177 | C | 14.9 | 113–177 | 83 | ||||
The most stable SS in which each base is unpaired. The six mutable sites in SS14.9 are in red, and mutable bases unpaired in structures other than SS14.9 are in green. See Fig. 3.
The 65 nt fold forming the SS.
SS14.9 pre
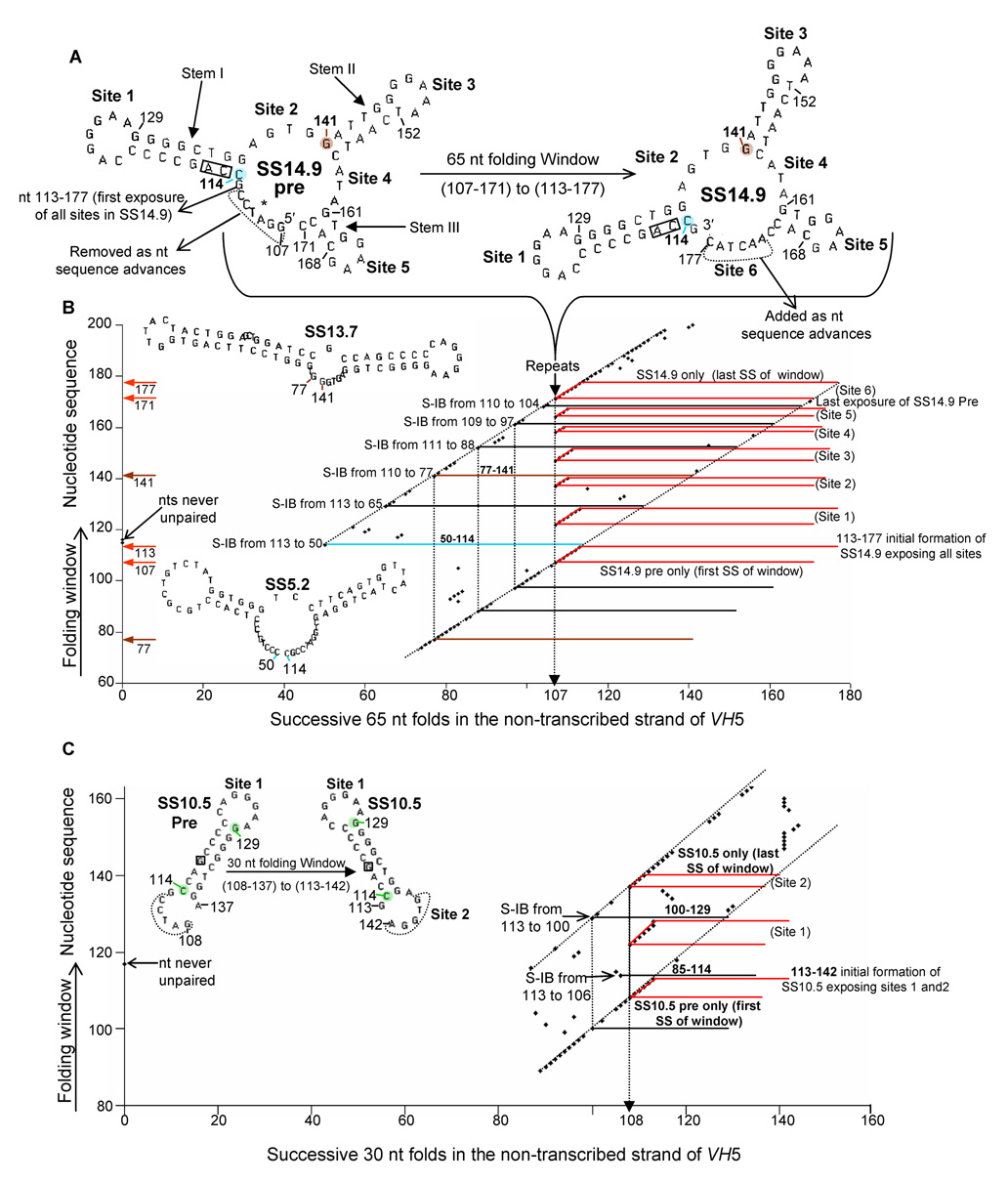
3.2 The process of “backtracking” underlying the repeated appearance and conversion of SS14.9 pre to SS14.9
In the formation of SS14.9 from its precursor, SS14.9 pre (Fig. 3A), each 65 nt fold beginning at nt 107 and proceeding to nt 113 is initiated at a successive base in the sequence (bolded black in Table 1). However, the C at nt 114 is paired, at the beginning of an extremely stable stem (boxed nts 115 and 116 are never unpaired subsequent to the initial formation of this stem). Therefore, when the folding window reaches paired base 114 in the sequence (Table 1), mfg must choose the most stable SS in which that C is unpaired. That fold, nts 50–114 (Fig. 3B; SS5.2 in blue), occurred earlier in the sequence at nt 50 (not shown). We call this phenomenon, in which a paired base initiates backtracking, “Stem-Induced Backtracking”, or S-IB. Conceptually, this complex process is best described by a graph (Figs. 3B and C) in which the nt sequence, 5′ to 3′, is shown on the ordinate, from bottom to top. The direction of the folding window is shown left to right. Thus, in Figure 3B, each 65 nt fold that occurs at each nt in the sequence (ordinate) is depicted in the horizontal direction (abscissa), and each point (diamond) is the first nt of the fold. The diagonal lines indicate the initiation of unfolded strands. Representative horizontal lines are shown, indicating the 65 nt length of each fold. Note that the same fold can occur more than once. For example, fold 77–141 (brown) is first seen initiated at nt 77; later in time, following S-IB, this fold occurs again at base 141. The resulting pattern of points initiating successive folds moves both upward, 5′ to 3′ (bottom to top), and to the right. However, the repeated formation and conversion of SS14.9 pre to SS14.9 becomes a “standing wave” that does not move. The extent to which a fold at any nt can backtrack depends upon the chosen window size. The stem (paired base) at which S-IB is initiated depends upon how far transcription, and thus the resulting folding window of the non-transcribed strand, has progressed. Folds which begin with each of the eight paired bases of the first stem (nts 114 through 121; Fig. 3A and B) result in eight backtracks, with respect to the location of the folding complex at nt 113, to form less stable SSs (Table 1). The first backtracked fold, nt 50–114 (SS5.2, blue), is an entire 65 nt window length 5′ to the position of the folding complex. The subsequent seven folds at Stem I are seen scattered between nts 114–121. When the folding window reaches the first unpaired base at each Site, mfg reinitiates folding at nt 107, as SS14.9 pre is the most stable SS in which all bases at all Sites are simultaneously unpaired. Thus, the formation and conversion of SS14.9 pre to SS14.9 occurs repeatedly at each Site of unpaired bases; the number of folds (always beginning with nt 107) equals the number of unpaired bases at each Site (Fig. 3A and B). As this process occurs at each unpaired base (30 times), the simultaneous exposure of unpaired bases for coordinated mutagenesis is maximized within a minimum sequence length. At each stem after Site 1 (nts 129, 141, 152, 161, and 168), S-IB continues to occur, but must stop all together when the folding window advances to the point at which no more unpaired bases exist at the 5′ end of SS14.9 pre (Fig. 3A). Thus, at the first unpaired C of Site 6 (nt 171), mfg reports all seven folds as the most stable in which that base is unpaired. At the last 3′ C, only one fold (113–177) is shown.
Fig. 3. Stem-Induced Backtracking during the conversion of SS14.9 pre to SS14.9.
(A) During transcription the window size for folding the non-coding strand moves 5′ to 3′ as SS14.9 is formed from its precursor. (B) As mfg must choose an unpaired base to initiate each fold, S-IB occurs at each paired base of each stem. Two examples of S-IB are highlighted, triggered at nt 114 preceding Site 1 (blue), and at nt 141, preceding Site three (brown). The latter fold first occurs at nt 77 (brown), prior to the initial formation of Stem 1 (seen Table 1). (C) A similar analysis showing S-IB using a 30 nt window during the conversion of SS10.5 pre to SS10.5
S-IB is not window size-sensitive, i.e., essentially the same mechanism is observed using a 30 nt window (Fig. 3C) as seen using a 65 nt window. However, three SSs are now required to include the six Sites in SS14.9 (Fig. 1C–E). The example shown is the conversion of SS10.5 pre with Site 1 to SS10.5 with Sites 1 and 2. Only one repeat of SS10.5 pre occurs (Site 1) before the last window of SS10.5 is formed (Fig. 3C).
The length of nts folded (window size) at any level of transcription would be expected to vary in vivo, especially at pause sites. As a single window size must be chosen for each analysis, mfg cannot reflect a more realistic, variable size for predicting SSs. However, as seen in Figure 3C (and see Supplementary data), window size is not critical to S-IB (see Discussion).
A similar analysis of S-IB in two inter-converting SSs of VH4 is described in Supplementary data.
3.3 Independent experimental evidence for the model of SHM and S-IB in VH5
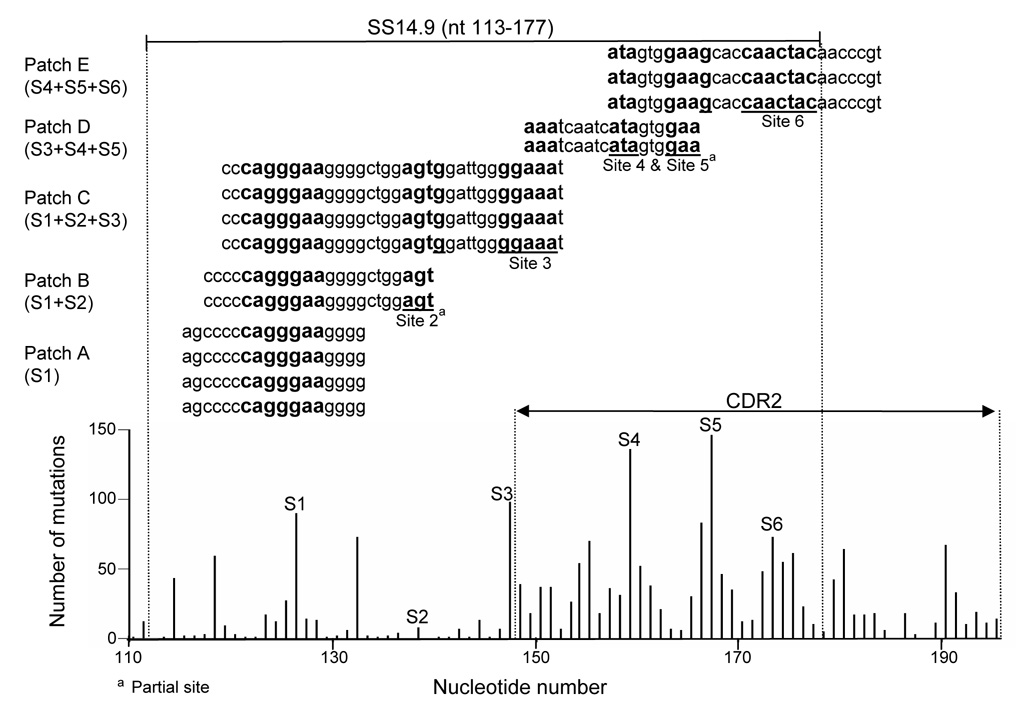
Ronai et al. (2007) isolated nuclei from splenocytes of immunized mice and prepared “chromatinized” DNA, to determine whether ssDNA is enriched in vivo in regions that undergo SHM during transcription in a constitutively hypermutating Ramos B cell line. Using a chemical assay (sodium bisulfite, which deaminates dCs in ssDNA), they described chromatin-associated ssDNA regions targeted for SHM in the VH5 gene. We therefore sought to locate the SS14.9 Sites (Fig. 2A) in the sequences containing ssDNA “patches” (generously provided by D. Ronai and M. D. Schraff). Table 2 and Table 3 summarize the occurrences of our mutable Sites (bolded) in those patch sequences. The nt sequence of each patch and each of the six Sites are also located with respect to mutation frequencies (Fig. 1A and Fig. 4). Thus, in the non-transcribed strand, the simultaneously-exposed mutable bases in SS14.9 are identical to the ssDNA sequence segments of Ronai et al. In the transcribed strand, the data of Ronai et al. [see Fig. 3C of (Ronai et al., 2007)] show only one ssDNA patch in CDR2 (which has the highest frequency of mutations), and we found only two occurrences of our Sites in patches of that strand, suggesting that mutations occur primarily in the non-transcribed strand.
Table 2.
SS14.9 Mutable Sites found in ssDNA patches of VH5
| Patch | Sites in Patches | Location of Site(s) (bold) in patches | No. of occurrences of each Site in patches |
|---|---|---|---|
| A | S1 | agcccccagggaagggg | 6 |
| B | S1 + S2 a | cccccagggaaggggctggagt a | 2 |
| C | S1 + S2 + S3 | cccagggaaggggctggagtggattggggaaat | 4 |
| D | S3 + S4 + S5a | aaatcaatcatagtggaa a | 2 |
| E | S4 + S5 + S6 | atagtggaagcaccaactacaacccgt | 3 |
Partial site
Table 3.
VH5 Mutable Sites Information
| Site | Total mutations at Site | Stem stability (−ΔG) a | All occurrences of each Site in a patch | No. S-IB repeats exposing all Sites b |
|---|---|---|---|---|
| S1 | 174 | 13.9 (5′) | 12 | 7 |
| S2 | 9 | 13.9 (3′) | 6 | 4 |
| S3 | 228 | 3.7 (5′) | 6 | 5 |
| S4 | 219 | 3.7 (3′) | 5 | 3 |
| S5 | 265 | 4.7 (5′) | 5 | 4 |
| S6 | 286 | 4.7 (3′) | 3 | 1 |
The 5′ side of each stem precedes the loop Sites and the 3′ side precedes Sites 2, 4, and 6.
See Figure 3B.
Fig. 4. Mutable Sites in SS14.9 located in ssDNA patches determined by chemical assay.
The location of the six mutable Sites in SS14.9 are shown (bolded) in five different ssDNA patches as defined by Ronai et al. (2007). The location of these Sites is also seen in the mutation frequency profile of the VH5 gene (Figure 2 and Table 2 and Table 3).
Site 1 (with seven unpaired bases) was the only Site located alone (six times) in a ssDNA patch, and was observed a total of 12 times in ssDNA patches (alone and with Sites 2 and 3) (Table 2 and Table 3). The diminishing occurrence, 5′ to 3′, of mutable Sites in the ssDNA patches (Table 2) correlates with the diminishing repeats of SS14.9 pre that expose all mutable Sites (Fig. 3B and Table 3). These correlations are consistent with a key role for stem stability (Fig. 3A and Table 2 and Table 3) in the initiation of S-IB, in which the maximum number of eight backtracks (nts 114–121) are followed by the most repeats (seven at S1). These repeats are triggered in response to the first and most stable stem (Stem I, Fig. 3A and Table 3). The total number of mutations at all sites is shown in Table 3 (see Discussion).
4. Discussion
4.1 Transcription-driven mutagenesis and S-IB
Evidence for mutagenesis in SSs and was pioneered by Ripley and Glickman (1983), and more recently recognized as the result of transcription-driven supercoiling that stabilizes SSs containing intrinsically mutable unpaired bases (Reimers et al., 2004; Schmidt et al., 2006; Wright, 2000; Wright et al., 2002; Wright et al., 2003; Wright, 2004; Wright et al., 2004; Wright et al, 2006). Hoede et al. (2006) investigated the influence of transcription-directed mutagenesis (TDM) on genome evolution, and demonstrated that the control of TDM through DNA SSs is under selection in the E. coli bacterial genome. In a recent analysis of SHM in the TP53 gene in B-cell chronic lymphocytic leukemia, Malcikova et al. (2008), found evidence for TDM in a number of genes. In two cases, the AID enzyme also contributed to (i.e, was superimposed upon) the hypermutation process, suggesting a reconciliation for the simultaneous existence of intrinsic and enzyme-based mutagenesis.
The formation of a SS from a specific ssDNA sequence, predicted by mfold (Markham and Zuker, 2005), is determined by free energy parameters governing superhelical strand separation transitions where torsional stress destabilizes the duplex and results in strand separation, ssDNA and secondary structure formation (Materials and Methods). However, to our knowledge, mfg is the only tool for predicting the successive formation and inter-conversion of the most stable, smaller SSs that presumably exist in vivo during transcription of sequences hundreds of nucleotides long. The assumptions underlying this algorithm are logical: each successive fold must be initiated by an unpaired base, and the most stable SS in which each base is unpaired will dominate the folding pathway. Although SS14.9 dominates and reappears most frequently, other SSs of lesser stability are formed, as the result of complex inter-conversions, the energetic properties of ssDNA, and changing window sizes that must occur. Use of this program has revealed a new mechanism of mutagenesis that incorporates and augments the underlying cause of TDM - base exposure - by repetitive formation of the most stable SS in which all unpaired bases are exposed. Repetitive SS formation is dependent upon SS stability (−ΔG) and upon relative stem stabilities within the dominant SS (Fig. 3 and see Supplementary data). Although the mechanism of pausing and S-IB predicted by mfg analysis cannot precisely simulate conditions in vivo, it could approximate the essential characteristics of these mechanisms triggered by high stability SSs.
4.2 Mechanisms of Pausing and Backtracking
The point in time during transcription at which pausing and S-IB occurs depends upon the location of the folding window, which moves 5′ to 3′ at a rate determined by transcription. Presumably, S-IB occurs in negatively supercoiled ssDNA in the wake of the transcription complex, and therefore has relatively little effect on the rate of transcription. However, supercoil-stabilized DNA structures in promoters are known to cause RNA polymerase pausing and block transcription (Bagga et al., 1990; Peck and Wang, 1985). Krohn et al. (1992) examined the effects of template topology on RNA polymerase pausing during in vitro transcription, and demonstrated a direct correlation between pause strength and the negative superhelical densities of the templates used. There are also a number of studies demonstrating that pause sites tend to occur at the base of a stem in SSs (Suo and Johnson, 1998; Weaver and DePamphilis, 1984). In the latter work the arrest of DNA synthesis in vitro was analyzed, and the polymerase stopped precisely at the base of the stem, regardless of the direction from which the enzyme approached. Thus, mfg simulations of folding DNA per se may have revealed a contributing cause of pausing in vitro and in vivo, i.e., new folds in ssDNA are inhibited by stems because each fold must be initiated with an unpaired base.
RNA polymerase arrest and backtracking have been explained by disengagement of the enzyme from the 3′ end of the transcript followed by backward movement along the DNA with concomitant reverse threading of the intact RNA through the enzyme, which could account for most polymerase pauses (Galburt et al., 2007; Komissarova and Kashlev, 1997). In a recent theoretical paper (Voliotis et al., 2008) transcriptional pausing was found to result in bursts of mRNA production, suggesting that transcriptional pauses may be a significant contributor to variability in rates of transcription.
With respect to pausing and backtracking in the non-transcribed strand of VH5, an appropriate comparison may be to the non-coding segment of human mitochondrial DNA (mtDNA). Replication fork arrest and stalling occurs frequently during transcription, and is thought to be mediated by hairpin SSs in D-loops which form a structural barrier (Bowmaker et al., 2003; Kaguni and Clayton, 1982). Pereira, et al. (Mol. Biol. Evolution, in press), recently used mfold to investigate the formation of SSs in mtDNA. A new cloverleaf-like SS (very similar to SS14.9) with an atypically high folding potential was predicted for a 93 bp stretch of the Control Region 5′ peripheral domain. Mutational heterogeneity was analyzed in a phylogenetic tree containing 2,137 human mtDNA sequences with more than 10,000 independent mutations, and these authors conclude that this structure (SS11.3) is responsible for the rate of base substitutions in hypervariable region I (HVRI). An mfg analysis of this sequence shows S-IB very similar to that in VH5, i.e., repeated folding of the most stable SS and exposure of unpaired bases triggered by the first, most stable stem in SS11.3 (unpublished data).
4.3 A model for SHM
Evidence suggests that the hypermutation mechanism, although site-specific, is random in terms of base-pair substitutions (MacLennan, 1994). Extremely high mutation frequencies (10−3) are apparently essential for producing the number of variations required to test, change and coordinate mutations in fitting antibody to antigen. The rate of VH and VL transcription is negligible in germ-line DNA, but variable-region gene rearrangements close the gap between enhancers and promoters to increase the rate of transcription approximately 10,000-fold. This in turn should increase supercoiling that stabilizes SSs, and therefore increases the availability of unpaired bases in these structures. Unpaired bases are thermodynamically unstable, and point mutations occur by known chemical mechanisms having finite, significant activation energies under physiological conditions (Drake et al., 1983; Singer and Kusmierek, 1982). Non-enzymatic protonation and deamination of cytosine in ssDNA occur 140-fold more frequently than in dsDNA (Frederico et al., 1990; Frederico et al., 1993; Lindahl, 1993; Lindahl and Nyberg, 1974). CpG sequences are methylated non-enzymatically by S-adenosylmethionine, rendering them 40-fold more susceptible to deamination than non-methylated CpG sequences. Two major non-enzymatic mutagenic events, the hydrolytic deamination of C and the oxidation of G, are estimated to occur 100–500 times per day in each human cell, and 2,000–10,000 purine bases turn over per day in each cell owing to the hydrolytic depurination and repair of unpaired bases in ssDNA. Thus, G and C are much more mutable by these mechanisms than A and T, and, because of its size, A is more likely than C or T to replace G at apurinic sites. It is therefore conceivable that the million-fold increase in mutation frequency is due to the approximate 10,000-fold increase in rate of transcription and exposure of unpaired bases in SSs. In that event, enzyme-based mutagenesis would subsequently be superimposed upon a mutation frequency of 10−3 per base per generation, and it would be difficult experimentally to distinguish between the two mutagenic mechanisms. However, transcription per se has been shown to result in a 20-fold increase in AID-induced mutations in E. coli (Ramiro et al., 2003), and recent studies of Ronai et al. (2007) have shown that the formation of ssDNA requires transcription but not AID. Although DNA repair proteins clearly modify the spectrum of mutations, they have relatively minor effects on mutation frequency (Frey et al., 1998; Kim et al., 1999; Li et al., 2006; Phung et al., 1998).
A dynamic model coupling transcription and mutation is proposed for intrinsic and superimposed enzyme-catalyzed mutagenesis over time during affinity maturation. Such a mechanism is ideal for coordinating mutagenesis in producing, repeatedly modifying, rejecting and finally selecting the vast number of gain-of-function mutations and high affinity antibodies required for humoral immunity. The primary SS dominating this model in VH5 is SS14.9, in which experimentally-determined mutation frequencies correlate with the extent to which a base is unpaired (% unpaired, Fig. 2D). Table 1 and Figure 2 depict a transitional, dynamic equilibrium in which 26 highly unpaired and mutable bases in SS14.9 have multiple opportunities for testing and modifying mutagenesis within changing populations of unpaired bases in different structures available to enzyme modification. For example, when SS12.7 (Fig. 2A) is formed, mutable unpaired bases (green) can coordinate mutagenesis with all bases at Sites 1, 2, and some bases at Site 3. At high levels of transcription, S-IB results in the repeated appearance of SS14.9 stabilized by supercoiled DNA, in effect providing a stationary yet dynamic platform for mutagenesis. This mutagenic mechanism offers multiple sources of variability for modifying mutagenic patterns and repeatedly receiving feedback for the fit of antibody to antigen during the course of affinity maturation. The degree of success with which the relevant mutant antibody increases its affinity for cognate foreign antigen monitors and guides the outcome of selection or rejection.
Independent data of Ronai et al. provide strong support for the proposed model by demonstrating (Fig. 4 and Table 2 and Table 3) that the mutable Sites in SS14.9 are actually located in ssDNA patches that occur ex vivo. The frequency with which specific patches of ssDNA were found in the non-transcribed strand does not correlate with mutation frequency (Fig. 4 and Table 2 and Table 3). The most striking correlation is between stem stability and the number of patches containing Site 1 (Table 3). Moreover, no other Site was found alone in any patch. The frequency of Site 1 in ssDNA patches, and the correlation between occurrences of a Site in a patch and the number of S-IB repeats (Table 3) are consistent with the mechanism of S-IB (Fig. 3). The data of Ronai et al. are also consistent with the 65 nt size of SS14.9, which can accommodate all six Sites. Four of their ssDNA patches include Sites 1, 2, and 3 (Table 2 and Table 3) in a 34 nt sequence. According to mfold, these three structures (Fig. 1C–E) cannot co-exist in a conformation of this size, i.e., the SS must be larger (data not shown). Also, Site 3 is found in ssDNA patches containing Sites 1 and 2 as well as in patches containing Sites 4 and 5; similarly, Sites 4 and 5 are also found with Site 6. These data are consistent with the presence of all Sites and patches within the same large structure, i.e., SS14.9.
The predictive value of mfg has been demonstrated by correlations between predicted and experimentally-determined mutation frequencies during transcription, in particular, for hypermutable codons of p53 (Wright et al., 2002; Wright et al., 2006), and by the predicted effects of transcription, promoter strength and supercoiling on base mutability. In the present study, the model of SHM is substantiated by three types of experimental data: (1) high mutation frequencies determined experimentally in VH5 occur in highly unpaired (% unpaired) bases predicted by mfg (Fig. 1A and F; Fig. 2D and Fig. 4); (2) unpaired mutable bases in SS14.9 are identical to those identified in ssDNA patches described by Ronai et al. during transcription and SHM in mutating V regions of VH5 in which DNA-protein complexes were preserved in the context of chromatin in human B cell lines and in primary mouse B cells; and (3) the frequency with which our mutable Sites are found in their ssDNA patches is consistent with the mechanism of S-IB, and not correlated with the concentration of ssDNA or mutation frequencies in a patch (Table 2 and Table 3). Thus, in effect, the ex vivo experimental data of Ronai et al. were predicted by the in silico model of mutagenesis in the non-transcribed strand of VH5 presented here. As the data of Ronai et al. come from a cell line that mutates constitutively, the resulting mutation frequency should in part reflect the intrinsic mutability of unpaired bases in ssDNA (Lindahl, 1993; Singer and Kusmierek, 1982).
The mutagenic mechanisms observed in this study are consistent with several characteristics of SHM. Since mutation frequency is directly dependent upon transcription frequency, the mechanism of mutagenesis would be expected to be essentially the same at all levels of transcription. Thus, base exposure (% unpaired) should, ideally, be high at all levels of transcription. This has been observed in VH5 (Fig. 1F) as well as in VH4, VH 186.2 and VH 94 (unpublished data). Also, the repeated exposure of unpaired bases via S-IB is usually initiated at the same stems at different levels of transcription. This is seen in VH5 (Fig. 3B and C) as well as in VH4 (Supplementary data). Maximizing variability and the efficiency of coordinated mutagenesis for high frequency gain-of-function mutations is the essence of SHM. These analyses reveal sets of contiguous unpaired bases located in loops of the single most stable SSs formed at each level of transcription, and S-IB results in the repeated, simultaneous exposure of all unpaired bases within a minimum sequence distance at all levels of transcription. Such a dynamic platform of ssDNA is an ideal target for enzyme-based diversification resulting in high affinity antibodies.
Supplementary Material
Supplementary data associated with this article can be found, in the online version,
Acknowledgments
We thank N. Papavasiliou for suggesting that ssDNA Sites in SS14.9 could be the same as the ssDNA patches described by D. Ronai and M. D. Schraff, whom we thank for providing the sequences of their ssDNA patches. For a critical review of this manuscript we are grateful to N. Papavasiliou, M. D. Schraff, J. Pfau, and D. Reschke. We thank A. Hunt for excellent technical assistance and N. Parrow for valuable information relevant to statistics. This research was funded by NIH grant RO1CA099242 and the Stella Duncan Memorial Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bachl J, Carlson C, Gray-Schopfer V, Dessing M, Olsson C. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J Immunol. 2001;166:5051–5057. doi: 10.4049/jimmunol.166.8.5051. [DOI] [PubMed] [Google Scholar]
- Bachl J, Olsson C. Hypermutation targets a green fluorescent protein-encoding transgene in the presence of immunoglobulin enhancers. Eur J Immunol. 1999;29:1383–1389. doi: 10.1002/(SICI)1521-4141(199904)29:04<1383::AID-IMMU1383>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Bagga R, Ramesh N, Brahmachari SK. Supercoil-induced unusual DNA structures as transcriptional block. Nucleic Acids Res. 1990;18:3363–3369. doi: 10.1093/nar/18.11.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto VM, Ramiro AR, Nussenzweig MC. Activation-induced deaminase: controversies and open questions. Trends Immunol. 2005;26:90–96. doi: 10.1016/j.it.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Benham CJ. Computation of DNA structural variability--a new predictor of DNA regulatory regions. Comput Appl Biosci. 1996;12:375–381. doi: 10.1093/bioinformatics/12.5.375. [DOI] [PubMed] [Google Scholar]
- Betz AG, Milstein C, Gonzalez-Fernandez A, Pannell R, Larson T, Neuberger MS. Elements regulating somatic hypermutation of an immunoglobulin kappa gene: critical role for the intron enhancer/matrix attachment region. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- Bowmaker M, Yang MY, Yasukawa T, Reyes A, Jacobs HT, Huberman JA, Holt IJ. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J Biol Chem. 2003;278:50961–50969. doi: 10.1074/jbc.M308028200. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslauer KJ, Frank R, Blocker H, Marky LA. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Dayn A, Malkhosyan S, Mirkin SM. Transcriptionally driven cruciform formation in vivo. Nucleic Acids Res. 1992;20:5991–5997. doi: 10.1093/nar/20.22.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW, Glickman BW, Ripley LS. Updating the theory of mutation. American Scientist. 1983;71:621–630. [Google Scholar]
- Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- Frederico LA, Kunkel TA, Shaw BR. Cytosine deamination in mismatched base pairs. Biochemistry. 1993;32:6523–6530. doi: 10.1021/bi00077a005. [DOI] [PubMed] [Google Scholar]
- Frey S, Bertocci B, Delbos F, Quint L, Weill JC, Reynaud CA. Mismatch repair deficiency interferes with the accumulation of mutations in chronically stimulated B cells and not with the hypermutation process. Immunity. 1998;9:127–134. doi: 10.1016/s1074-7613(00)80594-4. [DOI] [PubMed] [Google Scholar]
- Fukita Y, Jacobs H, Rajewsky K. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 1998;9:105–114. doi: 10.1016/s1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- Galburt EA, Grill SW, Wiedmann A, Lubkowska L, Choy J, Nogales E, Kashlev M, Bustamante C. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446:820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- Hoede C, Denamur E, Tenaillon O. Selection acts on DNA secondary structures to decrease transcriptional mutagenesis. PLoS Genet. 2006;2(11):1697–1701. doi: 10.1371/journal.pgen.0020176. e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo T, Nagaoka H, Shinkura R, Muramatsu M. AID to overcome the limitations of genomic information. Nat Immunol. 2005;6:655–661. doi: 10.1038/ni1218. [DOI] [PubMed] [Google Scholar]
- Kaguni LS, Clayton DA. Template-directed pausing in in vitro DNA synthesis by DNA polymerase a from Drosophila melanogaster embryos. Proc Natl Acad Sci U S A. 1982;79:983–987. doi: 10.1073/pnas.79.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Bozek G, Lo JC, Storb U. Different mismatch repair deficiencies all have the same effects on somatic hypermutation: intact primary mechanism accompanied by secondary modifications. J Exp Med. 1999;190:21–30. doi: 10.1084/jem.190.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc Natl Acad Sci U S A. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikov AS, Podtelezhnikov A, Vologodskii A, Mirkin SM. Large-scale effects of transcriptional DNA supercoiling in vivo. J Mol Biol. 1999;292:1149–1160. doi: 10.1006/jmbi.1999.3117. [DOI] [PubMed] [Google Scholar]
- Krohn M, Pardon B, Wagner R. Effects of template topology on RNA polymerase pausing during in vitro transcription of the Escherichia coli rrnB leader region. Mol Microbiol. 1992;6:581–589. doi: 10.1111/j.1365-2958.1992.tb01504.x. [DOI] [PubMed] [Google Scholar]
- Lebecque SG, Gearhart PJ. Boundaries of somatic mutation in rearranged immunoglobulin genes: 5′ boundary is near the promoter, and 3′ boundary is approximately 1 kb from V(D)J gene. J Exp Med. 1990;172:1717–1727. doi: 10.1084/jem.172.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Peled JU, Zhao C, Svetlanov A, Ronai D, Cohen PE, Scharff MD. A role for Mlh3 in somatic hypermutation. DNA Repair (Amst) 2006;5:675–682. doi: 10.1016/j.dnarep.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Lilley DM. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980;77:6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longerich S, Basu U, Alt F, Storb U. AID in somatic hypermutation and class switch recombination. Curr Opin Immunol. 2006;18:164–174. doi: 10.1016/j.coi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- MacLennan IC. Somatic mutation. From the dark zone to the light. Curr Biol. 1994;4:70–72. doi: 10.1016/s0960-9822(00)00017-8. [DOI] [PubMed] [Google Scholar]
- Malcikova J, Smardova J, Pekova S, Cejkova S, Kotaskova J, Tichy B, Francova H, Doubek M, Brychtova Y, Janek D, Pospisilova S, Mayer J, Dvorakova D, Trbusek M. Identification of somatic hypermutations in the TP53 gene in B-cell chronic lymphocytic leukemia. Mol Immunol. 2008;45:1525–1529. doi: 10.1016/j.molimm.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger MS, Rada C. Somatic hypermutation: activation-induced deaminase for C/G followed by polymerase eta for A/T. J Exp Med. 2007;204:7–10. doi: 10.1084/jem.20062409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- Peck LJ, Wang JC. Transcriptional block caused by a negative supercoiling induced structural change in an alternating CG sequence. Cell. 1985;40:129–137. doi: 10.1016/0092-8674(85)90316-2. [DOI] [PubMed] [Google Scholar]
- Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- Phung QH, Winter DB, Cranston A, Tarone RE, Bohr VA, Fishel R, Gearhart PJ. Increased hypermutation at G and C nucleotides in immunoglobulin variable genes from mice deficient in the MSH2 mismatch repair protein. J Exp Med. 1998;187:1745–1751. doi: 10.1084/jem.187.11.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C, Ehrenstein MR, Neuberger MS, Milstein C. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- Rahmouni AR, Wells RD. Direct evidence for the effect of transcription on local DNA supercoiling in vivo. J Mol Biol. 1992;223:131–144. doi: 10.1016/0022-2836(92)90721-u. [DOI] [PubMed] [Google Scholar]
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat Immunol. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- Reimers JM, Schmidt KH, Longacre A, Reschke DK, Wright BE. Increased transcription rates correlate with increased reversion rates in leuB and argH Escherichia coli auxotrophs. Microbiology. 2004;150:1457–1466. doi: 10.1099/mic.0.26954-0. [DOI] [PubMed] [Google Scholar]
- Ripley LS, Glickman BW. Unique self-complementarity of palindromic sequences provides DNA structural intermediates for mutation. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):851–861. doi: 10.1101/sqb.1983.047.01.097. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Diaz M. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J Immunol. 2004;172:3382–3384. doi: 10.4049/jimmunol.172.6.3382. [DOI] [PubMed] [Google Scholar]
- Ronai D, Iglesias-Ussel MD, Fan M, Li Z, Martin A, Scharff MD. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J Exp Med. 2007;204:181–190. doi: 10.1084/jem.20062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KH, Reimers JM, Wright BE. The effect of promoter strength, supercoiling and secondary structure on mutation rates in Escherichia coli. Mol Microbiol. 2006;60:1251–1261. doi: 10.1111/j.1365-2958.2006.05166.x. [DOI] [PubMed] [Google Scholar]
- Shen HM, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc Natl Acad Sci U S A. 2004;101:12997–13002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Potaman VN, Sinden RR, Lyubchenko YL. Structure and dynamics of supercoil-stabilized DNA cruciforms. J Mol Biol. 1998;280:61–72. doi: 10.1006/jmbi.1998.1855. [DOI] [PubMed] [Google Scholar]
- Singer B, Kusmierek JT. Chemical mutagenesis. Annu Rev Biochem. 1982;51:655–693. doi: 10.1146/annurev.bi.51.070182.003255. [DOI] [PubMed] [Google Scholar]
- Suo Z, Johnson KA. DNA secondary structure effects on DNA synthesis catalyzed by HIV-1 reverse transcriptase. J Biol Chem. 1998;273:27259–27267. doi: 10.1074/jbc.273.42.27259. [DOI] [PubMed] [Google Scholar]
- Teng G, Papavasiliou FN. Immunoglobulin Somatic Hypermutation. Annu Rev Genet. 2007;41:107–120. doi: 10.1146/annurev.genet.41.110306.130340. [DOI] [PubMed] [Google Scholar]
- Tumas-Brundage K, Manser T. The transcriptional promoter regulates hypermutation of the antibody heavy chain locus. J Exp Med. 1997;185:239–250. doi: 10.1084/jem.185.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voliotis M, Cohen N, Molina-Paris C, Liverpool TB. Fluctuations, pauses, and backtracking in DNA transcription. Biophys J. 2008;94:334–348. doi: 10.1529/biophysj.107.105767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver DT, DePamphilis ML. The role of palindromic and non-palindromic sequences in arresting DNA synthesis in vitro and in vivo. J Mol Biol. 1984;180:961–986. doi: 10.1016/0022-2836(84)90266-3. [DOI] [PubMed] [Google Scholar]
- Wright B, Reimers J, Schmidt K, Burkala E, Davis N, Wei P. Mechanisms of genotoxin-induced transcription and hypermutation in p53. Cancer Cell Int. 2006;6:27. doi: 10.1186/1475-2867-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright BE. A biochemical mechanism for nonrandom mutations and evolution. J Bacteriol. 2000;182:2993–3001. doi: 10.1128/jb.182.11.2993-3001.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright BE. Stress-directed adaptive mutations and evolution. Mol Microbiol. 2004;52:643–650. doi: 10.1111/j.1365-2958.2004.04012.x. [DOI] [PubMed] [Google Scholar]
- Wright BE, Reimers JM, Schmidt KH, Reschke DK. Hypermutable bases in the p53 cancer gene are at vulnerable positions in DNA secondary structures. Cancer Res. 2002;62:5641–5644. [PubMed] [Google Scholar]
- Wright BE, Reschke DK, Schmidt KH, Reimers JM, Knight W. Predicting mutation frequencies in stem-loop structures of derepressed genes: implications for evolution. Mol Microbiol. 2003;48:429–441. doi: 10.1046/j.1365-2958.2003.t01-1-03436.x. [DOI] [PubMed] [Google Scholar]
- Wright BE, Schmidt KH, Minnick MF. Mechanisms by which transcription can regulate somatic hypermutation. Genes Immun. 2004;5:176–182. doi: 10.1038/sj.gene.6364053. [DOI] [PubMed] [Google Scholar]
- Yang SY, Fugmann SD, Schatz DG. Control of gene conversion and somatic hypermutation by immunoglobulin promoter and enhancer sequences. J Exp Med. 2006;203:2919–2928. doi: 10.1084/jem.20061835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa K, Okazaki IM, Eto T, Kinoshita K, Muramatsu M, Nagaoka H, Honjo T. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 2002;296:2033–2036. doi: 10.1126/science.1071556. [DOI] [PubMed] [Google Scholar]
- Zheng NY, Wilson K, Jared M, Wilson PC. Intricate targeting of immunoglobulin somatic hypermutation maximizes the efficiency of affinity maturation. J Exp Med. 2005;201:1467–1478. doi: 10.1084/jem.20042483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data associated with this article can be found, in the online version,