Summary
The dendritic actin network generated by Arp2/3 complex in lamellipodia underlies formation of protrusions, directional sensing and migration. While the generation of this network is well studied, the mechanisms regulating network disassembly are poorly understood. We report that Coronin 1B disassembles Arp2/3-containing actin filament branches by inducing Arp2/3 dissociation. This activity is antagonized by Cortactin, a filament branch stabilizer. Consistent with this biochemical competition, depletion of both proteins partially rescues defects in lamellipodial dynamics observed upon depletion of either protein alone. Coronin 1B targets actin branches in a manner that is mutually exclusive with Arp2/3 complex and alters the branch angle. We conclude that Coronin 1B replaces Arp2/3 complex at actin filament branches as the dendritic network matures and drives the turnover of branched actin networks.
Introduction
The migration of cells is required for morphogenesis, wound healing, cellular immune response and establishes neuronal connections. Mis-regulated cell migration contributes to human disease, including metastasic cancers and compromised immune function. Dynamic cytoskeletal remodeling underlies cell migration, especially remodeling of actin filaments (Pollard and Borisy, 2003). Filament nucleation is a key step during dynamic actin network formation and is catalyzed by the Arp2/3 complex, a highly conserved filament nucleator (Goley and Welch, 2006). Arp2/3 complex nucleates actin filaments that branch from the sides of existing filaments and remains localized at branch junctions to maintain the dendritic structure of the network (Mullins et al., 1998; Svitkina and Borisy, 1999). On its own, Arp2/3 complex is inactive and requires accessory nucleation promoting factors (NPFs) to nucleate filaments (Pollard and Borisy, 2003). NPFs are grouped into two categories (Welch and Mullins, 2002). Type I NPFs, such as WASP and SCAR/WAVE, directly activate the Arp2/3 complex by inducing conformational changes in the complex and supplying the first actin monomer of the new filament. Type II NPFs, such as Cortactin, are weaker NPFs on their own, but potently synergize with Type I NPFs (Weaver et al., 2001).
Although branched filament formation by Arp2/3 complex is well studied, the mechanisms for disassembling branched filament networks are poorly understood. At the back of lamellipodia, ADF/Cofilin proteins (hereafter referred to as Cofilin) are thought to promote filament disassembly via severing and possibly enhanced depolymerization (Iwasa and Mullins, 2007; Svitkina and Borisy, 1999). Cofilin directly promotes debranching in vitro, although no data support this activity in vivo (Blanchoin et al., 2000). Recent data indicate that branches emanating from ATP-actin or ADP-Pi-actin containing filaments are more stable than those emanating from filaments containing ADP (Mahaffy and Pollard, 2006). ATP hydrolysis by the Arp2 subunit of yeast Arp2/3 complex is required for actin patch internalization and efficient spontaneous branch dissociation in vitro (Martin et al., 2006). In contrast, Cortactin stabilizes Arp2/3-dependent filament branches in vitro (Weaver et al., 2001). Depletion of Cortactin reduced dendritic spines in hippocampal neurons, decreased lamellipodial persistence and formation of invadopodia in cancer cells, impaired trans-epithelial migration, reduced receptor internalization, and pathogenic bacterial invasion (reviewed in Cosen-Binker and Kapus, 2006). All of these processes involve Arp2/3-dependent branched actin networks, but the contribution of Cortactin's branch stabilizing activity to these processes has not been assessed in vivo.
Coronins are a conserved family of actin-binding proteins that regulate cell motility in a variety of contexts (Uetrecht and Bear, 2006). Genetic deletion of Coronin 1A in mice leads to severe defects in the immune system including failure of T cell chemotaxis and homing to lymph nodes (Foger et al., 2006). Coronin 1B, a ubiquitously expressed isoform, influences both lamellipodial protrusion and whole-cell motility (Cai et al., 2007b). Consistent with results using yeast Coronin (Humphries et al., 2002), human Coronin 1B inhibits actin nucleation by Arp2/3 complex and this activity is regulated by phosphorylation at Serine 2 (Cai et al., 2005; Foger et al., 2006). Regulation of Coronin 1B by phosphorylation and dephosphorylation at Serine 2 is orchestrated by PKC and Slingshot 1L, respectively (Cai et al., 2007b). However, the mechanistic details of Coronin's inhibition of Arp2/3 complex are unknown. In this work, we describe the molecular antagonism between Coronin 1B and Cortactin at Arp2/3-containing actin branches.
Results
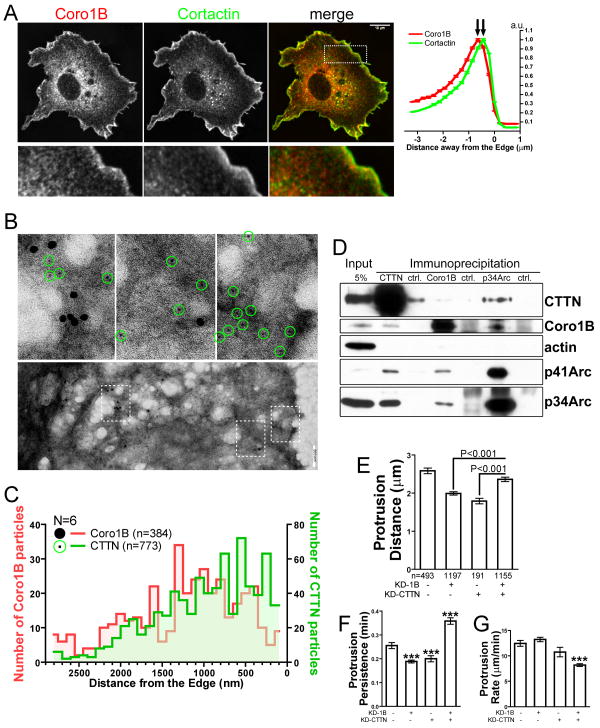
Coronin 1B and Cortactin are distinctly localized in lamellipodia and interact with different pools of Arp2/3 complex
To dissect the role of Coronin 1B in regulating Arp2/3 complex in vivo, we examined its localization relative to Cortactin. Immunolocalization for endogenous proteins reveals that Coronin 1B and Cortactin co-localize to a large extent in lamellipodia (Fig. 1A). Using a contour-based analysis method, we plotted the distribution of Coronin 1B and Cortactin intensity as a function of distance from the cell edge (Fig. 1A) (Cai et al., 2007a; Cai et al., 2007b). Coronin 1B and Cortactin overlap considerably, however, Cortactin consistently localized closer to the leading edge than Coronin 1B. Immunoelectron microscopy using different sized gold conjugates confirmed this result (Fig. 1B); the peak of Coronin 1B occurred ∼800 nm from the cell edge, while most Cortactin localized ∼500 nm from the cell edge (Fig. 1C). Thus, we conclude that Coronin 1B and Cortactin have distinct distributions in lamellipodia.
Figure 1. Coronin 1B and Cortactin localize differentially in lamellipodia and function antagonistically during protrusion.
A- Rat2 cells were immunolabeled for endogenous Coronin 1B and Cortactin. Normalized pixel intensities of Coronin 1B and Cortactin plotted as a function of distance from the cell edge (mean ± SEM). Scale bar = 10 μm.
B- Lower panels: Negative stained electron micrographs of lamellipodia in Rat2 cells immunolabeled for Coronin 1B (18-nm gold particles) and Cortactin (6-nm gold particles). Higher-mag views of boxed regions are presented in upper panels with 6-nm gold particles circled in green. Scale bar = 200 nm.
C- Distribution of distances from the cell edge of gold particles labeling Coronin 1B (red) and Cortactin (green). Student's t test of the distance from the edge is p<0.0001 (N=cells; n=particles counted).
D- Lysates of Rat2 cells immunoprecipitated with antibodies to Cortactin, Coronin 1B, p34Arc or control IgG (ctrl.) and immunoblotted with indicated antibodies.
E,F,G- Protrusion parameters of Rat2 cells expressing Coronin 1B shRNA (KD-1B), Cortactin shRNA (KD-CTTN), or both. Data presented are mean ± 95% CI from >7 cells in each category (n=number of protrusive events). Newman-Keuls multiple comparison test was used after one-way ANOVA to generate the p values.
Both Coronin 1B and Cortactin bind directly to Arp2/3 complex (Cai et al., 2007b; Weed et al., 2000). To determine if Coronin 1B, Cortactin and Arp2/3 complex exist in the same complex in vivo, we examined the interactions of these components by immunoprecipitation (Fig. 1D). Both Coronin 1B and Cortactin reciprocally co-immunoprecipitate with Arp2/3 complex, but the interaction between Coronin 1B and Cortactin is extremely weak, suggesting that these proteins interact with different pools of Arp2/3 complex.
Coronin 1B and Cortactin exert opposing activities on lamellipodial dynamics
The different cellular distributions and interactions with Arp2/3 complex for Coronin 1B and Cortactin prompted us to examine a functional interaction by comparing the effects of depleting each protein, both singly and in combination. Since both proteins are concentrated at the leading edge, we analyzed lamellipodial protrusion dynamics in depleted cells. As expected, depletion of either Coronin 1B or Cortactin reduces protrusion persistence (Fig. 1F) (Bryce et al., 2005; Cai et al., 2007b). Depletion of either protein also reduced protrusion distance (Fig. 1E). Interestingly, cells depleted of both Coronin 1B and Cortactin exhibit increased protrusion persistence and distance relative to cells depleted of only one of these proteins (Fig. 1E,F) but a protrusion rate lower than cells depleted of either protein (Fig. 1G). Thus, Coronin 1B and Cortactin exert opposing effects on some (but not all) parameters of lamellipodial dynamics. These data suggest that Coronin 1B and Cortactin may have antagonistic effects on actin dynamics.
Coronin 1B antagonizes Cortactin in vitro
Coronin 1B inhibits VCA-induced Arp2/3 nucleation activity, but high concentrations were required (IC50 ≈ 1 μM) (Cai et al., 2007b). We examined whether Coronin 1B also inhibits type II NPFs such as Cortactin that synergize with VCA (Fig. S2C). Coronin 1B potently inhibited actin nucleation induced by Cortactin and VCA in a dose-dependent manner (Fig. 2A, B). We estimate that Coronin 1B inhibits the Cortactin component of Cortactin/VCA-induced activation of Arp2/3 with a half-maximal concentration of 71 nM (Fig. 2B). In addition, Coronin 1B inhibits activation of Arp2/3 complex by Cortactin alone (Fig. S2E,F).
Figure 2. Coronin 1B antagonizes Cortactin in vitro and induces debranching.
A- Actin polymer concentration versus time in reactions containing 1.5μM actin (5% pyrene labeled), 20nM Arp2/3 complex, 1nM VCA, and Coronin 1B and Cortactin as indicated. Other curves: actin only (yellow), 1nM VCA & actin (grey) and Arp2/3 & actin (tan).
B- Maximal rates of actin assembly by Arp2/3 and VCA/Cortactin at various Coronin 1B concentrations. Data are normalized to the maximal assembly rate assembly by Arp2/3 and VCA without Cortactin. Data presented as mean ± SEM (N=4 independent experiments). One-phase exponential decay was fit to the data to calculate the IC50.
C- Actin (0.5μM, 30% Oregon green labeled) filaments growing in the presence of 13.3nM Arp2/3 and 13.3nM VCA ± 50nM Coronin 1B observed by TIRF. Scale bars = 4μm.
D- High-mag view of filament debranching observed in reactions from (C). Red arrows indicate actin branch disassembly events. Scale bars = 3μm.
E- Debranching frequency was calculated from TIRF movies as in (C) except that NEM-myosin was omitted to allow increased daughter filament detachment upon debranching. Data are presented as a box and whisker plot. Dunnett's multiple comparison test was used after one-way ANOVA to generate the p values. Data compiled from 3 independent experiments; number of debranching events indicated by n.
F- The number of actin branches/field (1400 μm2) was plotted as a function of time. Actin (0.5μM, 30% Oregon green labeled), 13.3nM Arp2/3, 13.3nM VCA, and indicated amounts of Cortactin and Coronin 1B were mixed immediately before the reactions started. Data are presented as mean ± SEM from 4 fields for each condition.
G- Schematic diagram of the second-wave debranching assay: 0.5 μM unlabeled G-actin (gray dots) with 13.3 nM Arp2/3 complex (blue) and 13.3 nM VCA (step 0). Actin polymerization proceeds for 10 min (step 1). Flow protein mixtures (a-h) containing 0.8 μM Capping protein and without G-actin, VCA or Arp2/3 complex into the reaction chambers for 2 min (step 2). Flush chamber with Rhodamine-phalloidin in imaging buffer to stop the reactions (step 3, red lines), and capture images.
H- Second-wave branch densities (a-f) were calculated from 18 fields (2 independent experiments) and presented in a box and whisker plot. Dunnett's multiple comparison test was used after one-way ANOVA to generate the p values (*, p< 0.05; ***, p<0.0001).
To determine the structural requirements for Coronin 1B's inhibition of Cortactin, we tested mutant Coronin 1B proteins. Coronin 1B R30D (which binds Arp2/3 complex but not F-actin) weakly inhibits Arp2/3 nucleation stimulated by Cortactin and VCA (half-maximal concentration of ∼11 μM) (Fig. S2D) and does not inhibit Arp2/3 activation by Cortactin alone (Fig. S2E,F). Coronin 1B R30D also does not inhibit VCA-dependent Arp2/3 complex activity (Fig. S2A). Thus, F-actin binding is essential for all of Coronin 1B's known biochemical activities. Coronin 1B S2D, a mutant defective in binding Arp2/3 (Cai et al., 2005; Cai et al., 2007b), had no effect on the synergistic activation of Arp2/3 by VCA and Cortactin or by Cortactin alone (Fig. S2D-F). Thus, Coronin 1B must bind both Arp2/3 and F-actin to antagonize Cortactin in vitro.
Coronin 1B destablizes actin filament branches and competes with branch stabilization by Cortactin
To clarify the mechanism of Coronin 1B's inhibition of Arp2/3 nucleation, we used total internal reflection microscopy (TIRF) to monitor branching actin polymerization in real time (Amann and Pollard, 2001). In the presence of 50 nM Coronin 1B, total actin polymer was reduced and branching was less extensive than in control reactions (Fig. 2C). To ensure that the decreased amount of actin filaments was not due to decreased rate of filament elongation at barbed ends, we quantified the filament elongation rate by kymography (Kuhn and Pollard, 2005). Coronin 1B did not alter the rate of filament elongation (Fig. S3A,B).
Examination of the TIRF movies revealed that the decreased branching observed in the presence of Coronin 1B was explained, in part, by instability of branches. In reactions without Coronin 1B, most branches remained stable throughout the course of the experiment (Fig. 2D, upper panels). In contrast, 50 nM Coronin 1B induced dissociation of daughter and mother filaments at a high frequency, suggesting that Coronin 1B destabilizes filament branches (Fig. 2D, Suppl. Movies 1-4).
We used two methods to quantify debranching. Since the length of the daughter filament is proportional to the time since branch formation, we estimated debranching frequency from daughter filament length measurements at the time of dissociation. Coronin 1B increased the frequency of debranching (Fig. 2E). In addition, we also counted actin branches formed over time in the presence of various concentrations of Coronin 1B and Cortactin. Addition of 42 nM Cortactin increased the rate of actin branch formation relative to that promoted by VCA alone (Fig. 2F). Addition of 56 nM Coronin 1B to reactions containing either VCA or 42nM Cortactin suppressed the rate of actin branch formation; this inhibition was reversed by increasing amounts of Cortactin. Conversely, increased amounts of Coronin 1B overcame the branch promoting/protection effect of Cortactin (Fig. S3D).
Since Cortactin both enhances nucleation by Arp2/3 complex and stabilizes actin filament branches (Weaver et al., 2001), we developed a second-wave debranching assay to further dissect Coronin 1B's effect on branching (Fig. 2G). In this assay, a pre-formed branched network intiated by Arp2/3 complex and VCA (first-wave) was incubated with various debranching/branch stabilization factors (second-wave) (treatments a - f, Fig. 2G,H) in the absence of G-actin, VCA and Arp2/3 to prevent formation of additional branched filaments. After two minutes, the reactions were stopped by flushing the chamber with rhodamine-phalloidin and the number of branched filaments per total filament length were counted. Coronin 1B decreased the density of actin branches (reactions b and c) and Cortactin increased the density of actin branches (reaction d). Furthermore, Coronin 1B reversed branch stabilization by Cortactin (reactions e and f). Together, these data indicate that Cortactin and Coronin 1B affect actin branches in a manner that is independent of the initial nucleation event.
Coronin 1B induces dissociation of Arp2/3 complex from sides of actin filaments
To further characterize the effects of Coronin 1B on branch stability, we used centrifugation to separate and quantify the fraction of Arp2/3 complex associated with actin filaments in nucleation reactions (Fig. 3A). Increasing amounts of Coronin 1B decreased the amount of Arp2/3 complex associated with F-actin; Cortactin overcame this effect of Coronin 1B and increased the amount of Arp2/3 complex associated with F-actin. Coronin 1B-induced Arp2/3 dissociation was independent of VCA activation since Coronin 1B also induced dissociation of Arp2/3 from the sides of pre-formed filaments (Fig. 3B-E). Cortactin competes with Coronin 1B-induced dissociation (Fig. 3E). Identical results were obtained with filaments nucleated by Spectrin-F-actin seeds (SAS), excluding a potential role of pointed end-Arp2/3 interactions in this assay.
Figure 3. Coronin 1B dissociates Arp2/3 complex from the sides of actin filaments.
A- Reactions contained 20 nM Arp2/3, 1 nM VCA, 1.5 μM actin and indicated amounts of Coronin 1B and/or Cortactin; polymerization proceeded 30 min at room temperature followed by sedimentation to pellet F-actin. Samples were separated by SDS-PAGE and Coomassie Blue stained or immunoblotted with indicated antibodies. Short/Long indicates duration of exposure of the immunoblot to film.
B,C,D- Pre-formed F-actin stabilized by phalloidin was mixed with 20 nM Arp2/3 complex and incubated at 4°C ON. Reactions were subjected to ultra-centrifugation to pellet F-actin and pellets suspended in phalloidin-containing MKEI-50 buffer. Samples were negative stainied and visualized by TEM. (B) reaction mixture before centrifugation; (C) supernatant; (D) resuspended pellet. Yellow arrowheads indicate Arp2/3 complex in association with actin filaments. Scale bars = 100 nm.
E- Actin (1.5 μM) filaments nucleated by Spectrin F-actin seeds (SAS) or spontaneously were pre-formed for 30 min and mixed with 50 nM Arp2/3 complex and the indicated concentrations of Coronin 1B and/or Cortactin for an additional 30 min. Reactions were incubated at room temperature and processed as in (A).
F- Actin (1.5 μM) filaments initiated by SAS were mixed with 25 nM Arp2/3 complex for 30 min at room temperature and then varying concentrations of Coronin 1B were added and reactions were incubated at 4°C overnight to reach equilibrium. Pellet samples were obtained and processed as in (A).
G- Reactions as in (F) with varying concentrations of Cortactin. Arp2/3 complex in pellet was quantified by densitometry and normalized to that in reactions lacking cortactin. Data from 7 independent experiments are presented as (mean ± SEM). One-phase exponential function was used to fit the data and calculate the EC50.
H- Experiments as in (F) with varying concentrations of Coronin 1B were analyzed as in (G). Data were compiled from the indicated number (N) of independent experiments. One-phase exponential decay was used to fit the data and calculate the IC50.
I,J- Actin (2 μM) filaments were pre-formed by spontaneous nucleation for 1 hr, mixed with indicated amounts of Coronin 1B and Cortactin, incubated at 4°C overnight to reach equilibrium and processed as in (A).
Quantitative immunoblotting indicated that Coronin 1B induced Arp2/3 dissociation from F-actin with a half-maximal concentration of 74 nM (Fig. 3F,H), while Cortactin increased the association of Arp2/3 with F-actin in a saturable manner with a half maximal concentration of 51 nM (Fig. 3G, S2B), consistent with its ability to stabilize actin branches. To test the molecular requirements for Coronin 1B's Arp2/3-dissociating activity, we examined different mutants of Coronin 1B. Coronin 1B S2D (defective in Arp2/3 binding) had ∼2 fold lower Arp2/3-dissociating activity, while Coronin 1B R30D (defective in F-actin binding) had ∼5 fold lower activity (Fig. 3H). Finally, a double mutant, Coronin 1B S2D, R30D (DD) did not dissociate Arp2/3 complex from F-actin (Fig. 3H). Thus, both Arp2/3 binding and F-actin binding are required for Coronin 1B to efficiently dissociate Arp2/3 complex from filaments. One possible explanation for the antagonism between Coronin 1B and Cortactin in these biochemical assays is that both proteins compete for a common binding site on F-actin. We performed actin co-sedimentation assays with varying amounts of Coronin 1B and fixed amounts of Cortactin or vise versa. Coronin 1B and Cortactin can simultaneously bind to actin filaments (Fig. 3I,J).
Coronin 1B localizes to actin branches both in vitro and in vivo
To characterize the effects of Coronin 1B on actin branches, we used negative staining and transmission electron microscopy (TEM) to visualize actin branches in the presence of Coronin 1B. The branch angle between mother and daughter filaments was not altered by Cortactin (Fig. 4A,B). In contrast, Coronin 1B perturbs the structure of branched filaments (Fig. 4A, yellow arrowheads), increasing the angle between the mother and daughter filaments (Fig. 4B).
Figure 4. Coronin 1B localizes to actin branches in vitro and in vivo.
A- Reactions containing 20 nM Arp2/3, 1 nM VCA and 1.5 μM actin were mixed with 2 μM phalloidin and incubated at 4°C overnight to reach equilibrium. An equal volume of 0.56 μM Coronin 1B, 0.63 μM Cortactin or buffer was added to each reaction 5 min before negative staining. Samples were visualized by TEM. Yellow arrowheads indicate abnormal actin branch structures. Scale bars = 100 nm.
B- Angles between the mother and daughter filaments were quantified and presented as mean ± 95% CI. Dunnett's multiple comparison test was used after one-way ANOVA to generate the p values (***, p<0.0001).
C- Actin (0.5 μM, 30% Oregon green labeled) was mixed 26 nM Arp2/3 complex and 37 nM VCA and actin branches were allowed to form in the flow chamber. After 20 min (time point 0 in the panel), 0.13 nM AlexaFluor-568 labeled Coronin 1B (labeled with ∼one dye molecule/protein) in imaging buffer was flowed into the chamber. Scale bars = 1 μm.
D- Reactions containing 20 nM Arp2/3, 1 nM VCA, 1.5 μM actin and either 150 nM Coronin 1B or 60 nM Cortactin were mixed with 1:100 dilution of primary antibodies, 1:25 dilution of colloidal gold secondary antibodies and 2 μM phalloidin and incubated at 4°C overnight. Samples were prepared by negative staining and visualized by TEM. Anti rabbit secondary antibodies were conjugated to 18-nm gold; anti-mouse secondary antibodies were conjugated to 6-nm gold. Stains: 1, polyclonal anti-Coronin 1B; 2, monoclonal anti-His tag (Coronin 1B-His); 3, polyclonal anti-p34Arc; 4, monoclonal anti-Cortactin; 5-6, monoclonal anti-Cortactin and polyclonal anti-p34Arc. Green circles highlight the 6-nm colloidal gold particles. Scale bars = 100 nm.
E- Samples were processed as in (D). Data compiled from 3 independent experiments. Chi squared test between the Cortactin-stained group (1) and the Coronin 1B-stained group (2) shows p = 0.0021. An expanded version of this table including staining controls is in the supplemental data section.
F- Platinum replica electron micrographs of lamellipodia in mouse embryo fibroblasts (MEF) immunolabeled for Coronin 1B with 18-nm gold (white dots). Expanded views of boxed regions (a-f) are presented in the lower panels; gold particles indicated by yellow arrowheads. Scale bar = 500 nm.
G,H- MEF cells were treated with either 0.05 μM or 0.1 μM cytochalasin D for 10 min and immunolabeled for Coronin 1B (G) or Arp2/3 complex (H). Yellow arrowheads indicate actin branches containing gold particles. Scale bars = 50 nm.
I- The distribution of Coronin 1B-labeled (red) or Arp2/3-labeled (green) actin branch angles. The 3D nature of platinum replica EM likely leads to a slight under-estimation of branch angle. Data compiled from 3 independent experiments. Student's t test of the branch angle shows p<0.001; (N = number of cells, n = number of branches).
Since Coronin 1B affects branch dynamics and alters the branch angle, we postulated that Coronin 1B must localize to actin branches. To test this idea, fluorescently labeled Coronin 1B was added to reactions containing pre-formed Arp2/3 branches. Fluorescent Coronin 1B preferentially localized to actin branches (Fig. 4C). Infrequently, we observed that actin branches disassembled after binding Coronin 1B (see Suppl. Movie 5).
To confirm the localization of Coronin 1B to branches, we used immunogold labeling in conjunction with negative staining and TEM. Immunogold labeling for Coronin 1B using two different antibodies preferentially decorated actin branches (Fig. 4D, 1-2). When 6 nm gold particles were used, we observed many gold particles at a single branch, suggesting that Coronin 1B may be multimeric at branches. As expected, Arp2/3 complex and Cortactin are also localized to actin branches (Fig. 4D, 3-4). We also examined the co-localization of Arp2/3, Cortactin and Coronin 1B at branches using double labeling with two different sized gold particles. Simultaneous labeling of Arp2/3 complex and Cortactin identified many branches containing both proteins (Fig. 4D, 5-6, 4E), consistent with reports that Cortactin remains associated with branches after nucleation (Egile et al., 2005). In contrast, with antibodies to Arp2/3 and Coronin 1B, we observed only one doubly-labeled branch out of nearly 1000 branches examined (Fig. 4E, Suppl. table 1). To exclude the possibility that the Coronin 1B and Arp2/3 antibodies did not simultaneously label branches due to steric hindrance, we tested alternate Arp2/3 complex antibodies. Again, no doubly-labeled branches were observed. In the absence of Arp2/3 complex, Coronin 1B does not induce the formation of end-to-side branches (data not shown). Thus, Arp2/3 complex and Coronin 1B are mutually exclusive at branches in vitro. We conclude that Coronin 1B targets pre-existing Arp2/3-containing branches and replaces Arp2/3 complex.
To determine if Coronin 1B localizes to branches in vivo, we used immunogold staining in conjunction with platinum replica electron microscopy (Svitkina and Borisy, 1999). Coronin 1B localized to actin branches in vivo (Fig. 4F,S6,8), as did Arp2/3 complex (Fig. 4H, S5,7). Coronin 1B also was detected at filament junctions containing multiple actin filaments. To better visualize single actin branches, we decreased the density of the actin network with low doses of cytochalasin D (Fig. 4G). In these conditions, Coronin 1B was localized at single filament branches. To determine the geometry of actin filament branches associated with either Arp2/3 complex or Coronin 1B, we measured the angle of labeled branches (Fig. 4I). Arp2/3-labeled junctions branch at an angle of 72.5° (mode) with a fairly normal Gaussian distribution of values, whereas the majority of Coronin1B-labeled junctions branch at larger angles (mode = 85°). Interestingly, a significant fraction of Coronin 1B-labeled branch junctions are more acute than Arp2/3 branches, suggesting that these branches may be more flexible. Thus, Coronin 1B localizes at the junctions of branched actin filaments in vivo and the majority of those junctions are oriented at larger angles than Arp2/3-containing branches.
Targeting active Coronin 1B to the plasma membrane reduces Arp2/3 branching and alters lamellipodial dynamics
To further investigate how Coronin 1B regulates dendritic actin networks, we targeted Coronin 1B to the plasma membrane where it is not normally located. Coronin 1B-GFP was engineered to contain a CAAX sequence to direct membrane targeting (WT-GAAX; Fig. 5A, B). Interestingly, we discovered that WT-GAAX becomes phosphorylated on Ser2, a site that negatively regulates Coronin 1B activity by decreasing the interaction between Coronin 1B and Arp2/3 complex (Fig. 5C) (Cai et al., 2005; Cai et al., 2007b). Phospho-WT-GAAX accumulates, in part, because membrane-targeted Coronin 1B cannot be dephosphorylated (Fig. S9).
Figure 5. Targeting of active Coronin 1B to the plasma membrane alters actin filament architecture and lamellipodial protrusion dynamics.
A- Schematic diagram of WT-GAAX; Coronin 1B S2A-GAAX is designated S2A-GAAX.
B- Rat2 cells expressing WT-GAAX were immunolabeled for p34Arc and F-actin. Scale bar = 5 μm.
C- Lysates from HEK293 cells expressing Coronin 1B-EGFP or WT-GAAX were immunoprecipitated with antibodies to GFP and immunoblotted with indicated antibodies. Cells were treated with or without 100 nM PMA for 30 minutes before lysis.
D- Rat2 cells expressing S2A-GAAX were immunolabeled as in (B).
E- The pixel intensity of p34Arc and F-actin were plotted vs. distance from the cell edge (mean ± SEM; N = number of cells). GFP fluorescence from both cell lines showed equal expression of WT-GAAX and S2A-GAAX (data not shown).
F- The ratio of p34Arc to F-actin intensity is plotted vs. distance from the cell edge (mean ± SEM).
G,H- Platinum replica electron micrographs of lamellipodia in Rat2 cells expressing WT-GAAX (H) or S2A-GAAX (G). Expanded views of boxed regions are presented on the right. Scale bars = 500 nm.
I- Actin filament densities of images as in panel G and H were quantified and plotted with distance away from the cell edge (mean ± SEM; N = number of cells).
J,K,L- Protrusion parameters of Rat2 cells expressing soluble Coronin 1B-GFP, WT-GAAX, or S2A-GAAX. Data are presented as mean ± 95% CI. Dunnett's multiple comparison test was used after one-way ANOVA to generate the p values against the control. >4 cells from each category were analyzed, n = number of protrusion events analyzed.
To circumvent this inhibitory phosphorylation, we targeted a non-phosphorylatable mutant of Coronin 1B (S2A-GAAX) to the plasma membrane (Fig. 5D, Fig. S9B). Arp2/3 complex staining in lamellipodia was reduced in cells expressing S2A-GAAX (Fig. 5D). We used a ratiometric variant of our contour-based quantitative immunofluorescence assay to measure the ratio of Arp2/3 to F-actin as a function of distance from the leading edge (Fig. 5E, F). The ratio of Arp2/3 to F-actin was reduced in cells expressing S2A-GAAX relative to that in cells expressing WT-GAAX or in uninfected control cells (data not shown). Platinum replicas were used to examine the architecture of actin filaments in the lamellipodia. Consistent with the immunofluorescent analysis, expression of S2A-GAAX reduces the density of the actin network in the lamellipodia relative to that observed in cells expressing WT-GAAX (Fig. 5G-I, S10).
To test the functional significance of Coronin 1B-induced changes in Arp2/3-branch density, we quantified lamellipodial dynamics in cells expressing soluble Coronin 1B-GFP or membrane-targeted Coronin 1Bs (Fig. 5J-L). Expression of soluble Coronin 1B-GFP did not alter lamellipodial dynamics. Expression of WT-GAAX induced a slight but significant increase in protrusion rate and a slight decrease in protrusion persistence. Expression of S2A-GAAX increased protrusion rate and decreased protrusion persistence, as well as significantly decreased protrusion distance (Fig. 5J-L). Thus, inappropriate localization of active Coronin 1B to the plasma membrane alters Arp2/3 branch density in the lamellipodia and affects protrusion dynamics at the leading edge.
Coronin 1B and Arp2/3 complex exhibit sequential spatio-temporal dynamics in lamellipodia
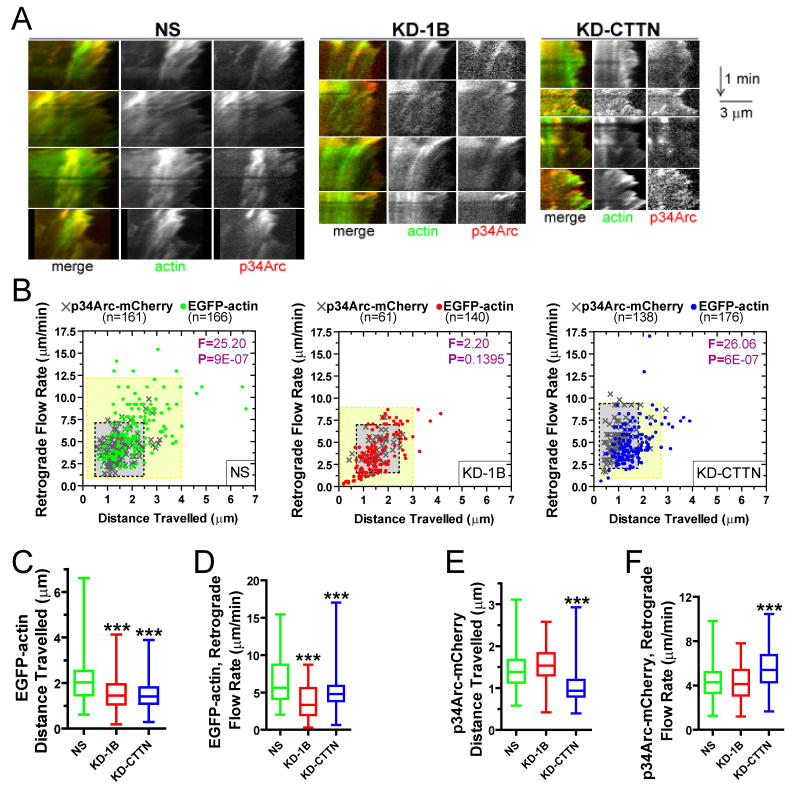
To study the relative spatial and temporal dynamics of actin, Coronin 1B, Cortactin and the Arp2/3 complex in lamellipodia, we used live-cell imaging of cells expressing functional fluorescent fusion proteins expressed in cells depleted of the endogenous proteins (Fig. S11). F-actin assembled in periodic bursts at the leading edge and, together with other actin-associated proteins, moved rearward by retrograde flow. Kymography was used to assess the rate of retrograde flow and distance travelled during retrograde flow for each protein. Actin, Cortactin, Coronin 1B and Arp2/3 complex exhibited similar retrograde flow rates (Fig. 6A, C), however, each of these proteins travelled a distinct distance from the leading edge during retrograde flow. Actin travelled the longest distance and Cortactin and Arp2/3 complex travelled the shortest distance. Coronin 1B travelled nearly twice as far from the leading edge as Cortactin and Arp2/3 complex (Fig. 6B). These data are consistent with previous observations that Coronin 1B extends deeper into the lamellipodia than either Arp2/3 complex or Cortactin (Cai et al., 2007a).
Figure 6. Coronin 1B, Cortactin an Arp2/3 complex have distinct dynamics in vivo.
A- Retrograde flow rate and distance travelled during retrograde flow of EGFP-tagged actin/actin-binding proteins presented as a scatter plot; >6 cells from each category were analyzed.
B- Distance traveled parameter from (A) presented as a box and whisker plot. (n = number of events analyzed)
C- Representative kymographs showing retrograde flow of EGFP tagged proteins: EGFP-actin, Cortactin-EGFP, p34Arc-EGFP and Coronin 1B-EGFP. Horizontal lines = 5 μm; vertical lines = 1 min. Yellow solid line illustrates distance travelled; the angle α was used to calculate the retrograde flow rate.
D- Normalized fluorescence intensity profiles of Coronin 1B-mCherry and p34Arc-EGFP expressed in Rat2 cells were extracted at varying distance from the cell edge. Three line profiles obtained 0, -0.4, -0.9 μm from the cell edge are shown; these data were used in (E) to calculate the correlation coefficient.
E- Line profiles were extracted as described in (D) for pair-wise combinations of labeled proteins. Each profile was used to calculate Pearson's correlation coefficient between the red and green channel. Results from >6 cells were grouped and plotted as a function of distance from the cell edge (mean ± SEM; N = number of protrusion events)
F- Kymographs were generated from a Rat2 cell expressing Cortactin-mCherry and p34Arc-EGFP (two examples shown). Left panel shows fluorescence intensity profiles of the two channels extracted from five lines as indicated on the kymograph. Horizontal lines = 1 μm; vertical lines = 1 minute.
G- Kymographs were generated from a Rat2 cell expressing Coronin 1B-mCherry and p34Arc-EGFP (two examples shown). Left panel shows fluorescence intensity profiles of the two channels extracted from five lines as indicated on the kymograph. The peak pixel intensity of both channels closest to the cell edge was traced and presented in the middle. Normalized fluorescence intensity profiles were extracted based on the distance from the cell edge. Profiles with 0, -0.4, -0.9 μm from the cell edge were plotted on the right, which are examples used in (H) for coefficient calculation.
H- Line profiles based on the distance to the cell edge were extracted as described in (G) from Rat2 cells expressing pair-wise combinations of fluorescent proteins. Each line profile with a given distance from the cell edge was used to calculate Pearson's correlation coefficient between the channels. Results from >6 cells were grouped together and plotted as a function of distance from the cell edge (mean ± SEM; N = number of flow events)
We also performed correlation analysis between Coronin 1B, Cortactin and Arp2/3 complex along the cell margin using dual-color live-cell imaging. Notably, Arp2/3 complex was distributed closer to the leading edge than Coronin 1B (Fig. 6D, S11A). To quantify these correlations, we adapted the contour-based pixel intensity extraction technique to calculate Pearson's correlation coefficient for pairwise combinations of proteins in different regions of the lamellipodium (Fig. 6E). Consistent with our biochemical experiments, the distributions of Coronin 1B and Cortactin are poorly correlated across the lamellipodia with the lowest value near the cell margin, while those of Cortactin and Arp2/3 complex are highly correlated across the entire lamellipodium (Fig. 6E).
To assess the spatio-temporal relationships for Arp2/3, Coronin 1B and Cortactin at a single location in the lamellipodium, we used kymographs generated from dual-color movies. As expected, Cortactin and Arp2/3 complex are highly correlated in both space and time (Fig. 6F, H). Interestingly, Coronin 1B and Arp2/3 exhibited sequential dynamics within the lamellipodium. Each Arp2/3 complex-rich region close to the membrane was gradually enriched with Coronin 1B concomitant with Arp2/3 complex depletion with distance from the leading edge (red/green line scans, Fig. 6G). Coronin 1B never accumulated without a pre-existing concentration of Arp2/3 complex, supporting the notion that Coronin 1B specifically targets Arp2/3-containing branches and replaces Arp2/3 at branched junctions. These qualitative observations are confirmed by Pearson's correlation analysis (Fig. 6H).
Depletion of Coronin 1B and Cortactin interfere with retrograde flow of actin and Arp2/3 complex
We also visualized the dynamics of actin and Arp2/3 complex in Coronin 1B- or Cortactin-depleted cells using two-color kymographs (Fig. 7A). In general, depletion of either protein reduced the distance travelled and the retrograde flow rate of actin (Fig. 7B-D). Thus, both Coronin 1B and Cortactin are required for normal dynamics of lamellipodial actin. Cortactin depletion decreased the distance Arp2/3 complex traveled rearward (Fig 7B, E, F), consistent with the in vitro branch stabilization activity of this protein. Interestingly, depletion of Cortactin increased the retrograde flow rate of Arp2/3, suggesting that Cortactin retards Arp2/3-actin branch movement in lamellipodia.
Figure 7. Depletion of either Coronin 1B or Cortactin affects the relative dynamics of Arp2/3 complex and actin in lamellipodia.
A- Kymographs showing retrograde flow of EGFP-actin and p34Arc-mCherry in cells expressing the indicated shRNA. Horizontal lines = 3 μm; vertical lines = 1 min.
B- Retrograde flow rate and distance traveled of EGFP-actin (dots) and p34Arc-mCherry (crosses) presented as scatter plots from cells expressing the indicated shRNAs; >16 cells from each category were analyzed. Yellow boxes represent the mean of actin for both parameters ± 2×SD (95% of data points); gray boxes represent the mean of p34Arc for both parameters ± 2×SD. Analysis of covariance was used to generate the F and P values for each condition. (n = number of flow events)
C,D- Distance traveled during retrograde flow of (B) presented as box and whisker plots. Dunnett's multiple comparison test was used after one-way ANOVA to generate the p values against the control (***, P<0.0001).
E,F- Retrograde flow rates for EGFP-actin and pArc34-mCherry of (B) was processed as in (C,D)
Several interesting features of the coordinate dynamics of actin and Arp2/3 complex in lamellipodia were noted. If all of the actin in lamellipodia were in the form of Arp2/3-containing branched actin filaments, then actin and Arp2/3 complex should exhibit similar dynamics. However, actin and Arp2/3 complex exhibit distinct distributions of retrograde flow rates and distances traveled in control cells (Fig. 7B, left panel). Surprisingly, in Coronin 1B-depleted cells, the dynamic properties of actin and the Arp2/3 complex are synchronized (Fig. 7B, middle panel), suggesting that Coronin 1B functions to remodel Arp2/3-containing actin networks into other forms. In contrast, depletion of Cortactin leads to divergence in the dynamic properties of actin and Arp2/3 (Fig. 7B, right panel). We tested the co-variance between Arp2/3 complex and actin dynamics to quantitatively access the effects of depleting Coronin 1B and Cortactin on kinematics of actin and Arp2/3 complex. Control cells and Cortactin-depleted cells exhibit divergent dynamics of Arp2/3 and actin, but Coronin 1B-depleted cells show no significant difference in the co-variance of these markers.
Discussion
In this work we show that Coronin 1B antagonizes Cortactin in vitro and in vivo. Furthermore, we show that Coronin 1B remodels Arp2/3-containing actin branches and plays an important role in coordinating actin dynamics within lamellipodia.
Antagonism between Coronin 1B and Cortactin
One of the most striking observations we made in this work is that Coronin 1B potently antagonizes Cortactin and vice versa. In lamellipodia, these proteins are localized in distinct patterns and interact with distinct pools of Arp2/3 complex, with little interaction between the pools. Coronin 1B and Cortactin compete in multiple biochemical assays in vitro and in functional analysis in vivo. This competition is not due solely to competition for binding to F-actin, but also involves Arp2/3 binding. Future studies will focus on defining the precise molecular interactions required for this competition. While the competition between Coronin 1B and Cortactin in vitro was efficient, results for cellular experiments suggest a more complex interaction between these proteins in vivo. The incomplete rescue observed upon double depletion of these proteins indicates that Coronin 1B and Cortactin each participate in additional functions within lamellipodia.
Coronin 1B remodels the Arp2/3-containing actin network
Remodeling of the Arp2/3-containing dendritic network by Coronin 1B may generate a second actin network with distinct properties. Our data from dual-color live-cell imaging indicate that Arp2/3 complex and actin have different dynamics within lamellipodia. Lamellipodial actin flows rearward to a greater extent than Arp2/3 complex in fibroblasts. These results are consistent with recent observations from Drosophila S2 cells obtained using speckle microscopy (Iwasa and Mullins, 2007). Intriguingly, depleting Coronin 1B leads to near perfect synchronization of Arp2/3 and actin dynamics in lamellipodia. Taken together with our biochemical data and observations of branch angles in vivo, we conclude that Coronin 1B remodels Arp2/3-containing branches into a new network that is free of Arp2/3 complex. Other studies have identified diverse and seemingly independent actin networks within lamellipodia (Ponti et al., 2004). Future studies will be required to delineate the role of Coronin 1B-mediated actin remodeling in generating diversity in lamellipodial actin networks.
What advantage might remodeling the lamellipodial actin network provide the cell? First, this mechanism allows efficient recycling of Arp2/3 complex for continued branched filament nucleation at the leading edge. Although the cytosolic concentration of Arp2/3 is high (Pollard et al., 2000), autocatalytic branching could locally deplete Arp2/3 complex and stall or delay dendritic network assembly. Replacing Coronin 1B at branch junctions would allow continued branch formation while maintaining existing branched filaments. Second, Coronin 1B provides the flexibility to either maintain branches or initiate branch disassembly on demand if Slingshot is recruited (Cai et al., 2007b). Finally, having both Cortactin-stabilized Arp2/3 branch junctions and Coronin 1B-branched filaments in lamellipodia allows differential coupling of the actin network to cellular structures such as nascent adhesions or trafficking vesicles.
Regulation of Coronin 1B branch remodeling activity
Branch remodeling by Coronin 1B is extremely potent in vitro, implying that a strict regulation of this activity likely operates in vivo. Our membrane targeting experiments revealed a possible mechanism for regulating branch remodeling. Previously, we showed that PKC phosphorylation of Ser2 on Coronin 1B reduced binding to Arp2/3 complex (Cai et al., 2005). Upon membrane targeting, Ser2 of Coronin 1B became phosphorylated and Arp2/3 interaction was strongly reduced, consistent with reports of a constitutively-active, plasma membrane-resident pool of PKC (Gallegos et al., 2006). Our pharmacological experiments revealed that WT-GAAX is hyper-phosphorylated at Ser2 because membrane-targeted Coronin1B could not be efficiently dephosphorylated by Slingshot 1L. This result indirectly suggests that activation of Cofilin by dephosphorylation also may not occur at the plasma membrane.
Membrane targeting of non-phosphorylatable Coronin 1B (S2A) reduced the density of Arp2/3 branches in the dendritic network. Localizing active Coronin 1B to the site where Arp2/3-containing actin branches are generated may inhibit stable branch formation and promote debranching, as observed using TIRF microscopy when nascent daughter filaments rapidly dissociated from mother filaments in the presence of Coronin 1B. Alternately, active Coronin 1B tethered at the membrane may inhibit filament nucleation. Regardless of the precise mechanism, this result illustrates the importance of regulating Coronin 1B spatially within the lamellipodial actin network. One question is whether synthetic membrane targeting is physiologically relevant. Interestingly, retention of Coronin 1A on phagosomes containing live M. tuberculosis bacteria blocks phagolysosome maturation, a processes that is dependent on actin polymerization (Anes et al., 2003; Ferrari et al., 1999). Future studies will examine how targeting of Coronins controls membrane-associated actin dynamics in normal and pathophysiological circumstances.
A working model for Coronin 1B function
Based on data presented here and on our previous work, we propose the following speculative model to explain the role of Coronin 1B in branched actin remodeling/disassembly (Fig. S12). Near the plasma membrane, activated Arp2/3 complex nucleates a new filament on the side of an existing filament, forming a branched junction. Cortactin associates with Arp2/3-containing branches, protecting them from spontaneous disassembly. Coronin 1B targets Arp2/3-containing actin branches in an antagonistic fashion with Cortactin. Targeting by Coronin 1B requires both F-actin and Arp2/3 binding for maximum efficiency and involves multimeric Coronin 1B complexes. Subsequently, Arp2/3 complex dissociates from the branch, leaving multimers of Coronin 1B in its place. Coronin 1B-branches have a larger, but more variable angle than Arp2/3-branches. Previously, we showed that Coronin 1B binds Slingshot 1L, the activating phosphatase of Cofilin (Cai et al., 2007b). Recruitment of Slingshot 1L by Coronin 1B brings it into close proximity to F-actin, an essential cofactor for its activation. Slingshot 1L activation leads to local dephosphorylation and activation of Cofilin. Active Cofilin promotes the release of inorganic phosphate (Pi) from actin filaments to which it binds. Since Coronin 1B binds with ∼50-fold lower affinity to ADP-F-actin than to ATP or ADP-Pi-F-actin (Cai et al., 2007a), local Pi release would lead to disassembly of Coronin 1B-containing branches. In addition, locally activated Cofilin may promote filament turnover via its severing activity in the vicinity of the recently disassembled branch. This model provides a mechanism for Coronin 1B function in promoting turnover of Arp2/3-branched actin networks. Although our studies have focused on lamellipodia, the mechanism of Coronin function revealed by these experiments likely applies to all Arp2/3-branched networks such as those found on endosomes, in dendritic spines or phagocytic cups.
Experimental Procedures
Descriptions of plasmid construction, cell culture and antibodies/reagents are in Supplemental Experimental Procedures. Immunoprecipitations were performed using standard conditions (Cai et al., 2007b); additional details are included in the Supplemental Experimental Procedures.
TIRF and other in vitro actin assays
Recombinant and native proteins were prepared as described (Cai et al., 2007b). TIRF-based actin branching assay was performed as previously described (Amann and Pollard, 2001; Kuhn and Pollard, 2005). Briefly, glass coverslips and parafilm spacers were used to prepare imaging chambers. NEM-inactivated myosin was used to immobilize actin filaments. Protein mixutres were diluted in freshly prepared TIRF imaging buffer (100 mM KCl, 2 mM MgCl2, 2 mM EGTA, 20 mM imidazole pH 7.0, 30 mM glucose, 1% 4000 cP methylcellulose, 200 mM DTT, 0.4 mM ATP, 40 μg/ml catalase, 200 μg/ml glucose oxidase) and imaged at 5- or 10-sec intervals by objective based TIRF microscopy. Under standard conditions, 0.5 μM actin (30% Oregon green labeled) was nucleated in the presence of 13.3 nM Arp2/3 and 13.3 nM VCA. To calculate debranching frequency, the length of the daughter filament was measured in ImageJ at the moment of its release from the mother filament. Debranching frequency = (barbed end elongation rate) / (de-branched filament length). Second-wave debranching was performed as described: First, reactions were initiated by mixing 0.5 μM G-actin with 13.3 nM Arp2/3 complex and 13.3 nM GST-VCA, and incubated for 10 min to form actin branches inside the imaging chamber. Proteins mixtures in TIRF imaging buffer (all including 0.8 μM capping protein) were flushed into the reaction chambers and incubated for 2 min. Control experiments confirmed that this procedure completely removed the residual VCA and G-actin from the first step and blocked further branched nucleation. Finally, Rhodamine-phalloidin (1.5 μM) was flushed into the chamber to stop the reactions and stain the actin filaments. Detailed methods for actin co-sedimentation-based assays (eg. Arp2/3 dissociation assay) and pyrene actin assembly assays are described in Supplemental Experimental Procedures.
Electron microscopy
Negative staining was performed using 2% (w/v) uranyl acetate of samples on glow discharged carbon grids (in vitro samples) or fibronectin-coated Colloidin grids (cells). Immunogold staining was performed using primary and secondary antibodies as described in the legends. The grids were examined in a Philips CM12 microscope. The images were taken at magnification 10,000× or 45,000× on Kodak SO-163 film, scanned at resolution 2500 dpi and adjusted for publication using Photoshop. Platinum replica EM was performed as described (Cai et al., 2007b). For immunogold labeling of platinum replicas, the extraction buffer was modified to preserve Coronin 1B staining as described in Supplemental Experimental Procedures.
Light microscopy
For immunofluorescent staining, the cells were fixed, stained and mounted as described (Cai et al., 2007b). Images were captured using a spinning disk confocal scan head (Yokogawa CSU-10) attached to an inverted microscope (IX-81, Olympus), a CCD camera (OCAR-ERG, Hamamatsu), and controlled by either Andor iQ or Slidebook software. Lamellipodial dynamics were analyzed using phase contrast kymography as described (Cai et al., 2007b). Retrograde flow analysis was performed using movies captured by spinning disk confocal microscopy (1-second interval, 1×1 binning for single channel signal; 2-second interval, 2×2 binning for dual-color signal). Kymographs were generated from these movies and analyzed to calculate the retrograde flow rate and the distance traveled as described in the legend.
Image and statistical analysis
Branch angle measurement was performed using ImageJ. Pearson's correlation coefficient was computed from regions of interest extracted using ImageJ and calculated based on normalized intensities in Excel. Other statistical analyses were performed using Prism.
Supplementary Material
Acknowledgments
We acknowledge F. Gertler, S. Rogers, R. Cheney and members of the Bear lab for critical comments on this manuscript, M. Welch for the p41Arc antibody and the substantial assistance of Jack Griffith in the LCCC Electron Microscopy core (supported by NIH CA16086 and GM31819). This work was supported by NIH grants to DAS (GM067222) and JEB (GM083035). JEB is also supported by the Sontag Foundation and the Carolina Center for Cancer Nanotechnology Excellence (NCI, 1U54CA119343).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amann KJ, Pollard TD. Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc Natl Acad Sci U S A. 2001;98:15009–15013. doi: 10.1073/pnas.211556398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anes E, Kuhnel MP, Bos E, Moniz-Pereira J, Habermann A, Griffiths G. Selected lipids activate phagosome actin assembly and maturation resulting in killing of pathogenic mycobacteria. Nat Cell Biol. 2003;5:793–802. doi: 10.1038/ncb1036. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD, Mullins RD. Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr Biol. 2000;10:1273–1282. doi: 10.1016/s0960-9822(00)00749-1. [DOI] [PubMed] [Google Scholar]
- Bryce NS, Clark ES, Leysath JL, Currie JD, Webb DJ, Weaver AM. Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr Biol. 2005;15:1276–1285. doi: 10.1016/j.cub.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Cai L, Holoweckyj N, Schaller MD, Bear JE. Phosphorylation of coronin 1B by protein kinase C regulates interaction with Arp2/3 and cell motility. J Biol Chem. 2005;280:31913–31923. doi: 10.1074/jbc.M504146200. [DOI] [PubMed] [Google Scholar]
- Cai L, Makhov AM, Bear JE. F-actin binding is essential for coronin 1B function in vivo. J Cell Sci. 2007a doi: 10.1242/jcs.007641. [DOI] [PubMed] [Google Scholar]
- Cai L, Marshall TW, Uetrecht AC, Schafer DA, Bear JE. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 2007b;128:915–929. doi: 10.1016/j.cell.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosen-Binker LI, Kapus A. Cortactin: the gray eminence of the cytoskeleton. Physiology (Bethesda) 2006;21:352–361. doi: 10.1152/physiol.00012.2006. [DOI] [PubMed] [Google Scholar]
- Egile C, Rouiller I, Xu XP, Volkmann N, Li R, Hanein D. Mechanism of filament nucleation and branch stability revealed by the structure of the Arp2/3 complex at actin branch junctions. PLoS Biol. 2005;3:e383. doi: 10.1371/journal.pbio.0030383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari G, Langen H, Naito M, Pieters J. A coat protein on phagosomes involved in the intracellular survival of mycobacteria. Cell. 1999;97:435–447. doi: 10.1016/s0092-8674(00)80754-0. [DOI] [PubMed] [Google Scholar]
- Foger N, Rangell L, Danilenko DM, Chan AC. Requirement for coronin 1 in T lymphocyte trafficking and cellular homeostasis. Science. 2006;313:839–842. doi: 10.1126/science.1130563. [DOI] [PubMed] [Google Scholar]
- Gallegos LL, Kunkel MT, Newton AC. Targeting protein kinase C activity reporter to discrete intracellular regions reveals spatiotemporal differences in agonist-dependent signaling. J Biol Chem. 2006;281:30947–30956. doi: 10.1074/jbc.M603741200. [DOI] [PubMed] [Google Scholar]
- Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- Humphries CL, Balcer HI, D'Agostino JL, Winsor B, Drubin DG, Barnes G, Andrews BJ, Goode BL. Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J Cell Biol. 2002;159:993–1004. doi: 10.1083/jcb.200206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa JH, Mullins RD. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr Biol. 2007;17:395–406. doi: 10.1016/j.cub.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaffy RE, Pollard TD. Kinetics of the formation and dissociation of actin filament branches mediated by Arp2/3 complex. Biophys J. 2006;91:3519–3528. doi: 10.1529/biophysj.106.080937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Welch MD, Drubin DG. Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nat Cell Biol. 2006;8:826–833. doi: 10.1038/ncb1443. [DOI] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetrecht AC, Bear JE. Coronins: the return of the crown. Trends Cell Biol. 2006;16:421–426. doi: 10.1016/j.tcb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Weaver AM, Karginov AV, Kinley AW, Weed SA, Li Y, Parsons JT, Cooper JA. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr Biol. 2001;11:370–374. doi: 10.1016/s0960-9822(01)00098-7. [DOI] [PubMed] [Google Scholar]
- Weed SA, Karginov AV, Schafer DA, Weaver AM, Kinley AW, Cooper JA, Parsons JT. Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J Cell Biol. 2000;151:29–40. doi: 10.1083/jcb.151.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Mullins RD. Cellular control of actin nucleation. Annu Rev Cell Dev Biol. 2002;18:247–288. doi: 10.1146/annurev.cellbio.18.040202.112133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.