Summary
PKR (protein kinase R) is a central component of the interferon antiviral defense pathway. Upon binding dsRNA, PKR undergoes autophosphorylation reactions that activate the kinase. PKR then phosphorylates eIF2α, thus inhibiting protein synthesis in virally-infected cells. Here, we define the mechanism of PKR activation using a series dsRNAs of increasing length. A minimal dsRNA of 30 bp is required to bind two PKR monomers and 30 bp is the smallest dsRNA that elicits autophosphorylation activity. Thus, the ability of dsRNAs to function as PKR activators is correlated with binding of two or more PKR monomers. Sedimentation velocity data fit a model where PKR monomers sequentially attach to a single dsRNA. These results support an activation mechanism where the role of the dsRNA is to bring two or more PKR monomers in close proximity to enhance dimerization via the kinase domain. This model explains the inhibition observed at high dsRNA concentrations and the strong dependence of maximum activation on dsRNA binding affinity. Binding affinities increase dramatically upon reducing the salt concentration from 200 to 75 mM NaCl and we observe that a second PKR can bind to the 20 bp dsRNA. Nonspecific assembly of PKR on dsRNA occurs stochastically without apparent cooperativity.
Keywords: analytical ultracentrifugation, protein-nucleic acid interactions, protein kinase, innate immunity, autophosphorylation
Introduction
Protein kinase R (PKR) is an interferon-induced kinase that plays a key role in the innate immunity response to viral infection.1;2 The enzyme is synthesized in a latent state but it is activated by binding dsRNA to undergo autophosphorylation at multiple serine, threonine and tyrosine residues.3 The most well characterized cellular substrate of PKR is the alpha subunit of eukaryotic initiation factor eIF2. Phosphorylation of eIF2á inhibits protein synthesis in virally-infected cells. Thus, production of dsRNA during viral infection4 results in PKR activation and subsequent inhibition of viral and host protein synthesis. The importance of PKR in antiviral defense is underscored by the large number of viruses that encode PKR inhibitors.5 PKR also functions in the control of cell growth and proliferation and as a tumor suppressor protein 6;7
PKR is a member of the family of stress-response kinases that includes PKZ, GCN2, HRI and PEK that inhibit translation by phosphorylation of eIF2á.8 These enzymes all contain a conserved catalytic domain linked to different regulatory domains. In PKR, the regulatory module consists of an N-terminal double-stranded RNA binding domain (dsRBD). The dsRBD consists of two tandem dsRNA binding motifs. Each of the motifs adopts the αβββα fold typically found in dsRNA binding modules with a short unstructured linker lying between the two folded units.9;10 The dsRBD recognizes dsRNA over dsDNA or RNA-DNA heteroduplexes but binds without sequence specificity.11 Like other protein kinases, the PKR catalytic domain is comprised of N-terminal and C-terminal lobes. A complex of the PKR kinase domain with eIF2α crystallized as a dimer with the interface formed by the kinase N-terminal lobes.12 Helix αC in the N-lobe of the kinase domain forms part of the dimer interface12 and conformational changes in this helix often regulate protein kinase activity13, suggesting that dimerization may allosterically modulate kinase activity. The ~ 80 residue linker region lying between the dsRBD and kinase is flexible or dynamic.14
PKR activation by dsRNA has been extensively studied and several molecular mechanisms have been proposed.15 In the autoinhibition model, the latent enzyme exists in a closed conformation mediated by interaction of the dsRBD with the kinase domain that blocks substrate access. Binding of dsRNA activates PKR by inducing a conformational change that relieves the latent enzyme of inhibition. In support of this mechanism, an interaction of the second dsRNA binding motif with the kinase domain has been reported based on NMR chemical shift perturbation experiments.10;16 However, several lines of evidence challenge the autoinhibition model. The strength of this interaction is too weak to form a stable complex.17 Equilibrium and kinetic analysis of nucleotide binding to PKR indicate that substrate accessibility of the kinase is not modulated by activation state.18 Sedimentation velocity19, AFM18 and NMR14 measurements all support an extended structure for PKR rather than a closed structure predicted by the autoinhibition model. Other models emphasize the role of dimerization in PKR activation.1;14;15;20 PKR is capable of dimerizing in the absence and the presence of dsRNA.19;21–23 Dimerization is sufficient to activate PKR in the absence of dsRNA.19 Fusion of a heterologous dimerization domain with the PKR kinase domain enhances autophosphorylation.24;25 A defining feature of PKR is the “bell-shaped” curve for activation where low concentrations of dsRNA activate but higher concentrations are inhibitory.26;27 These results can be rationalized in a model where low concentrations of dsRNA favor assembly of multiple proteins – possibly assembling as dimers – on a single dsRNA whereas higher dsRNA concentrations dilute PKR monomers onto separate molecules of dsRNA.28 Hybrid autoinhibition/dimerization models have also been proposed where dsRNA binding induces a conformation change in PKR that leads to protein dimerization and activation.14;29
The ability of dsRNA sequences to bind and activate PKR is dependent on length. The dsRBD of PKR can bind to sequences as short as 15–16 bp.11;30 As expected for a nonspecific interaction, the binding stoichiometries increase with dsRNA length.11;30;31 For sequences shorter than 30 bp, the binding stoichiometries and affinities conform to an overlapping ligand binding model where the domain binds to multiple faces of the dsRNA duplex and overlaps along the helical axis.30;32 It was originally reported that PKR activation requires a minimum dsRNA length of ~30 bp and reaches a maximum by 85 bp.27;33 However, recent studies have demonstrated PKR activation by 19–21 bp siRNAs containing 2 nt 3’ overhangs34;35 or even a blunt-ended 19 nt dsRNA.34
In the present study, we have addressed the molecular mechanism for PKR activation by dsRNA by systematically correlating binding stoichiometries and affinities with activation under controlled conditions. Nonspecific protein-nucleic acid interactions are labile and are readily perturbed by the separation of free and bound forms in conventional gel-shift and filter binding assays 32 Therefore, we have employed solution biophysical methods to define PKR binding to a series of dsRNAs of increasing length under conditions that can be directly related to the enzymatic activity assays. The results support a model where sequential binding of two or more PKR monomers on a single dsRNA leads to dimerization and activation.
Results
Correlation of dsRNA binding stoichiometry and activation
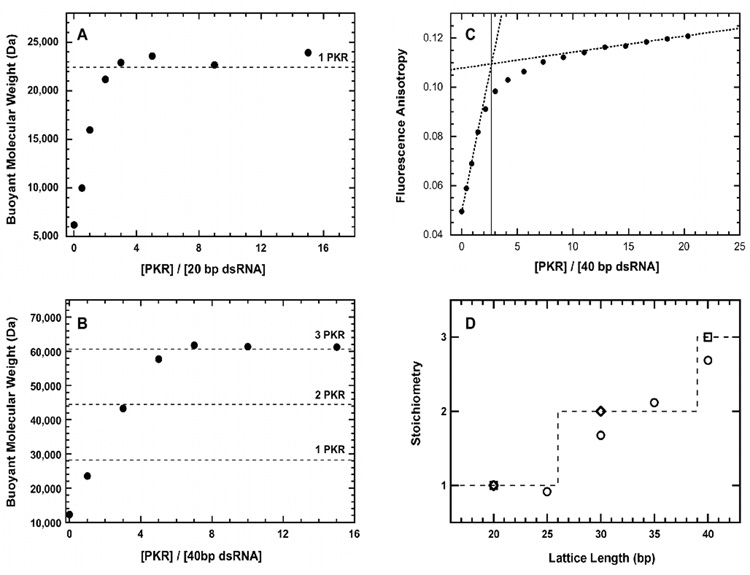
We have measured the PKR binding stoichiometries and enzymatic activation using a truncation series ranging from 20 bp to 40 bp in 5 bp increments. Our previous analysis of dsRBD binding to this panel of dsRNA sequences was performed in 75 mM NaCl.30;32 However, the current measurements were made at 200 mM NaCl because PKR aggregates upon binding dsRNAs of 30 bp or longer at lower salt concentrations (Lemaire and Cole, data not shown). Binding stoichiometries were measured using two independent approaches. Figure 1A shows a sedimentation equilibrium titration of PKR binding to a 20 bp dsRNA. The increase in the buoyant molecular weight at saturating concentrations of PKR corresponds to binding of one PKR / 20 bp dsRNA. As expected for nonspecific binding, the stoichiometry increases with RNA length and three PKR monomers bind to the 40 bp dsRNA (Figure 1B). The sedimentation equilibrium results were confirmed using fluorescence anisotropy titrations peformed under stoichiometric binding conditions where [dsRNA] >> Kd. Figure 1C shows that the equivalence point for binding of PKR to the fluorescein-labeled 40 bp dsRNA corresponds to a stoichiometry of 2.69 PKR / dsRNA, in fairly good agreement with the sedimentation data. Figure 1D summarizes the results of sedimentation and fluorescence stoichiometry assays for five dsRNAs ranging from 20–40 bp. Significantly, At least 30 bp are required for binding two PKRs.
Figure 1.
Stoichiometry of PKR binding to dsRNA. A) Sedimentation equilibrium titration of 20 bp dsRNA. Conditions: RNA concentration, 0.5 µM; rotor speed, 20,000 RPM; temperature, 20°C; wavelength, 260 nm. The dashed line indicates the predicted buoyant molecular weight of a 1:1 PKR-dsRNA complex. B) Sedimentation equilibrium titration of 40 bp dsRNA. Conditions: RNA concentration, 0.3 µM; rotor speed, 12,000 RPM; temperature, 20°C; wavelength, 260 nm. The dashed lines indicated the predicted buoyant molecular weights of 1:1, 2:1 and 3:1 PKR-dsRNA complexes. C) Fluorescence anisotropy titration of fluorescein-labeled 40 bp RNA. Conditions: RNA concentration, 5 µM; temperature, 20°C. Each data point represents the average of 3–4 measurements. A binding stoichiometry of 2.69 (solid line) was determined by linear extrapolations (dotted lines) of the rising and plateau regions of the titration. D) Summary of stoichiometries obtained by multiple techniques. Sedimentation equilibrium (□), fluorescence anisotropy (Δ) and sedimentation velocity (◊).
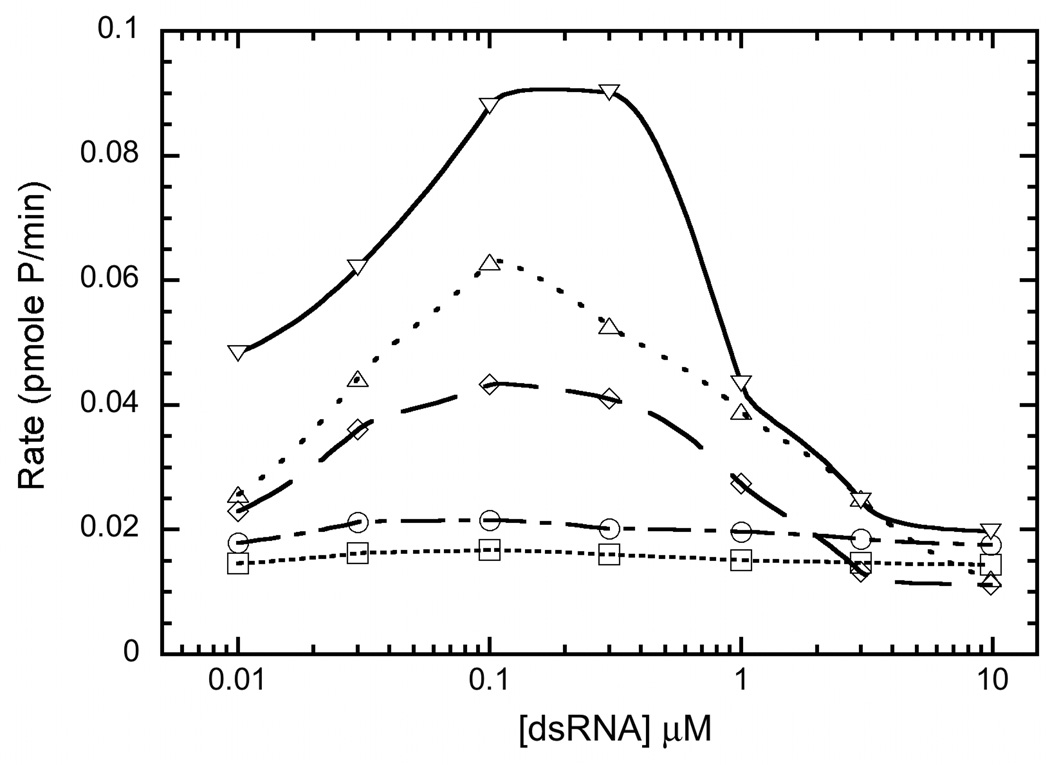
The dsRNA binding stoichiometries were correlated with enzymatic activation using autophosphorylation assays. Measurements were performed at a PKR concentration of 200 nM, which is below the concentrations where dsRNA-independent activation is observed.19 Figure 2 plots the rate of PKR autophosphorylation as a function of dsRNA concentration for sequences ranging from 20 to 40 bp. The 20–25 bp sequences failed to activate above background up to a concentration of 10 µM. The 30 bp sequence induced autophosphorylation, indicating that the ability of a dsRNA to activate is correlated with binding of two PKR monomers. Note that the earlier studies showing activating of PKR by 19–21 bp dsRNAs34;35 were performed at lower salt concentrations and PKR binding is strongly dependent on ionic strength (see below). As previously reported26;27, higher dsRNA concentrations inhibit, consistent with dilution of PKR dimers onto separately dsRNA molecules and the maximal activation level increases with dsRNA length. The maximum rates occur near 100–300 nM, which are close to the stoichiometric value of 2 PKR: RNA.
Figure 2.
Activation of PKR by dsRNA. (□), 25 bp RNA (○), 30 bp RNA (◊), 35 bp RNA (△) and 40 bp RNA (▽). PKR (0.2 µM) was preincubated with dsRNA for 10 minutes at 32°C in AU 200 buffer supplemented with 5 mM MgCl2. Autophosphorylation was initiated with 0.4 mM ATP containing 4 µCi [γ-32P] ATP. The reactions were quenched with addition of sample loading buffer after 20 min. The samples were analyzed by SDS PAGE and phosphorimaging.
Analysis of PKR binding to activating and nonactivating dsRNAs
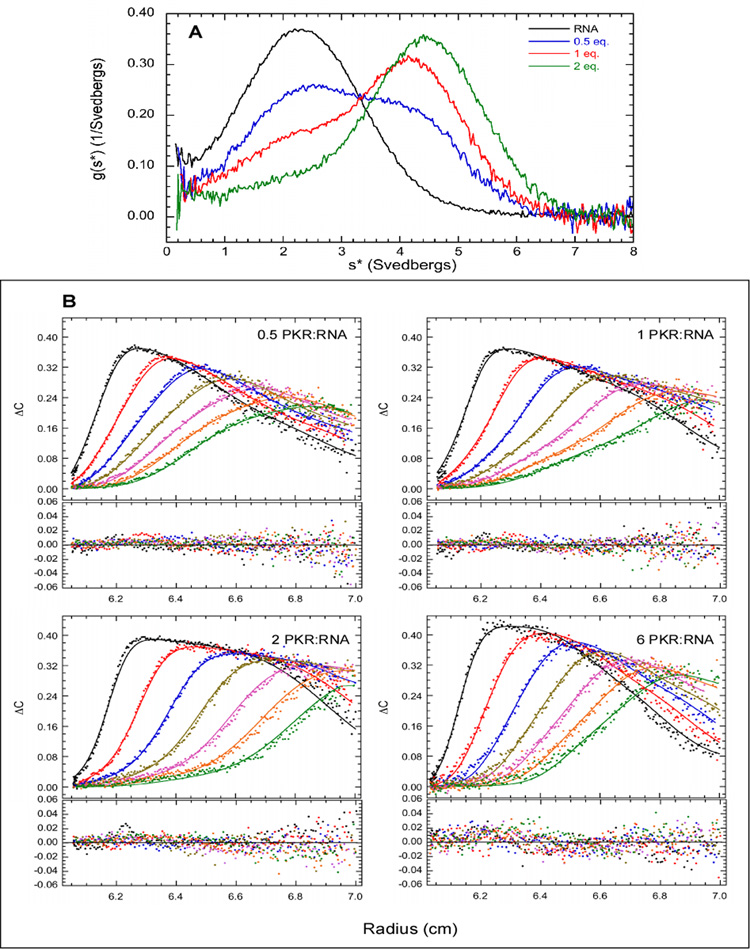
The 20 bp and 30 bp sequences were selected for detailed analysis of PKR binding by sedimentation velocity compare how PKR interacts with nonactivating and activating dsRNAs. Samples were prepared at multiple PKR:RNA ratios and measurements were performed using absorbance detection at 260 nm to detect free dsRNA and protein-RNA complexes. Initially, the data were analyzed using the time derivative method36 to define the binding mechanism. Figure 3A shows the normalized g(s*) distributions obtained for binding of PKR to the 20 bp dsRNA. In the absence of PKR, a single feature is observed near s = 2.5 S. The distribution fits well to a single species model with a molecular weight of 12.4 kDa, which agrees with the predicted value of 12.75 kDa for the 20 bp duplex. The corrected sedimentation coefficient of s20,w = 2.61 S corresponds to a frictional ratio of f/f0 = 1.38. The sedimentation coefficient is close to the value of s20,w of 2.37 S predicted from an earlier study of the hydrodynamic properties of short DNA and RNA duplexes,37 indicating that the 20 bp dsRNA adopts a typical duplex conformation in solution.
Figure 3.
Sedimentation velocity analysis of PKR binding to a 20 bp dsRNA. A) Normalized g(s*) distributions of 1 µM dsRNA (black), dsRNA + 0.5 eq. PKR (blue), dsRNA + 1 eq. PKR (red), dsRNA + 2 eq. PKR (green). The distributions are normalized by area. B) Global analysis of sedimentation velocity difference curves. The data were subtracted in pairs and four data sets at the indicated ratios of PKR: dsRNA were fit to 1:1 binding stoichiometry model. The top panels show the data (points) and fit (solid lines) and the bottom panels show the residuals (points). The best fit parameters are shown in Table 1. For clarity, only every 4th difference curve is shown. Conditions: rotor speed, 50,000 RPM; temperature, 20°C; wavelength, 260 nm; scan interval, 6 minutes.
Upon addition of PKR, the 2.5 S peak decreases in amplitude and a new feature develops at higher S, consistent with formation of an dsRNA-protein complex. The peak maximum of the new feature shifts continuously with increasing concentration of PKR, consistent with rapid exchange on the timescale of the sedimentation velocity experiment. Model – dependent analysis was performed by subtracting the sedimentation velocity traces in pairs for each sample to remove systematic noise and fitting the resulting difference curves to association models using the program SEDANAL.38 Figure 3B shows a fit of the same data depicted in Figure 3A to a simple 1:1 binding model,
| (1) |
where R refers to RNA and P refers to PKR. Based on the shift in peak position for the dsRNA-protein complex with concentration, a rapid equilibrium was assumed. The data fit well to this model with randomly-distributed residuals. The best fit Kd of 859 nM (Table 1) is relatively weak. The fit was not improved using an explicit kinetic model where the association and dissociation rates were treated at adjustable parameters (data not shown). The fitted sedimentation coefficient for the dsRNA-protein complex of 5.14 S corresponds to a frictional ratio of f/f0 =1.34.
Table 1.
PKR-dsRNA interaction parameters.a
| dsRNA | PKR | [NaCl] mM | Model | Kd1 | Kd2 | s(RP)c | s(RP2)c | RMSd |
|---|---|---|---|---|---|---|---|---|
| 20 bp | WTb | 200 | R+P ↔RP | 859 (746, 987) nM |
5.14 (5.03, 5.25) |
0.00961 | ||
| 30 bp | WT | 200 | R+P ↔RP RP+P ↔RP2 |
87 (49, 148) nM |
970 (690, 1450) nM |
4.85 (4.68, 5.09) |
6.88 (6.70, 7.13) |
0.00772 |
| 30 bp | WT | 200 | R+P ↔RP 2RP↔R2P2 |
932 nM | 1.65 uM | 7.52 | 5.31e | 0.0100 |
| 30 bp | R262D | 200 | R+P ↔RP RP+P ↔RP2 |
93 (51,157) nM |
1070 (620, 1730) nM |
4.94 (4.77, 5.14) |
7.32 (6.97, 7.76) |
0.00887 |
| 20 bp | WT | 75 | R+P ↔RP RP+P ↔RP2 |
15.0 (8.5, 25.3) nM |
228 (148, 351) nM |
4.48 (4.40, 4.57) |
6.60 (6.46,6.78) |
0.00967 |
| 20 bp | R262D | 75 | R+P ↔RP RP+P ↔RP2 |
44.3 (20.3, 125) nM |
386 (204, 1021) nM |
4.41 (4.19, 4.84) |
6.78 (6.52, 7.38) |
0.00960 |
Parameters obtained by global nonlinear least squares analysis of sedimentation velocity experiments. The values in parentheses represent the 95% joint confidence intervals.
Wild type.
Uncorrected sedimentation coefficient (Svedbergs).
Root mean square deviation of the fit in absorbance units.
For this model, the dimeric species is R2P2 not RP2.
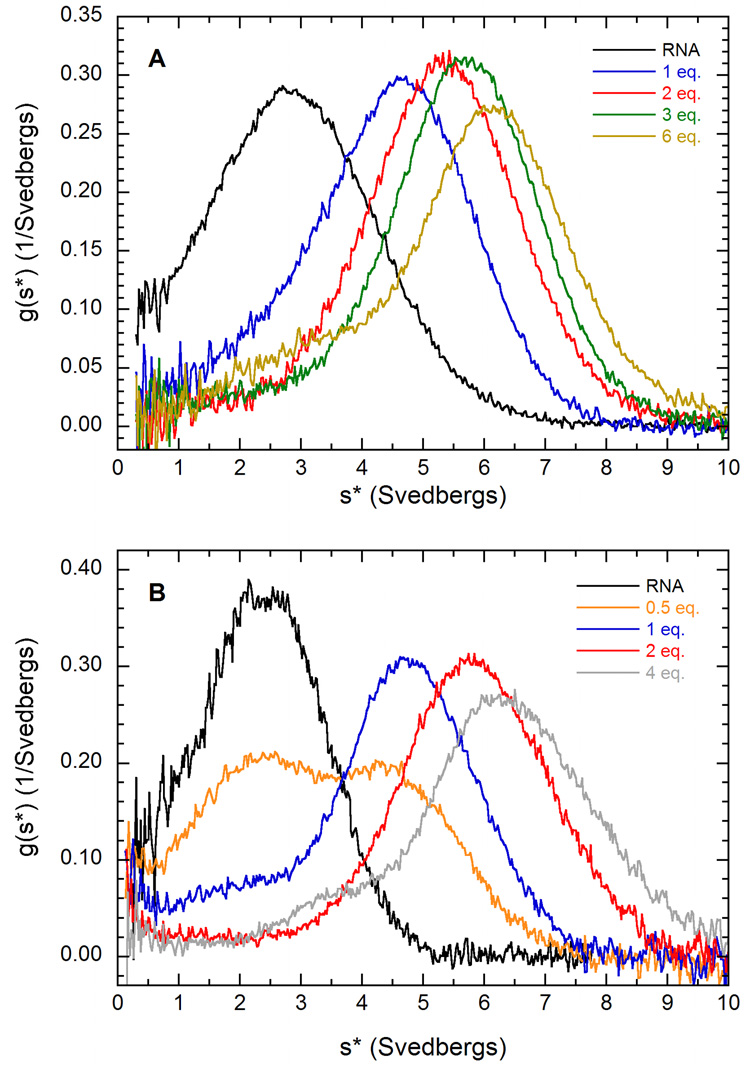
Consistent with binding of a second PKR, the g(s*) distributions for the activating 30 bp dsRNA shift more dramatically to higher s upon addition of PKR than was observed for the 20 bp sequence (figure 4A). The velocity data fit well to model 2 where two PKRs sequentially bound to the 30 bp dsRNA, with a low RMS deviation of 0.0079 (Table 1).
| (2) |
The data do not fit to model 1 involving a single PKR binding event (data not shown), confirming the stoichiometry data. The dissociation constant for binding the first PKR to the 30 bp dsRNA is about 10-fold lower than for the 20 bp dsRNA, indicating that PKR binds more strongly to the longer, activating dsRNA. This decrease in Kd for the longer sequence is consistent with the expected statistical effects for nonspecific protein – nucleic acid interactions.39 The macroscopic, stepwise binding constants measured here are products of an intrinsic binding constant and a statistical factor describing the number of microscopic configurations. The greater number of configurations for longer sequences leads to stronger binding affinities. Similarly, the increase in Kd for the second PKR binding relative to the first is associated with the reduced degrees of freedom on the lattice and does not imply negative cooperativity. The sedimentation coefficients for the RP and RP2 complexes correspond to asymmetric species with f/f0 = 1.57 and 1.50, respectively.
Figure 4.
g(s*) analysis of PKR binding to dsRNA. A) PKR binding to a 30 bp dsRNA in 200 mM NaCl. Normalized g(s*) distributions of 0.75 µM dsRNA (black), dsRNA + 1 eq. PKR (blue), dsRNA + 2 eq. PKR (red), dsRNA + 3 eq. PKR (green), dsRNA + 6 eq. PKR (tan). B) PKR binding to a 20 bp dsRNA in 75 mM NaCl. Normalized g(s*) distributions of 1 µM dsRNA (black), dsRNA + 0.5 eq. PKR (orange), dsRNA + 1 eq. PKR (blue), dsRNA + 2 eq. PKR (red), dsRNA + 4 eq. PKR (grey). Conditions: rotor speed, 40,000 RPM; temperature, 20°C; wavelength, 260 nm; scan interval, 5 minutes.
We have considered an alternative model14;29 where PKR forms a 1:1 complex with an activating RNA that subsequently dimerizes, leading to formation of an 2:2 complex:
| (3) |
In this model, binding of RNA to the dsRNA binding domain of PKR enhances protein dimerization via the interface localized on the kinase domain N-lobe. The sedimentation data do not fit well to this model with a large increase of the RMS deviations relative to model 2 (Table 1) and systematic deviations in the residuals (data not shown). Also, the best fit value for the sedimentation coefficient of the RP complex is greater than that of the R2P2 complex, which is not physically reasonable. Model 3 was also tested using a PKR mutant, R262D which disrupts an essential salt bridge in the dimer interface and eliminates self-association.40 In contrast to wild type PKR, the sedimentation coefficient of R262D PKR does not increase with protein concentration over a range of 0.1 to 1.8 mg/ml, confirming that dimerization has been eliminated or greatly reduced (data not shown). The parameters governing binding to the 30 bp dsRNA are not significantly perturbed by this mutation (Table 1), indicating that protein-protein interactions do not affect binding to this activating dsRNA. These results are inconsistent with model 3.
We have examined the effects of salt concentration on PKR – dsRNA interactions in order to facilitate comparison of the present results with our earlier studies of the dsRBD of PKR performed in 75 mM NaCl30 and to relate the biophysical studies to earlier studies in the literature where activation assays were performed at lower salt. Figure 4B shows a g(s*) analysis of sedimentation velocity titration of the 20 bp dsRNA with PKR. Interestingly, the distributions closely correspond to those observed at 200 mM NaCl (figure 3A). Upon addition of two equivalents, the peak shifts to ~ 6 S, which is higher than the fitted sedimentation coefficient of the dsRNA-PKR complex formed at 200 mM NaCl. In fact, the g(s*) more closely resemble the distributions observed for PKR binding to the 30 bp dsRNA at 200 mM NaCl (figure 4A). These results suggest that the decrease in salt concentration is inducing a second PKR to bind to the 20 bp dsRNA. Sedimentation equilibrium measurements confirm that two PKRs bind to this dsRNA at 75 mM NaCl (data not shown). The velocity data fit well to the sequential binding model (equation 2). The value of Kd1 of 15 nM is about 60-fold lower at 75 mM than at 200 mM NaCl, indicating a very strong salt dependence. As was observed for the 30 bp dsRNA, the second PKR binds more weakly with Kd2 ~ 230 nM. Assuming a similar salt dependence for the second PKR binding to the 20 bp dsRNA implies Kd2 would approach 15 µM in 200 mM NaCl. Such weak binding would not be readily detectable. The sedimentation coefficients of the RP and RP2 species correspond to asymmetric species with values of f/f0 of 1.58 and 1.51, which are very close to the corresponding complexes formed with the 30 bp dsRNA at 200 mM NaCl. As observed in the case of the 30 bp dsRNA, the R262D mutation also does not affect PKR binding to the 20 bp dsRNA in 75 mM NaCl.
Model for dsRNA activation of PKR
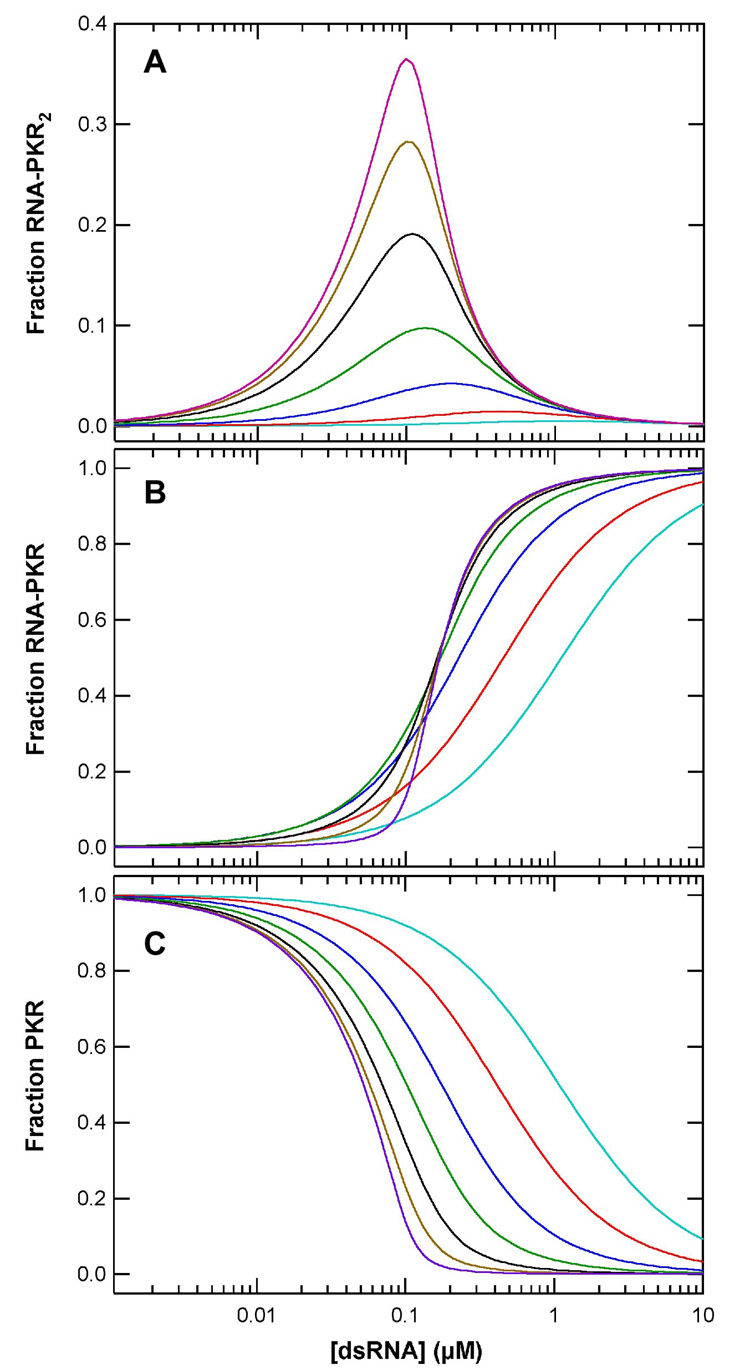
Our data support a sequential binding model for PKR activation by dsRNA where the autophosphorylation rates are proportional to the concentration of dsRNA species containing multiple PKR monomers. We have simulated of dsRNA activity titrations based on model 2 with the constraint that Kd2 = 10 × Kd1, as observed experimentally. Figure 5A shows the fraction of PKR in RNA-protein complexes containing two bound PKRs as function of RNA concentration over a range of binding affinities. Owing to the complexity of the autophosphorylation kinetics, it is difficult to quantitatively correlate these simulations with experimental activity data and we have not considered binding of more than two PKR ligands, as occurs for the longer RNAs. However, the simulations do reproduce the qualitative features of the activation profiles. As observed in figure 2, the simulated curves exhibit a maximum at intermediate dsRNA concentrations corresponding to the stoichiometric equivalence point where [dsRNA] = [PKR]/2. Significantly, the peak amplitudes decreases with reduced binding affinity. Figure 5B shows that inactive dsRNA-PKR complexes effectively competes with the active dsRNA-PKR2 species at RNA concentrations above stoichiometric.
Figure 5.
Simulation of PKR activation by dsRNA. A) Fraction of PKR present in the RNA-PKR2 species. B) Fraction of PKR present in the RNA-PKR species. C) Fraction of free PKR. Activation titrations were simulated using a model where two PKR monomers sequentially bind a dsRNA. The binding equilibria were numerically simulated using IGOR Pro (Wavemetrics Inc.) for [PKR] = 200 nM. A ratio of Kd2/Kd1 = 10 was assumed with the following values of Kd1: 1 nM (purple), 3.3 nM (tan), 10 nM (black), 33 nM (green), 100 nM (blue), 330 nM (red), 1 µM (acqua).
Discussion
Our studies of PKR interactions with a series of dsRNAs of increasing length demonstrate that the ability of sequences of 30 bp or longer to function as PKR activators is correlated with binding of two or more PKR monomers. Sedimentation velocity binding data fit a model where PKR monomers sequentially attach to a single dsRNA. Taken together, these results support an activation mechanism where the role of the dsRNA is to bring two or more PKR monomers in close proximity to enhance dimerization via the kinase domain. PKR dimerization is sufficient to induce PKR to undergo autophosphorylation, resulting in an active enzyme capable of phosphorylating eIF2α.19 Simulations based on this model reproduce the bell shaped activation curve (figure 2 and references 27;33). The model also predicts a strong dependence of maximal activation level on dsRNA binding affinity. This effect explains why larger dsRNAs elicit stronger PKR activations since the binding affinity increases with sequence length. Binding affinity effects also account for the lack of activation observed for the short 20 bp dsRNA in the present study. Although this sequence is capable of binding two PKRs and thus would be expected to function as an activator, the affinity is too low in 200 mM NaCl. Consistent with the strong enhancement of binding affinity at low salt, activation by siRNAs and dsRNAs as short as 19–21 bp34;35 has been reported when assayed at lower salt concentrations. The frictional ratios of the dsRNA-PKR and dsRNA-PKR2 complexes formed with the 30 bp dsRNA in 200 mM NaCl are very close to the corresponding complexes with the 20 bp sequence in 75 mM NaCl, indicating that they have similar overall shapes.
As illustrated by the crystal structure of the second dsRNA binding motif of the Xlrbpa protein complexed with dsRNA,41 the motif interacts with successive minor, major and minor grooves, spanning ~16 bp. Thus, to account for binding of two PKRs to the 20 bp dsRNA it is necessary to consider an overlapping ligand binding model where binding occurs on multiple faces of the dsRNA duplex, where the dsRBMs overlap along the helical axis.32 Previously, we demonstrated that this model accounts for the stoichiometries and affinities for dsRBD binding to sequences ranging from 15–30 bp with a site size of 12 bp and a minimum overlap of 4 bp.30;32 NMR mapping experiments indicated that dsRBM1 plays the dominant role in dsRBD binding to short dsRNAs.30 Three dsRBD bind to the 20 bp dsRNA in 75 mM NaCl with Kds of 11, 210, and 780 nM.32 Thus, the affinities of PKR and dsRBD are similar, suggesting a similar binding interface, but the presence of the kinase domain and linker region results in steric hindrance which prevents a third PKR from binding and increases the minimal overlap.
It is noteworthy that PKR assembly on dsRNA occurs without apparent cooperativity. Although aggregation of PKR upon binding larger dsRNAs in low salt buffer preclude a detailed analysis of the dependence of stoichiometry and affinity on sequence length, the fact that Kd2 ~ 10 × Kd1 implies that statistical factors far outweigh cooperativity and suggests that binding is essentially noncooperative. In addition, the similar dsRNA affinities for wild type and R262D PKR indicate that protein dimerization does not contribute measurably to the free energy for binding of the second PKR. Thus, the nonspecific binding to dsRNA is governed by statistical factors and dimerization that occurs upon binding to dsRNA must be driven by the same weak interactions that govern dimerization in the absence of dsRNA.
The analysis of PKR activation by simple dsRNA sequences provides insight into the regulation of PKR by viral RNAs. HIV TAR RNA is reported to function as a PKR activator.42 Although TAR has a complex secondary structure comprised of a stem-loop with three bulges interrupting the double-stranded region, the PKR activation properties of TAR resemble simple activating dsRNAs. PKR activation by TAR follows a bellshaped curve where inhibition is observed at high RNA concentration42 and TAR is capable of binding two PKRs.11;21;43 Thus, it seems likely that TAR activation is also mediated by sequential binding and dimerization of PKR. An alternative model has been proposed for PKR activation by the HIV TAR RNA in which PKR binding to this sequence enhances protein dimerization, leading to formation of a 2:2 TAR:PKR complex.14 However, this model is not compatible with our sedimentation velocity analysis of PKR binding to simple dsRNAs and also does account for the inhibition that occurs at high RNA concentrations.
Materials and Methods
All reagents used were reagent grade purchased from Fisher Scientific except as noted. Unphosphorylated PKR was expressed and purified as previously described.19 The R262D PKR mutant was created using Quik-Change (Stratagene, Inc.) and was verified by DNA sequencing. Synthetic oligoribonucleotides were obtained from Dharmacon, Inc. and prepared as previously described 30 The dsRNA duplexes ranged from 20 to 40 bp in 5 bp increments with the sequences shown below.
For fluorescence anisotropy experiments, dsRNAs were prepared with one strand containing a 5’-fluorescein label. Unless otherwise indicated, experiments were conducted at 20°C in a buffer containing 20 mM HEPES, 200 mM NaCl, 0.1 mM EDTA, 0.1 mM TCEP, pH 7.5 (AU200).
Sedimentation equilibrium analysis of dsRNA binding stoichiometries was performed using absorption optics with a Beckman-Coulter XL-I analytical ultracentrifuge as previously described.30 Fluorescence anisotropy data were collected using a Jobin Yvon FluoroMax-3 fluorimeter at 20°C using a 1 ×0.2 cm microcuvette. Titrations were recorded with sequential addition of PKR, allowing about 4 minute equilibration between additions.
Quantitative autophosphorylation assays were carried out by pre-incubating 0.2 µM PKR with variable dsRNA concentrations (0.01–10 µM) in AU200 buffer containing 5 mM MgCl2 at 32°C for 10 min. Phosphorylation was initiated with 0.4 mM ATP containing 4 μCi [γ-32P] ATP and allowed to proceed for 20 min except when otherwise stated. The reaction was quenched with sample loading buffer (Invitrogen) and analyzed by SDS-PAGE electrophoresis and phosphor imaging.
Sedimentation velocity analysis of protein-RNA binding affinities was carried out at 20°C and 50,000 RPM. Samples were loaded into two channel aluminum-epon double-sector cells equipped with quartz windows. Extinction coefficients, molecular masses (M), protein partial specific volume (v̄) and solvent densities (ρ) were calculated using SEDNTERP.44 Initial analysis was performed using DCDT+45 to obtain g(s*) distributions. Distributions were normalized by area (absorbance) for presentation. Multiple datasets were globally fit to alternative hetero-association models using SEDANAL.38 Joint confidence intervals were obtained using the F-statistic to define a statistically-significant increase in the variance46 upon adjusting each parameter from its best-fit value.
Acknowledgements
This work was supported by grant number AI-53615 from the NIH to J.L.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaufman RJ. The double stranded RNA-activated protein kinase PKR. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2000. pp. 503–528. [Google Scholar]
- 2.Toth AM, Zhang P, Das S, George CX, Samuel CE. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:369–434. doi: 10.1016/S0079-6603(06)81010-X. [DOI] [PubMed] [Google Scholar]
- 3.Su Q, Wang S, Baltzis D, Qu LK, Wong AH, Koromilas AE. Tyrosine phosphorylation acts as a molecular switch to full-scale activation of the eIF2{alpha} RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U S A. 2006;103:63–68. doi: 10.1073/pnas.0508207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langland JO, Cameron JM, Heck MC, Jancovich JK, Jacobs BL. Inhibition of PKR by RNA and DNA viruses. Virus Res. 2006;119:100–110. doi: 10.1016/j.virusres.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 6.Koromilas AE, Roy S, Barber GN, Katze MG, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 7.Meurs E, Galabru J, Barber GN, Katze MG, Hovanessian AG. Tumor supressor function of interferon-induced double-stranded RNA activated protein kinase. Proc. Natl. Acad. Sci. USA. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dever TE. Translation initiation: adept at adapting. Trends Biochem. Sci. 1999;24:398–403. doi: 10.1016/s0968-0004(99)01457-7. [DOI] [PubMed] [Google Scholar]
- 9.Nanduri S, Carpick BW, Yang Y, Williams BR, Qin J. Structure of the double-stranded RNA binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nanduri S, Rahman F, Williams BRG, Qin J. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 2000;19:5567–5574. doi: 10.1093/emboj/19.20.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bevilacqua PC, Cech TR. Minor-groove recognition of doublestranded RNA by the double-stranded RNA-binding domain of the RNA-activated protein kinase PKR. Biochemistry. 1996;35:9983–9994. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- 12.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 13.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 14.McKenna SA, Lindhout DA, Kim I, Liu CW, Gelev VM, Wagner G, Puglisi JD. Molecular framework for the activation of RNA-dependent protein kinase. J. Biol. Chem. 2007;282:11474–11486. doi: 10.1074/jbc.M700301200. [DOI] [PubMed] [Google Scholar]
- 15.Cole JL. Activation of PKR: an open and shut case? Trends Biochem. Sci. 2007;32:57–62. doi: 10.1016/j.tibs.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gelev V, Aktas H, Marintchev A, Ito T, Frueh D, Hemond M, Rovnyak D, Debus M, Hyberts S, Usheva A, Halperin J, Wagner G. Mapping of the auto-inhibitory interactions of protein kinase R by nuclear magnetic resonance. J. Mol. Biol. 2006;364:352–363. doi: 10.1016/j.jmb.2006.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson E, Cole JL. Domain Stabilities in Protein Kinase R (PKR): Evidence for Weak Interdomain Interactions. Biochemistry. 2008;47:4887–4897. doi: 10.1021/bi702211j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemaire PA, Tessmer I, Craig R, Erie DA, Cole JL. Unactivated PKR exists in an open conformation capable of binding nucleotides. Biochemistry. 2006;45:9074–9084. doi: 10.1021/bi060567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemaire PA, Lary J, Cole JL. Mechanism of PKR activation: dimerization and kinase activation in the absence of double-stranded RNA. J. Mol. Biol. 2005;345:81–90. doi: 10.1016/j.jmb.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 20.Robertson HD, Mathews MB. The regulation of the protein kinase PKR by RNA. Biochimie. 1996;78:909–914. doi: 10.1016/s0300-9084(97)86712-0. [DOI] [PubMed] [Google Scholar]
- 21.Carpick BW, Graziano V, Schneider D, Maitra RK, Lee X, Williams BRG. Characterization of the solution complex between the interferon-induced double-stranded RNA-activated protein kinase and HIV-I trans-activating region RNA. J. Biol. Chem. 1997;272:9510–9516. doi: 10.1074/jbc.272.14.9510. [DOI] [PubMed] [Google Scholar]
- 22.Langland JO, Jacobs BL. Cytosolic double-stranded RNA-dependent protein kinase is likely a dimer of partially phosphorylated Mr=66,000 subunits. J. Biol. Chem. 1992;267:10729–10736. [PubMed] [Google Scholar]
- 23.Patel RC, Stanton P, McMillan NM, Williams BR, Sen GC. The interferon-inducible double-stranded RNA-activated protein kinase self-associates in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 1995;92:8283–8287. doi: 10.1073/pnas.92.18.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ung TL, Cao C, Lu J, Ozato K, Dever TE. Heterologous dimerization domains functionally substitute for the double-stranded RNA binding domains of the kinase PKR. EMBO J. 2001;20:3728–3737. doi: 10.1093/emboj/20.14.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vattem KM, Staschke KA, Wek RC. Mechanism of activation of the double-stranded-RNA-dependent protein kinase, PKR: role of dimerization and cellular localization in the stimulation of PKR phosphorylation of eukaryotic initiation factor-2 (eIF2) Eur. J. Biochem. 2001;268:3674–3684. doi: 10.1046/j.1432-1327.2001.02273.x. [DOI] [PubMed] [Google Scholar]
- 26.Hunter T, Hunt T, Jackson RJ, Robertson HD. The characteristics of inhibition of protein synthesis by double-stranded ribonucleic acid in reticulocyte lysates. J. Biol. Chem. 1975;250:409–417. [PubMed] [Google Scholar]
- 27.Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kostura M, Mathews MB. Purification and activation of the doublestranded RNA-dependent eIF-2 kinase DAI. Mol. Cell. Biol. 1989;9:1576–1586. doi: 10.1128/mcb.9.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu S, Kaufman RJ. A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J. Biol. Chem. 1997;272:1291–1296. doi: 10.1074/jbc.272.2.1291. [DOI] [PubMed] [Google Scholar]
- 30.Ucci JW, Kobayashi Y, Choi G, Alexandrescu AT, Cole JL. Mechanism of interaction of the double-stranded RNA (dsRNA) binding domain of protein kinase R with short dsRNA sequences. Biochemistry. 2007;46:55–65. doi: 10.1021/bi061531o. [DOI] [PubMed] [Google Scholar]
- 31.Schmedt C, Green SR, Manche L, Taylor DR, Ma Y, Mathews MB. Functional characterization of the RNA-binding domain and motif of the double-stranded RNA-dependent protein kinase DAI (PKR) J. Mol. Biol. 1995;249:29–44. doi: 10.1006/jmbi.1995.0278. [DOI] [PubMed] [Google Scholar]
- 32.Ucci JW, Cole JL. Global Analysis of Nonspecific Protein-Nucleic Interactions by Sedimentation Equilibrium. Biophys. Chem. 2004;108:127–140. doi: 10.1016/j.bpc.2003.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minks MA, West DK, Benvin S, Baglioni C. Structural requirements of double-stranded RNA for the activation of 2'-5'-Oligo(A) polymerase and protein kinase of interferon-treated HeLa cells. J. Biol. Chem. 1979;254:10180–10183. [PubMed] [Google Scholar]
- 34.Puthenveetil S, Whitby L, Ren J, Kelnar K, Krebs JF, Beal PA. Controlling activation of the RNA-dependent protein kinase by siRNAs using sitespecific chemical modification. Nucleic Acids Res. 2006;34:4900–4911. doi: 10.1093/nar/gkl464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 36.Stafford WF. Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal. Biochem. 1992;203:295–301. doi: 10.1016/0003-2697(92)90316-y. [DOI] [PubMed] [Google Scholar]
- 37.Bonifacio GF, Brown T, Conn GL, Lane AN. Comparison of the electrophoretic and hydrodynamic properties of DNA and RNA oligonucleotide duplexes. Biophys J. 1997;73:1532–1538. doi: 10.1016/S0006-3495(97)78185-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stafford WF, Sherwood PJ. Analysis of heterologous interacting systems by sedimentation velocity: curve fitting algorithms for estimation of sedimentation coefficients, equilibrium and kinetic constants. Biophys. Chem. 2004;108:231–243. doi: 10.1016/j.bpc.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 39.Cole JL. Analysis of heterogeneous interactions. Methods Enzymol. 2004;384:212–232. doi: 10.1016/S0076-6879(04)84013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dey M, Cao C, Dar AC, Tamura T, Ozato K, Sicheri F, Dever TE. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell. 2005;122:901–913. doi: 10.1016/j.cell.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 41.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maitra R, McMillan NAJ, Desai S, McSwiggen J, Hovanessian AG, Sen G, Williams BRG, Silverman RH. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology. 1994;204:823–827. doi: 10.1006/viro.1994.1601. [DOI] [PubMed] [Google Scholar]
- 43.Kim I, Liu CW, Puglisi JD. Specific recognition of HIV TAR RNA by the dsRNA binding domains (dsRBD1-dsRBD2) of PKR. J. Mol. Biol. 2006;358:430–442. doi: 10.1016/j.jmb.2006.01.099. [DOI] [PubMed] [Google Scholar]
- 44.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S, Rowe A, Horton J, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 45.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal. Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 46.Johnson ML, Faunt LM. Parameter estimation by least-squares methods. Methods Enzymol. 1992;210:1–37. doi: 10.1016/0076-6879(92)10003-v. [DOI] [PubMed] [Google Scholar]