Abstract
Transforming growth factor (TGF)-β induces various cellular responses principally through Smad-dependent transcriptional regulation. Activated Smad complexes cooperate with transcription factors in regulating a group of target genes. The target genes controlled by the same Smad-cofactor complexes are denoted a synexpression group. We found that an Id-like helix-loop-helix protein, human homologue of Maid (HHM), is a synexpression group-restricted regulator of TGF-β signalling. HHM suppressed TGF-β-induced growth inhibition and cell migration but not epithelial–mesenchymal transition. In addition, HHM inhibited TGF-β-induced expression of plasminogen activator inhibitor-type 1 (PAI-1), PDGF-B, and p21WAF, but not Snail. We identified a basic-helix-loop-helix protein, Olig1, as one of the Smad-binding transcription factors affected by HHM. Olig1 interacted with Smad2/3 in response to TGF-β stimulation, and was involved in transcriptional activation of PAI-1 and PDGF-B. HHM, but not Id proteins, inhibited TGF-β signalling-dependent association of Olig1 with Smad2/3 through physical interaction with Olig1. HHM thus appears to regulate a subset of TGF-β target genes including the Olig1-Smad synexpression group. HHM is the first example of a cellular response-selective regulator of TGF-β signalling with clearly determined mechanisms.
Keywords: HHM, Olig1, Smad, synexpression group, TGF-β
Introduction
The growth factors are a group of proteins that mediate intercellular communication through regulation of cell growth and differentiation, and thus have important functions in maintaining homoeostasis of multicellular organisms. Aberrant activation of growth factor signalling results in various diseases, including malignant tumour. Control of growth factor signalling has thus been considered one of the most attractive targets in the treatment of malignant tumours (Cohen, 2002).
However, transforming growth factor (TGF)-β should be considered separately from other growth factors for the following reasons (Blobe et al, 2000). TGF-β suppresses the proliferation of epithelial cells and certain carcinoma cells, but promotes proliferation of fibroblasts and glioma cells. TGF-β also promotes apoptosis of most types of cells, but induces cell survival under certain conditions (Ehata et al, 2007). In addition, TGF-β promotes epithelial–mesenchymal transition (EMT), cell migration, and extracellular matrix production. Therefore, whether comprehensive suppression of TGF-β signalling promotes or suppresses progression of malignant tumours depends on various factors, including tumour origin, pathological type, and microenvironment.
Transforming growth factor-β binds to types I and II serine/threonine kinase receptors and transduces intracellular signals through Smad proteins (Miyazawa et al, 2002; Derynck and Zhang, 2003; Shi and Massagué, 2003). Upon phosphorylation by type I receptors, receptor-regulated Smads (R-Smads; Smad2 and 3) form heteromeric complexes with common-partner Smad (Co-Smad; Smad4) and translocate into the nucleus. In the nucleus, the activated Smad complexes cooperate with other transcription factors to elicit specific transcriptional regulation, as the affinity of the activated Smad complex for the Smad-binding element (SBE) is insufficient to support association with endogenous promoters of target genes except those with multiple SBE clusters. TGF-β-induced gene responses are thus classified by groups of genes that are jointly controlled by a given Smad–cofactor combination (Shi and Massagué, 2003). A group of genes that are simultaneously regulated by a common Smad–cofactor complex is denoted a ‘synexpression group'. Such gene responses orchestrate the successful maintenance of homoeostasis, and aberrant regulation of such responses may lead to various diseases.
Human homologue of Maid (HHM) was originally identified as a protein structurally related to mouse maternal Id-like molecule (Maid) (Hwang et al, 1997; Terai et al, 2000). HHM is also termed GCIP (Grap2 cyclin D interacting protein), CCNDBP1 (cyclin D-type binding protein 1), and DIP1 (D-type cyclin interacting protein) (Xia et al, 2000; Yao et al, 2000). As HHM has a helix-loop-helix (HLH) domain, but lacks a basic DNA-binding domain, it is structurally similar to Id proteins. HHM appears to exert opposite effects on cell cycle progression depending on cellular context (Sonnenberg-Riethmacher et al, 2007), and the pathophysiological functions of HHM have not been fully determined.
In this study, we found that HHM disrupts the physical interaction of specific transcription factors with R-Smads to inhibit TGF-β signalling in a synexpression group-restricted manner. We also identified a novel TGF-β effector, oligodendrocyte transcription factor 1 (Olig1) (Zhou et al, 2000), as one of the Smad-binding transcription factors affected by HHM. In contrast to Id proteins, which interact with ubiquitously expressed basic-helix-loop-helix (bHLH) transcription factors, HHM interacts with the tissue-specific bHLH transcription factor Olig1 and regulates Smad-dependent transcription. Our findings raise the possibility of control of TGF-β signalling in a cellular response-specific manner through targeting of synexpression groups and add a novel dimension to understanding of the regulation of growth factor signalling.
Results
HHM attenuates TGF- signalling through suppression of Smad-dependent transcriptional activity
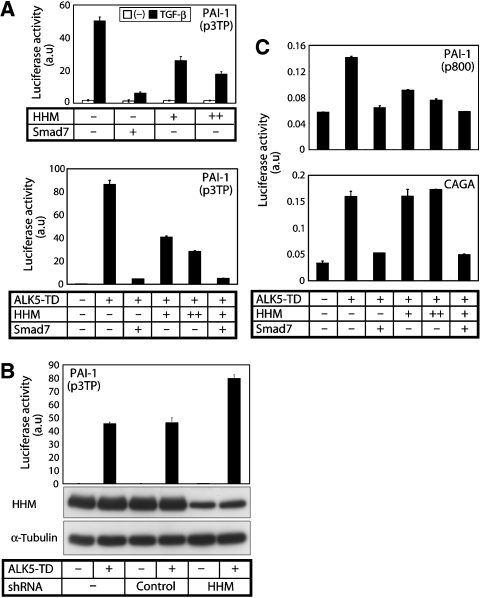
We first examined whether HHM affects TGF-β signalling by luciferase reporter assay. Exogenous HHM inhibited the transactivation of p3TP-Lux induced by TGF-β ligand or by constitutively active TGF-β type I receptor (ALK5-TD) (Figure 1A), whereas knockdown of endogenous HHM enhanced signalling (Figure 1B). These findings indicate that HHM is a negative regulator of TGF-β signalling.
Figure 1.
HHM attenuates TGF-β signalling through suppression of Smad-dependent transcriptional activity. (A) Effects of HHM on the transactivation of p3TP-Luc induced by TGF-β ligand (1 ng/ml) or constitutively active TβR-I (ALK5-TD) in NIH3T3 cells. Error bars represent s.d. Smad7 was used as a positive control. (B) Effects of HHM knockdown by shRNA on the transactivation of p3TP-Luc induced by constitutively active TβR-I (ALK5-TD) in HeLa cells (top panel). Error bars represent s.d. Expression of HHM protein was determined by anti-HHM antibody (middle panel). The lower panel shows expression level of tubulin protein as a loading control. (C) Effects of HHM on the transcriptional activity of p800-Luc in NMuMG cells (upper panel) and (CAGA)12-MLP-Luc in NIH3T3 cells (lower panel) induced by ALK5-TD. Error bars represent s.d. Smad7 was used as a positive control.
We next examined the effects of HHM on other luciferase reporter constructs (Figure 1C). Although HHM inhibited transactivation of p800-Luc, it failed to affect the transactivation of (CAGA)12-MLP-Luc. These findings suggest that HHM inhibits Smad signalling at the transcriptional level. Consistent with this fact, HHM did not affect phosphorylation (data not shown) as well as nuclear accumulation of Smad2 and 3 induced by TGF-β (Supplementary Figure S1), and it failed to interact with Smad2 or 3 (data not shown). Notably, these three reporter constructs, p3TP-Lux, p800-Luc, and (CAGA)12-MLP-Luc, are all derived from the promoter region of the human plasminogen activator inhibitor-type 1 (PAI-1) gene. p800-Luc contains the −800/+75 region of the PAI-1 promoter (Keeton et al, 1991) and p3TP-Lux contains the −740/−636 region of the PAI-1 promoter together with three repeats of the TPA-responsive element derived from the human collagenase gene (Wrana et al, 1992). Activation of these reporter constructs appears to be driven by cooperation between Smad proteins and other transcription factor(s). In contrast, (CAGA)12-MLP-Luc contains only 12 tandem repeats of the Smad binding ‘CAGA' boxes, and activation of it is exclusively driven by the activated Smad complex (Dennler et al, 1998). These findings suggest that HHM does not affect the function of Smad proteins, but instead those of other component(s) of the transcriptional complex, leading to the inhibition of TGF-β signalling.
HHM inhibits TGF- signalling in a cell response-specific manner
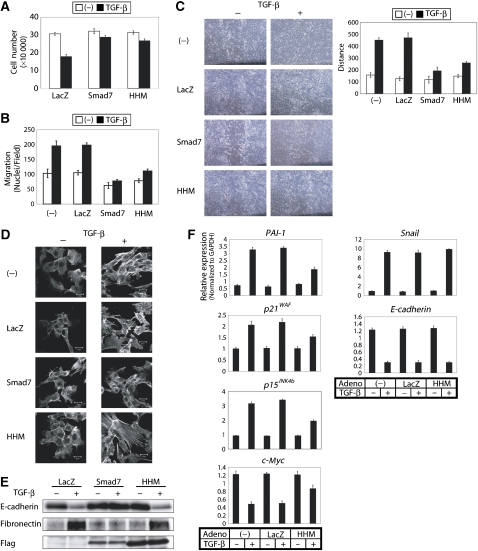
We next examined the effects of HHM on various TGF-β-induced cell responses in NMuMG mouse mammary epithelial cells, using adenoviruses carrying HHM cDNA (Ad-HHM), Smad7 cDNA (Ad-Smad7), and LacZ cDNA (Ad-LacZ). Ad-Smad7 and Ad-LacZ were used as positive and negative controls, respectively. First, we examined the sensitivity to TGF-β-induced growth inhibition. HHM-infected cells exhibited resistance to TGF-β-induced growth inhibition (Figure 2A) as well as to downregulation of c-myc expression (Figure 2F). Second, we examined the effect of HHM on TGF-β-induced stimulation of cell migration. In a chamber assay, TGF-β-induced cell migration was clearly suppressed by HHM (Figure 2B). Similarly, in a wound healing assay, HHM-infected cells exhibited delay in the closure of the scratched area (Figure 2C). However, we found that TGF-β induced EMT in HHM-infected cells as in LacZ-infected cells but not in Smad7-infected cells, as assessed by actin reorganization (Figure 2D) and epithelial or mesenchymal marker expression (Figure 2E) (Zavadil and Böttinger, 2005). Similarly, HHM attenuated TGF-β-induced growth inhibition in human keratinocyte cell line, HaCaT, and inhibited TGF-β-induced cell migration in human lung adenocarcinoma cell line, A549, whereas it did not affect TGF-β-induced EMT in A549 cells (Supplementary Figure S2). HHM thus inhibits TGF-β signalling in a cell response-specific manner, involving antagonism of the TGF-β-induced growth inhibition and migration, but not TGF-β-induced EMT.
Figure 2.
HHM inhibits TGF-β signalling in a cell response-specific manner. (A) HHM attenuated TGF-β-induced growth inhibition. NMuMG cells infected with Ad-LacZ, Ad-Smad7, or Ad-HHM were seeded and treated with or without TGF-β (1 ng/ml). Cell numbers were counted 6 days after stimulation. Error bars represent s.d. (B) HHM inhibited TGF-β-induced cell migration in chamber assay. Error bars represent s.d. (C) HHM inhibited TGF-β-promoted wound closure in wound healing assay. The result was quantified as shown in the graph on the right side. Migration of wound edges was measured at three random points on the photograph. (D) HHM failed to affect TGF-β-induced actin reorganization. Cells were stimulated with TGF-β (1 ng/ml) for 24 h, and TRITC-phalloidin staining was then performed. (E) Effects of HHM on the expressions of E-cadherin and fibronectin. Cells were infected with Ad-LacZ, Ad-Smad7, or Ad-HHM, and cultured with or without TGF-β for 24 h. Proteins were extracted for immunoblotting. The bottom panel shows protein expression of Smad7 and HHM determined by anti-Flag antibody. (F) Effects of overexpression of HHM on the expressions of target genes of TGF-β, PAI-1, p21WAF, p15INK4b, c-myc, Snail, and E-cadherin. NMuMG cells were infected with Ad-LacZ or Ad-HHM, and then stimulated with 1 ng/ml TGF-β for 1 h (except for E-cadherin; 24 h). Expression of each gene was determined by quantitative real-time PCR analyses. Values were normalized to the amount of GAPDH mRNA. Error bars represent s.d.
We also confirmed that HHM inhibited expression of only a subset of TGF-β target genes in NMuMG cells. The representative data are listed in Figure 2F and Supplementary Figure S3. TGF-β target genes whose induction was suppressed by HHM included PAI-1, p21WAF, p15INK4b, and c-myc, but not Snail or E-cadherin.
Identification of Olig1 as a binding partner of HHM
The above findings suggest that HHM inhibits the transcription of a subset of TGF-β-induced genes and thus affects TGF-β signalling in a cell response-specific manner. The targets of HHM appear to be transcription factors that cooperate with the activated Smad complex.
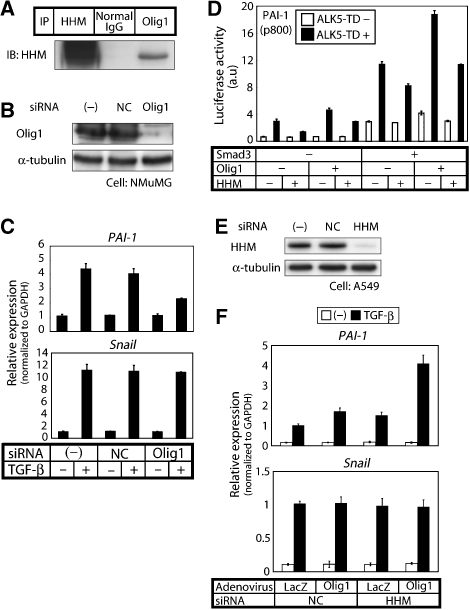
To elucidate the mechanism underlying these specific inhibitory effects of HHM, we explored transcription factors that cooperate with Smads and that are affected by HHM. We performed glutathione S-transferase (GST) pull-down using NMuMG cell nuclear extract. The proteins that bound to GST-HHM, but not to GST, were identified by mass spectrometry analysis (Hellman, 2000), and one of the proteins was a tissue-specific bHLH transcription factor, Olig1 (Zhou et al, 2000). We confirmed the interaction between endogenous HHM and endogenous Olig1 by co-immunoprecipitation assay in NMuMG cells (Figure 3A).
Figure 3.
Olig1 upregulates expression of PAI-1, but not Snail, in concert with Smad2/3, and their synergistic effects are antagonized by HHM. (A) Physical interaction of endogenous HHM with endogenous Olig1 in NMuMG cells. Cell lysates were subjected to immunoprecipitation with anti-Olig1 antibody, anti-HHM antibody as a positive control, or normal mouse IgG as a negative control, followed by immunoblotting with anti-HHM. (B) Knockdown of Olig1 in NMuMG cells. NMuMG cells were transfected with control or Olig1 siRNA duplex, and the expression of Olig1 protein was determined by immunoblotting. Expression of endogenous α-tubulin is also shown as a loading control. NC, negative control siRNA. (C) Effects of Olig1 knockdown on the expression of target genes of TGF-β, PAI-1, and Snail. NMuMG cells were transfected with control or Olig1 siRNA and then stimulated with 1 ng/ml TGF-β for 1 h. Expressions of PAI-1 (upper panel) and Snail (lower panel) were examined by quantitative real-time PCR analyses. Values were normalized to the amount of GAPDH mRNA. Error bars represent s.d. (D) Cooperation of Smad3 with Olig1 in transactivation of p800-Luc. Cells were co-transfected with the p800-Luc reporter construct and various combinations of indicated cDNAs, and luciferase activities in cell lysates were determined. Error bars represent s.d. (E) Knockdown of HHM in A549 cells. A549 cells were transfected with control or HHM siRNA duplex, and expression of HHM protein was determined by immunoblotting. Expression of endogenous α-tubulin is also shown as a loading control. (F) A549 cells were transfected with control or HHM siRNA duplex. Twenty-four hours after transfection, cells were infected with Ad-LacZ or Ad-HHM and cultured for another 24 h. Cells were then treated with or without TGF-β (1 ng/ml) for 1 h before harvest, and then expression of PAI-1 and Snail was examined by quantitative real-time PCR analyses. Error bars represent s.d.
Olig1 is a transcription factor that cooperates with Smad2/3 and is antagonized by HHM in the induction of PAI-1
We next knocked down endogenous Olig1 by small interfering RNAs (siRNAs; Figure 3B) and examined TGF-β-induced gene expression (Figure 3C) in NMuMG cells. The induction of PAI-1 expression by TGF-β was partially suppressed, whereas basal PAI-1 expression was not altered. In contrast, Snail induction was not significantly affected. We also examined expression of other target genes of TGF-β and found that Smad7 and procollagen 1 were affected by siRNA for Olig1, whereas p21WAF, p15INK4b, c-myc, and E-cadherin were not affected (Supplementary Figure S4). p21WAF, p15INK4b, and c-myc appear to be regulated by other transcription factors that interact with HHM. To assess the effects of Olig1 and HHM on whole TGF-β target genes, we performed oligonucleotide microarray analysis. Among 318 TGF-β target genes, 49 genes were affected by HHM. Among these 49 genes, 30 genes were affected by Olig1, whereas 19 genes were not affected (Supplementary Table S1).
As Olig1 was shown to be involved in the induction of PAI-1 by TGF-β, we examined whether Olig1 functions synergistically with Smad2/3. As shown in Figure 3D, Olig1 modestly enhanced the transactivation of p800-Luc, but striking enhancement by Olig1 was observed when Smad3 was co-transfected. Similar results were obtained for Smad2 (data not shown). Moreover, co-transfection of HHM cancelled the synergistic effects between Olig1 and Smad3. Similar results were obtained when we used p3TP-Luc or Smad7-Luc instead of p800-Luc (data not shown).
We further investigated the effects of HHM on the synergistic action of Smads and Olig1 in endogenous gene expression. Olig1 enhanced PAI-1 expression synergistically with TGF-β signalling, and their synergistic effect was more salient when HHM was knocked down (Figure 3E and F). On the other hand, TGF-β-induced Snail expression was not significantly affected by Olig1 and HHM.
These findings indicate that Olig1 upregulates expression of target genes including PAI-1 in concert with R-Smads, and that HHM attenuates their synergistic effects and consequently downregulates expression of a subset of target genes.
Olig1 interacts with Smad2/3 in a signalling-dependent manner
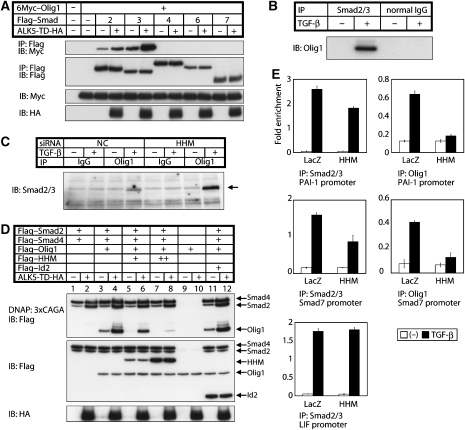
We also examined interactions between Olig1 and Smad proteins. Olig1 interacted with Smad2 and Smad3 in a signalling-dependent manner, whereas it failed to interact with Smad4, 6, and 7 (Figure 4A). We confirmed that endogenous Olig1 is associated with endogenous Smad2/3 in glioma cell line, U373MG, in response to TGF-β stimulation (Figure 4B and C). In addition, stronger binding of endogenous Olig1 to endogenous Smad2/3 was observed when HHM was knocked down (Figure 4C). Olig1 interacts with Smad3 through the MH2 domain (Supplementary Figure S5A), whereas Smad3 interacts with Olig1 through the bHLH and the C-terminal regions (Supplementary Figure S5B).
Figure 4.
Olig1 is associated with Smad2/3, and their cooperative action on chromosome is abrogated by HHM. (A) TGF-β signalling-dependent association of Olig1 with Smad2 and Smad3. Physical interactions of Flag–Smads with 6Myc–Olig1 were examined by immunoprecipitation with anti-Flag antibody followed by immunoblotting with anti-Myc antibody in transfected HEK293T cells. TGF-β signalling was induced by ALK5-TD. (B) Physical interaction of endogenous Olig1 with endogenous Smad2 and Smad3 in U373MG cells. U373MG cells were treated with or without TGF-β (2.5 ng/ml) for 3 h before harvest. Cell lysates were subjected to immunoprecipitation with anti-Smad2/3 antibody, or normal mouse IgG as a negative control, followed by immunoblotting with anti-Olig1 antibody. (C) Stronger binding between endogenous Olig1 and Smad2/3 when HHM was knocked down. U373MG cells were transfected with control or HHM siRNA duplex and treated with or without TGF-β (2.5 ng/ml) for 3 h before harvest. Cell lysates were subjected to immunoprecipitation with anti-Olig1 antibody, or normal mouse IgG as a negative control, followed by immunoblotting with anti-Smad2/3 antibody. Smad3 is dominantly detected by this antibody in U373MG cells. (D) Interaction of Olig1 with the activated Smad complex bound to DNA is abrogated by HHM. HEK293T cells were transfected as indicated. Cell lysates were subjected to DNA-affinity precipitation assay using biotinylated 3 × CAGA as a probe. (E) Association of Olig1–Smad complex with the PAI-1 and Smad7 promoter regions is abrogated by HHM. ChIP analysis was performed using U373MG cells infected with Ad-LacZ or Ad-HHM. Cells were treated with TGF-β (2.5 ng/ml) for 1 h and harvested. Eluted DNAs were subjected to quantitative real-time PCR analysis. Values were normalized to the amount of the first intron of hypoxanthine phosphoribosyltransferase 1. Error bars represent s.d. Primer sequences are listed on the Supplementary Table S2.
HHM inhibits interaction of Olig1 with the activated Smad complexes bound to DNA
We next examined effects of HHM on the interaction between Olig1 and the activated Smad complexes bound to DNA (Figure 4D) using a DNA-affinity precipitation method (Nishihara et al, 1999). In the absence of TGF-β signalling, Smad4 was precipitated mainly by 3 × CAGA probe, with weak precipitation of Smad2 also observed. In the presence of TGF-β signalling, Smad2 was efficiently precipitated through complex formation with Smad4 (Figure 4D, lanes 1 and 2). Olig1 was precipitated in Smad- and TGF-β-signalling-dependent manners (Figure 4D, lanes 3, 4, 9, and 10), indicating that the precipitated Olig1 was associated with the activated Smad complexes.
When HHM was co-transfected with Olig1, Smad2, and Smad4, co-precipitation of Olig1 was abrogated depending on the amount of HHM (Figure 4D, lanes 5–8). These findings suggest that HHM interferes with the interaction between Olig1 and the activated Smad complex and, as a consequence, HHM exhibits inhibitory effects on transcription induced by Olig1–Smad complex.
For further investigation, the association of Olig1–Smad complex with the promoter regions of TGF-β target genes was examined by chromatin immunoprecipitation (ChIP) assay (Figure 4E). Anti-Smad2/3 antibody or anti-Olig1 antibody immunoprecipitated the DNA fragments of the PAI-1 and Smad7 promoters (containing SBEs and E-boxes) in response to TGF-β. The recruitment of Smad2/3 and Olig1 to these promoters was attenuated in the HHM-overexpressed cells. The weaker attenuation of the recruitment of Smad2/3 suggests that Olig1–Smad complex does not bind to all of the Smad-binding elements in the PAI-1 or Smad7 promoters. We also examined the recruitment of Smad2/3 to the leukaemia inhibitory factor (LIF) promoter. LIF expression is induced by TGF-β in U373MG cells (Bruna et al, 2007), and this induction was not affected by Olig1 or HHM (Supplementary Figure S6). TGF-β-induced recruitment of endogenous Smad2/3 to the LIF promoter was not attenuated in the HHM-overexpressed U373MG cells (Figure 4E). The binding of Olig1 to the LIF promoter was around the background level (data not shown).
These results indicate that Olig1 and R-Smads interact with each other on chromosomes and yield synergistic upregulation of TGF-β-target gene expression when Olig1-binding sequence(s) and Smad-binding sequence(s) reside in close vicinity.
The HLH region of HHM has unique effects in inhibiting interaction of Olig1 with Smad3
As HHM has an HLH region that lacks an adjacent basic domain, its function may be similar to those of Id family proteins. Id proteins interact with bHLH transcription factors through their HLH region and interfere with dimerization of bHLH transcription factors (Norton, 2000). Similarly, HHM and Olig1 interact with each other through their HLH region (Supplementary Figure S5C and D). We also found that the HLH region of HHM is required for inhibition because only the constructs containing the HLH region effectively inhibited the transactivation of p800-Luc (Supplementary Figure S5E).
Id2 was reported previously to interact with Olig1 (Samanta and Kessler, 2004). We therefore examined co-precipitation of Olig1 and Smads in the presence of Id2 (Figure 4D, lanes 11 and 12). Although Id2 was expressed at the same level as HHM, Id2 did not inhibit the interaction between Olig1 and Smads. These findings suggest that the HLH region of HHM has unique effects in disruption of the interaction of Olig1 with Smad3.
Olig1 enhances TGF--induced proliferation of glioma cells through upregulation of PDGF-B, whereas HHM exhibits the opposite effect
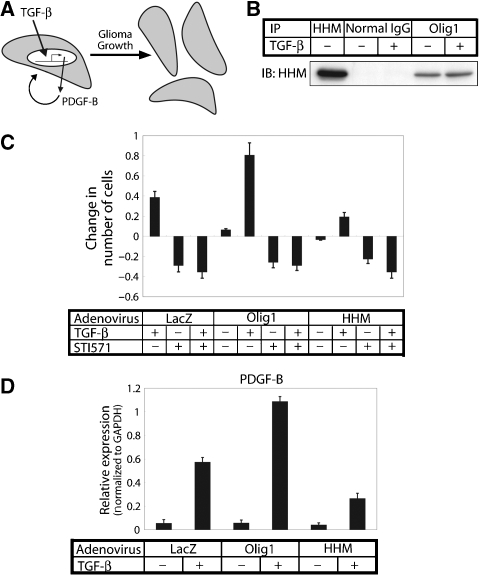
We further attempted to demonstrate the effects of HHM on the TGF-β-induced cellular responses mediated by the Olig1–Smad complex. Olig1 has been suggested recently to have an important function in glioma growth (Ligon et al, 2007). In addition, TGF-β has been reported to be one of the key cytokines in glioma cell proliferation through direct induction of PDGF-B expression (Figure 5A; Bruna et al, 2007). These reports prompted us to examine the functions of Olig1 and HHM in TGF-β-induced glioma growth.
Figure 5.
HHM antagonizes the TGF-β-induced proliferation of glioma cells accelerated by Olig1. (A) Model of promotion of the growth of glioma by TGF-β through induction of PDGF-B exerting an effect through an autocrine mechanism (Bruna et al, 2007). (B) Physical interaction of endogenous Olig1 and HHM in U373MG cells. U373MG cells were treated with or without TGF-β (1 ng/ml) for 3 h before harvest. Cell lysates were subjected to immunoprecipitation with anti-Olig1 antibody, anti-HHM antibody as a positive control, or normal mouse IgG as a negative control, followed by immunoblotting with anti-HHM. (C) Olig1 promotes TGF-β-induced proliferation of glioma cells, whereas HHM suppresses it. U373MG cells were infected with Ad-LacZ, Ad-Olig1, or Ad-HHM and seeded. After 24 h, the cells were treated with TGF-β (1 ng/ml) and/or STI571 (5 μM), and cell number was counted 72 h after treatment. Results were expressed as the change from control (untreated cells). Error bars represent s.e. (D) TGF-β-induced expression of PDGF-B in Ad-Olig1- or Ad-HHM-infected cells. Expression of PDGF-B was examined by quantitative real-time PCR analyses after infected U373MG cells were treated with TGF-β (1 ng/ml) for 3 h. Values were normalized to the amount of GAPDH mRNA. Error bars represent s.d.
We first confirmed interaction of endogenous Olig1 with endogenous HHM (Figure 5B). Their interaction was not dependent on TGF-β signalling. We then examined the effect of Olig1 on proliferation of malignant glioma cell line U373MG in response to TGF-β. Expression of Olig1 resulted in enhancement of TGF-β-induced proliferation of U373MG cells, which was antagonized by STI571 (Gleevec), an inhibitor of PDGF receptor kinase (Figure 5C). Olig1 also enhanced TGF-β-induced upregulation of PDGF-B expression (Figure 5D). These findings suggest that the effect of Olig1 was mediated through induction of PDGF-B. In contrast, TGF-β-induced proliferation of U373MG cells as well as induction of PDGF-B were suppressed by HHM (Figure 5C and D). HHM thus exhibited effects opposite to those of Olig1 on TGF-β-induced proliferation of U373MG cells.
Next, we knocked down the expression of endogenous Olig1 or HHM in U373MG cells by microRNA introduced by lentivirus vector (Figure 6A). The cell proliferation induced by TGF-β was partially abrogated in the cells that expressed microRNA targeting Olig1 (Figure 6B). In contrast, TGF-β more potently induced proliferation of the cells expressing microRNA targeting HHM, and this induction was completely blocked by STI571, as in the cells expressing negative-control microRNA. Furthermore, induction of PDGF-B by TGF-β was suppressed in the U373MG cells in which the Olig1 microRNA was expressed, whereas TGF-β more potently upregulated PDGF-B expression in the cells expressing the HHM microRNA (Figure 6C).
Figure 6.
Effects of endogenous Olig1 and HHM on the growth of U373MG glioma cells in vitro and in vivo. (A) Knockdown of Olig1 and HHM using microRNA. U373MG cells were infected with lentivirus-encoding control microRNA, Olig1 microRNA, or HHM microRNA. Expression of Olig1 and HHM proteins were determined by immunoblot. Expression of α-tubulin was determined as a loading control. NC, negative-control microRNA. (B) In vitro proliferation assay of U373MG glioma cells in which Olig1 or HHM was knocked down. Results were expressed as the change from control (untreated cells). Error bars represent s.e. (C) TGF-β-induced expression of PDGF-B in U373MG glioma cells in which Olig1 or HHM was knocked down. mRNAs were prepared from cells treated with TGF-β (1 ng/ml) for 3 h. Expression of PDGF-B, Olig1, or HHM was determined by quantitative real-time PCR analyses. Values were normalized to the amount of GAPDH mRNA. Error bars represent s.d. (D) Association of Olig1 and Smad2/3 with the PDGF-B promoter region. ChIP analysis was performed using U373MG cells infected with lentivirus-encoding control microRNA, Olig1 microRNA, or HHM microRNA. Cells were treated with TGF-β (2.5 ng/ml) for 1 h and harvested. Eluted DNAs were subjected to quantitative real-time PCR analysis. Values were normalized to the amount of the first intron of hypoxanthine phosphoribosyltransferase 1. Error bars represent s.d. Primer sequences are listed on the Supplementary Table S2. (E) In vivo growth of U373MG cells in which Olig1 or HHM was knocked down by infection with lentivirus-encoding control microRNA, Olig1 microRNA, or HHM microRNA. Values are plotted as the mean tumour volume±s.e. of nine tumours per condition. Panels on the right side show the results of histological examination of the samples. Tumour tissues dissected at 3 weeks after transplantation appeared encapsulated and exhibited no significant variation histologically. Tissue sections were stained with haematoxylin–eosin or Azan. Scale bars: 60 μm.
These findings suggest that Olig1 is involved in TGF-β-induced PDGF-B expression, which promotes proliferation of the glioma cell line U373MG, whereas HHM exhibits effects counteracting those of Olig1.
Olig1 associates with the PDGF-B promoter region in response to TGF-, and HHM facilitates their dissociation
Smad3 was shown previously to associate with the PDGF-B promoter region in U373MG cells in response to TGF-β (Bruna et al, 2007). To examine the recruitment of Olig1 to the PDGF-B promoter in vivo, we performed ChIP assay in U373MG cells. Anti-Smad2/3 antibody or anti-Olig1 antibody immunoprecipitated the DNA fragment (approximately −336 to −36 from the transcription initiation site, containing SBE and E-boxes) of the PDGF-B promoter in response to TGF-β (Figure 6D). When Olig1 was knocked down, the co-precipitation of the DNA fragment was substantially reduced. These findings suggest that Olig1 associates with the PDGF-B promoter synergistically with the Smad complex, and thus upregulates PDGF-B expression. We also found that TGF-β-dependent interaction of the PDGF-B promoter with Smad2/3 and Olig1 was enhanced in U373MG cells with HHM microRNA. HHM thus facilitated the dissociation of Olig1 and Smad2/3 from the PDGF-B promoter, resulting in the suppression of TGF-β-induced PDGF-B induction.
HHM negatively regulates in vivo growth of glioma, whereas Olig1 is required for this growth
To examine the functions of endogenous Olig1 and HHM in in vivo growth of glioma, we subcutaneously implanted U373MG cells into nude mice. Knockdown of Olig1 in U373MG cells greatly attenuated the in vivo growth of U373MG cells compared with control cells, whereas knockdown of HHM caused more rapid growth of these cells (Figures 6E). These in vivo growth data are thus consistent with those for in vitro proliferation.
In summary, Olig1 is required for glioma proliferation, which was negatively regulated by HHM through modulation of TGF-β signalling.
Discussion
Transforming growth factor-β induces a variety of cellular responses principally through Smad-dependent transcriptional regulation. Cytoplasmic as well as nuclear proteins have been identified as regulators of Smad signalling, most of which regulate it in a comprehensive manner. One exception to this is YY1, which affects the expression of PAI-1, but not of p21WAF, p15INK4b, or c-myc (Kurisaki et al, 2003). Although YY1 interacts with the MH1 domain of Smad4 to interfere with the Smad4–DNA interaction, the mechanism underlying the selective suppression has not been fully elucidated. In this study, we report that an Id-like protein, HHM, is a synexpression group-restricted regulator of Smad signalling, which exerts an effect through abrogation of physical interaction between Smad2/3 and certain bHLH transcription factors, including Olig1. HHM is the first example of a cell response-specific regulator of Smad signalling with clearly determined mechanisms.
Dual functions of Maid in vivo
Maid was first identified as a maternally transcribed gene product that has a structural feature of dominant-negative HLH proteins (Hwang et al, 1997). Mouse Maid is actively transcribed in eggs and zygotes, but it is also widely expressed in adults. HMM is a human homologue of mouse Maid (Terai et al, 2000). Subsequently, it was shown to interact with a leukocyte-specific adaptor protein Grap2 and cyclin D, and it is also termed GCIP, CCNDBP1, and DIP1 (Xia et al, 2000; Yao et al, 2000). However, the physiological functions of HHM have not been clearly determined.
Maid exerts opposite effects on cell proliferation and tumorigenesis, depending on cellular context. In Maid-deficient mice, cell proliferation after partial hepatectomy was suppressed compared with that in wild-type mice (Sonnenberg-Riethmacher et al, 2007). In addition, HHM expression is elevated in early stages of hepatocarcinogenesis (Takami et al, 2005). These findings suggest positive regulatory functions of Maid in cell proliferation. In contrast, Maid-deficient mice more frequently develop liver tumours (Sonnenberg-Riethmacher et al, 2007), and hepatic overexpression of HHM in transgenic mice decreases susceptibility to chemical hepatocarcinogenesis (Ma et al, 2006). These findings suggest a tumour suppressor function of Maid in the liver.
In this study, we observed that HHM positively regulates the proliferation of cells whose growth is inhibited by TGF-β (NMuMG and HaCaT cells), whereas it negatively regulates the proliferation of cells whose growth is promoted by TGF-β (U373MG cells). Although HHM was reported previously to promote (Takami et al, 2005) or inhibit cell cycle progression (Xia et al, 2000; Ma et al, 2007), the Smad-signalling-mediated mechanism in cell proliferation appears to be dominant, at least in these cell lines.
HHM has a mode of action distinct from that of prototypical HLH proteins Ids
Human homologue of Maid shares a structural feature with Id proteins in having an HLH domain but lacking a basic domain responsible for the interaction with DNA. It has therefore been expected that Maid functions in a manner analogous to Id proteins (Hwang et al, 1997). However, we found that Maid inhibits bHLH proteins in a manner distinct from that of Id proteins.
Basic-helix-loop-helix transcription factors are classified into two groups, that is, those ubiquitously expressed (class I), including E12/47, and those expressed in a tissue-specific manner (class II), including Olig1 (Massari and Murre, 2000). They are thought to function in dimeric forms because DNA-binding activities in their monomeric form are not strong enough and, in general, heterodimeric forms are favoured. Id proteins are known to inhibit heterodimerization of bHLH proteins through predominant binding to class I bHLH proteins (Norton, 2000). We also observed that Olig1 heterodimerizes with E12/47 (Supplementary Figure S7A), and Id2 interferes with this heterodimerization principally through binding to E12/47 (Supplementary Figure S7B), although Id2 can directly interact with Olig1 (Samanta and Kessler, 2004). Thus, the inhibitory effects of Id proteins should be comprehensive, as Id proteins target common components of heterodimers of bHLH transcription factors.
In contrast, when HHM inhibits the formation of the complex composed of class II bHLH transcription factor and Smad2/3, HHM principally targets class II bHLH proteins, yielding more specific inhibitory effects. Id2 failed to disrupt the interaction between Olig1 and Smad2/3, probably because the affinity between Id2 and Olig1 was not sufficient to produce dissociation of the Olig1–Smad2/3 complex. HHM also inhibits heterodimerization between Olig1 and E12/47, although it fails to interact with E12/47 (Supplementary Figure S7A and B). Direct interaction with class II proteins appears to be a preferred mechanism in HHM-mediated inhibition of functions of the class II bHLH proteins (Supplementary Figure S7C).
Function of the Olig1-Smad synexpression group in TGF- signalling
Olig1 was first identified as an oligodendrocyte lineage-specific bHLH transcription factor (Zhou et al, 2000) and subsequently shown to be required for maturation of oligodendrocyte progenitors (Arnett et al, 2004; Xin et al, 2005). However, Olig1 is expressed in cells other than oligodendrocytes (Supplementary Figure S8) and has been implicated in malignant tumours including oligodendroglioma (Lu et al, 2001) and non-small cell lung carcinoma (Brena et al, 2007).
In a neurosphere implantation model of malignant glioma, in which neural progenitor cells from p16INK4a/p19ARF-null mouse embryos were used after manipulation to express constitutively active EGF receptor mutant, Olig1-null background resulted in the delay of glioma formation (Ligon et al, 2007). Olig1 thus also appears to have an important function in the progression of glioma, although the mechanism remains to be determined. Here, we demonstrated that Olig1 is involved in TGF-β-induced proliferation of glioma cells through induction of PDGF-B.
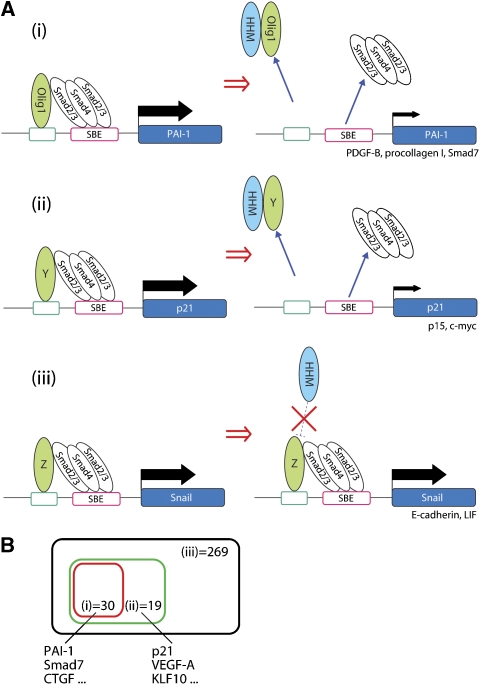
The Olig1-Smad synexpression group includes PAI-1, PDGF-B, procollagen 1, and Smad7, but not p21WAF, p15INK4b, c-myc, Snail, E-cadherin, or LIF. According to our findings, TGF-β-target genes can be classified as follows (Figure 7A): genes of the Olig1-Smad synexpression group that are suppressed by HHM (Figure 7A(i)), genes of the other synexpression groups that are suppressed by HHM (Figure 7A(ii)), and genes that are not suppressed by HHM (Figure 7A(iii)). Figure 7B is the diagrammatic representation of this classification with the results of DNA microarray analysis (Supplementary Table S1). The Olig1-Smad synexpression group appears to include target genes involved in fibrosis, but not those mediating the anti-proliferative or EMT-inducing effects of TGF-β. Further investigation is needed to establish the pathophysiological functions of the Olig1-Smad synexpression group. Other Smad-binding transcription factors antagonized by HHM appear to regulate target genes involved in the anti-proliferative effect of TGF-β. Identification of such targets of HHM is a task for the future.
Figure 7.
(A) Model of synexpression group-restricted inhibition of TGF-β signalling by HHM. On activation of TGF-β signalling, Smads form complexes with transcription factors, including Olig1 in the nucleus, and interact with the promoter region of target genes. HHM binds to a subset of the transcription factors including Olig1 (i) and Y (ii), but not Z (iii) and inhibits their physical interaction with Smads, which results in the dissociation of both the Smad complex and the partner transcription factors from the promoter regions. Y, unidentified Smad-binding transcription factors that interact with HHM; Z, Smad-binding transcription factors that do not interact with HHM. (B) Diagram representing data obtained from the microarray analysis of gene expression profiles of NMuMG cells transfected with negative-control siRNA, HHM siRNA, or Olig1 siRNA and treated for 1 h with TGF-β (1 ng/ml). Black square, TGF-β-up regulated genes; green square, HHM-regulated genes; red square, Olig1-regulated genes. Raw data are shown in Supplementary Table S1.
Synexpression group-specific regulators of TGF- signalling as molecular targets
Our present findings open the way for the control of each synexpression group through inhibiting formation of complexes composed of transcription factors and activated Smad2/3, which enables cellular response-specific regulation of TGF-β signalling. Recently, inhibitors of the TGF-β type I receptor as well as neutralizing antibodies to TGF-β ligands have been developed and used to inhibit metastasis of certain cancers (Yingling et al, 2004). As TGF-β has important functions in the maintenance of homoeostasis, transcription factors that cooperate with activated Smad complexes in the regulation of transcription of TGF-β-target genes should be promising molecular targets with reduced side effects.
Materials and methods
Cell culture
NIH3T3, HaCaT, HEK293T, NMuMG, and U373MG cells were obtained from the American Type Culture Collection, and A549 cells were from Cell Resource Center for Biomedical Research, Institute of Development, Aging and Cancer, Tohoku University. All cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum, 50 U/ml penicillin, and 50 μg/ml streptomycin. For the culture of NMuMG cells, insulin (10 μg/ml) was supplemented. For the culture of U373MG cells, sodium pyruvate (1 mM) and non-essential amino acids (0.1 mM) were supplemented. HEK293FT cells were purchased from Invitrogen and maintained according to a standard protocol.
Luciferase assay
Luciferase assay was performed as described previously (Goto et al, 2007) using TGF-β-responsive reporters, (CAGA)12-MLP-Luc (Dennler et al, 1998), p3TP-Luc (Wrana et al, 1992), p800-Luc (Keeton et al, 1991), and Smad7-Luc (Nagata et al, 2006). Values were normalized to Renilla luciferase activity under the control of thymidine kinase promoter.
RNA interference
To generate short hairpin RNA (shRNA) constructs, oligonucleotides corresponding to HHM-pSUPER and NC-pSUPER (see Supplementary Table S2 for sequences) were annealed, followed by ligation into the pSUPER vector, which was digested with BglII/HindIII. siRNAs (see Supplementary Table S2 for sequences) were introduced into NMuMG cells using HiPerFect transfection reagent (Qiagen) according to the manufacturer's instructions. The final concentration of siRNAs used was 5 nM. MicroRNA constructs against Olig1 and HHM were cloned into the pcDNA6.2-GW/EmGFP-mir vector (Invitrogen) after annealing the oligonucleotides (see Supplementary Table S2 for sequences). miR-neg (negative control) was provided by Invitrogen.
Antibodies
The antibodies used were as follows: anti-Flag M2 (Sigma-Aldrich); anti-Myc 9E10 (Pharmingen); anti-haemagglutinin (HA) 3F10 (Roche Diagnostics); anti-α-tubulin DM1A (Sigma-Aldrich); anti-E-cadherin 610182 (BD Transduction Laboratories); anti-fibronectin (Calbiochem); anti-Olig1 (R&D Systems); and normal mouse IgG (Santa Cruz Biotechnology) as a negative control for immunoprecipitation using anti-Olig1 antibody. Anti-HHM antibody was prepared by immunizing rabbits with bacterially expressed HHM (full-length).
DNA transfection, cell lysis, immunoprecipitation, and immunoblotting
HEK293T cells were transiently transfected using FuGENE6 transfection reagent and incubated for 24 h before analysis. Cells were lysed with a buffer containing 1% Nonidet P-40, 20 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM PMSF, 1% aprotinin, and 5 mM EDTA. For immunoprecipitation, cleared lysates were incubated with anti-Flag antibody for 1 h at 4 °C, or with anti-Olig1 antibody or mouse normal IgG overnight at 4 °C. Proteins in immunoprecipitates or cleared cell lysates were subjected to SDS–PAGE and transferred to Fluoro Trans W membrane (Pall). Immunoblotting was performed using the indicated antibodies.
Adenoviruses
Flag-tagged full-length Olig1 and HHM cDNAs were cloned into a pENTR vector (Invitrogen) and introduced into the adenoviral genome through recombination between pENTR vector and the pAd/CMV/V5-DEST vector using LR Clonase (Invitrogen). HEK293A cells were infected with pAd/CMV/Olig1 or pAd/CMV/HHM after its linearization with PacI. Viral particles were isolated by three freeze–thaw cycles and amplified by reinfection to HEK293A cells.
Cell proliferation assay
Cells were seeded in triplicate at a density of 5 × 104 cells per well in 12-well plates and cultured for 24 h in the growth medium for each cell line. After treatment with TGF-β (1 or 2.5 ng/ml) and/or STI571 (5 μM, Novartis) for the indicated periods, cells were trypsinized and then counted. STI571 stock solution was prepared as described previously (Matsuyama et al, 2003).
Quantitative real-time PCR
Quantitative real-time reverse transcription–PCR was performed as described previously (Goto et al, 2007). All samples were run in triplicate in each experiment. The primers used are listed in Supplementary Table S2. Values were normalized to that for glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Phalloidin staining
To allow direct fluorescence of the actin cytoskeleton, cells were fixed in 3.7% paraformaldehyde in phosphate-buffered saline (PBS), permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature, and subsequently stained with 0.25 mM tetramethylrhodamine B isocyanate (TRITC)-conjugated phalloidin (Sigma-Aldrich). Fluorescence was examined by confocal laser scanning microscopy (Carl Zeiss).
Protein identification
Selected protein-containing bands were excised from silver-stained SDS–PAGE gels and digested in-gel with trypsin (Hellman, 2000). Proteins were identified from the obtained peptide mass spectra using NCBInr sequence database with the aid of PROFOUND (http://prowl.rockefeller.edu/prowl-cgi/profound.exe).
DNA-affinity precipitation
Cell lysates were prepared and DNA-affinity precipitation assay was then performed as described previously (Nishihara et al, 1999).
GST pull-down
Equal amounts of GST, GST-Olig1, or GST-HHM mutants were adsorbed to glutathione-Sepharose beads and incubated with normalized amounts of lysates from HEK293T cells expressing Flag-tagged protein. The beads were washed three times in NETN buffer (0.1% Nonidet P-40, 50 mM Tris–HCl (pH 8.0), 150 mM NaCl, and 1 mM EDTA). Bound proteins were analysed by immunoblotting.
Lentivirus
To create entry clones, cDNAs encoding GFP and microRNA constructs against Olig1 or HHM were transferred from pcDNA6.2-GW/EmGFP-mir into pDONR221 (Invitrogen) using BP clonase (Invitrogen). To create lentivirus constructs, they were transferred from entry clones into lentivirus vector (CSII-EF-RfA) using LR clonase (Invitrogen).
For the production of defective lentivirus vectors, HEK293FT cells were transfected using Lipofectamine2000 (Invitrogen) with three plasmids: vector construct, VSV-G- and Rev-expressing construct (pCMV-VSV-G-RSV-Rev), and packaging construct (pCAG-HIVgp). The culture supernatants were collected 48 h after transfection, and viral particles were concentrated by centrifugation. For lentiviral infection, 1 × 105 U373MG cells per well in 6-well plates were infected with lentivirus particles. At 48 h after infection, infection efficiency was examined on the basis of GFP expression, and 95–100% U373MG cells were confirmed to be positive for GFP.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as described previously (Suzuki et al, 2004). Following reverse crosslinking, DNA was treated with proteinase K and purified using a PCR purification kit (Qiagen). DNA was eluted in 30 μl of TE, and used for PCR analysis or quantitative real-time PCR. PCR primers are listed in the Supplementary Table S2.
In vivo proliferation assay
A total of 1 × 106 U373MG cells in 50 μl of serum-free DMEM and 50 μl Matrigel (BD Biosciences) were injected subcutaneously into male Balb/c nu/nu mice (5 weeks old). Tumours were measured externally every week until week 3, and tumour volume was approximated using the equation volume=(a × b2) π/6, where a and b are the lengths of the major and minor axes, respectively. All animal experimental protocols were performed in accordance with the policies of the Animal Ethics Committee of University of Tokyo.
Microarray analysis
NMuMG cells were transfected with negative-control siRNA, Olig1 siRNA, or HHM siRNA and treated with or without TGF-β for 1 h. Total RNAs were prepared with RNeasy (Qiagen) and used to conduct oligonucleotide microarray analysis using GeneChip Mouse Genome 430 2.0 Array (Affymetrix) according to the manufacturer's instructions.
Supplementary Material
Supplementary Figures S1
Supplementary Figures S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Information
Acknowledgments
We are grateful to Dr Takeshi Imamura for valuable discussion and Keiko Yuki and Yasuyuki Morishita for skilled technical assistance. This work was supported by KAKENHI (Grant-in-aids for Scientific Research) and Global COE Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
- Arnett HA, Fancy SP, Alberta JA, Zhao C, Plant SR, Kaing S, Raine CS, Rowitch DH, Franklin RJ, Stiles CD (2004) bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science 306: 2111–2115 [DOI] [PubMed] [Google Scholar]
- Blobe GC, Schiemann WP, Lodish HF (2000) Role of transforming growth factor β in human disease. N Engl J Med 342: 1350–1358 [DOI] [PubMed] [Google Scholar]
- Brena RM, Morrison C, Liyanarachchi S, Jarjoura D, Davuluri RV, Otterson GA, Reisman D, Glaros S, Rush LJ, Plass C (2007) Aberrant DNA methylation of OLIG1, a novel prognostic factor in non-small cell lung cancer. Plos Med 4: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruna A, Darken RS, Rojo F, Ocaña A, Peñuelas S, Arias A, Paris R, Tortosa A, Mora J, Baselga J, Seoane J (2007) High TGFβ-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell 11: 147–160 [DOI] [PubMed] [Google Scholar]
- Cohen P (2002) Protein kinases—the major drug targets of the twenty-first century? Nat Rev Drug Discov 1: 309–315 [DOI] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM (1998) Direct binding of Smad3 and Smad4 to critical TGFβ-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J 17: 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang Y (2003) Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 425: 577–584 [DOI] [PubMed] [Google Scholar]
- Ehata S, Hanyu A, Hayashi M, Aburatani H, Kato Y, Fujime M, Saitoh M, Miyazawa K, Imamura T, Miyazono K (2007) TGF-β promotes survival of mammary carcinoma cells through induction of anti-apoptotic transcription factor DEC1. Cancer Res 67: 9694–9703 [DOI] [PubMed] [Google Scholar]
- Goto K, Kamiya Y, Imamura T, Miyazono K, Miyazawa K (2007) Selective inhibitory effects of Smad6 on bone morphogenetic protein type I receptors. J Biol Chem 282: 20603–20611 [DOI] [PubMed] [Google Scholar]
- Hellman U (2000) Sample preparation by SDS/PAGE and in-gel digestion. EXS 88: 43–54 [DOI] [PubMed] [Google Scholar]
- Hwang SY, Oh B, Füchtbauer A, Füchtbauer EM, Johnson KR, Solter D, Knowles BB (1997) Maid: a maternally transcribed novel gene encoding a potential negative regulator of bHLH proteins in the mouse egg and zygote. Dev Dyn 209: 217–226 [DOI] [PubMed] [Google Scholar]
- Keeton MR, Curriden SA, van Zonneveld AJ, Loskutoff DJ (1991) Identification of regulatory sequences in the type 1 plasminogen activator inhibitor gene responsive to transforming growth factor β. J Biol Chem 266: 23048–23052 [PubMed] [Google Scholar]
- Kurisaki K, Kurisaki A, Valcourt U, Terentiev AA, Pardali K, ten Dijke P, Heldin CH, Ericsson J, Moustakas A (2003) Nuclear factor YY1 inhibits transforming growth factor β- and bone morphogenetic protein-induced cell differentiation. Mo Cell Biol 23: 4494–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, Depinho RA, Anderson DJ, Stiles CD, Rowitch DH (2007) Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron 53: 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Park JK, Noll E, Chan JA, Alberta J, Yuk D, Alzamora MG, Louis DN, Stiles CD, Rowitch DH, Black PM (2001) Oligodendrocyte lineage genes (OLIG) as molecular markers for human glial brain tumors. Proc Natl Acad Sci USA 98: 10851–10856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Stafford LJ, Li D, Luo J, Li X, Ning G, Liu M (2007) GCIP/CCNDBP1, a helix-loop-helix protein, suppresses tumorigenesis. J Cell Biochem 100: 1376–1386 [DOI] [PubMed] [Google Scholar]
- Ma W, Xia X, Stafford LJ, Yu C, Wang F, LeSage G, Liu M (2006) Expression of GCIP in transgenic mice decreases susceptibility to chemical hepatocarcinogenesis. Oncogene 25: 4207–4216 [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C (2000) Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S, Iwadate M, Kondo M, Saitoh M, Hanyu A, Shimizu K, Aburatani H, Mishima HK, Imamura T, Miyazono K, Miyazawa K (2003) SB-431542 and Gleevec inhibit transforming growth factor-β-induced proliferation of human osteosarcoma cells. Cancer Res 63: 7791–7798 [PubMed] [Google Scholar]
- Miyazawa K, Shinozaki M, Hara T, Furuya T, Miyazono K (2002) Two major Smad pathways in TGF-β superfamily signalling. Genes Cells 7: 1191–1204 [DOI] [PubMed] [Google Scholar]
- Nagata M, Goto K, Ehata S, Kobayashi N, Saitoh M, Miyoshi H, Imamura T, Miyazawa K, Miyazono K (2006) Nuclear and cytoplasmic c-Ski differently modulate cellular functions. Genes Cells 11: 1267–1280 [DOI] [PubMed] [Google Scholar]
- Nishihara A, Hanai JI, Imamura T, Miyazono K, Kawabata M (1999) E1A inhibits transforming growth factor-β signaling through binding to Smad proteins. J Biol Chem 274: 28716–28723 [DOI] [PubMed] [Google Scholar]
- Norton JD (2000) ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci 113: 3897–3905 [DOI] [PubMed] [Google Scholar]
- Samanta J, Kessler JA (2004) Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development 131: 4131–4142 [DOI] [PubMed] [Google Scholar]
- Shi Y, Massagué J (2003) Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113: 685–700 [DOI] [PubMed] [Google Scholar]
- Sonnenberg-Riethmacher E, Wüstefeld T, Miehe M, Trautwein C, Riethmacher D (2007) Maid (GCIP) is involved in cell cycle control of hepatocytes. Hepatology 45: 404–411 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Yagi K, Kondo M, Kato M, Miyazono K, Miyazawa K (2004) c-Ski inhibits the TGF-β signaling pathway through stabilization of inactive Smad complexes on Smad-binding elements. Oncogene 23: 5068–5076 [DOI] [PubMed] [Google Scholar]
- Takami T, Terai S, Yokoyama Y, Tanimoto H, Tajima K, Uchida K, Yamasaki T, Sakaida I, Nishina H, Thorgeirsson SS, Okita K (2005) Human homologue of maid is a useful marker protein in hepatocarcinogenesis. Gastroenterology 128: 1369–1380 [DOI] [PubMed] [Google Scholar]
- Terai S, Aoki H, Ashida K, Thorgeirsson SS (2000) Human homologue of maid: a dominant inhibitory helix-loop-helix protein associated with liver-specific gene expression. Hepatology 32: 357–366 [DOI] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Cárcamo J, Zentella A, Doody J, Laiho M, Wang XF, Massagué J (1992) TGFβ signals through a heteromeric protein kinase receptor complex. Cell 71: 1003–1014 [DOI] [PubMed] [Google Scholar]
- Xia C, Bao Z, Tabassam F, Ma W, Qiu M, Hua S, Liu M (2000) GCIP, a novel human grap2 and cyclin D interacting protein, regulates E2F-mediated transcriptional activity. J Biol Chem 275: 20942–20948 [DOI] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu FF, Gow A, Lu QR (2005) Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci 25: 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Doki Y, Jiang W, Imoto M, Venkatraj VS, Warburton D, Santella RM, Lu B, Yan L, Sun XH, Su T, Luo J, Weinstein IB (2000) Cloning and characterization of DIP1, a novel protein that is related to the Id family of proteins. Exp Cell Res 257: 22–32 [DOI] [PubMed] [Google Scholar]
- Yingling JM, Blanchard KL, Sawyer JS (2004) Development of TGF-β signalling inhibitors for cancer therapy. Nat Rev Drug Discov 3: 1011–1022 [DOI] [PubMed] [Google Scholar]
- Zavadil J, Böttinger EP (2005) TGF-β and epithelial-to-mesenchymal transitions. Oncogene 24: 5764–5774 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Wang S, Anderson DJ (2000) Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron 25: 331–343 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1
Supplementary Figures S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Information