Abstract
In yeast, three small nucleolar RNAs (snoRNAs) are essential for the processing of pre-ribosomal RNA—U3, U14 and snR30—whereas 72 non-essential snoRNAs direct site-specific modification of pre-rRNA. We applied a quantitative screen for alterations in the pre-ribosome association to all 75 yeast snoRNAs in strains depleted of eight putative helicases implicated in 40S subunit synthesis. For the modification-guide snoRNAs, we found no clear evidence for the involvement of these helicases in the association or dissociation of pre-ribosomes. However, the DEAD box helicase Rok1 was required specifically for the release of snR30. Point mutations in motif I, but not in motif III, of the helicase domain of Rok1 impaired the release of snR30, but this was less marked than in strains depleted of Rok1, and resulted in a dominant-negative growth phenotype. Dissociation of U3 and U14 from pre-ribosomes is also dependent on helicases, suggesting that release of the essential snoRNAs might differ mechanistically from release of the modification-guide snoRNAs.
Keywords: ribosome biogenesis, RNA helicase, snoRNA
Introduction
During ribosome synthesis in Saccharomyces cerevisiae, the 35S pre-ribosomal RNA is processed into the mature 18S, 5.8S and 25S rRNAs through a complex sequence of endo- and exonucleolytic cleavages and base modifications (Fromont-Racine et al, 2003; Tschochner & Hurt, 2003; Granneman & Baserga, 2004). The most numerous modifications are 2′-O-methylation and pseudouridylation at sites selected by base-pairing to box C/D and box H/ACA small nucleolar RNAs (snoRNAs), respectively (Kiss-László et al, 1996; Ganot et al, 1997; Bachellerie et al, 2002). All snoRNA-directed rRNA modifications are individually non-essential, and the corresponding snoRNAs are similarly non-essential for the processing of pre-rRNA, ribosome synthesis and viability (Kiss, 2002). By contrast, three snoRNAs—the box C/D snoRNAs U3 and U14, and the box H/ACA snoRNA snR30—are essential for viability and early rRNA cleavages at sites A0–A2 (Li et al, 1990; Hughes & Ares, 1991; Beltrame & Tollervey, 1995; Liang & Fournier, 1995; Morrissey & Tollervey, 1997). U3 and snR30 are not believed to function as modification guides. U14 does direct 2′-O-methylation, but this activity is distinct from its essential function in the processing of pre-rRNA.
More than 180 proteins are required for ribosome biogenesis in S. cerevisiae, including 19 putative RNA helicases. One of these is the DEVH box helicase Mtr4, which acts as a cofactor for the nuclear exosome complex and is required for the maturation of 5.8S rRNA. The remaining helicases are all members of the DEAD/H box family. On the basis of the pre-rRNA processing defects in mutant strains, seven helicases are specifically required for 40S synthesis and nine for 60S synthesis, whereas Has1 is involved in both pathways (reviewed by Rocak & Linder, 2004; Bleichert & Baserga, 2007). Prp43 is also required for the biogenesis of both subunits and, in addition, functions in the splicing of pre-mRNA (Lebaron et al, 2005; Combs et al, 2006; Leeds et al, 2006); it was not analysed here. Little is known about the detailed function of any helicase during ribosome synthesis, but all except Dbp3 and Dbp7 are essential for viability, showing that they do not generally have redundant functions.
The snoRNAs direct pre-rRNA modification through site-specific base pairing (Kiss, 2002). Particularly in the case of the box C/D snoRNAs, this can involve extended, perfect complementarity of up to 19 nucleotides, which is predicted to be stable under physiological conditions, suggesting that protein cofactors are required to promote association with, and dissociation from, the pre-rRNAs. The many putative helicases were obvious candidates for this activity and Dbp4 was shown to be required for the release of U14 and snR41 from pre-ribosomes (Kos & Tollervey, 2005), whereas Has1 was implicated in the release of both U14 and U3 (Liang & Fournier, 2006).
Here, we report a systematic analysis of all yeast snoRNAs by using quantitative PCR (qPCR), assessing changes in pre-ribosome association following depletion of eight DEAD/H box RNA helicases that are required for synthesis of the 40S ribosomal subunit (Dbp4, Dbp8, Dhr1, Dhr2, Fal1, Has1, Rok1 and Rrp3). Our results confirm the requirement for Dbp4 and Has1 in the release of the snoRNA U14 from pre-ribosomes. We further report that the release of snR30 from pre-ribosomes is specifically dependent on the helicase Rok1. Markedly, however, none of the modification guide snoRNAs showed such strong alteration in any helicase mutant, suggesting that their binding and release involve unanticipated mechanisms.
Results
A screen for the quantification of yeast snoRNAs
With the aim of systematically testing the hypothesis that snoRNAs require specific RNA helicases for their release from association with the pre-rRNA, we developed a screen based on qPCR. This was modified from the technique of Ro et al (2006), and allowed the detection and quantification of all yeast snoRNAs in strains depleted of individual helicases.
To allow depletion of the essential RNA helicases, the genomic copies of DBP4, DBP8, DHR1, DHR2, HAS1, ROK1 and RRP3 were tagged with an amino-terminal 3HA tag and placed under the control of a tetoff promoter, which is repressed in response to doxycycline. For FAL1, the GAL1 promoter was used, as integration of the tet promoter was unsuccessful. For each strain, the time course of the response to doxycycline or glucose repression was determined by growth curves, and depletion was analysed by Western blots (data not shown) to allow the optimal depletion time point for analysis to be determined. Cells were collected one generation before detection of growth inhibition to reduce the likelihood of indirect effects. Depletion times used were 4 h for Dbp4, 6 h for Rrp3 and Fal1, 7 h for Dhr1, Has1 and Rok1, and 11 h for Dhr2 and Dbp8. Cells were lysed and soluble material was fractionated by centrifugation through sucrose gradients. RNA was extracted from pools of fractions containing either unbound snoRNAs or pre-ribosomal complexes and processed as shown in Fig 1A.
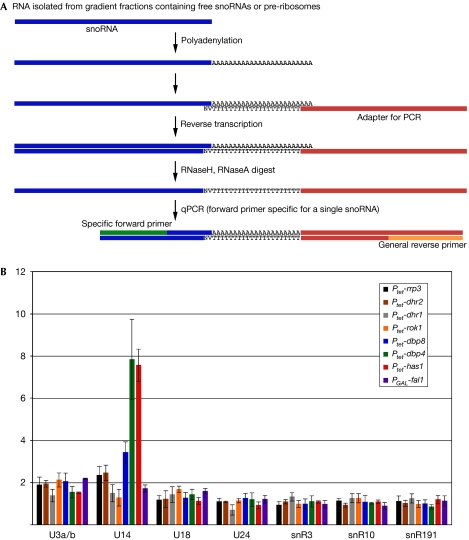
Figure 1.
A quantitative screen to analyse the levels of all yeast snoRNAs. (A) Scheme to illustrate sample preparation and analysis (adapted from Ro et al, 2006; see main text and Methods for details). (B) Levels of free and pre-ribosome-bound snoRNAs were analysed on depletion of eight putative RNA helicases required for the biogenesis of the small ribosomal subunit. Soluble material from lysates of wild-type and depleted cells was centrifuged in sucrose gradients, and fractions containing either free or pre-ribosome-bound snoRNAs were pooled (see Fig 3). RNA was extracted and analysed as shown in (A). Data from depletions were normalized to wild-type samples processed in parallel. Average ratios of pre-ribosome bound to unbound snoRNAs in the indicated depletion strains shown were calculated from at least three independent experiments. Error bars represent standard error. U14 accumulates on pre-ribosomes on depletion of Dbp4 (green bar) and Has1 (red); a minor accumulation is observed for depletion of Dbp8 (blue). Ptet, tetracycline-repressible promoter; snoRNAs, small nucleolar RNAs.
The RNA present in each fraction was polyadenylated in vitro and reverse transcribed with an oligo(dT) probe carrying an adapter. Each qPCR reaction used one snoRNA-specific primer, together with a common primer complementary to the adapter region introduced during reverse transcription (for primers, see the supplementary Table online). To validate the technique we initially analysed a subset of snoRNAs, including U14 that had previously been shown to require the helicases Dbp4 and Has1 for release from pre-ribosomes (Kos & Tollervey, 2005; Liang & Fournier, 2006). Our data confirmed the accumulation of U14 in the pre-ribosomal fraction in the absence of Dbp4 (Fig 1B, green) or Has1 (red). We did not, however, observe an effect on pre-ribosomal snR41 levels on depletion of Dbp4 with the strain background and time points used here (data not shown). A minor effect on the association of U14 with pre-ribosomes was also seen following depletion of Dbp8 (Fig 1B, blue).
Release of U3 was reported previously to be reduced by depletion of Has1 for 20 h (Liang & Fournier, 2006). We performed our analyses at shorter depletion time points (7 h in the case of Has1) and detected similar accumulation of pre-rRNA-associated U3 on depletion of several helicases. U3 is known to have multiple binding sites on 35S pre-rRNA and to interact with several proteins in the SSU processome, and several helicases might therefore participate in its release.
Release of snR30 requires Rok1
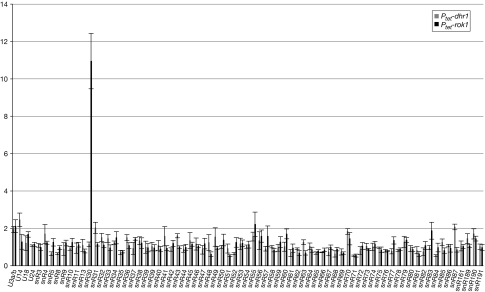
The results obtained with a subset of snoRNAs verified the method and therefore we screened the full set of 75 yeast snoRNAs for changes in pre-ribosome association in each of the eight helicase mutants (supplementary Fig S1 online; the primary qPCR data set is available on request from the authors). Depletion of Rok1 caused substantial accumulation of snR30 on pre-ribosomes (Fig 2). No other snoRNA showed statistically significant alterations following the depletion of Rok1. Surprisingly, such strong alteration in pre-ribosome association was not observed for any of the 72 modification-guide snoRNAs in any helicase mutant tested.
Figure 2.
Depletion of the putative RNA helicase Rok1 leads to accumulation of snR30 on pre-ribosomes. Rok1 and Dhr2 were depleted and the levels of free and pre-ribosome-bound pools of all yeast snoRNAs were analysed by using qPCR. Data are presented as described for Fig 1B. On depletion of Rok1, but not Dhr2, snR30 accumulates on pre-ribosomes. Ptet, tetracycline-repressible promoter; qPCR, quantitative PCR; snoRNAs, small nucleolar RNAs.
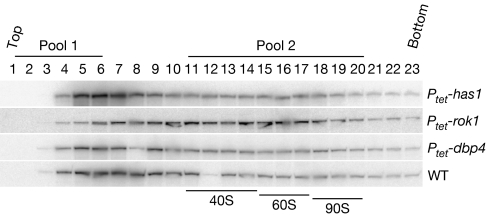
The requirement for Rok1 in the release of snR30 from pre-ribosomes was confirmed by Northern blotting of the RNA isolated from individual gradient fractions (Fig 3). Depletion of only Rok1 caused a shift of snR30 to gradient fractions with higher density. No clear changes in the abundance of snoRNA were detected by using qPCR following depletion of any helicase tested (data not shown), and this was confirmed for a subset of snoRNAs by Northern blotting (supplementary Fig S2 online). Changes in the ratio of pre-ribosome-bound compared with free snoRNAs therefore reflect their altered distribution.
Figure 3.
snR30 is retained on pre-ribosomes following the depletion of Rok1. Northern blot analysis of sucrose gradient fractions loaded with soluble material from either wild-type (WT) cells or after helicase depletion. Strains were grown in a medium containing doxycycline for helicase depletion. After lysis, soluble material was fractionated on sucrose gradients, RNA was isolated and analysed by Northern blot for the distribution of snR30. In the WT, as well as Has1 and Dbp4 depletion strains, snR30 is present largely in the top fractions of the gradient, whereas it shifts into higher molecular weight complexes on depletion of Rok1. Pools are indicated as the pre-ribosomal fractions (pool 2) and the gradient fractions containing the largest unbound fraction across all snoRNAs (pool 1). Pools were defined by comparison to the gel, which was stained before Northern transfer. Pool 1 contains material sedimenting well above the 40S ribosomal subunits. Pool 2 contains 40S and 60S ribosomes, together with the 90S pre-ribosomes. The 40S and 60S peaks were identified by ethidium staining of the gel. The 90S peak was identified by hybridization of an identical gradient with a probe directed against the ITS1 region of the 35S pre-rRNA. Ptet, tetracycline-repressible promoter.
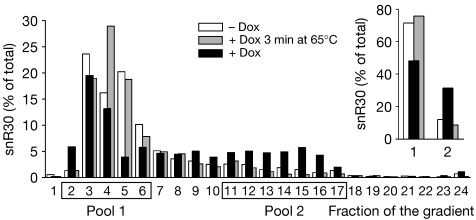
To confirm that snR30 present in lower gradient fractions was base-paired to large RNA species, we tested this association in deproteinized extracts (Fig 4; supplementary Fig S3 online; Tollervey, 1987; Kos & Tollervey, 2005). Cell lysates from the Ptet-rok1 strain grown in the presence or absence of doxycycline were deproteinized by treatment with proteinase K and loaded onto sucrose gradients. As a control, the doxycycline-treated lysate was incubated at 65°C for 3 min to disrupt base-paired interactions. Depletion of Rok1 (+Dox samples) increased the proportion of snR30 that sedimented in the lower fractions of the gradient, which contained complexes of high molecular weight. Treatment at 65°C largely released snR30 into the upper gradient fractions. No alteration was observed for the other snoRNAs tested, U3 and U14 (supplementary Fig S3 online). As expected, 5.8S rRNA sedimented in the lower gradient fractions owing its association with 25S rRNA, and was predominantly released by heat treatment (supplementary Fig S3 online). We conclude that the depletion of Rok1 increases the amount of snR30 that is base-paired to high molecular weight pre-rRNA.
Figure 4.
snR30 shows increased association with high molecular weight RNA following the depletion of Rok1. Northern blot analysis of 10–30% sucrose gradient fractions loaded with deproteinized RNA from Ptet-rok1 cells expressing Rok1 (−Dox samples) or after helicase depletion (+Dox samples). For deproteinization, lysates were treated with proteinase K in a buffer containing 1% lithium dodecyl sulphate for 3 h at 4°C, before loading on sucrose gradients. To dissociate RNA–RNA interactions, +Dox samples were, in addition, incubated at 65°C for 3 min before loading the sucrose gradient. Dox, doxycycline; Ptet, tetracycline-repressible promoter.
Mutation of motif I in Rok1 inhibits snoRNA release
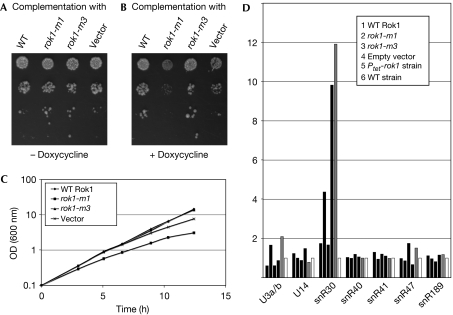
To determine whether the putative helicase activity of Rok1 is required for the release of snR30, point mutations were generated in the conserved motifs I (rok1-m1) and III (rok1-m3, SAT domain). Wild-type and mutant Rok1 constructs were expressed from the ROK1 promoter in vector pRS415 in the Ptet-rok1 strain. For complementation analysis, ten-fold serial dilutions of cultures were spotted on plates lacking doxycycline (Fig 5A; permissive conditions for Ptet-rok1 expression) or containing doxycycline (Fig 5B; repressive conditions). Growth was also tested in liquid culture (Fig 5C). The strain expressing wild-type Rok1 grew well in the presence of doxycycline, whereas growth of the vector control was impaired (Fig 5B,C). The rok1-m1 mutant showed a dominant-negative effect relative to the empty vector control, whereas rok1-m3 did not show a growth phenotype following depletion of endogenous Rok1.
Figure 5.
Analysis of Rok1 mutants for growth complementation and snR30 distribution. (A–C) Growth complementation analysis of Rok1 with mutations in motifs I (rok1-m1) and III (rok1-m3). In a background in which the genomic ROK1 gene is under control of the tetracycline-repressible promoter (Ptet-rok1), growth complementation was tested with plasmid-derived wild-type (WT), mutant Rok1 or empty plasmid (vector). Dilutions of cultures were spotted on plates (A) without doxycycline or (B) under Ptet-rok1 depletion conditions on plates containing doxycycline. Growth of the strains was also analysed in liquid culture (C). Although the rok1-m3 mutant can complement for growth, the mutation in motif I (rok1-m1) results in a slight dominant-negative effect. (D) Analysis of snoRNA accumulation on pre-ribosomes in the Rok1 complementation strains described in (A–C) shown in black bars, the Ptet-rok1 strain (grey) and wild type in white (bars 1–6 for each snoRNA). The complementation strains are shown in the same order as in (A–C). Average ratios of pre-ribosome-bound to unbound snoRNAs from two separate experiments are shown. Although plasmid-derived wild-type Rok1 and rok1-m3 can largely rescue the phenotype of snR30 accumulation, the empty vector cannot; the rok1-m1 mutant shows an intermediate phenotype. OD, optical density.
Analysis of the levels of pre-ribosome-bound compared with free snoRNAs showed an accumulation of snR30 on pre-ribosomes in the strain expressing rok1-m1 (Fig 5D), although this accumulation was less strong than in the vector control lacking Rok1. Accumulation of snR30 in pre-ribosomes was not seen in the rok1-m3 mutant, and no dominant-negative effects of rok1-m1 were observed.
We conclude that the ATPase activity of Rok1 is required for the efficient release of snR30 from pre-ribosomes.
Discussion
By using qPCR, we screened for changes in the levels of pre-ribosome-bound snoRNAs on depletion of each of the putative RNA helicases involved in 40S biogenesis. These analyses confirmed the functional interactions of Dbp4 and Has1 with U14 (Kos & Tollervey, 2005; Liang & Fournier, 2006). In addition, we found that Rok1 is required for the release of snR30 from pre-ribosomes.
To test the requirements for the putative helicase activity of Rok1, we analysed mutants in conserved helicase domains: the motif I (rok1-m1) and the SAT domain or motif III (rok1-m3). Expression of Rok1-m1 resulted in a dominant-negative growth defect, whereas Rok1-m3 supported near wild-type growth during depletion of endogenous Rok1. Dominant-negative effects were associated previously with mutations in motif I, the ATP binding and ATPase domain, in other helicases required for ribosome biogenesis (Bernstein et al, 2006; Granneman et al, 2006). By contrast, mutations in motif III, which is predicted to link the ATPase and helicase activities (Pause & Sonenberg, 1992; Plumpton et al, 1994; Bernstein et al, 2006; Granneman et al, 2006), generally show less severe growth phenotypes. Expression of the Rok1-m1 protein only partly rescued the release of snR30 from pre-ribosomes in the Rok1-depleted strain. These results show that the ATPase activity of Rok1 is not essential for the release of snR30 from pre-ribosomes, but it is important for optimal activity. Conceivably, binding of Rok1 to the pre-rRNA in close proximity to snR30 might assist its displacement in the absence of ATPase activity. Box H/ACA snoRNAs form two short base-paired sequences with the pre-rRNA, which are generally shorter than the sequences bound by box C/D snoRNAs (Kiss, 2002). The binding site of snR30 on 18S has not been reported but, as a member of the H/ACA snoRNA family, its release might be less dependent on helicase activity than U14.
Perhaps the most striking result from these analyses is the discrepancy between the strong accumulation seen for essential snoRNAs and the minor effects observed for several of the 72 modification-guide snoRNAs tested in strains depleted of any of the eight DEAD/H box RNA helicases investigated. As all the helicases tested here are individually essential for viability, they are presumably not simply functionally redundant. A proposed mechanism for the DEAD box RNA helicases in RNA duplex unwinding envisaged that the catalytic domains could act within a certain radius from the binding site (Yang et al, 2007). This suggested that several helicases might overlap in their ‘activity radius'. In this case, snoRNAs binding to pre-rRNA in regions that are accessible to more than one helicase might only show minor effects on the depletion of each individual helicase. However, clustering analyses of the raw data from our qPCR analysis did not yield robust results supporting this hypothesis (data not shown), and setting a more stringent threshold resulted in insufficient data for clustering. The clustering data suggested that Rok1 and Dhr1 showed the greatest similarities. However, a strain in which both ROK1 and DHR1 were depleted under tet-control showed no clear cumulative effects on pre-ribosomal snoRNA levels (data not shown). Thus, the mild alterations that were observed for several different snoRNAs provided no evidence that snoRNAs with pre-rRNA-binding sites in the same region of the 18S rRNA would show increases in the same helicase mutants.
We propose that the main function of the helicases involved in the pathway is the structural remodelling of pre-ribosomal intermediates, rather than acting on individual snoRNAs. It can be noted that the snoRNAs that do require helicases for their release from pre-ribosomes, including snR30, are essential for viability and are required for the initial steps in the processing of 35S pre-rRNA (Kos & Tollervey, 2005; Liang & Fournier, 2006). By contrast, dissociation of none of the non-essential modification-guide snoRNAs was associated with a specific helicase. This suggests that there might be significant differences in the mechanisms of dissociation of these two classes of snoRNAs. We speculate that the act of modification triggers destabilization of the base pairing, presumably involving some other type of activity.
Methods
Yeast strains and growth complementation. The strains used are listed in the supplementary Table online. Genomic integration of the tetracycline regulated promoter and the GAL1 promoter was carried out as described previously (Longtine et al, 1998). Yeast strains were grown in YPD medium for helicase depletions, containing 2 μg ml−1 doxycycline (Sigma-Aldrich, St Louis, MO, USA) for the repression of tet. For growth complementation, the ROK1 open reading frame (including 352 nt upstream and 449 nt downstream) was cloned into the XhoI–SmaI sites of pRS415 for expression from its own promoter. The mutants rok1-m1 (K172L) and rok1-m3 (S312A and T314A) were generated by site-directed mutagenesis. Plasmids were transformed into the BY4741 strain with genomic ROK1 under control of a tetoff-regulated promoter. Growth was measured on plates or in liquid culture containing 0.67% yeast nitrogen base, 2% glucose and 0.5% ammonium sulphate, supplemented with the required amino acids and in the presence of 10 μg ml−1 doxycycline for genomic depletion.
Sucrose gradient analyses, RNA preparation and qPCR. Cells were collected and lysed as described previously (Kos & Tollervey, 2005), and lysates were loaded on top of 12 ml 10–45% sucrose gradients and centrifuged for 16 h at 25 k.r.p.m. in a SW40Ti rotor. A portion of 500 ml fractions was collected and RNA was isolated as described previously (Sambrook et al, 1989). For qPCR analysis, RNA from fractions containing either pre-ribosome-bound (pool 2) or unbound snoRNAs (pool 1) was processed as shown in Fig 1A and as described in the supplementary information online. For deproteinization of RNA, lysates were treated with 2 mg ml−1 proteinase K in a buffer containing 200 mM NaCl and 1% LDS for 3 h at 4°C (Tollervey, 1987), and separated on 10–30% sucrose gradients.
Northern blotting. Northern transfer and hybridization were performed as described previously (Sambrook et al, 1989). Hybridization probes used are listed in the supplementary Table online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org)
Supplementary Material
Supplementary Information
Acknowledgments
We thank A. Wächter and E. Schleiff for help with the statistical analysis, K. Kotovic and S. Innocente for support with the qPCR set-up, and C. Schneider for a critical reading of the manuscript. This project was supported by the Wellcome Trust and a Federation of European Biochemical Societies Long-Term Fellowship to M.T.B.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bachellerie JP, Cavaille J, Huttenhofer A (2002) The expanding snoRNA world. Biochimie 84: 775–790 [DOI] [PubMed] [Google Scholar]
- Beltrame M, Tollervey D (1995) Base-pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J 14: 4350–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KA, Granneman S, Lee AV, Manickam S, Baserga SJ (2006) Comprehensive mutational analysis of yeast DEXD/H box RNA helicases involved in large ribosomal subunit biogenesis. Mol Cell Biol 26: 1195–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F, Baserga SJ (2007) The long unwinding road of RNA helicases. Mol Cell 27: 339–352 [DOI] [PubMed] [Google Scholar]
- Combs DJ, Nagel RJ, Ares M Jr, Stevens SW (2006) Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol Cell Biol 26: 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F (2003) Ribosome assembly in eukaryotes. Gene 313: 17–42 [DOI] [PubMed] [Google Scholar]
- Ganot P, Bortolin ML, Kiss T (1997) Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809 [DOI] [PubMed] [Google Scholar]
- Granneman S, Baserga SJ (2004) Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res 296: 43–50 [DOI] [PubMed] [Google Scholar]
- Granneman S, Bernstein KA, Bleichert F, Baserga SJ (2006) Comprehensive mutational analysis of yeast DEXD/H box RNA helicases required for small ribosomal subunit synthesis. Mol Cell Biol 26: 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JMX, Ares MJ (1991) Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J 10: 4231–4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T (2002) Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148 [DOI] [PubMed] [Google Scholar]
- Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T (1996) Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Kos M, Tollervey D (2005) The putative RNA helicase Dbp4p is required for release of the U14 snoRNA from pre-ribosomes in Saccharomyces cerevisiae. Mol Cell 20: 53–64 [DOI] [PubMed] [Google Scholar]
- Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, Henry Y (2005) The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol Cell Biol 25: 9269–9282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds NB, Small EC, Hiley SL, Hughes TR, Staley JP (2006) The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol Cell Biol 26: 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HV, Zagorski J, Fournier MJ (1990) Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol 10: 1145–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W-Q, Fournier MJ (1995) U14 base-pairs with 18S rRNA: a novel snoRNA interaction required for rRNA processing. Genes Dev 9: 2433–2443 [DOI] [PubMed] [Google Scholar]
- Liang XH, Fournier MJ (2006) The helicase Has1p is required for snoRNA release from pre-rRNA. Mol Cell Biol 26: 7437–7450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Morrissey JP, Tollervey D (1997) U14 small nucleolar RNA makes multiple contacts with the pre-ribosomal RNA. Chromosoma 105: 515–522 [DOI] [PubMed] [Google Scholar]
- Pause A, Sonenberg N (1992) Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J 11: 2643–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumpton M, McGarvey M, Beggs JD (1994) A dominant negative mutation in the conserved RNA helicase motif ‘SAT' causes splicing factor PRP2 to stall in spliceosomes. EMBO J 13: 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S, Park C, Jin J, Sanders KM, Yan W (2006) A PCR-based method for detection and quantification of small RNAs. Biochem Biophys Res Commun 351: 756–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocak S, Linder P (2004) DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5: 232–241 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Tollervey D (1987) A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J 6: 4169–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner H, Hurt E (2003) Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol 13: 255–263 [DOI] [PubMed] [Google Scholar]
- Yang Q, Del Campo M, Lambowitz AM, Jankowsky E (2007) DEAD-box proteins unwind duplexes by local strand separation. Mol Cell 28: 253–263 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information