Abstract
Accumulating evidence suggests that multiple complex mechanisms may be involved, simultaneously or complementarily, in the emergence and development of multidrug resistance (MDR) in various cancers. Cell adhesion-mediated MDR is one such mechanism. In the present study, we initially observed increased cell adhesion to extracellular matrix proteins by the MDR human breast tumor cell line MCF-7/MX compared to its parental cells. We then used a strategy that combined antibody-based screening technique and mass spectrometry-based proteomics to identify membrane proteins that contribute to the enhanced adhesion of MCF-7/MX cells. Using MCF-7/MX cells as immunogen, we isolated a mouse monoclonal antibody, 9C6, that preferentially reacts with MCF-7/MX cells over the parental MCF-7 cells. The molecular target of 9C6 was identified as cytokeratin 8 (CK8), which was found to be overexpressed on the cell surface of MCF-7/MX cells. We further observed that down-regulation of cell surface levels of CK8 through siRNA transfection significantly inhibited MCF-7/MX cell adhesion to fibronectin and vitronectin. In addition, anti-CK8 siRNA partially reversed the MDR phenotype of MCF-7/MX cells. Taken together, our results suggest that alterations in the expression level and cellular localization of CK8 may play a significant role in enhancing the cellular adhesion of MDR MCF-7/MX cells.
Introduction
Multidrug resistance (MDR) is the phenomenon whereby tumor cells acquire cross-resistance to a variety of structurally and functionally unrelated drugs. After cytotoxic chemotherapy, MDR occurs almost universally in various tumors and becomes a major obstacle to successful cancer treatment. The complex multimodal mechanisms involved in MDR have been extensively investigated and include overexpression of a family of ATP-binding cassette (ABC) transporters, such as P-glycoprotein (ABCB-1), multidrug resistance-associated protein 1 (ABCC-1), and breast cancer resistance protein (BCRP; ABCG-2), changes in topoisomerase II activity, and altered expression of apoptosis-associated proteins and drug binding proteins [1–4]. Recently, accumulating evidence suggests that the extracellular microenvironment may also influence the drug response and acquisition of drug resistance in cancer cells [5,6]. Notably, cell adhesion has been demonstrated to modulate drug response and prevent cell death, implicating the interaction of cell-cell or cell-extracellular matrix as a potentially important determinant in the emergence of drug resistance [7,8].
Indeed, increasing volumes of data stress that various molecular mechanisms are concomitantly activated during cytotoxic drug exposure and may complementarily and/or cooperatively contribute to MDR phenotypes. Schneider et al. established a human breast cancer cell line, MCF-7/MX, which was selected against mitoxantrone and is cross-resistant to several cytotoxic agents, including mitoxantrone, topotecan, and daunorubicin [9–11]. Expression of BCRP is significantly up-regulated and is considered as the primary, but not the only, contributing factor to drug resistance in MCF-7/MX cells [10]. Initially, we found that the capacity of MCF-7/MX cells to adhere to the extracellular matrix was increased compared to the parental cells. Therefore, we wondered whether the drug resistance of MCF-7/MX cells is concomitantly associated with cell adhesion- mediated MDR. To test this hypothesis, we first tried to identify novel membrane molecules that participate in the enhanced adhesion of MCF-7/MX cells. We used a mass spectrometry (MS)-based proteomic approach to identify changes in membrane components between MCF-7/MX and parental cells.
However, owing to their inherently hydrophobic nature and low abundance of membrane proteins, the success of direct differential proteomics analysis to separate and identify membrane proteins is limited [12,13]. Thus, we adopted an alternative strategy that combines comparative antibody screening to identify the target antigen with MS sequencing to identify differentially expressed proteins. We immunized mice with MCF-7/MX cells and generated several monoclonal antibodies that preferentially reacted with MCF-7/MX compared to parental MCF-7 cells. One of the antibodies, 9C6, bound to a unique epitope on cytokeratin 8 (CK8), which is found to be overexpressed in the drug-resistant MCF-7/MX cells compared to the drug-sensitive parental cells. Down-regulation of CK8 expression through siRNA transfection resulted in reduced cell adhesion to the extracellular matrix and in partial reversal of the MDR phenotype in MCF-7/MX cells. Our results suggest that membrane CK8 plays a significant role in the enhanced cell adhesion capacity of MCF-7/MX cells.
Materials and Methods
Cell Culture
The human breast cancer cell line MCF-7 and the mitoxantroneselected MDR cell line MCF-7/MX were kindly provided by Dr. E. Schneider (Wadsworth Center, NY). All cell lines were grown as a monolayer culture in Dulbecco's modified Eagle's medium (GIBCO, Grand Island, NY) supplemented with 10% fetal calf serum (GIBCO) at 37°C and 5% CO2. The drug-resistant cell line was cultured in mitoxantrone (Sigma, St. Louis, MO) at 800 ng/ml.
Comparative Screening of Hybridomas
BALB/c mice were immunized with MCF-7/MX cells (2 x 107) through intraperitoneal injections, which were repeated three times at 15-day intervals, until a positive test was obtained by ELISA. After a final intrasplenic booster injection with MCF-7/MX cells (1 x 105), splenocytes were harvested from mice with the highest antibody titers and fused to myeloma SP2/0 cells to generate hybridoma clones. Comparative screening of hybridoma clone culture supernatants was tested by ELISA. Briefly, parental MCF-7/WT and MDR MCF-7/MX cells (5 x 104 cells per well) were seeded in 96-well plates and cultured overnight. The cells were fixed in prechilled 0.1% glutaraldehyde (50% solution; Amresco, Solon, Ohio) for 15 minutes at 4°C and blocked with 1% BSA in PBS overnight at 4°C. The hybridoma supernatant (100 µl) was added to each well and incubated for 2 hours at room temperature. The plates were then incubated with a horseradish peroxidase (HRP)-conjugated anti-mouse IgG antibody, followed by incubation with orthophenylenediamine (Sigma) substrate. The plates were read at 490 nm using an ELISA reader (Dynatech, Guernsey, UK). Sera from nonimmunized mice were used as negative controls. Hybridoma clones producing antibodies that showed preferential binding to MCF-7/MX compared to MCF-7/WT cells were selected and subcloned three times by limiting dilution. For large-scale antibody production, hybridoma clones of interest were expanded and intraperitoneally injected into BALB/c mice. Ascites were harvested, and antibodies were affinity-purified using a Protein G-coupled Sepharose column according to the manufacturer's protocol (Amersham Bioscience, Piscataway, NJ).
Immunoprecipitation and Western Blot
For immunoprecipitation, whole cell lysates were prepared in modified RIPA buffer [50 mM Tris-HCl, 150 mM NaCl, 0.25% SDS, 1% Triton X-100, 0.25% sodium deoxycholate, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol (DTT)] containing protease inhibitor cocktail (Sigma). Cell lysates containing 800 to 1000 µg of total protein (at ∼1 mg/ml) were first incubated with 1 µg of isotype control IgG antibody together with 20 µl of Protein A agarose suspension (Sigma) with gentle shaking for 1 hour at 4°C. After centrifugation, the preabsorbed supernatants were incubated with 5 µg of 9C6 at 4°C for 2 hours followed by 50 µl of Protein A-agarose suspension overnight at 4°C with gentle shaking. The beads were then pelleted through brief centrifugation and washed three times with lysis buffer, and the proteins were then eluted by boiling the sample in 2x SDS-PAGE loading buffer for 5 minutes. Sera from nonimmunized mice were used as negative controls.
For immunoblot analysis, lysates containing 50 µg of total protein were resolved by SDS-PAGE, transferred to nitrocellulose membranes (Millipore, Billerica, MA), and blocked overnight at 4°C with 5% skim milk. The membrane was incubated with various primary antibodies at room temperature, followed by HRP-conjugated secondary antibodies. All signals were visualized using SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL) according to the instructions of the manufacturer. The following antibodies were used for immunoblot analysis: mouse monoclonal antibody 9C6 (0.5 µg/ml); ABCG2 (BXP-21, 1:1000; Alexis, San Diego, CA); CK8 (C51, 1:1000); and GAPDH (6C5, 1:1000; Santa Cruz Biotechnology, Santa Cruz, CA).
MALDI-TOF/TOF MS and MS/MS Analyses
The immunoprecipitated proteins were separated by 10% SDS-PAGE gel and visualized by Coomassie Brilliant Blue staining. Bands demonstrating a prominent differentiation between drug-sensitive and drug-resistant cells were excised. In-gel digestion with trypsin was performed using the following protocol. Briefly, the bands were cut into tiny pieces, washed twice with ultrapure water, incubated with 50 mM NH4HCO3 in 50% acetonitrile (enough to immerse the gel particles), and sonicated for 10 minutes. The solution was removed, and the gel pieces were incubated in acetonitrile for 15 minutes at room temperature. The gel pieces were then vacuum dried and reduced with 10 mM DTT in 25 mM NH4HCO3 for 1 hour at 56°C. The DTT solution was then replaced with the same volume of 55 mM iodoacetamide in 25 mM NH4HCO3, followed by incubation for 45 minutes at room temperature in the dark. The gel pieces were washed with 100 µl of 25 mM NH4HCO3 for 10 minutes while vortexing and dehydrated with 100 µl of 25 mM NH4HCO3 in 50% acetonitrile. The liquid phase was removed, and the gel pieces were again dried. The dried gel pieces were reswelled in 50 mM NH4HCO3 buffer containing trypsin (K/R) for 45 minutes at 4°C and then incubated at 4°C overnight. Trypsinization was stopped with 5% trifluoroactic acid, and the supernatant was assessed using a 4700 Proteomics Analyzer (Applied Biosystems, Framingham, CA) operated in positive ion reflector mode with a m/z range of 700 to 4000. Spectra were obtained by averaging 1000 acquired spectra in MS mode or 3000 acquired spectra in MS/MS mode. The MS/MS spectra of selected peptides from collision-induced dissociation were obtained at an acceleration voltage of 1 keV and approximately 1 x 106 torr collision gas. The 4000 series explorer software 3.0 (Applied Biosystems) was used for peak list generating with default parameters. Data were analyzed with GPS explorer 3.5 (Applied Biosystems) using Mascot Version 1.9.05 for peak identification against the NCBInr human protein database (released on May 28, 2007; 4880906 sequences, 1738906834 residues). The search criteria permitted one missed cleavage, at least four matching peptide masses, a mass tolerance of <50 ppm for precursor ions, a mass tolerance of <0.2 Da for fragment ions, and a minimum signal-to-noise ratio of 10.0. Variable modifications, including carbamidomethyl modification and oxidation, were assigned to cysteine and methionine residues, respectively. The peptide sequences were manually reviewed for fidelity and consensus with known proteins. Significant matches were identified based on an expected Mascot value of <5% for protein identification, equivalent to statistically significant search scores of >95%. Finally, the molecular weights of the proteins were compared to the values estimated by gel electrophoresis.
Subcellular Fractions
Confluent cell monolayer were lysed in a hypotonic buffer (10 mM HEPES pH 7.4, 25 mM KCl, 5 mM MgCl2, 5 mM EDTA) and centrifuged at 200g for 10 minutes. The resulting pellet consisted of nuclei and unhomogenized cells. The postnuclear lysates were centrifuged at 20,000g for 20 minutes. The supernatant was designated as the cytoplasmic fraction (C), whereas the resulting pellet was enriched in plasma membrane. The plasma membrane fraction was solubilized in 1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% SDS. After another centrifugation at 20,000g for 10 minutes, the supernatant and pellet were collected as the detergent-soluble membrane fraction (M–S) and detergent-insoluble membrane fraction (M–P), respectively. Whole cell lysates (W) were prepared in modified RIPA buffer or in 1x SDS-PAGE loading buffer containing 2% SDS, 5% 2-mercaptoethanol, and 25% glycerol in 0.05 M Tris-HCl buffer (pH 6.8). Protease inhibitor cocktail was included at each stage of preparation. Before SDS-PAGE, each fraction was adjusted to an equal protein concentration using the BCA Protein Assay Kit (Pierce).
RNA Interference
CK8 siRNA and BCRP siRNA, each consisting of a pool of three target-specific 20- to 25-nt siRNA designed to knockdown gene expression, were purchased from Santa Cruz Biotechnology and Invitrogen (Carlsbad, CA), respectively. The Stealth Select RNAi Set was used to reduce off-target and nonspecific effects. A nonspecific siRNA was also purchased from Invitrogen and used as the negative control.
MCF-7/MX cells (2.5 x 105 per well) were cultured in six-well plates for 24 hours and transfected with 100 nM of the CK8 siRNA or BCRP siRNA per well using LipofectAMINE 2000 (Invitrogen) according to the manufacturer's instructions. Control cells were incubated with transfection reagent and the control nonspecific siRNA. To confirm subsequent CK8 and BCRP knockdown, whole cell lysates were prepared and subjected to Western blot analysis.
Immunofluorescence and Confocal Microscopy
Cells were grown on eight-well glass slides (Laboratory-Tek Brand Products, Naperville, IL). To stain the plasma membrane, cells were incubated with 2 µM CM-DiI (Invitrogen) for 5 minutes at 37°C and then for an additional 15 minutes at 4°C. After two washes with PBS, the cells were fixed with 4% (w/v) paraformaldehyde in PBS at 37°C for 10 minutes and then blocked for 1 hour with PBS containing 10% normal goat serum. The cells were then further incubated with either clone 9C6 or C51, a commercially available anti-CK8 antibody (Santa Cruz Biotechnology), for 1 hour at 37°C, followed by incubation with an FITC-conjugated goat anti-mouse secondary antibody for 30 minutes at room temperature. Finally, DAPI (Sigma) was applied at a final concentration of 1 µM in PBS for 10 minutes to counterstain the nuclei. All images were obtained using a confocal fluorescence microscope (Leica, Nussloch, Germany).
Cell Adhesion Assay
Ninety-six-well plates were first coated with either 10 or 20 µg/ml of fibronectin (FN; Sigma-Aldrich, St. Louis, MO) or 5 or 10 µg/ml of vitronectin (VN; BD Biosciences, San Diego, CA) in PBS at 4°C overnight. Immediately before addition of the cells, the plates were blocked with 0.5% BSA for 30 minutes at room temperature and washed three times with PBS. All cells were harvested with 5 mM EDTA in PBS and resuspended in DMEM containing 2% fetal bovine serum at a concentration of 5 x 105 (MCF-7 and MCF-7/MX) or 1 x 105 (HEK293) cells/ml. Then, 0.1 ml of cell suspension was added to each well. The plates were incubated for 1 hour (MCF-7 and MCF-7/MX) or 30 minutes (HEK293) at 37°C and 5% CO2 and washed twice with PBS to remove unbound cells. The adherent cells were fixed with 3% formaldehyde and stained with 30% ethanol in PBS containing 0.5% crystal violet for 15 minutes. After two gentle washes in PBS, 0.1 ml of DMSO was added to lyse the cells, and the absorbance at 570 nm was read using a microtiter plate reader. All data are presented as the mean ± SD of three determinations. Statistical analysis was performed using a 2-tailed Student's t test.
Cytotoxicity Assay
MCF-7/MX cells were transfected with siRNA as described above, seeded in 96-well plates in triplicate at a density of 2000 cells per well, and cultured overnight. The cells were incubated at 37°C in 5% CO2 for 48 hours with different drugs at the concentrations indicated. Cell cytotoxicity was assayed using the optimized sulforhodamine B (SRB; Sigma) method as previously described [14]. Drug concentrations producing 50% cell growth inhibition (IC50) were determined using curve-fitting analyses with Prism software (GraphPad Software, San Diego, CA) using the following sigmoid dose-response model equation: Y = Bottom + (Top - Bottom) / {1 + 10[(LogEC50 - X)]}. The IC50 values were calculated based on five independent experiments for each cell line. Statistical significance was evaluated using the Mann-Whitney U test with SPSS software (Version 16.0; SPSS, Chicago, IL).
Results
Cell-Extracellular Matrix Adhesion Is Enhanced in MCF-7/MX Cells
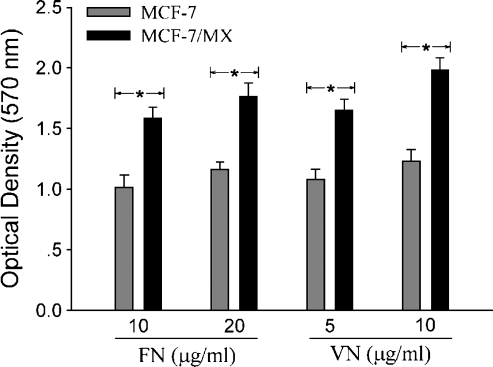
We first compared the adhesive activity of drug-resistant and drugsensitive cells. As shown in Figure 1, MCF-7/MX cells demonstrated significantly greater cellular attachment to both FN and VN compared to the parent cells. This suggests a discrepancy in cell adhesion between drug-resistant and drug-sensitive cells.
Figure 1.
Comparison of cell-extracellularmatrix adhesion in MCF-7/MX and drug-sensitive cells. Drug-sensitive MCF-7 and MDR MCF-7/MX cells (5 x 104 cells per well plated in 96-well plates in DMEM containing 2% FBS) were incubated in FN- or VN-coated plates for 1 hour at 37°C and 5% CO2. Cell attachment was quantitated using the crystal violent adhesion assay as described in the Materials and Methods section. *P < .01, MCF-7/MX versus MCF-7.
Selective Screening of 9C6, a Monoclonal Antibody That Binds Preferentially to MDR MCF-7/MX Cells
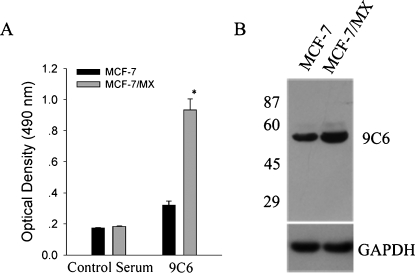
To identify cell surface molecules that are selectively up-regulated in MDR MCF-7/MX cells compared to parent MCF-7 cells, we generated several monoclonal antibodies demonstrating preferential binding to MDR cells over parent cells by immunizing mice with MCF-7/MX cells. Clone 9C6 was selected for further studies based on its strong and selective reactivity with MCF-7/MX cells (Figure 2A). Western blot analysis using whole cell lysate demonstrated that 9C6 binds to a single protein band of approximately 53 kDa in both MCF-7 and MCF-7/MX cells (Figure 2B). As expected, the expression level of the protein recognized by 9C6 was much higher inMDR cells.
Figure 2.
Identification of 9C6, a monoclonal antibody that preferentially binds to MDR MCF-7/MX cells. (A) Binding of the 9C6 hybridoma culture supernatant to parental drug-sensitive MCF-7 cells and MDR MCF-7/MX cells by ELISA. Nonimmunized mouse serum was used as a negative control. Data are presented as the mean ± SD of three independent experiments. *P < .05 versus MCF-7. (B) Western blot analysis of whole cell lysates using 9C6. Molecular mass standards (in kDa) are shown on the left.
Identification of the Differential Expression of CK8 by MALDI-TOF/TOF MS
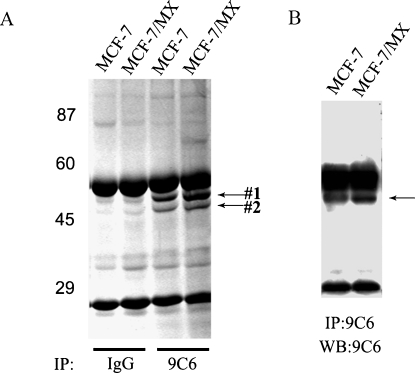
Immunoprecipitation was performed with 9C6 using whole cell lysates from both MCF-7 and MCF-7/MX cells, and immunocomplexes were resolved by SDS-PAGE analysis. Commassie blue staining of the gel revealed two unique protein bands of approximately 53 and 48 kDa, respectively (Figure 3A). Only upper band #1, however, was recognized by 9C6 when the gel was subjected to Western blot analysis (Figure 3B). The two protein bands were excised and analyzed by MALDI-TOF/TOF MS, followed by database interrogation. The MS/MS data obtained for five peptides from the 53-kDa band matched CK8 based on a total of 301 ion scores (accession number gi|49256423 in the NCBI database; Table 1). In contrast, the MS/MS data obtained for four peptides from the 48-kDa band matched cytokeratin 18 based on 208 ion scores (accession number gi|61357801 in the NCBI database; data not shown). It is known that CK8 and CK18 form a heterotypic network in a wide variety of epithelial cells. Thus, CK18 was identified here by coimmunoprecipitation with 9C6, further confirming CK8 to be the specific target recognized by 9C6.
Figure 3.
Separation of proteins recognized specifically by 9C6. The 9C6 and control IgG antibodies were used for immunoprecipitation with whole lysates from MCF-7 and MDR MCF-7/MX cells. The precipitated proteins were separated by SDS-PAGE and visualized by Commassie blue staining (A) or by Western blot analysis using 9C6 (B). The IgG heavy chains (∼56 kDa) and light chains (∼25 kDa) were also identified in the gel and blot. Arrows indicate the positions of the protein bands that were subjected to mass spectrometry analysis.
Table 1.
Identification of CK8 by Mass Spectrometry.
| Calculated Mass | Observed Mass | ± da | Start Sequence | End Sequence | Sequence |
| 911.4217 | 911.3864 | -0.0353 | 24 | 32 | SYTSGPGSR |
| 1079.5116 | 1079.4653 | -0.0463 | 265 | 273 | AQYEDIANR |
| 1208.5939 | 1208.5443 | -0.0497 | 303 | 312 | TKTEISEMNR |
| 1277.7101 | 1277.7314 | 0.0214 | 382 | 392 | LALDIEIATYR |
| 1341.7484 | 1341.6884 | -0.0601 | 317 | 328 | LQAEIEGLKGQR |
| 1344.6754 | 1344.6198 | -0.0556 | 329 | 341 | ASLEAAIADAEQR |
| 1352.6766 | 1352.6122 | -0.0644 | 187 | 197 | TEMENEFVLIK |
| 1405.8049 | 1405.7451 | -0.0598 | 382 | 393 | LALDIEIATYRK |
| 1412.6838 | 1412.6245 | -0.0593 | 274 | 285 | SRAEAESMYQIK |
| 1419.7478 | 1419.7799 | 0.0321 | 214 | 225 | LEGLTDEINFLR |
| 1428.6787 | 1428.6119 | -0.0668 | 274 | 285 | SRAEAESMYQIK |
| 1480.7716 | 1480.7017 | -0.0699 | 187 | 198 | TEMENEFVLIKK |
| 1508.7777 | 1508.7146 | -0.0631 | 186 | 197 | RTEMENEFVLIK |
| 1797.8323 | 1797.7588 | -0.0735 | 199 | 213 | DVDEAYMNKVELESR |
| 1847.8051 | 1847.8541 | 0.049 | 134 | 148 | SNMDNMFESYINNLR |
| 1956.0396 | 1955.9664 | 0.0733 | 329 | 347 | ASLEAAIADAEQRGELAIK |
| 2003.9062 | 2003.8374 | 0.0688 | 134 | 149 | SNMDNMFESYINNLRR |
| 2034.0576 | 2033.9888 | -0.0688 | 159 | 176 | LKKLEAELGNMQGLVEDFK |
| 2109.0129 | 2108.9451 | -0.0678 | 234 | 252 | ERLQSQISDTSVVLSMDNSR |
#1 band in SDS-PAGE was cut, digested by trypsin, and analyzed by mass spectrometry. Measured masses from MALDI-TOF, theoretical masses from NCBInr human protein database, their difference (±da), and the corresponding sequence in CK8 were shown for each matched peptide. Sequences underlined were also obtained by MS/MS. After MALDI-TOF MS and database searching, 19 tryptic peptides matched with theoretical masses, leading to a sequence coverage of 38.3%.
Characterization of Membrane CK8 Expression in MDR MCF-7/MX Cells
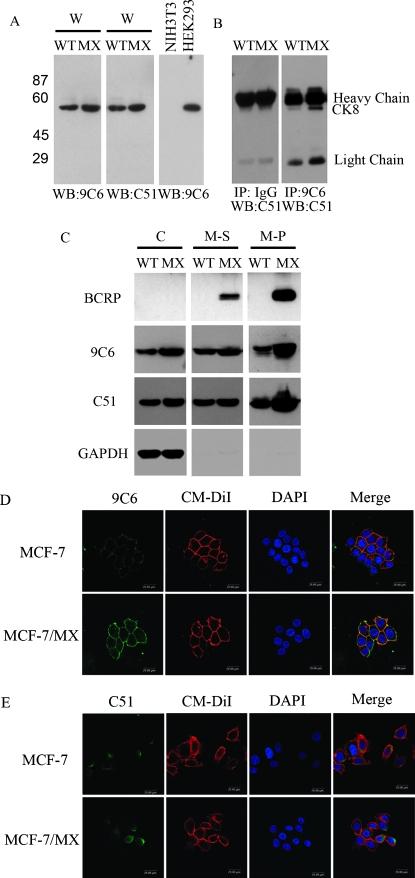
To confirm the proteomic results described above and provide further evidence that the protein recognized by 9C6 was indeed CK8, Western blot analysis was repeated using a commercial monoclonal antibody, C51, which has been shown to react specifically with CK8. As shown in Figure 4A, both the C51 and 9C6 antibodies yielded identical patterns of immunoreactivity with the 53-kDa protein when blotted against whole cell lysate. As a control, neither antibody detected any protein bands when applied to blots of whole cell lysate from NIH3T3 cells, which are known to not express CK8 (Figure 4A). In a parallel experiment, whole cell lysate was first immunoprecipitated with 9C6 or a control IgG antibody, followed by blot analysis with C51. As shown in Figure 4B, the 53-kDa protein was only observed in the 9C6-precipitated sample, further confirming the binding specificity of 9C6 for CK8 protein.
Figure 4.
Increased expression of CK8, both intracellularly and on the cell surface, of MDR MCF-7/MX cells. (A) Western blot analysis of whole cell lysates (W) with 9C6 or commercial the anti-CK8 antibody, C51. A protein band of the same molecular weight was identified by both antibodies. NIH3T3 and HEK293 cell lines were used as negative and positive control, respectively. (B) Western blot analysis of immunocomplexes precipitated by 9C6 or a control IgG using C51 as the blotting agent. Note that 9C6 yielded a single protein band at ∼53 kDa, whereas no specific protein band was identified in the control IgG sample. IgG heavy chains (∼55 kDa) and light chains (∼25 kDa) were also identified in the blots. WT indicates parent MCF-7 cells; MX, MCF-7/MX cells. (C) Western blot analysis of the subcellular protein fractions from MCF-7 and MCF-7/MX cells. C indicates cytoplasmic fraction; M–S, detergent-soluble portion of the plasma membrane fraction; M–P, detergent-insoluble pellet from the plasma membrane fraction. BCRP and GAPDH were used to assess the purity of the membrane and cytoplasmic fractions, respectively. (D–E) Immunofluorescence staining of cell membrane CK8. Cell surface CK8 expression was detected (in green) using 9C6 (D) or C51 (E) in fixed, nonpermeabilized MCF-7 and MCF-7/MX cells. The plasma membrane was further stained with CM-DiI (red), whereas the nuclei were stained with DAPI (blue). Note that surface CK8 staining in MCF-7/MX cells using 9C6 was significantly stronger than that in MCF-7 cells, whereas no obvious membrane CK8 staining was observed when using C51. The merged images clearly demonstrate costaining (yellow) in the MCF-7/MX cell membrane with both 9C6 and CM-DiI.
The expression pattern of CK8 in different subcellular fractions was then compared between drug-sensitive and drug-resistant cells (Figure 4C). As expected, BCRP expression was significantly elevated in MDR MCF-7/MX cells and was located exclusively in the plasma membrane fractions (M–S and M–P). Consistent with an earlier observation, CK8 expression, as detected by both 9C6 and C51, was significantly increased in the MDR cells, especially in the detergent-insoluble portion (M-P) of the plasma membrane fraction. Similar to the Western blot analysis, immunofluorescence confocal microscopy confirmed the distinct membrane staining patterns of CK8 between MCF-7/MX and MCF-7 cells. As shown in Figure 4D, 9C6 exhibited much stronger plasma membrane staining in MCF-7/MX cells compared to that in MCF-7 cells, further confirming the surface overexpression of CK8 in MDR cells. Upon cell membrane permeabilization, 9C6 yielded clear cytoplasmic staining in both MCF-7/MX and MCF-7 cells (data not shown). In contrast, C51, while demonstrating clear cytoplasmic staining in both permeabilized MCF-7/MX and MCF-7 cells, failed to bind to the plasma membrane of MCF-7/MX cells (Figure 4E). It seems apparent that the binding epitope(s) on CK8 recognized by 9C6 and C51 are different; unlike 9C6, C51 may bind to an epitope that is not exposed on the cell surface.
Increased Cell Surface CK8 Expression Caused Enhanced Cell Adhesion in MDR MCF-7/MX Cells
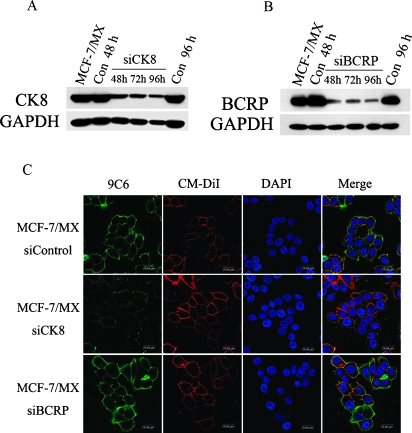
To determine whether increased CK8 expression in MCF-7/MX cells is involved in their enhanced adhesion, a siRNA-silencing approach was used to down-regulate the expression of CK8. MCF-7/MX cells were transfected with anti-CK8 siRNA. The optimal concentration of both anti-CK8 siRNA for cell transfection was determined based on a series of titration experiments to be 100 nM (data not shown). As shown in Figure 5A, transfection of anti-CK8 siRNA resulted in significant down-regulation of the expression of the respective proteins 48 to 96 hours after transfection. As expected, the control siRNA did not show any effects on CK8 expression 48 and 96 hours after transfection. Most importantly, in addition to the reduced expression in whole cell lysate, immunofluorescence staining showed that the surface expression of CK8 inMCF-7/MX cells was also significantly down-regulated after transfection with anti-CK8 siRNA (Figure 5C). In contrast, the siRNA-mediated down-regulation of BCRP had no effect on the cell surface expression of CK8 in MCF-7/MX cells (Figure 5, B and C). Taken together, these results confirm both the efficacy and specificity of our siRNA silencing approach.
Figure 5.
Modulation of CK8 and BCRP expression by specific siRNA. (A–B) Analysis of siRNA-mediated knockdown of CK8 or BCRP protein expression in MCF-7/MX cells by Western blot. RNAi interference was performed using (A) anti-CK8 siRNA and (B) anti-BCRP siRNA at a concentration of 100 nM. CK8 or BCRP expression at the indicated time points (48, 72, and 96 hours) after siRNA transfection was assessed by Western blot analysis using C51 and BXP-21. (C) Immunofluorescence staining of surface CK8 on MCF-7/MX cells after siRNA transfection. MCF-7/MX cells treated with anti-CK8 siRNA or anti-BCRP siRNA were incubated with 9C6 for 1 hour at 37°C, followed by staining with an FITC-conjugated goat anti-mouse antibody for 30 minutes at room temperature (green). The plasma membrane was further stained with CM-DiI (red), and the nuclei were stained with DAPI (blue). Note that anti-BCRP siRNA transfection did not result in a decrease in the surface CK8 expression of MCF-7/MX cells.
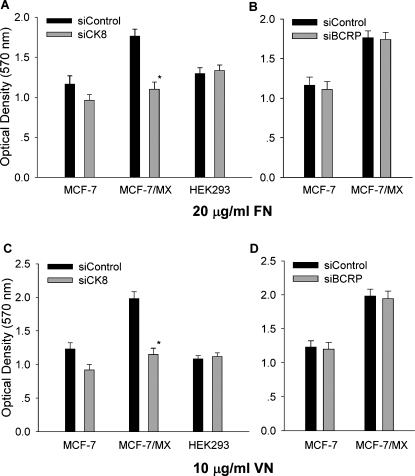
We next assessed the possible role of membrane CK8 in cell adhesion by specifically down-regulating membrane CK8 by RNA interference. Down-regulation of CK8 expression effectively reduced cell-FN and cell-VN adhesion in MCF-7/MX cells; however, a less obvious change in drug-sensitive cells was also observed (Figure 6, A and C). Because the expression levels of CK8 in both the cell membrane and cytoplasm were reduced by suppression of cytoplasmic CK8, it was necessary to further investigate whether the increased cell adhesion was only affected by membrane CK8 rather than by cytoplasmic CK8. Thus, the same assessment was carried out in adenovirus-transformed human embryonic kidney cells (HEK293), which demonstrate strong expression of CK8 only in the cytoplasm, using 9C6 (data not shown). Remarkably, CK8 siRNA did not change the adhesive properties HEK293 cells, although the expression of intracellular CK8 was significantly knocked down. These results suggest that membrane CK8, but not cytoplasmic CK8, played an important role in enhancing the attachment of drug-resistant MCF-7/MX cells to FN and VN. In addition, siRNA-mediated down-regulation of BCRP expression had no effect on the adhesion of MCF-7/MX cells (Figure 6, B and D).
Figure 6.
Membrane CK8 mediates enhanced cell adhesion of MDR MCF-7/MX cells to FN and VN. (A and C) MCF-7, MCF-7/MX, and HEK293 cells were transfected with anti-CK8 siRNA or a control siRNA. After 48 hours, the cells were harvested and analyzed using the cell adhesion assay by coating plates with FN (20 µg/ml) or VN (10 µg/ml). Note that anti-CK8 siRNA significantly inhibited the cell adhesion of MCF-7/MX cells to FN and VN to a comparable level exhibited by parent MCF-7 cells. In contrast, anti-CK8 siRNA showed only a marginal effect on MCF-7 cells and did not affect the attachment of HEK293 cells. (B and D) MCF-7 and MCF-7/MX cells were transfected with anti-BCRP siRNA. Note that anti-BCRP siRNA have no effects on the cell-extracellular matrix adhesion in both cell lines. Data are shown as the mean ± SD of triplicate determinations.
CK8 Partially Contributed to Drug Resistance in MDR in MCF-7/MX Cell
The effect of siRNA-mediated down-regulation of CK8 and BCRP expression on the MDR phenotype of MCF-7/MX cells was examined. Drug sensitivity of the various transfected tumor cells to three cytotoxic agents, mitoxantrone, topotecan, and daunorubicin, was investigated. As shown in Table 2, MDR MCF-7/MX cells showed 1112-, 322-, and 3.7-fold increased resistance to mitoxantrone, topotecan, and daunorubicin, respectively, compared to its parental MCF-7 cells. Down-regulation of CK8 expression by siRNA significantly increased drug sensitivity of MCF-7/MX cells by approximately 40.2%, 33.0%, and 62.2%, to mitoxantrone, topotecan, and daunorubicin, respectively (P < .01 in all three cases).Meanwhile, down-regulation of BCRP expression in MCF-7/MX cells by siRNA transfection resulted in a more drastic drug sensitization: cell resistance to mitoxantrone, topotecan, and daunorubicin was reduced by 76.8%, 63.1%, and 100%, respectively (all P < .01; Table 2).
Table 2.
Effects of siRNA Transfection on Chemosensitivity of MCF-7/MX Cells.
| Cells | IC50 (RR)*,† | ||
| Mitoxantrone (nM) | Topotecan (nM) | Daunorubicin (nM) | |
| MCF-7 | 22.13 ± 1.60 (1) | 54.99 ± 5.82 (1) | 116.53 ± 10.98 (1) |
| MCF-7/MX | 24,610 ± 1085 (1112) | 17,759 ± 1265 (322) | 427.13 ± 15.41 (3.7) |
| siRNA-Control | 22,491 ± 982 (1016) | 16,867 ± 1978 (306) | 433.73 ± 4.48 (3.7) |
| siRNA-CK8 | 13,450 ± 1087 (608)‡ | 11,311 ± 658 (205)‡ | 163.80 ± 32.39 (1.4)‡ |
| siRNA-BCRP | 5232 ± 348 (236)‡ | 5201 ± 733 (95)‡ | 122.17 ± 17.10 (1.0)‡ |
IC50 values (mean ± SD) were calculated from five independent experiments performed in triplicates.
RR indicates relative resistance (x-fold compared to that of MCF-7 cells).
P < .01 compared to the control siRNA-treated cells.
Discussion
In our initial study, we have found a salient discrepancy in cell adhesion between MDR MCF-7/MX cells and drug-sensitive MCF-7/WT cells, but the mechanism remains unclear. To examine the possible mechanisms that contribute to the enhanced adhesion of MDR cells, we set out to investigate potential membrane proteins that demonstrate differential expression in MDR breast cancer cells. By immunizing mice with an MDR MCF-7/MX cell line, we obtained an antibody that specifically reacts with CK8. The antibody preferentially binds to the surface of MCF-7/MX cells compared to their parental MCF-7 cells, providing direct evidence of the cell surface overexpression of CK8 in MDR MCF-7/MX cells. It has been a subject of debate whether cell surface expression of CK8 under various experimental conditions represents a true phenomenon associated with cell transformation or just an artifact of study methods. In our preliminary experiment, we failed to identify cell surface CK8 through N-terminal biotinylation of intact cells (data not shown). This is consistent with the observations of Riopel et al. [15]. Conversely, a number of reports have clearly demonstrated that CK8 is expressed on the surface of a number of cancer cell lines, especially MCF-7 cells [16,17]. For example, by combining cell surface iodination and two-dimensional electrophoresis methods, Godfroid et al. [16] demonstrated that CK8, CK18, and CK19 are expressed on the surface of several established human breast cancer cell lines, including MCF-7 cells. Ultrastructural immunocytochemistry further confirmed that cytokeratins were localized to the blebs formed by the cell membrane. Similarly, CK8 was shown to localize to the plasma membrane of cancer cells but was absent on the cell membrane of other healthy human tissues, excluding hepatocytes, based on confocal laser scanning microscopy [17].
Several potential mechanisms have been proposed for the aberrant expression of CK8 on the tumor cell surface, including lipid binding followed by translocation to the outer membrane [18], penetration and projection through the plasma membrane as part of a protein complex [19,20], and noncovalent association, or secondary binding, to the cell membrane after proteolytic release from cells into the extracellular space [21–23]. Overproduction of cytokeratins by transformed cells may also lead to increased cell surface expression because of insufficient incorporation into intermediate filaments [24]. It is also noteworthy that a truncated form of CK8 was identified in colorectal cancer cells (but not in normal colon cells), suggesting that malignant cells may possess CK8 degradation pathways that differ from those of normal cells [25].
What are the exact physiological and pathological roles of cell surface-expressed CK8 remains a question to be clearly elucidated. Several reports have demonstrated that CK8 is able to bind plasminogen and promote its activation through tissue-type plasminogen activator on the cancer cell surface, suggesting that cell surface CK8 may function as a major plasminogen receptor and contribute to cancer invasion and metastasis [19,26,27]. Here, we report the positive correlation between membrane CK8 and cell adhesion in MDR cancer cells, adding further comprehensive information to the function of membrane CK8. We demonstrated that cell surface overexpression of CK8 led to increased attachment of MCF-7/MX cells to FN and VN, and this effect could be completely reversed by anti-CK8 siRNA transfection. To date, studies examining BCRP function have focused on its efflux of cytotoxic agents, and no such correlation of BCRP expression with cell adhesion has been reported. Consistent with this, we did not detect any effects of BCRP on cell-extracellular matrix adhesion in MCF-7/MX cells, although BCRP is also highly expressed on the cell surface of MCF-7/MX cells.
There is a great deal of evidence suggesting that cell-cell and cell-extracellular matrix adhesion could mediate drug resistance through different mechanisms in cancer cells [28–30]. For example, attachment of tumor cells to FN through certain integrins has been shown to protect the cells from drug-induced apoptosis and confer an MDR phenotype [29,31]. Furthermore, overexpression of PECAM-1 in lymphoid cancers has been shown to confer resistance to apoptosis induced by DNA-damaging chemotherapeutic agents, and decreased PECAM-1 expression could render cancer cells more susceptible to chemotherapy [30]. Similarly, an anti-E-cadherin antibody has been found to increase the chemosensitivity of MCF-7 cells, which express a high level of E-cadherin [32]. Therefore, we further examined the role of overexpressed membrane CK8 in drug resistance to investigate whether adhesion-mediated drug resistance is also exhibited by MDR MCF-7/MX cells. It is intriguing that reduced cell surface CK8 expression by CK8 siRNA transfection partially restored the sensitivity of MCF-7/MX cells to all three chemotherapeutic agents tested (Table 2), clearly supporting the notion that membrane CK8 may be involved in cellular protection against chemotherapeutic agents in MDR MCF-7/MX cells; although the BCRP-mediated MDR phenotype is still a major contributor. It is likely that CK8 enhances adhesion between cells and cell-extracellular matrix proteins through its increased surface expression and, as a result, mediates drug resistance. However, it is also plausible that the up-regulation of CK8 may be the result of prolonged exposure of MCF-7 cells to mitoxantrone, a secondary effect of cellular response to the drug, rather than the primary cause of resistance to the compound. Further, although a number of studies have unequivocally demonstrated increased CK8 expression in MDR tumor cells, there was at least one report showing that CK8 was underexpressed in both adriamycin- and paclitaxel-resistant breast tumor cells [33]. More in-depth mechanistic studies are clearly warranted to further elucidate the relevance and the roles of CK8 expression in the development of MDR in tumor cells.
In summary, we demonstrated that CK8 expression is up-regulated on the cell surface of the MDR human breast cancer cell line, MCF-7/MX. Down-regulation of CK8 in MCF-7/MX cells using the siRNA approach significantly reduced tumor cell adhesion to extracellular matrix proteins and partially reversed the MDR phenotype in tumor cells. Taken together, our results suggest that tumor surface-expressed CK8, through enhancing cell-cell matrix adhesion, may represent a complementary mechanism of MDR in MCF-7/MX cells. Further understanding of the molecular mechanisms involved in the emergence and maintenance of MDR is essential for the development of novel and more effective agents for efficient diagnosis and therapy for cancer.
Acknowledgments
The authors thank E. Schneider (Wadsworth Center, NY) for the generous gifts of the human breast cancer cell line MCF-7 and the mitoxantrone-selected MDR cell line MCF-7/MX.
Abbreviations
- MDR
multidrug resistance
- BCRP
breast cancer resistance protein
- CK8
cytokeratin 8
- WT
wild type
- MX
mitoxantrone
- SRB
sulforhodamine B
- FN
fibronectin
- VN
vitronectin
- HEK
human embryonic kidney
- MALDI-TOF MS
Matrix-assisted laser desorption/deionization-time-of-flight mass spectrometry
Footnotes
This research was supported by National Natural Science Foundation of China (Nos. 30572203, 30570772, 30400405), Natural Science Foundation of Tianjin (No. 05YFJZ°C01200), and National High Technology Research and Development Program (the 863 Program) of the Chinese Government (No. 2002AA2Z346D).
References
- 1.Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 2.Stavrovskaya AA. Cellular mechanisms of multidrug resistance of tumor cells. Biochemistry (Mosc) 2000;65:95–106. [PubMed] [Google Scholar]
- 3.Park DC, Yeo SG, Wilson MR, Yerbury JJ, Kwong J, Welch WR, Choi YK, Birrer MJ, Mok SC, Wong KK. Clusterin interacts with paclitaxel and confer paclitaxel resistance. Neoplasia. 2008;10:964–972. doi: 10.1593/neo.08604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberti A, La Sala D, Cinti C. Multiple genetic and epigenetic interacting mechanisms contribute to clonally selection of drug-resistant tumors: current views and new therapeutic prospective. J Cell Physiol. 2006;207:571–581. doi: 10.1002/jcp.20515. [DOI] [PubMed] [Google Scholar]
- 5.Hazlehurst LA, Landowski TH, Dalton WS. Role of the tumor microenvironment in mediating de novo resistance to drugs and physiological mediators of cell death. Oncogene. 2003;22:7396–7402. doi: 10.1038/sj.onc.1206943. [DOI] [PubMed] [Google Scholar]
- 6.Thews O, Gassnery B, Kelleher DK, Schwerdty G, Gekley M. Impact of extracellular acidity on the activity of P-glycoprotein and the cytotoxicity of chemotherapeutic drugs. Neoplasia. 2006;8:143–152. doi: 10.1593/neo.05697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White DE, Rayment JH, Muller WJ. Addressing the role of cell adhesion in tumor cell dormancy. Cell Cycle. 2006;5:1756–1759. doi: 10.4161/cc.5.16.2993. [DOI] [PubMed] [Google Scholar]
- 8.Cimbora-Zovko T, Ambriović-Ristov A, Loncarek J, Osmak M. Altered cell-cell adhesion in cisplatin-resistant human carcinoma cells: a link between betacatenin/plakoglobin ratio and cisplatin resistance. Eur J Pharmacol. 2007;558:27–36. doi: 10.1016/j.ejphar.2006.11.077. [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa M, Schneider E, Dixon KH, Horton J, Kelley K, Morrow C, Cowan KH. Reduced intracellular drug accumulation in the absence of P-glycoprotein (mdrl) overexpression in mitoxantrone-resistant MCF-7 breast cancer cells. Cancer Res. 1992;52:6175–6181. [PubMed] [Google Scholar]
- 10.Ross DD, Yang WD, Abruzzo LV, Dalton WS, Schneider E, Lage H, Dietel M, Greenberger L, Cole SPC, Doyle LA. Atypical multidrug resistance: breast cancer resistance protein messenger RNA expression in mitoxantrone-selected cell lines. J Natl Cancer Inst. 1999;91:429–433. doi: 10.1093/jnci/91.5.429. [DOI] [PubMed] [Google Scholar]
- 11.Yang CJ, Horton JK, Cowan KH, Schneider E. Cross-resistance to camptothecin analogues in a mitoxantrone-resistant human breast carcinoma cell line is not due to DNA topoisomerase I alterations. Cancer Res. 1995;55:4004–4009. [PubMed] [Google Scholar]
- 12.Wallin E, Von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rabilloud T. Use of thiourea to increase the solubility of membrane proteins in two-dimensional. Electrophoresis. 1998;19:758–760. doi: 10.1002/elps.1150190526. [DOI] [PubMed] [Google Scholar]
- 14.Papazisis KT, Geromichalos GD, Dimitriadis KA, Kortsaris AH. Optimization of the sulforhodamine B colorimetric assay. J Immunol Methods. 1997;208:151–158. doi: 10.1016/s0022-1759(97)00137-3. [DOI] [PubMed] [Google Scholar]
- 15.Riopel CL, Butt I, Omary MB. Method of cell handling affects leakiness of cell surface labeling and detection of intracellular keratins. Cell Motil Cytoskeleton. 1993;26:77–87. doi: 10.1002/cm.970260108. [DOI] [PubMed] [Google Scholar]
- 16.Godfroid E, Geuskens M, Dupressoir T, Parent I, Szpirer C. Cytokeratins are exposed on the outer surface of established human mammary carcinoma cells. J Cell Sci. 1991;99:595–607. doi: 10.1242/jcs.99.3.595. [DOI] [PubMed] [Google Scholar]
- 17.Gires O, Andratschke M, Schmitt B, Mack B, Schaffrik M. Cytokeratin 8 associates with the external leaflet of plasma membranes in tumour cells. Biochem Biophys Res Commun. 2005;328:1154–1162. doi: 10.1016/j.bbrc.2005.01.074. [DOI] [PubMed] [Google Scholar]
- 18.Asch HL, Mayhew E, Lazo RO, Asch BB. Lipids noncovalently associated with keratins and other cytoskeletal proteins of mouse mammary epithelial cells in primary culture. Biochim Biophys Acta. 1990;1034:303–308. doi: 10.1016/0304-4165(90)90056-3. [DOI] [PubMed] [Google Scholar]
- 19.Hembrough TA, Li L, Gonias SL. Cell-surface cytokeratin 8 is the major plasminogen receptor on breast cancer cells and is required for the accelerated activation of cell-associated plasminogen by tissue-type plasminogen activator. J Biol Chem. 1996;271:25684–25691. doi: 10.1074/jbc.271.41.25684. [DOI] [PubMed] [Google Scholar]
- 20.Vidrich A, Gilmartin ME, Mitchell J, Freedberg IM. Postsynthetic modifications of epithelial keratins. Ann N Y Acad Sci. 1985;455:354–370. doi: 10.1111/j.1749-6632.1985.tb50422.x. [DOI] [PubMed] [Google Scholar]
- 21.Chan R, Rossitto PV, Edwards BF, Cardiff RD. Presence of proteolytically processed keratins in the culture medium of MCF-7 cells. Cancer Res. 1986;46:6353–6359. [PubMed] [Google Scholar]
- 22.Hembrough TA, Kralovich KR, Li L, Gonias SL. Cytokeratin 8 released by breast carcinoma cells in vitro binds plasminogen and tissue-type plasminogen activator and promotes plasminogen activation. Biochem J. 1996;317:763–769. doi: 10.1042/bj3170763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou CF, Riopel CL, Rott LS, Omary MB. A significant soluble keratin fraction in “simple” epithelial cells. Lack of an apparent phosphorylation and glycosylation role in keratin solubility. J Cell Sci. 1993;105:433–444. doi: 10.1242/jcs.105.2.433. [DOI] [PubMed] [Google Scholar]
- 24.Ditzel HJ, Garrigues U, Andersen CB, Larsen MK, Garrigues HJ, Svejgaard A, Helistrom I, Helistrom KE, Jensenius JC. Modified cytokeratins expressed on the surface of carcinoma cells undergo endocytosis upon binding of human monoclonal antibody and its recombinant Fab fragment. Proc Natl Acad Sci USA. 1997;94:8110–8115. doi: 10.1073/pnas.94.15.8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishibori H, Matsuno Y, Iwaya M, Osada T, Kubomura N, Iwamatsu A, Kohno H, Sato S, Kitajima M, Hirohashi S. Human colorectal carcinomas specifically accumulate Mr 42,000 ubiquitin-conjugated cytokeratin 8 fragments. Cancer Res. 1996;56:2752–2757. [PubMed] [Google Scholar]
- 26.Hembrough TA, Vasudevan J, Allietta MM, Glass WF, Gonias SL. A cytokeratin 8-like protein with plasminogen-binding activity is present on the external surfaces of hepatocytes, HepG2 cells and breast carcinoma cell lines. J Cell Sci. 1995;108:1071–1082. doi: 10.1242/jcs.108.3.1071. [DOI] [PubMed] [Google Scholar]
- 27.Gonias SL, Hembrough TA, Sankovic M. Cytokeratin 8 functions as a major plasminogen receptor in select epithelial and carcinoma cells. Front Biosci. 2001;6:D1403–D1411. doi: 10.2741/gonias. [DOI] [PubMed] [Google Scholar]
- 28.Shain KH, Dalton WS. Cell adhesion is a key determinant in de novo multidrug resistance (MDR): new targets for the prevention of acquired MDR. Mol Cancer Ther. 2001;1:69–78. [PubMed] [Google Scholar]
- 29.Hazlehurst LA, Argilagos RF, Emmons M, Boulware D, Beam CA, Sullivan DM, Dalton WS. Cell adhesion to fibronectin (CAM-DR) influences acquired mitoxantrone resistance in U937 cell. Cancer Res. 2006;66:2338–2345. doi: 10.1158/0008-5472.CAN-05-3256. [DOI] [PubMed] [Google Scholar]
- 30.Bergom C, Goel R, Paddock C, Gao C, Newman DK, Matsuyama S, Newman PJ. The cell-adhesion and signaling molecule PECAM-1 is a molecular mediator of resistance to genotoxic chemotherapy. Cancer Biol Ther. 2006;5:1699–1707. doi: 10.4161/cbt.5.12.3467. [DOI] [PubMed] [Google Scholar]
- 31.Wu RC, Wang Z, Liu MJ, Chen DF, Yue XS. Beta2-integrins mediate a novel form of chemoresistance in cycloheximide-induced U937 apoptosis. Cell Mol Life Sci. 2004;61:2071–2082. doi: 10.1007/s00018-004-4165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura T, Kato Y, Fuji H, Horiuchi T, Chiba Y, Tanaka K. E-cadherin-dependent intercellular adhesion enhances chemoresistance. Int J Mol Med. 2003;12:693–700. [PubMed] [Google Scholar]
- 33.Chuthapisith S, Layfield R, Kerr ID, Hughes C, Eremin O. Proteomic profiling of MCF-7 breast cancer cells with chemoresistance to different types of anti-cancer drugs. Int J Oncol. 2007;30:1545–1551. [PubMed] [Google Scholar]