Abstract
Hec1 (Highly Expressed in Cancer 1) is one of four proteins of the outer kinetochore Ndc80 complex involved in the dynamic interface between centromeres and spindle microtubules. Its overexpression is seen in a variety of human tumors and correlates with tumor grade and prognosis. We show here that the overexpression of Hec1 in an inducible mouse model results in mitotic checkpoint hyperactivation. As previously observed with overexpression of the Mad2 gene, hyperactivation of the mitotic checkpoint leads to aneuploidy in vitro and is sufficient to generate tumors in vivo that harbor significant levels of aneuploidy. These results underscore the role of chromosomal instability as a result of mitotic checkpoint hyperactivation in the initiation of tumorigenesis.
Keywords: aneuploidy, cancer, chromosome instability
Whole chromosome instability has long been thought to play a role in the tumorigenic process (1). Abnormal chromosome numbers are observed frequently in tumor samples, particularly in solid tumors, which often correlates with tumor grade and prognosis. This observation has triggered a search for the molecular mechanisms that guide proper chromosome segregation and their abnormalities as mediators of tumorigenesis.
Research in the past two decades has led to an understanding of the molecular mechanisms ensuring that in each mitosis daughter cells receive one, and only one, copy of each pair of sister chromatids (2). At the completion of DNA replication in S phase, each replicated chromatid lies topologically linked to its sister by a large ring structure comprised of cohesins. After G2 and prometaphase, only the centromeric cohesins remain and the mitotic spindle begins to form from polar centrosomes. The alignment of sister chromatid pairs at the metaphase plate, a prerequisite for correct segregation, is achieved by bipolar attachment of microtubules to each chromatid pair at the site of a large dynamic scaffold of ≈100 proteins assembled around centromeric DNA known as the kinetochore (3). Broadly speaking, the kinetochore consists of an inner plate containing the centromeric DNA characterized by CenpA-containing nucleosomes and an outer plate where the microtubule interface lies (4). Among the protein complexes known to directly interact with the kinetochore microtubule fiber plus (+) ends is the conserved Ndc80 complex (5, 6), which together with the KNL-1 protein and the Mis-12 complex form the KMN (KNL-1/Mis-12/Ndc80) network responsible for the kinetochore–microtubule interaction. The Ndc80 tetrameric complex is formed by the interaction of Hec1 (Highly Expressed in Cancer 1/also called Kinetochore-associated protein 2, Kntc2; the human homologue of Ndc80) and Nuf2 at one end and Spc24 and Spc25 at the other, creating a rod-shaped structure with globular domains at the extremities (7–10). The N-terminal globular domains of Hec1 and Nuf2 interact with each other and have moderate microtubule binding activity, whereas the C-terminal globular domains of Spc24 and Spc25 are thought to link to the kinetochore outer plate (6). These two globular domains are connected by an overlapping coiled-coil domain that lies at an angle to the microtubule fiber. Multiple Ndc80 rod complexes are thought to bind around each kinetochore fiber, creating a kinetochore–microtubule interface that can slide as the plus end assembles and disassembles. This movement, propelled by the action of plus and minus end-directed motor proteins that also bridge the kinetochore with the microtubule, is responsible for the chromosome dynamics seen during chromosome congression before metaphase and during poleward movement in anaphase and telophase (for an extensive review, see ref. 11).
In addition to its role in microtubule binding by the kinetochore, the Ndc80 complex is essential for the recruitment of the mitotic checkpoint proteins Mad1, Mad2, and Mps1 to the kinetochore (12). Initial studies using RNAi to deplete Hec1 suggested a Mad2-dependent checkpoint arrest independent of its localization to the kinetochore (12). Subsequent studies have shown that such partial reduction of Hec1 results in weak kinetochore–microtubule attachments. Mad1 and Mad2 are easily stripped by microtubules in these unstable kinetochore–microtubule attachments, but the remaining low levels of Mad1 and Mad2 at kinetochores are still capable of inducing a prometaphase block. In fact, more complete depletions of Hec1 appear to completely inhibit mitotic checkpoint function (13), supporting a model whereby Hec1 is required for the recruitment of Mad1 and Mad2 to the kinetochore and hence to execute the mitotic checkpoint.
A number of recent studies have shown that abnormalities in chromosome segregation during mitosis are not only correlated with tumorigenesis, but might in fact act as initiators of the process. Animals heterozygous for Mad2 have been shown to develop tumors with increased frequency compared with control littermates (14). In addition, BubR1 heterozygosity increases tumor burden in a model of colon adenocarcinoma (15), and animals heterozygous for the microtubule motor protein CENP-E also have an increased incidence of spontaneous tumor formation (16). Conversely, overexpression of Mad2, a feature more commonly found in human tumors, has been shown to lead to tumorigenesis in an inducible mouse model (17). Expression analysis of human tumors shows misregulation of a number of mitotic checkpoint genes (18, 19). Although it remains a possibility that the tumor phenotypes observed in mouse models of mitotic checkpoint misregulation are caused by unknown functions of these proteins, the weight of evidence from in vivo studies together with expression analysis data and in vitro work seems to point to a causal link between chromosome instability and tumor initiation.
Human Hec1 was originally identified as a retinoblastoma-interacting protein in a yeast two-hybrid screen (20). Subsequent studies have shown that, although its expression is ubiquitous in dividing cells and absent in quiescent and differentiated cells, its levels are greatly increased in cell lines upon transformation (21). Furthermore, a recent study using immunohistochemical analysis of pathological samples found high levels of this protein in a panel of lung tumors where, in addition, these levels correlated with tumor grade and prognosis (22).
Given the finding that Hec1 levels correlate with tumor grade and prognosis and that the Ndc80 complex is essential for recruitment of mitotic checkpoint proteins to the kinetochore (12, 13), we decided to explore the possibility that Hec1 overexpression has a causal role in tumor formation. We generated an inducible mouse model of Hec1 overexpression and show that it is sufficient to initiate tumorigenesis in all likelihood by stabilization of Mad2, hyperactivation of the mitotic checkpoint, and acquisition of chromosome instability.
Results
Generation of an Antibody Specific to Murine Hec1.
To aid in the characterization of Hec1-induced phenotypes an anti-Hec1 antibody against a GST-mouse Hec1 fusion protein was generated and affinity-purified. The purified anti-Hec1 antibody reacted in Western blot assays with a 77-kDa protein in HeLa cells and a 79-kDa protein in 3T3 mouse cells, both in accordance with their predicted molecular masses (Fig. 1A). We refer to this antibody preparation as panHec1. The panHec1 antibody was able to immunoprecipitate a band of the expected size as seen by immunoprecipitation–Western blotting [Fig. 1B and supporting information (SI) Fig. S1A] that was competed away with the fusion protein used in its generation (data not shown). Moreover, immunofluorescence analysis in normal murine embryonic fibroblasts revealed a staining pattern similar to that described for Hec1 in HeLa cells (23): a diffuse signal was observed in interphase cells (data not shown), and during mitosis Hec1 localized to the kinetochores (Fig. 1C) from prophase to late anaphase. Staining at the centrosomes [as previously reported for HeLa (23)] was also observed. Purified centrosomes from HeLa cells were also found to contain Hec1 protein together with the centrosomal markers γ-tubulin and PLK1 (Fig. S1B). These data confirm that Hec1, in addition to being present at the kinetochore, is a component of the centrosome. When Hec1 was depleted from HeLa cells using siRNA, a high percentage of cells could be seen to contain multiple centrosomes that generated multipolar spindles (Fig. S1C), indicating an important role for Hec1 in centrosome copy number maintenance. In addition and as reported (24), a mitotic block after Hec1 depletion was also observed that was associated with a failure of chromosome congression at the metaphase plate (Fig. S1 C and D).
Fig. 1.
Generation and characterization of a panHec1 antibody. (A) HeLa or 3T3 cell extracts were separated by SDS/PAGE and Western-blotted with the anti-hHec1 (Lower) or the panHec1 antibody (Upper). (B) The panHec1 antibody immunoprecipitated a band of the expected size in both HeLa and murine 3T3 cells. The anti-hHec1 antibody used as a positive control was raised in mouse. HC, heavy chain. (C) Immunofluorescence with purified anti-panHec1 antibody in WT MEFs. White arrows indicate centrosomes, and yellow arrows indicate kinetochores. (Magnification: C, × 63.)
Generation of Mice Carrying an Inducible Hec1 Gene.
To determine whether the Hec1 overexpression observed in many human cancers can by itself induce tumors, transgenic mice containing a regulatable mouse Hec1 were generated. A 1.9-kb DNA fragment containing the mouse Hec1 coding sequence was subcloned in-frame with an HA epitope to facilitate transgene detection into the pTRE-2 vector (Fig. 2A). In addition to these elements, the final construct contained seven direct repeats of the tet operator sequence (tetO7) and the SV40 polyadenylation site.
Fig. 2.
Generation and characterization of pTRE-Hec1 overexpression vector. (A) Diagram of the transgene construct used to overexpress Hec1. TetO, tetracycline operator; HA, hemagglutinin; poly(A), SV40 gene polyadenylation sequence. (B) Empty vector (pTRE) or pTRE-HAmHec1 were transfected in HeLa-Tet-Off cells, and fold increments of doxycycline added to repress transgene expression. (C) (Left) Immunofluorescence of exogenous mHec1 stained both kinetochores and centrosomes of a mitotic cell. panHec1 antibody, red; DNA, blue. (Center) panHec1 stained the centrosomes of an interphase cell. (Right) Immunofluorescence of the competed antibody. White arrows indicate centrosomes, and yellow arrows indicate kinetochores. (D) PCR of tail DNA confirming the presence of mHec1 transgene in different founders and Southern blot of genomic DNA using a transgene-specific probe. C: control. (Magnification: C, × 63.)
To verify that the construct was regulatable and that the inclusion of the HA epitope did not interfere with Hec1 localization, the TetO-Hec1 vector was transfected into HeLa cells carrying the tetracycline transactivator (tTA), which is repressed by doxycycline. Overexpression of mHec1 in this cell line was indeed doxycycline repressible (Fig. 2B). Exogenous Hec1 localized to the kinetochore of mitotic chromosomes, as described for the endogenous Hec1 (Fig. 2C Left) and could also be detected at the centrosome in mitotic and nonmitotic cells when overexpressed at high levels (Fig. 2C Left and Center). Both centrosomal and kinetochore staining were absent when the immunizing peptide was used in competition with the panHec1 antibody (Fig. 2C Right). Similar results were obtained when probing with an antibody to the HA tag (Fig. S2A).
Injection of the 3.2-kb TetO-Hec1 transgene into fertilized F2 eggs obtained from mating of C57BL/6J × CBA/J F1 mice produced 82 pups that were then analyzed for the presence of the transgene by PCR analysis of tail-purified DNA (Fig. 2D Upper) and verified by Southern blot analysis using a probe of 420 bp that was specific for the exogenous Hec1 (Fig. 2D Lower). To generate mice that overexpressed Hec1 in a manner that could be repressed or induced by doxycycline, TetO-Hec1 mice were bred with mice that expressed the tTA or rtTA transactivators under the CMV promoter (17, 25), respectively. To induce Hec1 expression in the rtTA system, bitransgenic animals were fed pellets embedded with doxycycline after weaning. For tTA animals, normal food was administered and only uninduced control animals were maintained on a doxycycline diet.
In Vivo Expression of the TetO-Hec1 Transgene.
To verify transgene expression in vivo, RT-PCR was performed on RNA samples from several tissues from control or bitransgenic TetO-Hec1/CMV-rtTA (Tet-On) animals after 4 weeks of transgene induction. A majority of analyzed tissues expressed the transgene, albeit at different levels (Fig. 3A Center). The expression level of TetO-Hec1 was very high in proliferating tissues such as testis, spleen, and intestine, intermediate in kidney and lung, and very low in liver and brain. To evaluate the repressibility of the system in vivo, tissues were collected from littermates that had been fed with doxycycline-embedded food for the same period and then changed to normal food for 3 extra weeks. As shown in Fig. 3A Right, no exogenous Hec1 expression was detected in any tissue except for residual amounts in testis.
Fig. 3.
In vivo expression of TetO-Hec1. (A) RT-PCR from different tissues of nontransgenic mice and TetO-Hec1/CMV-rtTA mice exposed to doxycycline from 4 weeks to harvest at 8 weeks or exposed to doxycycline for 4 weeks and then to normal food for 3 weeks. PCRs were carried out in the presence (Top) and absence (Middle) of reverse transcription. Amplification of GADPH mRNA confirmed the presence of RNA in all samples (Bottom). (B) Western blot analysis showing Hec1 protein expression in testis (Left), kidney (Center), and MEFs (Right) from nontransgenic animals (N Tg) or TetO-Hec1/CMV-rtTA exposed to doxycycline (ON) or released from it (OFF). (C) Immunofluorescence of G2-M MEFs showing HAmHec1 localization to spindle poles and kinetochores (DNA, blue; mHec, red; γ-tubulin, green). (D) Diffuse localization of mHec1 in induced MEFs by immunofluorescence in G1-S cells. White arrows indicate centrosomes. (E) Mitotic index of asynchronous cells and cells synchronized by serum starvation. At least 2,000 cells of each condition were counted. (Magnification: C and D, × 63.)
To analyze the induction of Hec1 protein in these animals, protein extracts were prepared from several tissues and assayed by Western blot using the panHec1 antibody. Overexpression of Hec1 protein was detected in two tissues tested, testis and kidney (Fig. 3B Left and Center) in which the exogenous protein comigrated with endogenous murine Hec1. Similar results were seen in protein extracts from TetO-Hec1/CMV-tTA (Tet-Off) animals (data not shown). These results demonstrate the doxycycline-dependent inducibility of Hec1 overexpression in the double transgenic system.
Characterization of TetO-Hec1 Overexpression in Primary Murine Embryonic Fibroblasts (MEFs).
MEFs were derived from TetO-Hec1/CMV-rtTA crosses and maintained in media supplemented with doxycycline to induce Hec1 overexpression. Protein extracts prepared from early-passage MEFs were analyzed by Western blot with the panHec1 antibody. Fig. 3B Right shows increased levels of Hec1 in the presence of doxycycline compared with the endogenous levels or nontransgenic MEFs derived from littermates. In addition, we could induce expression of the transgene in a time- and dose-dependent fashion (data not shown). To determine Hec1 localization after overexpression, immunofluorescence staining of asynchronously growing MEFs treated with doxycycline was performed. As expected, mHec1 localized to the kinetochores and centrosomes/spindle poles in mitotic cells, the latter demonstrated by colocalization with γ-tubulin (Fig. 3C). In addition, a diffuse staining pattern with some centrosome localization was seen in nonmitotic cells (Fig. 3D). Similar results were observed when an HA antibody was used and this signal was similarly eliminated when an HA peptide was included for competition (Fig. S2A).
Primary TetO-Hec1/CMV-rtTA MEFs grew well in culture and did not display overt proliferative differences when compared with uninduced cells (data not shown). Moreover, cell cycle profiles assessed by flow cytometry were indistinguishable between WT and bitransgenic MEFs. To determine whether Hec1 overexpression leads to an increase in mitotic cells that would be missed by conventional flow cytometry analysis, the mitotic fraction in asynchronous cells and cells synchronized by serum starvation was scored by conventional microscopy. Hec1-overexpressing MEF cultures had higher mitotic fractions compared with controls in both cases (Fig. 3E). Thus, moderate levels of Hec1 overexpression do not affect the proliferative characteristics of primary MEFs but do result in a small, but significant, accumulation of cells in mitosis.
Overexpression, mutation, or down-regulation of kinetochore and mitotic checkpoint proteins has recently been shown to lead to chromosomal instability and aneuploidy in vitro (16, 17, 26). To assess whether Hec1 overexpression could also lead to alterations in whole chromosome segregation, chromosome counts of metaphase spreads generated from early-passage MEFs were performed. In cells overexpressing moderate levels of Hec1 after 72 h of induction, statistically significant differences were found in chromosome numbers between induced and uninduced samples or when induced samples were compared with WT and single transgenic controls (Fig. 4A). This difference in aneuploidy was observed in the 2N-containing but not the 4N-containing populations. Interestingly, chromosome bridges and lagging chromosomes were frequently seen in Hec1-overexpressing MEFs but not in the uninduced populations (Fig. 4B). Unlike the case with Mad2 overexpression, chromosome breaks in metaphases from Hec1-overexpressing cells were never observed (data not shown). Having shown that Hec1, in addition to being present at the kinetochore, is also found at centrosomes, the spindle and spindle poles in Hec1-overexpressing cells were examined. The number of spindle poles (as determined by staining with the centrosomal marker γ-tubulin) was not significantly different between control and induced MEFs (data not shown). Nevertheless, abnormal spindle figures (as visualized with a β-tubulin antibody) were observed in Hec1-overexpressing MEFs but never in control cells (Fig. 4C).
Fig. 4.
Characterization of Hec1-overexpressing primary cells. (A) Percentage of aneuploidy in the 2N and 4N population in TetO-Hec1-overexpressing (n = 128) and control MEFs (n = 78). (B) Evidence of lagging chromosomes and chromosome bridges in TetO-Hec1/CMV-rtTA-induced MEFs. (C) Example of a normal spindle in control cells and aberrant spindles in TetO-Hec1/CMV-rtTA-induced MEFs. (D) Western blot analysis of in vitro-stimulated splenic lymphocytes isolated from a nontransgenic and two different TetO-Hec1/CMV-rtTA mice in the presence of doxycycline showing the expression of Hec1 and the stabilization of Mad2 and Securin. (Magnification: C, × 63.)
Effects of the Overexpression of Hec1 in Primary Lymphocytes.
To determine the effect of Hec1 overexpression on progression through mitosis in synchronized cells, analyses were performed in primary lymphocytes isolated from spleens of control and TetO-Hec1/CMV-rtTA animals. Primary lymphocytes are preferable to fibroblasts in this situation as they enter the cell cycle synchronously upon stimulation. Such tight synchronization is difficult to obtain in fibroblasts. After addition of ionomycin and phorbol 12-myristate 13-acetate to induce entry into a synchronous cell cycle, lymphocytes at different time points were collected. Doxycycline was added to the culture medium to induce Hec1 expression in lymphocytes derived from the double transgenics. As predicted, Hec1 levels were elevated in double transgenic lymphocytes. Surprisingly, when Mad2 expression was measured as an indicator of mitotic checkpoint activation, it was also highly elevated and did not cycle as in the control lymphocytes, but remained constant throughout the cell cycle (Fig. 4D). As expected for a mitotic arrest seen with high Mad2 levels, TetO-Hec1 lymphocytes also showed elevated Securin levels that were relatively constant throughout the cycle. A less dramatic effect (perhaps caused by less synchrony) was observed in similar experiments performed in serum-starved MEFs released into G1 by the addition of serum (Fig. S2B).
These results indicate that Hec1 overexpression results in an overactive mitotic checkpoint, the likely cause of the accumulation of mitotic cells observed above (Fig. 3E).
Hec1 Overexpression Induces Tumor Formation in Different Tissues.
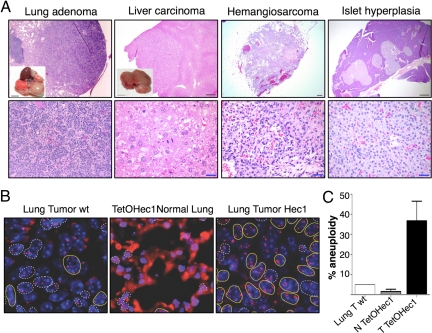
Hec1 expression has been shown to be elevated in tumor cell lines compared with normal cell lines or untransformed controls (21). To determine whether Hec1 overexpression can initiate tumorigenesis in vivo, we followed a cohort of 24 TetO-Hec1/CMV-rtTA mice on doxycycline and 23 TetO-Hec1/CMV-tTA mice that were induced after weaning. Control cohorts (n = 69) included nontransgenic, single transgenic CMV-tTA, or CMV-rtTA and mice containing the TetO-Hec1 transgene alone. Double transgenics in the uninduced setting were also included in the control cohort. No significant differences in tumor onset were found between the different genotypes included in the control cohort (data not shown). Animals were aged to 14–18 months and examined for the development of spontaneous tumors (Fig. 5). As shown in Table 1, lung adenomas were found in 12.8% of Hec1-overexpressing mice, whereas only 1.4% of the control mice had similar tumors, a difference that is highly statistically significant (P < 0.001). Mice that overexpress Hec1 also had a significant increase in liver tumors (25.5%) compared with control animals (11.6%). In addition to these lesions, a hemangiosarcoma and nontumor lesions such as a hyperplastic spleen (data not shown) and a hyperplastic fatty pancreas accompanied by hyperplasia of the islets of Langerhans were found in mice that overexpressed Hec1 (Fig. 5A). Interestingly, tumor spectrum and latency was comparable to that of animals that overexpress Mad2 (described in ref. 17). In line with the chromosomal instability seen in vivo, FISH analysis using a chromosome 12-specific probe revealed marked aneuploidy in the lung adenomas overexpressing Hec1 (Fig. 5B). Neither normal lungs overexpressing Hec1, nor a lung adenoma found in a WT mouse showed significant levels of aneuploidy (Fig. 5C). Abnormal chromosome numbers were seen in the hepatocellular carcinomas (data not shown), although the tendency of murine hepatocytes toward polyploidy precludes us from ascribing this finding to Hec1 overexpression alone. Importantly, tumors were found in both the CMV-tTA and CMV-rtTA systems, and in mice derived from different founders, excluding a strain-specific effect.
Fig. 5.
Hec1 overexpression induces tumor formation in vivo. (A) H&E staining of indicated tumors found in Hec1 transgenic animals taken at low (Upper) or high (Lower) magnification. (Insets) Macroscopic pictures of some of the tumors are shown. (Scale bars: Inset, 1 cm; black bar, 300 μm; blue bar, 30 μm.) (B) FISH images of lung sections from a WT tumor, a nontumor Hec1 lung, and a lung tumor from a Hec1-overexpressing animal showing aneuploidy in the later. DNA is shown in blue, and FISH paint probe to chromosome 12 is in red. Yellow circles mark aneuploid cells, and white circles are diploid cells. (C) Percentage of aneuploidy in a spontaneous WT lung tumor, normal lung overexpressing Hec1, and lung tumors from Hec1 mice. At least 80 cells were counted per condition. (Magnification: C, × 63.)
Table 1.
Comparison between Hec1 and Mad2 overexpressing mice
| Tumor type | Hec1 mice |
Mad2 mice |
||
|---|---|---|---|---|
| Incidence, % | Latency, weeks | Incidence, % | Latency, weeks | |
| Lung adenoma | 12.8 | 67 | 35 | 78 |
| Hepatocellular adenoma | 25.5 | 60 | 25 | 75 |
| Sarcoma | 2.12 | 59 | 5 | 31 |
| Intestinal tumor | 0 | 12.5 | 99 | |
| Lymphoma | 0 | 7.5 | 86 | |
| Prostate tumor | 0 | 5 | 75 | |
Hec1 mice scored at 56–72 weeks. Mad2 mice scored at humane end point.
Finally, we sought to determine whether tumors in the Hec1-overexpressing animals had an overactive mitotic checkpoint. Western blot analysis of tumor tissue from Hec1 overexpressors showed an increase in Mad2 levels compared with control tissues (Fig. S2C), suggesting that Hec1 is also leading to mitotic checkpoint overactivation in vivo.
These data demonstrate that Hec1 overexpression can initiate tumorigenic events in vivo and that it is associated with hyperactivation of the mitotic checkpoint, an event known to cause tumors in mice.
Discussion
Chromosomal instability has long been suggested to be a driving force for tumor initiation and/or progression. This notion largely stems from correlations between abnormal mitotic figures, aneuploidy, tumor grade and prognosis in pathological samples. In addition, mutation analysis and transcriptional data reinforce this notion by showing that a large number of genes known to play a part in the proper control of mitosis are misregulated in tumors. Further still, for the case of some mitotic regulatory genes that are involved in chromosome segregation such as Hec1 and Mad2, high expression levels correlate with tumor grade and prognosis in a variety of human tumors (22, 27–30). Nevertheless, whether these correlations are indicative of a causative role in tumor initiation is being explored. It remains possible that initial oncogenic insults together with loss of tumor suppressors are the main driving force for the formation of early lesions and subsequently with the generation of genomic instability (point mutations, microsatellite instability, or whole chromosomal instability) these lesions progress to full-blown cancers.
In this study we show that inducible overexpression of Hec1, a gene known to be overexpressed in a variety of tumors and whose expression correlates with tumor grade and prognosis (22, 30), can, in an otherwise WT animal, initiate tumorigenesis in multiple organs. The overexpression of Hec1 was also shown to correlate with overactivation of the mitotic checkpoint and widespread aneuploidy in primary fibroblasts and aneuploidy in the resulting tumors. This finding adds to an increasing number of studies showing that single-gene misregulation of the components of the mitotic cycle can contribute to tumor initiation.
Importantly, for the mitotic checkpoint genes known to be implicated in tumors and later validated in murine models, both partial inactivation and overactivation of the checkpoint seem to promote tumorigenesis. Although complete loss of Mad2 is cell lethal both in human and mouse cells (14, 31), heterozygous animals develop lung tumors with long latencies (32). Similarly, CenpE heterozygous animals also develop lung tumors (16) and heterozygous BubR1 animals develop colon adenocarcinomas when carcinogenesis is induced (33) or in a background of an APC (min) mutation (15). Conversely, Mad2 overexpression (more frequently seen in human tumor samples) is sufficient to initiate tumorigenesis in an inducible mouse model in a variety of tissues (17). These data are consistent with the likely possibility that chromosome instability, either caused by premature or delayed separation of sister chromatids, is the initiating event in these cancers. However, the possibility that other events mediated by these proteins contributes to tumor formation cannot be ruled out.
Hec1 is a core component of the outer kinetochore whose function is intricately involved in establishing appropriate microtubule attachments (5). However, cell culture studies have shown that complete loss of Hec1 at the kinetochore results in an inability to recruit the mitotic checkpoint proteins Mad1, Mad2, and Mps1 (12, 24). We show here that overexpression of Hec1 leads to activation of the mitotic checkpoint, as determined by up-regulation and stabilization of Mad2 and Securin. Whether the tumorigenic phenotype observed is a consequence of the elevated levels of Mad2 or some other function at the centrosome (where Hec1 is also localized) remains to be determined. Interestingly, however, there are striking similarities between Hec1 overexpression and Mad2 overexpression. The checkpoint overactivation seen with Hec1 overexpression results in aneuploidy, as has also previously been shown with Mad2 overexpression. Furthermore, tumors arising in Mad2-overexpressing mice contain abnormal chromosomes, and substantial aneuploidy and tetraploidy as also shown here for Hec1, strengthening the causal link between chromosomal instability and tumorigenesis. Nevertheless, Mad2 overexpression also leads to numerous chromosome breaks and interstitial deletions, neither of which were found in the case of Hec1 overexpression. In addition, Mad2 overexpression induces aneuploidy in both 2N- and 4N-containing populations, whereas Hec1 overexpression does so only in the 2N population. These differences may be related to levels of mitotic checkpoint hyperactivation or separate effects of Mad2 overexpression. As mentioned, Hec1 has moderate microtubule binding activity on its own, so it remains a possibility that very high levels of Hec1 saturate the Ndc80 complex binding sites on kinetochore fibers, thus leading to microtubules that bind to the kinetochore with reduced efficiency. Alternatively, as suggested by the presence of abnormal spindles, it is possible that overexpression of Hec1 is interfering with an as-yet-uncharacterized role at the centrosome and/or spindle assembly. Both of these potential mechanisms would activate the mitotic checkpoint and thereby also explain the increases seen in Mad2 levels upon induction of the Hec1 gene in synchronized lymphocytes. Whether Mad2 overexpression is required for the Hec1-induced tumor phenotype can now be explored genetically. We have shown that overexpression of an outer kinetochore protein, Hec1, whose overexpression is correlated with tumor severity, can by itself initiate the oncogenic process.
Methods
Generation of Hec1 Transgenic Mice.
Murine Hec1 cDNA was amplified with specific primers in-frame with the 5′ HA epitope and subcloned into the pTRE2 vector (Clontech) that contains the tetracycline operator and the SV40 polyadenylation sequence. Restriction digest and sequencing were used to verify correct clones. The construct was linearized and injected into fertilized F2 eggs obtained from mating of C57BL/6J × CBA/J F1 mice and newborn pups genotyped as described.
Animal Husbandry, Genotyping, and Southern Blotting.
Mice were kept in pathogen-free housing under Research Animal Resources Center-approved Memorial Sloan-Kettering Cancer Center institutional guidelines. CMV-tTA and CMV-rtTA mice have been described (17, 25). Doxycycline was administered by feeding mice with doxycycline-embedded pellets (625 ppm; Harlan-Teklad). TetO-Hec1 mice were genotyped by PCR on tail purified DNA by using the following primers: Hec1-Forward, 5′-GTCGAGTAGGCGTGTACGG-3 and Hec1-Reverse, 5′-AAGTGGTCTTGGGTCCTTGA-3′. For Southern blotting, a probe specific for the transgene and including part of the pTRE promoter, the HA epitope, and the 5′end of mHec cDNA was used.
Cell Culture, Transfections, and siRNA.
All cells were cultured at 37°C in a humidified atmosphere in the presence of 5% CO2–95% air. Transfection of HeLa-Tet-Off cells was performed with Lipofectamine (Invitrogen) following the manufacturer's instructions. For siRNA, oligonucleotides corresponding to the Hec1 ORF described by Martin-Lluesma et al. (12) were generated by Dharmacon, and transfections were carried out by using 10 μl of a 10-mmol RNA solution. Generation of MEFs and lymphocytes, FACS, and karyotyping are described in SI Text.
RNA Preparation and RT-PCR.
RNA was isolated with the RNeasy kit (Qiagen) and treated with DNaseI (Ambion) to eliminate any contaminating DNA. RT-PCRs were performed with SuperScript III (Invitrogen) according to the manufacturer's instructions.
Antibody Preparation and Immunoblotting.
The panHec1-specific antiserum was raised in rabbits against a GST-fusion protein containing 174 aa of the murine Hec1 sequence (amino acides 235–409). The antiserum was purified by affinity chromatography using protein-Sepharose columns as described (34). Protein expression was assessed by immunoblotting using 40 μg of total cell or tissue lysates, following standard protocols (35). Blots were probed with the purified panHec1 antibody, anti-hHec1 (Abcam; mouse monoclonal, clone 9G3), HA (Roche), Mad2 (BD Transduction Laboratories), Securin (Neomarkers), PLK1 (Bethyl Laboratories), γ-tubulin, and actin (Sigma). HRP-conjugated anti-mouse or anti-rabbit (Amersham) were used as secondary antibodies, and proteins were visualized by using the ECL detection system (Amersham).
Supplementary Material
Acknowledgments.
We thank the members of R.B.'s laboratory for helpful discussions of the project; S. Curelariu, Y. Chin, and C. Coker for technical assistance. M. Leversha and C. Kalyani for help with FISH analysis; K. La Perle for assistance with pathology; E. Hernando for continuous encouragement and helpful discussions; and A. Pandiella for support. R.S. was supported by the Charles Haskell Revson Foundation. J.-M.S. is supported by a Breast Cancer Research Program Predoctoral Traineeship Award from the Department of Defense (Congressionally Directed Medical Research Program). R.B. is supported by the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803504105/DCSupplemental.
References
- 1.Bharadwaj R, Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004;23:2016–2027. doi: 10.1038/sj.onc.1207374. [DOI] [PubMed] [Google Scholar]
- 2.Wassmann K, Benezra R. Mitotic checkpoints: From yeast to cancer. Curr Opin Genet Dev. 2001;11:83–90. doi: 10.1016/s0959-437x(00)00161-1. [DOI] [PubMed] [Google Scholar]
- 3.Ciferri C, Musacchio A, Petrovic A. The Ndc80 complex: Hub of kinetochore activity. FEBS Lett. 2007;581:2862–2869. doi: 10.1016/j.febslet.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 5.DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 6.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 7.Bharadwaj R, Qi W, Yu H. Identification of two novel components of the human NDC80 kinetochore complex. J Biol Chem. 2004;279:13076–13085. doi: 10.1074/jbc.M310224200. [DOI] [PubMed] [Google Scholar]
- 8.Ciferri C, et al. Architecture of the human ndc80–hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem. 2005;280:29088–29095. doi: 10.1074/jbc.M504070200. [DOI] [PubMed] [Google Scholar]
- 9.Hori T, Haraguchi T, Hiraoka Y, Kimura H, Fukagawa T. Dynamic behavior of Nuf2–Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J Cell Sci. 2003;116:3347–3362. doi: 10.1242/jcs.00645. [DOI] [PubMed] [Google Scholar]
- 10.McCleland ML, et al. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore–microtubule attachment. Curr Biol. 2004;14:131–137. doi: 10.1016/j.cub.2003.12.058. [DOI] [PubMed] [Google Scholar]
- 11.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 12.Martin-Lluesma S, Stucke VM, Nigg EA. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- 13.Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- 15.Rao CV, et al. Colonic tumorigenesis in BubR1+/−ApcMin/+ compound mutant mice is linked to premature separation of sister chromatids and enhanced genomic instability. Proc Natl Acad Sci USA. 2005;102:4365–4370. doi: 10.1073/pnas.0407822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Sotillo R, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehman NL, et al. Oncogenic regulators and substrates of the anaphase promoting complex/cyclosome are frequently overexpressed in malignant tumors. Am J Pathol. 2007;170:1793–1805. doi: 10.2353/ajpath.2007.060767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan B, et al. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res. 2006;12:405–410. doi: 10.1158/1078-0432.CCR-05-0903. [DOI] [PubMed] [Google Scholar]
- 20.Zheng L, Chen Y, Riley DJ, Chen PL, Lee WH. Retinoblastoma protein enhances the fidelity of chromosome segregation mediated by hsHec1p. Mol Cell Biol. 2000;20:3529–3537. doi: 10.1128/mcb.20.10.3529-3537.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Riley DJ, Chen PL, Lee WH. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol Cell Biol. 1997;17:6049–6056. doi: 10.1128/mcb.17.10.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayama S, et al. Activation of CDCA1-KNTC2, members of centromere protein complex, involved in pulmonary carcinogenesis. Cancer Res. 2006;66:10339–10348. doi: 10.1158/0008-5472.CAN-06-2137. [DOI] [PubMed] [Google Scholar]
- 23.Lin YT, Chen Y, Wu G, Lee WH. Hec1 sequentially recruits Zwint-1 and ZW10 to kinetochores for faithful chromosome segregation and spindle checkpoint control. Oncogene. 2006;25:6901–6914. doi: 10.1038/sj.onc.1209687. [DOI] [PubMed] [Google Scholar]
- 24.DeLuca JG, et al. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr Biol. 2003;13:2103–2109. doi: 10.1016/j.cub.2003.10.056. [DOI] [PubMed] [Google Scholar]
- 25.Furth PA, et al. Temporal control of gene expression in transgenic mice by a tetracycline-responsive promoter. Proc Natl Acad Sci USA. 1994;91:9302–9306. doi: 10.1073/pnas.91.20.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shepard JL, et al. A mutation in separase causes genome instability and increased susceptibility to epithelial cancer. Genes Dev. 2007;21:55–59. doi: 10.1101/gad.1470407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garber ME, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernando E, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka K, et al. Mitotic checkpoint protein hsMAD2 as a marker predicting liver metastasis of human gastric cancers. Jpn J Cancer Res. 2001;92:952–958. doi: 10.1111/j.1349-7006.2001.tb01186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van't Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 31.Michel L, Benezra R, Diaz-Rodriguez E. MAD2-dependent mitotic checkpoint defects in tumorigenesis and tumor cell death: A double-edged sword. Cell Cycle. 2004;3:990–992. [PubMed] [Google Scholar]
- 32.Michel LS, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 33.Dai W, et al. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 2004;64:440–445. doi: 10.1158/0008-5472.can-03-3119. [DOI] [PubMed] [Google Scholar]
- 34.Harlow E, Lane D. Antibodies: A Laboratory Manual. New York: Cold Spring Harbor Lab Press; 1988. [Google Scholar]
- 35.Diaz-Rodriguez E, Montero JC, Esparis-Ogando A, Yuste L, Pandiella A. Extracellular signal-regulated kinase phosphorylates tumor necrosis factor α-converting enzyme at threonine 735: A potential role in regulated shedding. Mol Biol Cell. 2002;13:2031–2044. doi: 10.1091/mbc.01-11-0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.