Abstract
We previously identified a relatively high frequency of B-cell proliferations along with simultaneous T-cell receptor γ-chain gene (TRG) and immunoglobulin heavy chain gene (IGH) rearrangements in a series of angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma, unspecified. Here, we report on a series of 74 peripheral T-cell lymphoma (PTCL) cases composed entirely of specific PTCL subtypes, including 28 cases of ALK+ anaplastic large-cell lymphoma (ALCL), 35 cases of ALK− ALCL, and 11 cases that represent other specific PTCL subtypes. We performed IGH and TRG gene rearrangement studies and in situ hybridization for Epstein-Barr virus (EBV) to determine the frequency of IGH clonality and to investigate the relationship between EBV, clonality, and associated B-cell proliferations. Using BIOMED-2 PCR assays, we detected TRG clones in 64 of 74 (86%) cases and IGH clones in 6 of 74 (8%) cases, with all IGH-positive cases exhibiting a concurrent TRG clone. Despite the detection of occasional IGH clones, there was no correlation between IGH clonality and EBV, and B-cell proliferations were not identified in any of the cases. These findings suggest that other factors contribute to IGH clonality and demonstrate that, in the absence of an associated B-cell proliferation, IGH clonality occurs infrequently (8%) in specific PTCL subtypes.
Peripheral T-cell lymphoma (PTCL) is an uncommon malignancy that accounts for less than 10% of non-Hodgkin lymphomas worldwide. By current World Health Organization criteria, the diagnosis and classification of PTCL is based on the combination of clinical, histologic, immunophenotypic, and genetic findings.1 However, the diagnosis of PTCL is difficult even for experienced pathologists, given the infrequency of cases, the often unusual histologic features, and perhaps most importantly, the lack of good immunophenotypic markers to assess for clonality in T-lineage neoplasms.
In cases of suspected PTCL, pathologists often evaluate for clonality and assess for lineage by performing PCR-based assays for clonal rearrangement in both the T-cell receptor γ-chain gene (TRG) and the immunoglobulin heavy chain gene (IGH). Although PCR-based assays for TRG typically detect clones in 80 to 90% of PTCL cases, several studies have shown that IGH clones are also detected relatively frequently (9 to 16%).2,3,4,5,6 The detection of an IGH clone, with or without a concurrent TRG clone, complicates an already challenging diagnosis.
Recently, we investigated the etiology of IGH clones in PTCL by studying a large series of two of the most common subtypes of PTCL, angioimmunoblastic T-cell lymphoma (AILT) and PTCL-unspecified (PTCL-U).7 A subset of cases were complicated by associated B-cell proliferations, a finding that we and others have described as an atypical infiltrate of B cells that is often associated with Epstein-Barr virus (EBV).8,9,10,11 Using multiplex PCRs developed in a European collaborative study (BIOMED-2), we detected TRG clones in approximately 80% of cases and IGH clones in approximately 35% of cases, with the majority of IGH clones detected along with simultaneous TRG clones.
Interestingly, a positive IGH clone correlated, at least in part, with the presence of a B-cell proliferation, suggesting that B-cell proliferations contribute to IGH clonality. However, alternate explanations for the detection of simultaneous TRG and IGH clones include those that are technical in nature and so-called lineage infidelity. In this context, lineage infidelity refers to recombination of both TRG and IGH in the same clone. However, this phenomenon is more common in immature hematolymphoid neoplasms and occurs only rarely (<5%) in mature B- and T-cell non-Hodgkin lymphomas.12,13,14,15
Regardless of the etiology, our findings raised several important questions. First, it remains unclear whether IGH clonality can occur in all PTCL subtypes or whether it is limited to AILT and PTCL-U, which represent the only two subtypes in which B-cell proliferations have been reported. If IGH clonality were limited to AILT and PTCL-U, this would suggest that B-cell proliferations are responsible for IGH clonality. Alternatively, although B-cell proliferations have not been described in other subtypes, this possibility has not been thoroughly investigated using the combination of immunohistochemical stains and assays for clonality and EBV. If IGH clones occur in other PTCL subtypes, they may be the manifestation of subtle or incipient B-cell proliferations that have been overlooked thus far.
To address these issues, we collected a series of 74 PTCL cases composed of subtypes other than AILT and PTCL-U, which corresponds to specific subtypes in which B-cell proliferations have not been reported. This series includes 28 cases of ALK+ anaplastic large-cell lymphoma (ALCL), 35 cases of ALK− ALCL, and 11 additional cases representing other specific PTCL subtypes. We assayed all cases for IGH and TRG clonality, and we performed the experiments under “blinded” conditions to minimize experimental and interpretive bias. In addition, we evaluated cases for the presence of B-cell proliferations using immunohistochemical stains, and we performed in situ hybridization for EBV to investigate the relationship between EBV, IGH clonality, and possible B-cell proliferations.
Materials and Methods
Cases
Twenty-eight cases of ALK+ ALCL, 35 cases of ALK− ALCL, 3 cases of subcutaneous panniculitis-like T-cell lymphoma, 3 cases of enteropathy-type T-cell lymphoma, 1 case of hepatosplenic T-cell lymphoma, 4 cases of T-cell large granular lymphocytic leukemia (T-LGL), and 24 cases of precursor B-cell acute lymphoblastic leukemia/lymphoma (pre-B-ALL) were selected from the Laboratory of Hematopathology, Stanford University Medical Center (Stanford, CA). Cases were received between January 1, 1995 and December 31, 2006, and diagnostic classification for all cases in this study was based on current World Health Organization criteria.1 Cases of CD30+, ALK− PTCL that lacked the morphologic features of ALCL as defined by the World Health Organization criteria were classified as PTCL-unspecified and excluded from the study. Cases were selected based on the availability of archived paraffin-embedded tissue or frozen bone marrow cells (decalcified specimens were excluded). The selected materials represented cases in which no previous DNA-based clonality study had been performed, and these cases have not been included in any previously published report, including our recent study on AILT and PTCL-U.7 All cases showed greater than 10% involvement, and the majority showed >50% involvement. Use of tissue for this study was approved by the Stanford University Panel on Medical Human Subjects [Protocol ID 79034; Institutional Review Board number 348 (Panel 1)].
Histology and Immunohistochemistry
Histologic sections were prepared from formalin-fixed, paraffin-embedded tissue by cutting 3- to 4-μm-thick sections and staining with H&E. Sections were stained for immunohistochemistry on either a BenchMark instrument (Ventana Medical Systems, Tucson, AZ) or Autostainer (Dako, Carpinteria, CA) using the biotin-avidin technique with diaminobenzidine as the chromogen.16 All cases were stained with at least one B- and one T-cell marker. The following mouse monoclonal antibodies were used: AB75 (anti-CD2; Vision BioSystem, Norwell, MA), IF6 (anti-CD4; Vision BioSystem), 4C7 (anti-CD5; Vision BioSystem), CD7-272 (anti-CD7; Vision BioSystem), C8/144B (anti-CD8; Dako), 56C6 (anti-CD10; Vision BioSystem), MMA (anti-CD15; Ventana), L26 (anti-CD20; Dako), Ber-H2 (anti-CD30; Dako), MY10 (anti-CD34; Becton Dickinson, San Jose, CA), L60 (anti-CD43; Becton Dickinson), ROA6 (anti-CD45RO; Zymed, South San Francisco, CA), JCV117 (anti-CD79a; Dako), ALK1 (anti-ALK; Dako), E029 (anti-EMA; Dako), 24 (anti-PAX5; BD Transduction, Lexington, KY), GrB-7 (anti-granzyme; Dako), and 2G9 (anti-T-cell intracellular antigen-1; Immunotech, Quebec, Canada). The following rabbit polyclonal antibodies were used: anti-CD3 (Cell Marque, Hot Springs, AR) and anti-TdT (Supertechs, Bethesda, MD). Antigen retrieval was performed by using automated heat pretreatment (Ventana) for AB75, IF6, 4C7, CD7-272, C8/144B, 56C6, L26, Ber-H2, MY10, L60, JCV117, ALK1, and anti-CD3 or by using manual heat-induced epitope retrieval (Dako) with an ethylene diamine tetraacetic acid buffer for E029 or citrate buffer for GrB-7, 2G9, 24, and anti-TdT. For MMA and ROA6, staining was performed without antigen retrieval using the BenchMark instrument (Ventana).
Flow Cytometry
Bone marrow aspirate specimens were diluted with 1 volume of Dulbecco's PBS (Sigma, St. Louis, MO), vortexed for 15 seconds, and incubated at 37°C for 20 minutes. After incubation, the mixture was centrifuged at 500 × g for 5 minutes, and the supernatant was discarded. The cells were resuspended in PBS with 20% fetal calf serum (HyClone, Logan, UT), washed three times in PBS, and diluted in PBS to 104 cells/μL for staining. An aliquot of cells was resuspended in PBS with 20% fetal calf serum and 20% dimethyl sulfoxide (Sigma) and archived at −70°C for subsequent clonality assays.
For detection of surface antigens, the following four-color antibody combinations were used (fluorescein isothiocyanate/phycoerythrin/peridinin-chlorophyll-protein/allophycocyanin): CD5/CD38/CD45/CD19, lambda/kappa/CD45/CD19, CD20/CD10/CD45/CD19, CD8/CD3/CD45/CD4, CD16/CD3/CD45/CD56, CD57/CD8/CD45/CD3, CD7/CD3/CD45/CD2, TCRαβ/TCRγδ/CD45/CD3, and CD38/CD56/CD45/CD34. Antibodies to CD2 (S5.2), CD3 (SK7), CD4 (SK3), CD5 (L17F12), CD7 (M-T701), CD8 (SK1), CD10 (HI10A), CD34 (8G12), CD38 (HB7), CD45 (2D1), CD56 (NCAM16.2), CD57 (HNK-1), TCRαβ (WT31), TCRγδ (11F2), kappa (TB28-2), and lambda (1-155-2) were obtained from Becton Dickinson. For staining, 100 μl of cells was mixed with antibodies at the manufacturer's recommended concentrations and incubated at 4°C for 20 minutes in the dark. After staining, the cells were treated with PharmLyse (Pharmingen, San Diego, CA) according to the manufacturer's protocol and resuspended in PBS for flow cytometry.
For detection of intracellular antigens, the following three-color antibody combinations were used (fluorescein isothiocyanate/phycoerythrin/peridinin-chlorophyll-protein): MPO/CD79A/CD45 and TdT/CD3/CD45. Antibodies to TdT (HT1 + HT4 + HT8 + HT9) and CD79A (HM47) were obtained from Beckman Coulter (Fullerton, CA), antibody to MPO (H-43-5) was obtained from Dako, and antibody to CD3 (SK7) was obtained from Becton Dickinson. For membrane permeabilization, 50 μl of cells was treated with Fix & Perm (Caltag, Burlingame, CA) according to the manufacturer's protocol. After permeabilization, cells were mixed with antibodies at the manufacturer's recommended concentrations and incubated at room temperature for 20 minutes in the dark. Stained cells were resuspended in PBS for flow cytometry.
Flow cytometry was performed on a four-color FACSCalibur instrument (Becton Dickinson), and data were acquired and analyzed using CellQuest software (Becton Dickinson). Specific cell populations were identified and gated based on forward versus orthogonal light scatter, CD45 expression versus orthogonal light scatter, and a variety of antigen expression patterns. Populations were evaluated for antigen expression by comparing the fluorescence intensity versus that of the same population stained with an isotype control.
Isolation of DNA
DNA was obtained from either formalin-fixed, paraffin- embedded (FFPE) tissue blocks or archived bone marrow aspirates that were each assigned a unique three-digit number. The corresponding case identities were maintained anonymous from the experimenters and authors until final clonality results were assigned for every case. For FFPE tissue, DNA was obtained by cutting four to eight 20-μm-thick sections and deparaffinizing by extracting three times in 1.0 ml of xylene (Fisher Scientific, Pittsburgh, PA). The extracted tissue was washed two times in 1.0 ml of 100% ethanol (Gold Shield Chemical Co., Hayward, CA) and then dried at 65°C. The tissue was resuspended in 2 volumes (50 to 200 μl) of a mixture of 4× PCR buffer II (Applied Biosystems, Foster City, CA), 0.1% sodium dodecyl sulfate, and 0.6 mg/ml proteinase K (Stratagene, La Jolla, CA) and incubated at 65°C overnight. Samples were further purified using a DNeasy Tissue kit (Qiagen, Valencia, CA) according to the manufacturer's protocols and eluting the DNA in 100 to 200 μl of DNase/RNase-free water (Invitrogen, Carlsbad, CA). For bone marrow aspirates, DNA from 1 to 2 × 106 cells was isolated using a QIAamp DNA Blood mini kit (Qiagen) according to the manufacturer's protocols and eluting the DNA in 50 to 100 μl of DNase/RNase-free water (Invitrogen).
TRG and IGH Clonality Studies
Cases were evaluated for TRG and IGH clonality as described previously7 using commercially available PCR-based kits (InVivoScribe Technologies, San Diego, CA) based on a European collaborative study (BIOMED-2 Concerted Action).17 The PCRs were performed according to the manufacturer's protocols. Briefly, a 5-μl aliquot of DNA sample was added to 45 μl of each reaction mixture and 1.25 units of AmpliTaq Gold (Applied Biosystems). Amplification was performed using a PTC-200 thermocycler (M&J Research, Waltham, MA) by initially heating at 95°C for 7 minutes, followed by 35 cycles of 95°C for 45 seconds, 60°C for 45 seconds, and 72°C for 90 seconds. The final step was incubation at 72°C for 10 minutes.
After amplification, 1 μl of PCR product was added to 10 μl of Hi-Di Formamide (Applied Biosystems) and 1 μl of ROX-500 internal size standard (Applied Biosystems). The mixture was then denatured at 95°C for 5 minutes, chilled on ice for 5 minutes, and resolved by capillary electrophoresis on an ABI 3100 instrument using performance optimized polymer-4 (Applied Biosystems). The data were stored electronically and analyzed using GeneScan software (Applied Biosystems). Printed electropherograms were reviewed independently by two separate pathologists (B.T.T. and D.A.A.) who were blinded to the identity of specimens. Criteria for assigning a positive or negative result were similar to those used for clinical specimens at the Stanford Molecular Pathology laboratory. Briefly, the products of a primer set were considered clonal if one or two distinct peaks were present within the expected size range. In some cases, a polyclonal background was present in addition to the distinct peak(s). To be considered clonal, the height of at least one distinct peak had to exceed that of the polyclonal background by at least twofold. When a single primer set yielded three or more distinct peaks within the expected size range, the results for that primer set were classified as oligoclonal, regardless of whether a polyclonal background was present or absent. Cases in which one or more primer sets yielded clonal products were scored as positive. Cases in which all of the primer sets yielded polyclonal, oligoclonal, and/or undetectable products were scored as negative. Assays yielding a positive IGH study were scored as positive only if a repeat assay yielded identically sized products to the original study.
Control specimens included clonal DNA isolated from cell lines (InVivoScribe), polyclonal DNA isolated from human tonsil specimens (InVivoScribe), a sensitivity control composed of clonal DNA diluted in polyclonal DNA, and a blank composed of DNase/RNase-free water (Invitrogen). Multiple polyclonal DNA specimens, each isolated from different patients (InVivoScribe), consistently yielded polyclonal signals and no clonal signals. For a run of clonality studies to be accepted into the data set, a positive clone had to be detectable from the sensitivity control, which was prepared at 2 or 3% (expressed as micrograms clonal DNA per microgram polyclonal DNA) for TRG or IGH assays, respectively. Individual DNA specimens had to yield detectable products of at least 300 nucleotides from the control amplification tube, which contains primers to amplify the human leukocyte antigen-DQ gene. In our validation of the InVivoScribe TRG and IGH kits, we were able to detect clones in 80 to 90% of known B- and T-cell neoplasms, and amplification of the provided polyclonal control DNA (InVivoScribe) or DNA isolated from paraffin-embedded reactive tonsil specimens in our laboratory yielded only polyclonal results.
In Situ Hybridization for EBV
EBV-encoded small RNA was detected from 3- to 4-μm-thick, formalin-fixed, paraffin-embedded tissue sections by in situ hybridization using a Ventana BenchMark instrument running a standardized program incorporating deparaffinization, hybridization to the Inform EBV-encoded small RNA probe cocktail, and staining with ISH iVIEW nitro blue tetrazolium (Ventana). Cases were evaluated for EBV-labeled cells under visible light microscopy, and the number of cells with blue-colored nuclei was visually estimated at 0, <1, 1 to 10, >10 to 100, or >100 per medium power field using a ×15 ocular and ×20 objective lens on a BX50 microscope (Olympus, Tokyo, Japan). The minimum number of labeled nuclei required for a nonzero score was three per section, which is more than that typically seen in a lymph node or tonsil removed from a healthy individual without evidence of immunodeficiency (B.T.T. and R.A.W., unpublished observations). For 30 of the 49 cases studied for EBV, a control hybridization was performed with an oligo-deoxythymidine probe and successfully detected polyadenylated-mRNA in all 30 cases. For 19 of the 49 cases, sufficient material was not available for a control, but the proportion of EBV-positive cases was similar for the 30 cases with controls (30%) versus the 19 cases without controls (32%). In addition, an external positive control consisting of an EBV-positive malignancy (NK/T-cell lymphoma or nasopharyngeal carcinoma) was run with each set of hybridizations.
Statistical Analysis
The data were analyzed using the software program JMP-IN version 5.1 (SAS Institute, Cary, NC). Age was entered as a continuous numerical parameter, and all other data were entered as categorical parameters. Differences in age between categorically defined groups were evaluated by t-test, and statistical significance was defined as P < 0.05. Correlations between categorical parameters were evaluated by Pearson χ2 test and likelihood ratio test, and the lower P was recorded.
Results
Cases
We collected a series of 74 cases of PTCL from the Stanford hematopathology laboratory. This series is composed of 28 cases of ALK+ ALCL, 35 cases of ALK− ALCL, 3 cases of subcutaneous panniculitis-like T-cell lymphoma, 1 case of hepatosplenic T-cell lymphoma, 4 cases of T-cell large granular lymphocytic leukemia (T-LGL), and 3 cases of enteropathy-type T-cell lymphoma. We also collected 24 cases of pre-B-ALL to serve as a positive control for IGH clonality. All cases were classified according to current World Health Organization criteria based on the combination of histologic, immunophenotypic, and clinical findings.1
The patient demographics and anatomic sites of cases are summarized in Table 1. Each case was selected from a different patient, and the vast majority of cases represented the primary diagnosis. The majority of pre-B-ALL cases were from pediatric patients, and the mean age of pre-B-ALL patients was significantly lower than that of ALK+ (P < 0.0368) and ALK− ALCL patients (P < 0.0001). For ALCL cases, the mean age of ALK+ patients was significantly lower than that of ALK− patients (P < 0.0001). Comparison between the diagnostic groups showed no significant difference in gender composition, although males slightly outnumbered females among ALK+ and ALK− ALCL patients, and females slightly outnumbered males among pre-B-ALL patients.
Table 1.
Patient Demographics of Collected Cases
| Diagnosis | Cases (n) | Men [n (%)] | Women [n (%)] | Mean age (years) | Site (n of cases) |
|---|---|---|---|---|---|
| ALK+ ALCL | 28 | 16 (57) | 12 (43) | 27 | Lymph node (23), soft tissue (2), leg (1), psoas (1), L1 vertebrae (1) |
| ALK− ALCL | 35 | 21 (60) | 14 (40) | 54 | Lymph node (25), abdomen (1), eye (1), stomach (1), mesentery (2), soft tissue (3), retroperitoneum (2) |
| SPTCL | 3 | 1 (33) | 2 (67) | 50 | Abdomen (1), palm (1), leg (1) |
| HSTCL | 1 | 1 (100) | 0 (0) | 65 | Liver (1) |
| T-LGL | 4 | 2 (50) | 2 (50) | 56 | Bone marrow (2), spleen (1), tonsil (1) |
| ETCL | 3 | 0 (0) | 3 (100) | 59 | Small bowel (2), stomach (1) |
| Total PTCL | 74 | 41 (55) | 33 (45) | 44 | Lymph node (48), bone marrow (2), spleen (1), leg (2), tonsil (1), stomach (2), small bowel (2), liver (1), abdomen (2), retroperitoneum (2), eye (1), psoas (1), mesentery (2), soft tissue (5), L1 vertebrae (1), palm (1) |
| Pre-B-ALL | 24 | 10 (42) | 14 (58) | 16 | Lymph node (13), bone marrow (3), skin (2), testis (1), ovary (1), mediastinum (1), nasal (1), periorbital (1), thigh (1) |
ETCL, Enteropathy-type T-cell lymphoma; HSTCL, hepatosplenic T-cell lymphoma; SPTCL, subcutaneous panniculitis-like T-cell lymphoma.
The ALK+ and ALK− ALCL cases showed similar histologic features with large atypical cells with pleomorphic, often kidney-shaped nuclei and abundant cytoplasm with occasional eosinophilic inclusions. The immunophenotypic findings of the ALCL cases are summarized in Table 2, and representative histologic, immunohistochemical, and EBV in situ studies are shown for an ALK+ and ALK− case in Figure 1, a–e and f–i, respectively. The majority of cases showed a predominance of large atypical CD30+ cells, consistent with the common variant of ALCL. However, three ALK+ cases showed mostly small- to medium-sized, CD30+ and ALK+ cells and were classified as the small cell variant.
Table 2.
Immunophenotypic Findings of ALCL Cases
| Positive cases/cases studied [n/n(%)] |
||||||||
|---|---|---|---|---|---|---|---|---|
| Diagnosis | Cases (n) | CD2 | CD3 | CD15 | CD43 | CD30 | ALK | EMA |
| ALK+ ALCL | 28 | 8/18 (44) | 4/25 (16) | 0/7 (0) | 11/17 (65) | 28/28 (100) | 28/28 (100) | 8/11 (73) |
| ALK− ALCL | 35 | 9/14 (64) | 16/32 (50) | 1/12 (8) | 16/23 (70) | 35/35 (100) | 0/35 (0) | 4/13 (31) |
| Total | 63 | 17/32 (53) | 20/57 (35) | 1/19 (5) | 27/40 (68) | 63/63 (100) | 28/63 (44) | 12/24 (50) |
Figure 1.
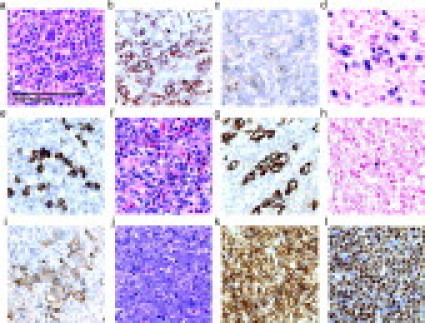
Histology of representative ALCL and pre-B-ALL cases. All panels shown at identical magnification. a–e: ALK+ ALCL stained with H&E (a), immunohistochemistry with anti-CD2 (b), anti-CD30 (c), in situ hybridization for EBV (d), or anti-ALK (e). f–i: ALK− ALCL stained with H&E (f), anti-CD3 (g), in situ hybridization for EBV (h), or anti-CD30 (i). j–l: pre-B-ALL stained with H&E (j), anti-CD79a (k), or anti-TdT (l). Images were captured with a Nikon Eclipse E1000 microscope (Nikon, Tokyo, Japan) using a 60×/0.95 objective lens (Nikon) and a Spot CCD camera (Diagnostic Instruments, McHenry, IL) using Spot software version 4.6 for image acquisition.
For the other PTCL subtypes, the three subcutaneous panniculitis-like T-cell lymphoma cases showed a dense lymphoid infiltrate involving subcutaneous tissue with atypical CD3+, CD8+ cells forming rings around adipocytes. The single hepatosplenic T-cell lymphoma case was a liver biopsy with numerous CD3+, CD4−, CD8− lymphocytes in a characteristic sinusoidal pattern. The three enteropathy-type T-cell lymphoma cases were characterized by an infiltrate of large atypical CD3+ lymphoid cells involving gastrointestinal mucosa, with the adjacent epithelium showing findings consistent with celiac spruce. For the four T-LGL cases, three were composed of CD3+, CD4−, CD8+ neoplastic cells that expressed one or more cytotoxic markers, including granzyme, T-cell intracellular antigen-1, and/or CD57. One T-LGL case was composed of CD3+, CD4−, CD8− T-cells expressing the γ/δ T-cell receptor and was classified as a γ/δ T-LGL.
All of the pre-B-ALL cases were composed of numerous immature lymphoid cells (Figure 1, j–l). For 21 of 24 cases, specimens from extramedullary sites were chosen to obtain nondecalcified, formalin-fixed, paraffin-embedded tissue from anatomic sites that were similar to those of the PTCL cases (Table 1). Three pre-B-ALL cases, along with two of the T-LGL cases, were obtained from archived bone marrow aspirates. All of the pre-B-ALL cases exhibited a CD79a+, TdT+, CD3− immunophenotype by immunohistochemical stains and/or flow cytometry.
IGH Clonality Studies
We assayed cases for clonal rearrangements in the immunoglobulin heavy chain gene (IGH) using a multiplex PCR based on a European BIOMED-2 collaborative study.17 The PTCL and pre-B-ALL cases were assayed concurrently, and the diagnoses of all cases were kept anonymous until clonality results were assigned for every case. The rationale for performing these studies in a “blinded” manner was to minimize any experimental or interpretive bias. In addition, the pre-B-ALL cases, which were expected to yield IGH clones, served as an internal positive control.
Representative IGH PCR data for an ALK− ALCL are shown in Figure 2, and the results for all cases are summarized in Table 3. Overall, IGH clones were detected in only 6 of 74 (8%) PTCL cases, including 3 of 28 (11%) ALK+ ALCL cases, 3 of 35 (9%) ALK− ALCL cases, and 0 of 11 other PTCL cases. By contrast, IGH clones were detected in 23 of 24 (96%) pre-B-ALL cases, and the frequency of IGH clonality for pre-B-ALL cases was significantly higher than that of PTCL cases (P < 0.0001).
Figure 2.

Electropherograms of PCRs for IGH rearrangements. The x axis indicates DNA length in nucleotides with expected product sizes above the axis. The y axis indicates relative fluorescence units. Shown are amplification products from a case of ALK− ALCL. a, b, and c represent different multiplex PCRs that use primers that bind to the FR1 (a), FR2 (b), or FR3 (c) region of IGH. This case was interpreted as positive with clonal peaks (arrows) detected by FR1, FR2, and FR3 primers.
Table 3.
Clonality by PCR for IGH by Diagnosis
| Positive cases detected by specific primer sets [n (% cases studied)] |
|||||||
|---|---|---|---|---|---|---|---|
| Diagnosis | Cases studied (n) | Positive cases [n (%)] | FR1 | FR2 | FR3 | DH1-6 | DH7 |
| ALK+ ALCL | 28 | 3 (11) | 0 (0) | 1 (4) | 1 (4) | 2 (7) | 1 (4) |
| ALK− ALCL | 35 | 3 (9) | 1 (3) | 1 (3) | 2 (6) | 1 (3) | 0 (0) |
| SPTCL, HSTCL, T-LGL, and ETCL | 11 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Total PTCL | 74 | 6 (8) | 1 (1) | 2 (3) | 3 (4) | 3 (4) | 1 (1) |
| Pre-B-ALL | 24 | 23 (96) | 17 (71) | 23 (96) | 19 (79) | 6 (25) | 1 (4) |
FR primer sets amplified complete rearrangements inIGH between variable segment (VH), diversity segment (DH), and joining segment (JH). DH primer sets amplified incomplete rearrangements between DH and JH. ETCL, Enteropathy-type T-cell lymphoma; HSTCL, hepatosplenic T-cell lymphoma; SPTCL, subcutaneous panniculitis-like T-cell lymphoma.
In regard to the six ALCL cases with IGH clones, all six showed concurrent TRG clones, and five of six showed a CD3+, CD20− immunophenotype. The remaining case was negative for multiple B- and T-cell markers, including CD3 and CD20, and only CD30, CD43, and CD45 were expressed by the lymphoma cells. The combined morphologic and immunophenotypic findings were interpreted as supportive of the so-called null cell type of ALK− ALCL.
Also listed in Table 3 are the frequencies of clonal detection by specific primer sets in the PCR. The framework region (FR) primers, which amplify a complete VH-DH-JH rearrangement, detected more clones than the DH primers, which amplify an incomplete DH-JH rearrangement. For a given case, clonality was typically demonstrated by more than one primer set, which is summarized in Table 4. Of the 29 total IGH clones, 18 were detected simultaneously by the FR1, FR2, and FR3 primers, consistent with amplification of a complete rearrangement on one IGH allele by all three FR primers. In 5 of these 18 cases, the DH1-6 or DH7 primers also detected a clone, consistent with amplification of an incomplete rearrangement on the other IGH allele. Overall, 11 of the 29 total IGH clones were detected by the DH1-6 or DH7 primers, and in 8 of these 11 cases, clonality was also demonstrated by at least one FR primer.
Table 4.
Clonality by PCR for IGH by Specific Primer Set Combination
| Cases [n (% of cases studied)] |
|||
|---|---|---|---|
| Primer sets yielding IGH clones | ALK+ ALCL | ALK− ALCL | Pre-B-ALL |
| DH1-6 | 1 (4) | 1 (3) | 0 (0) |
| DH7 | 1 (4) | 0 (0) | 0 (0) |
| FR1, FR2, FR3 | 0 (0) | 1 (3) | 12 (48) |
| FR1, FR2, FR3, DH1-6 | 0 (0) | 0 (0) | 4 (16) |
| FR1, FR2, FR3, DH7 | 0 (0) | 0 (0) | 1 (4) |
| FR2 | 0 (0) | 0 (0) | 3 (12) |
| FR2, DH1-6 | 0 (0) | 0 (0) | 1 (4) |
| FR2, FR3 | 0 (0) | 0 (0) | 1 (4) |
| FR2, FR3, DH1-6 | 1 (4) | 0 (0) | 1 (4) |
| FR3 | 0 (0) | 1 (3) | 0 (4) |
| Negative | 25 (89) | 32 (91) | 1 (4) |
| Total | 28 (100) | 35 (100) | 24 (100) |
Interestingly, the frequency of detection by single versus multiple primer sets differed between ALCL and pre-B-ALL cases (Table 4). Of the 6 ALCL cases with IGH clones, 4 were detected by only one primer set, and in 3 of these 4 cases, the clone was detected exclusively by the DH1-6 or DH7 primers. Only 2 ALCL cases were detected by multiple primer sets, with one ALK+ case detected by the FR2, FR3, and DH1-6 primers and one ALK− case detected by the FR1, FR2, and FR3 primers (Figure 2, a, b, and c, respectively). By contrast, 20 of the 23 pre-B-ALL cases with IGH clones were detected by multiple primer sets, and only 3 were detected by a single primer set. None of the pre-B-ALL cases were detected solely by the DH1-6 or DH7 primers.
TRG Clonality Studies
We assayed for clonal rearrangements in the T-cell receptor γ-chain gene (TRG) using a multiplex PCR based on the same European BIOMED-2 collaborative study as described previously.17 The TRG assays were also performed under blinded conditions, and as an added measure, the TRG and IGH studies were performed independently. For example, assays for TRG were performed and interpreted without any knowledge of the IGH results, and vice versa.
Representative TRG PCR data for a pre-B-ALL and ALK− ALCL case are shown in Figure 3, a and b, respectively, and the results for all cases are summarized in Table 5. Overall, TRG clones were detected in 64 of 74 (86%) PTCL cases, including 24 of 28 (86%) ALK+ ALCL cases, 30 of 35 (86%) ALK− ALCL cases, and 10 of 11 (91%) other PTCL cases. For pre-B-ALL cases, TRG clones were detected in 16 of 24 (67%) cases, and the frequency of TRG clonality for pre-B-ALL cases was significantly less than that of PTCL cases (P < 0.03). Further statistical analyses yielded no significant correlation between TRG clonality and IGH clonality or EBV. However, a positive TRG clone correlated with older age (P = 0.045) and male sex (P = 0.009).
Figure 3.
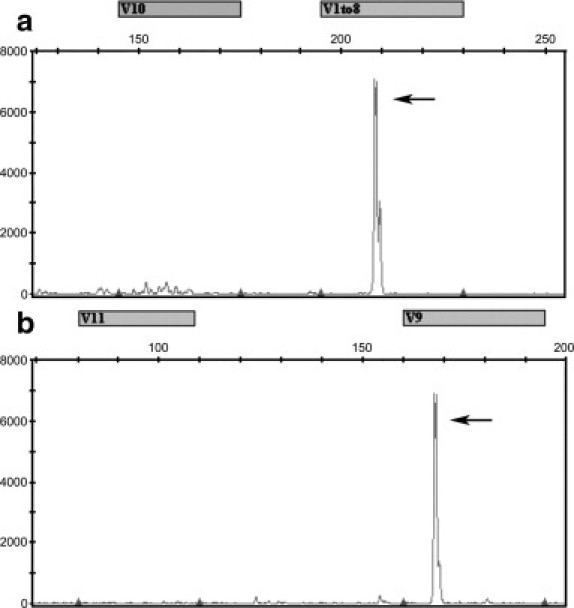
Electropherograms of PCRs for TRG rearrangements. The x axis indicates DNA length in nucleotides with expected product sizes above the axis. The y axis indicates relative fluorescence units. a: A case of pre-B-ALL with a clonal peak (arrow) detected in the multiplex PCR that uses primers to detect rearrangements involving Vγ10 or Vγ1-8 of TRG. b: An ALK− ALCL case with a clonal peak (arrow) detected in the multiplex PCR that uses primers to detect rearrangements involving Vγ11 or Vγ9 of TRG.
Table 5.
Clonality by PCR for TRG by Diagnosis
| Positive cases detected by specific primer sets [n (% of cases studied)] |
||||||
|---|---|---|---|---|---|---|
| Diagnosis | Cases studied (n) | Positive cases [n (%)] | Vγ1-8 | Vγ9 | Vγ10 | Vγ11 |
| ALK+ ALCL | 28 | 24 (86) | 14 (50) | 10 (36) | 10 (36) | 2 (7) |
| ALK− ALCL | 35 | 30 (86) | 19 (54) | 12 (34) | 12 (34) | 3 (9) |
| Other PTCL | 11 | 10 (91) | 8 (73) | 8 (73) | 5 (45) | 4 (36) |
| SPTCL | 3 | 3 (100) | 3 (100) | 3 (100) | 2 (67) | 1 (33) |
| HSTCL | 1 | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 0 (0) |
| T-LGL | 4 | 3 (75) | 2 (50) | 3 (75) | 1 (25) | 1 (25) |
| ETCL | 3 | 3 (100) | 2 (67) | 2 (67) | 2 (67) | 2 (67) |
| Total PTCL | 74 | 64 (86) | 41 (55) | 30 (41) | 27 (37) | 9 (12) |
| Pre-B-ALL | 24 | 16 (67) | 12 (50) | 6 (25) | 2 (8) | 2 (8) |
ETCL, Enteropathy-type T-cell lymphoma; HSTCL, hepatosplenic T-cell lymphoma; SPTCL, subcutaneous panniculitis-like T-cell lymphoma.
Also listed in Table 5 are the number of clones detected by specific primer sets in the multiplex PCR, which target rearrangements between specific variable genes Vγ and the joining segment Jγ of TRG. For a given case, clonality was typically demonstrated by more than one primer set, which is summarized in Table 6 for all cases, and in Table 7, which provides details on the six PTCL cases with concurrent TRG and IGH clones. Overall, rearrangements in Vγ1-8 were the most common, detected in 53% of cases, followed by rearrangements in Vγ9, Vγ10, and Vγ11, detected in 37, 30, and 11% of cases, respectively.
Table 6.
Clonality by PCR for TRG by Specific Primer Set Combination
| Cases [n (% of total PTCL or Pre-B-ALL cases studied)] |
||||||||
|---|---|---|---|---|---|---|---|---|
| Rearrangement(s) | ALK+ ALCL | ALK− ALCL | ETCL | HSTCL | SPTCL | T-LGL | Total PTCL | Pre-B-ALL |
| Vγ1-8 | 6 | 9 | 0 | 1 | 0 | 0 | 16 (22) | 7 (29) |
| Vγ1-8, Vγ10 | 4 | 2 | 0 | 0 | 0 | 0 | 6 (8) | 2 (8) |
| Vγ1-8, Vγ10, Vγ11 | 0 | 0 | 1* | 0 | 0 | 0 | 1 (1) | 0 (0) |
| Vγ1-8, Vγ11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 (0) | 1 (4) |
| Vγ1-8, Vγ9 | 3 | 4 | 1 | 0 | 1 | 1 | 10 (14) | 2 (8) |
| Vγ1-8, Vγ9, Vγ10 | 1* | 3* | 0 | 0 | 1* | 0 | 5 (7) | 0 (0) |
| Vγ1-8, Vγ9, Vγ10, Vγ11 | 0 | 0 | 0 | 0 | 1* | 1* | 2 (3) | 0 (0) |
| Vγ1-8, Vγ9, Vγ11 | 0 | 1* | 0 | 0 | 0 | 0 | 1 (1) | 0 (0) |
| Vγ10 | 2 | 5 | 0 | 0 | 0 | 0 | 7 (10) | 0 (0) |
| Vγ10, Vγ11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 (1) | 0 (0) |
| Vγ11 | 1 | 2 | 0 | 0 | 0 | 0 | 3 (4) | 0 (0) |
| Vγ9 | 4 | 2 | 0 | 0 | 0 | 1 | 7 (10) | 3 (13) |
| Vγ9, Vγ10 | 2 | 2 | 0 | 0 | 0 | 0 | 4 (5) | 0 (0) |
| Vγ9, Vγ10, Vγ11 | 0 | 0 | 1* | 0 | 0 | 0 | 1 (1) | 0 (0) |
| Vγ9, Vγ11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 (0) | 1 (4) |
| Negative | 4 | 5 | 0 | 0 | 0 | 1 | 10 (14) | 8 (33) |
Cases in which clonality was demonstrated by three or more primer sets. ETCL, Enteropathy-type T-cell lymphoma; HSTCL, hepatosplenic T-cell lymphoma; SPTCL, subcutaneous panniculitis-like T-cell lymphoma.
Table 7.
Results and Demographics for PTCL Cases with Simultaneous IGH and TRG Clones
| Diagnosis | Age (years) | Sex | Site | TRG clonality results | IGH clonality results |
|---|---|---|---|---|---|
| ALK+ ALCL | 26 | F | Posterior auricular lymph node | Vγ1-8 | FR2, FR3, DH1-6 |
| ALK+ ALCL | 26 | F | Axillary lymph node | Vγ1-8, Vγ10 | DH7 |
| ALK+ ALCL | 22 | F | Inguinal lymph node | Vγ1-8, Vγ9 | DH1-6 |
| ALK− ALCL | 54 | M | Chest wall mass | Vγ11 | DH1-6 |
| ALK− ALCL | 69 | F | Supraclavicular lymph node | Vγ1-8 | FR3 |
| ALK− ALCL | 54 | M | Axillary lymph node | Vγ10 | FR1, FR2, FR3 |
In regard to the pattern of variable gene usage, there were few differences between the diagnostic groups (Table 5). For example, the Vγ1-8 primers detected the most clones across all diagnostic groups, whereas the Vγ11 primers detected the least. However, the frequency of Vγ10 primer detection varied significantly by diagnosis, with Vγ10 rearrangements detected in 27 of 74 (36%) PTCL cases but only in 2 of 24 (8%) pre-B-ALL cases (P = 0.004). An additional finding that was unique to PTCL was the detection of TRG clonality by more than two primer sets (Table 6). Of the 64 PTCL cases with TRG clones, 8 cases were detected by three primer sets, and 2 cases were detected by all four primer sets. By contrast, none of the pre-B-ALL cases were detected by more than two primer sets.
EBV Studies
To investigate the role of EBV in IGH clonality, we performed in situ hybridization for EBV-encoded small RNA on 49 of the 74 (66%) PTCL cases, which were selected based on the availability of remaining tissue. Representative in situ hybridizations for an ALK+ ALCL and ALK− ALCL are shown in Figure 1, and the results for all cases are summarized in Table 8. A positive result, which was defined as more than three EBV-labeled cells per section, was observed in 15 of 49 (31%) cases, including 6 of 21 (29%) ALK+ ALCL cases, 8 of 24 (33%) ALK− ALCL cases, and 1 of 4 (25%) other PTCL cases. For most EBV-positive cases, labeled nuclei were scattered and rare, which is unlikely to represent EBV within the lymphoma cells (Figure 1h). However, one ALK+ ALCL case and two ALK− ALCL cases showed unanimous EBV labeling of large cells in a pattern consistent with an EBV-positive lymphoma (Figure 1d).
Table 8.
EBV Studies by in Situ Hybridization for EBER
| EBV-labeled cells per medium-power field (n) |
|||||||
|---|---|---|---|---|---|---|---|
| Diagnosis | Cases studied (n) | Positive cases [n (%)] | 0 | Less than 1 | 1 to 10 | Greater than 10 to 100 | Greater than 100 |
| ALK+ ALCL | |||||||
| With IGH clone | 3 | 0 (0) | 3 | 0 | 0 | 0 | 0 |
| Without IGH clone | 18 | 6 (33) | 12 | 4 | 1 | 0 | 1* |
| Total | 21 | 6 (29) | 15 | 4 | 1 | 0 | 1 |
| ALK− ALCL | |||||||
| With IGH clone | 2 | 0 (0) | 2 | 0 | 0 | 0 | 0 |
| Without IGH clone | 22 | 8 (36) | 14 | 4 | 1 | 1 | 2* |
| Total | 24 | 8 (33) | 16 | 4 | 1 | 1 | 2 |
| SPTCL, HSTCL, T-LGL, and ETCL | 4 | 1 (25) | 3 | 0 | 1 | 0 | 0 |
| Total PTCL | 49 | 15 (31) | 34 | 8 | 3 | 1 | 3 |
One SPTCL, one HSTCL, one T-LGL, and one ETCL were tested for EBV, and only the T-LGL case, which did not have an IGH clone, showed EBV-labeled cells. EBER, EBV-encoded small RNA; ETCL, enteropathy-type T-cell lymphoma; HSTCL, hepatosplenic T-cell lymphoma; SPTCL, subcutaneous panniculitis-like T-cell lymphoma.
Cases in which EBV-labeled cells corresponded to the neoplastic cells.
Despite the detection of EBV in 15 of 49 (31%) PTCL cases, the presence of the virus did not correlate with IGH clonality. Of the 6 PTCL cases with IGH clones, which included 3 ALK+ ALCL cases and 3 ALK− ALCL cases, 5 cases were studied by in situ hybridization, and all were negative for EBV. Likewise, of the 15 PTCL cases that contained EBV-positive cells, all were negative for IGH clonality. Taken together, these findings suggest that EBV does not contribute to IGH clonality, and further analysis yielded no significant correlation between EBV and age, gender, ALK expression, or TRG clonality.
Discussion
Our main goal was to determine whether IGH clones occur in cases of PTCL that lack associated B-cell proliferations. To address this question, we performed clonality studies on a series of ALCL and other specific PTCL subtypes in which B-cell proliferations have not been reported. In addition, we concurrently assayed 24 cases of pre-B-ALL, which is a neoplasm known to commonly show both IGH and TRG rearrangements, and we performed these studies in a blinded manner. Our intent was to minimize any experimental or interpretive bias by eliminating a priori knowledge of the diagnosis. For example, if a case could be identified as a PTCL, interpretation of the data may favor the expected results, namely a positive TRG clone and a negative IGH clone.
Under these blinded conditions, we detected IGH clones in only 6 of 74 (8%) PTCL cases, consistent with previous reports on large series of PTCL (9 to 16%).2,3,4,5,6 By contrast, we detected IGH clones in 23 of 24 (96%) pre-B-ALL cases, and similarly high rates of detection have been reported for pre-B-ALL.18,19,20 In regard to PTCL, the frequency of IGH clonality differed significantly from our previous report, in which we detected IGH clones in 33% of AILT cases and 35% of PTCL-U cases (P < 0.004).7 We attribute this difference to the distinct composition of cases, with 20% of previous cases showing demonstrable B-cell proliferations. Collectively, our findings suggest that IGH clonality occurs infrequently (<10%) in most PTCL subtypes, with the exceptions of AILT and PTCL-U. This finding was also observed in a recent BIOMED-2 study, in which IGH clones were detected in 10% of PTCL cases, but the majority of cases with IGH clones were classified as AILT or PTCL-U.21
Regarding the etiology of IGH clones, it remains our hypothesis that B-cell proliferations are largely responsible for this finding, at least in AILT and PTCL-U. However, IGH clonality can also occur in other PTCL subtypes, even in the absence of an associated B-cell proliferation or EBV. We investigated other possible etiologies, including pseudoclonality, which refers to PCR amplification of a single cell that results in a nonreproducible product.22,23 However, repeat testing of all positive IGH clones yielded reproducible products, making pseudoclonality unlikely.
Other explanations include lineage infidelity and solitary germinal centers. Proof of lineage infidelity requires single cell analysis, which others have achieved by performing PCR on single cells isolated by microdissection.6,24 However, the technical complexity of these experiments limits the number of cases that can be studied, and although evaluation for lineage infidelity is beyond the scope of this report, it remains a possible cause of IGH clonality. Several studies have shown that a single germinal center isolated by microdissection can yield an IGH clone by PCR.25,26 Therefore, we re-examined all of our PTCL cases for germinal centers. However, none of the cases with IGH clones contained solitary or multiple germinal centers, and there was no correlation between germinal centers and IGH clonality.
It should be noted that three of the six IGH clones in PTCL were detected exclusively by the DH1-6 or DH7 primers. There are several explanations for this finding, which was also observed in a recent BIOMED-2 study on PTCL.21 Because B-cell maturation requires complete VH-DH-JH rearrangement of at least one IGH allele,27 a clone detected solely by DH primers must be the result of DH-primed amplification of an incomplete DH-JH rearrangement on one allele and the inability of the FR primers to amplify a complete VH-DH-JH rearrangement on the other allele. Lack of amplification may be caused by translocation or mutation of the FR primer binding sites, and among mature B-cell lymphomas, isolated DH primer detection is most common in those that have undergone somatic hypermutation.28 If these events also occur in PTCL, this would suggest that IGH clonality is caused by germinal center or postgerminal center B cells. Another intriguing possibility is EBV-mediated survival and clonal expansion of a B cell that has acquired a large deletion or nonsense mutation in its functional IGH allele. This phenomenon, also known as EBV rescue of a “crippled” B cell, occurs in classical Hodgkin lymphoma29 and posttransplant lymphoproliferative disorders.30,31,32 However, in our study, all of the cases with IGH clones lacked EBV. An entirely alternate explanation for isolated DH primer detection involves lineage infidelity, with a sole DH-JH rearrangement on one IGH allele. In this context, a complete rearrangement on the other allele is not a developmental requirement because the events occur in a neoplastic T cell rather than a B cell.
The assays for TRG clonality also provided results that were consistent with the reported literature. For PTCL cases, TRG clones were detected in 64 of 74 (86%) cases, which included 24 of 28 (86%) ALK+ ALCL cases, 30 of 35 (86%) ALK− ALCL cases, and 10 of 11 (91%) other PTCL cases (Table 4). These frequencies are similar to those of previous reports2,33,34 and of recent studies using the BIOMED-2 TRG PCR.21,35,36 Although few reports include large numbers of ALCL, Brüggemann et al21 detected TRG clones in 24 of 34 (71%) ALK+ cases and 8 of 9 (89%) ALK− cases in a BIOMED-2 study. For the pre-B-ALL cases, we detected TRG clones in 16 of 24 (67%) cases, and this frequency is similar to comparable studies on pre-B-ALL.18,20,37
We also evaluated TRG variable gene usage by analyzing which specific primer sets yielded a clonal product (Table 5), and the frequencies of specific variable gene rearrangements were similar to our previous study.7 Rearrangements in Vγ1-8 were most common, occurring alone or in combination with other variable genes in 53 of 80 (66%) TRG clones (Table 5). By corollary, 27 of 80 (34%) TRG clones would have been missed if the Vγ1-8 primers were used alone. In general, the pattern of variable gene usage was similar across diagnostic groups, with the exception of Vγ10 rearrangements, which were frequent in PTCL, but nearly absent in pre-B-ALL. We previously reported that Vγ10 rearrangements are relatively common in AILT and PTCL-U,7 and others have shown Vγ10 rearrangements are virtually absent in pre-B-ALL.38
For most TRG clones, one or two rearrangements were detected, consistent with one or two rearranged TRG alleles, respectively (Table 5). However, of the 80 TRG clones, 8 showed three variable gene rearrangements, and 2 showed four variable gene rearrangements (Table 6). Interestingly, all of the cases with more than two rearrangements were PTCL cases. Because diploid cells have two TRG alleles, explanations for this finding include a second minor clone or oligoclones, genome instability, and aneuploidy. Regardless of the etiology, we and others have previously shown that more than two TRG rearrangements are detected by PCR in a minority (∼10%) of histologically proven PTCL.7,21,39 In this study, more than two rearrangements were detected in 10 of 74 (14%) PTCL cases, accounting for 10 of the 64 (16%) TRG clones in PTCL.
Taken together, the TRG and IGH assays provided correct lineage assignment for the majority of PTCL cases; however, simultaneous TRG and IGH clones were detected in 6 of 74 (8%) cases, resulting in ambiguous lineage. Although an IGH clone was not detected without a concurrent TRG clone, the potential for incorrect lineage assignment is readily apparent. Therefore, for cases of suspected PTCL, we recommend using DNA-based clonality studies primarily to provide support for a neoplastic process, and any unexpected results should be evaluated within the context of the histopathologic findings. Although an IGH clone may suggest a diagnosis of AILT or PTCL-U, IGH clonality can also occur in other PTCL subtypes, and this finding should not negate the diagnosis established by the combination of morphologic, immunophenotypic, and clinical findings.
Acknowledgements
We thank Lisa Ma for excellent technical assistance and Anet James for assistance with figure composition.
Footnotes
Supported by the Stanford Department of Pathology.
References
- 1.Jaffe ES, Harris NL, Stein H, Vardiman JW. In: Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. Kleihues P, Sobin LH, editors. IARC Press; Lyon, France: 2001. pp. 111–114. and 189–235. [Google Scholar]
- 2.Diss TC, Watts M, Pan LX, Burke M, Linch D, Isaacson PG. The polymerase chain reaction in the demonstration of monoclonality in T cell lymphomas. J Clin Pathol. 1995;48:1045–1050. doi: 10.1136/jcp.48.11.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia MJ, Martinez-Delgado B, Granizo JJ, Benitez J, Rivas C. IgH. TCR-gamma, and TCR-beta gene rearrangement in 80 B- and T-cell non-Hodgkin's lymphomas: study of the association between proliferation and the so-called “aberrant” patterns. Diagn Mol Pathol. 2001;10:69–77. doi: 10.1097/00019606-200106000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Noorali S, Pervez S, Moatter T, Soomro IN, Kazmi SU, Nasir MI, Smith JL. Characterization of T-cell non-Hodgkin's lymphoma and its association with Epstein-Barr virus in Pakistani patients. Leuk Lymphoma. 2003;44:807–813. doi: 10.1080/1042819031000067747. [DOI] [PubMed] [Google Scholar]
- 5.Thériault C, Galoin S, Valmary S, Selves J, Lamant L, Roda D, Rigal-Huguet F, Brousset P, Delsol G, Al Saati T. PCR analysis of immunoglobulin heavy chain (IgH) and TcR-gamma chain gene rearrangements in the diagnosis of lymphoproliferative disorders: results of a study of 525 cases. Mod Pathol. 2000;13:1269–1279. doi: 10.1038/modpathol.3880232. [DOI] [PubMed] [Google Scholar]
- 6.Vergier B, Dubus P, Kutschmar A, Parrens M, Ferrer J, de Mascarel A, Merlio JP. Combined analysis of T cell receptor gamma and immunoglobulin heavy chain gene rearrangements at the single-cell level in lymphomas with dual genotype. J Pathol. 2002;198:171–180. doi: 10.1002/path.1192. [DOI] [PubMed] [Google Scholar]
- 7.Tan BT, Warnke RA, Arber DA. The frequency of B- and T-cell gene rearrangements and Epstein-Barr virus in T-cell lymphomas: a comparison between angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma, unspecified with and without associated B-cell proliferations. J Mol Diagn. 2006;8:466–475. doi: 10.2353/jmoldx.2006.060016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higgins JP, van de Rijn M, Jones CD, Zehnder JL, Warnke RA. Peripheral T-cell lymphoma complicated by a proliferation of large B cells. Am J Clin Pathol. 2000;114:236–247. doi: 10.1309/72CM-KAXF-66DE-4XVA. [DOI] [PubMed] [Google Scholar]
- 9.Lome-Maldonado C, Canioni D, Hermine O, Delabesse E, Damotte D, Raffoux E, Gaulard P, Macintyre E, Brousse N. Angio-immunoblastic T cell lymphoma (AILD-TL) rich in large B cells and associated with Epstein-Barr virus infection: a different subtype of AILD-TL? Leukemia. 2002;16:2134–2141. doi: 10.1038/sj.leu.2402642. [DOI] [PubMed] [Google Scholar]
- 10.Xu Y, McKenna RW, Hoang MP, Collins RH, Kroft SH. Composite angioimmunoblastic T-cell lymphoma and diffuse large B-cell lymphoma: a case report and review of the literature. Am J Clin Pathol. 2002;118:848–854. doi: 10.1309/VD2D-98ME-MB3F-WH34. [DOI] [PubMed] [Google Scholar]
- 11.Zettl A, Lee SS, Rudiger T, Starostik P, Marino M, Kirchner T, Ott M, Muller-Hermelink HK, Ott G. Epstein-Barr virus-associated B-cell lymphoproliferative disorders in angloimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma, unspecified. Am J Clin Pathol. 2002;117:368–379. doi: 10.1309/6UTX-GVC0-12ND-JJEU. [DOI] [PubMed] [Google Scholar]
- 12.van Dongen JJ, Wolvers-Tettero IL. Analysis of immunoglobulin and T cell receptor genes. Part II: possibilities and limitations in the diagnosis and management of lymphoproliferative diseases and related disorders. Clin Chim Acta. 1991;198:93–174. doi: 10.1016/0009-8981(91)90247-a. [DOI] [PubMed] [Google Scholar]
- 13.Smith LJ, Curtis JE, Messner HA, Senn JS, Furthmayr H, McCulloch EA. Lineage infidelity in acute leukemia. Blood. 1983;61:1138–1145. [PubMed] [Google Scholar]
- 14.Greaves MF, Chan LC, Furley AJ, Watt SM, Molgaard HV. Lineage promiscuity in hemopoietic differentiation and leukemia. Blood. 1986;67:1–11. [PubMed] [Google Scholar]
- 15.McCulloch EA. Lineage infidelity or lineage promiscuity? Leukemia. 1987;1:235. [PubMed] [Google Scholar]
- 16.Bindl JM, Warnke RA. Advantages of detecting monoclonal antibody binding to tissue sections with biotin and avidin reagents in Coplin jars. Am J Clin Pathol. 1986;85:490–493. doi: 10.1093/ajcp/85.4.490. [DOI] [PubMed] [Google Scholar]
- 17.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, van Krieken JH, Droese J, Gonzalez D, Bastard C, White HE, Spaargaren M, Gonzalez M, Parreira A, Smith JL, Morgan GJ, Kneba M, Macintyre EA. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98–3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 18.Brumpt C, Delabesse E, Beldjord K, Davi F, Cayuela JM, Millien C, Villarese P, Quartier P, Buzyn A, Valensi F, Macintyre E. The incidence of clonal T-cell receptor rearrangements in B-cell precursor acute lymphoblastic leukemia varies with age and genotype. Blood. 2000;96:2254–2261. [PubMed] [Google Scholar]
- 19.Szczepański T, Langerak AW, Wolvers-Tettero IL, Ossenkoppele GJ, Verhoef G, Stul M, Petersen EJ, de Bruijn MA, van't Veer MB, van Dongen JJ. Immunoglobulin and T cell receptor gene rearrangement patterns in acute lymphoblastic leukemia are less mature in adults than in children: implications for selection of PCR targets for detection of minimal residual disease. Leukemia. 1998;12:1081–1088. doi: 10.1038/sj.leu.2401071. [DOI] [PubMed] [Google Scholar]
- 20.van der Velden VH, Szczepanski T, Wijkhuijs JM, Hart PG, Hoogeveen PG, Hop WC, van Wering ER, van Dongen JJ. Age-related patterns of immunoglobulin and T-cell receptor gene rearrangements in precursor-B-ALL: implications for detection of minimal residual disease. Leukemia. 2003;17:1834–1844. doi: 10.1038/sj.leu.2403038. [DOI] [PubMed] [Google Scholar]
- 21.Brüggemann M, White H, Gaulard P, Garcia-Sanz R, Gameiro P, Oeschger S, Jasani B, Ott M, Delsol G, Orfao A, Tiemann M, Herbst H, Langerak AW, Spaargaren M, Moreau E, Groenen PJ, Sambade C, Foroni L, Carter GI, Hummel M, Bastard C, Davi F, Delfau-Larue MH, Kneba M, van Dongen JJ, Beldjord K, Molina TJ. Powerful strategy for polymerase chain reaction-based clonality assessment in T-cell malignancies: Report of the BIOMED-2 Concerted Action BHM4 CT98-3936. Leukemia. 2007;21:215–221. doi: 10.1038/sj.leu.2404481. [DOI] [PubMed] [Google Scholar]
- 22.Elenitoba-Johnson KS, Bohling SD, Mitchell RS, Brown MS, Robetorye RS. PCR analysis of the immunoglobulin heavy chain gene in polyclonal processes can yield pseudoclonal bands as an artifact of low B cell number. J Mol Diagn. 2000;2:92–96. doi: 10.1016/S1525-1578(10)60622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoeve MA, Krol AD, Philippo K, Derksen PW, Veenendaal RA, Schuuring E, Kluin PM, van Krieken JH. Limitations of clonality analysis of B cell proliferations using CDR3 polymerase chain reaction. Mol Pathol. 2000;53:194–200. doi: 10.1136/mp.53.4.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallardo F, Pujol RM, Bellosillo B, Ferrer D, Garcia M, Barranco C, Planaguma M, Serrano S. Primary cutaneous B-cell lymphoma (marginal zone) with prominent T-cell component and aberrant dual (T and B) genotype: diagnostic usefulness of laser-capture microdissection. Br J Dermatol. 2006;154:162–166. doi: 10.1111/j.1365-2133.2005.06947.x. [DOI] [PubMed] [Google Scholar]
- 25.Iijima T, Inadome Y, Noguchi M. Clonal proliferation of B lymphocytes in the germinal centers of human reactive lymph nodes: possibility of overdiagnosis of B cell clonal proliferation. Diagn Mol Pathol. 2000;9:132–136. doi: 10.1097/00019606-200009000-00002. [DOI] [PubMed] [Google Scholar]
- 26.Zhou XG, Sandvej K, Gregersen N, Hamilton-Dutoit SJ. Detection of clonal B cells in microdissected reactive lymphoproliferations: possible diagnostic pitfalls in PCR analysis of immunoglobulin heavy chain gene rearrangement. Mol Pathol. 1999;52:104–110. doi: 10.1136/mp.52.2.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alt FW, Yancopoulos GD, Blackwell TK, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans PA, Pott C, Groenen PJ, Salles G, Davi F, Berger F, Garcia JF, van Krieken JH, Pals S, Kluin P, Schuuring E, Spaargaren M, Boone E, Gonzalez D, Martinez B, Villuendas R, Gameiro P, Diss TC, Mills K, Morgan GJ, Carter GI, Milner BJ, Pearson D, Hummel M, Jung W, Ott M, Canioni D, Beldjord K, Bastard C, Delfau-Larue MH, van Dongen JJ, Molina TJ, Cabecadas J. Significantly improved PCR-based clonality testing in B-cell malignancies by use of multiple immunoglobulin gene targets: Report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia. 2007;21:207–214. doi: 10.1038/sj.leu.2404479. [DOI] [PubMed] [Google Scholar]
- 29.Kanzler H, Kuppers R, Hansmann ML, Rajewsky K. Hodgkin and Reed-Sternberg cells in Hodgkin's disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med. 1996;184:1495–1505. doi: 10.1084/jem.184.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bräuninger A, Spieker T, Mottok A, Baur AS, Kuppers R, Hansmann ML. Epstein-Barr virus (EBV)-positive lymphoproliferations in post-transplant patients show immunoglobulin V gene mutation patterns suggesting interference of EBV with normal B cell differentiation processes. Eur J Immunol. 2003;33:1593–1602. doi: 10.1002/eji.200323765. [DOI] [PubMed] [Google Scholar]
- 31.Timms JM, Bell A, Flavell JR, Murray PG, Rickinson AB, Traverse-Glehen A, Berger F, Delecluse HJ. Target cells of Epstein-Barr-virus (EBV)-positive post-transplant lymphoproliferative disease: similarities to EBV-positive Hodgkin's lymphoma. Lancet. 2003;361:217–223. doi: 10.1016/S0140-6736(03)12271-4. [DOI] [PubMed] [Google Scholar]
- 32.Capello D, Cerri M, Muti G, Berra E, Oreste P, Deambrogi C, Rossi D, Dotti G, Conconi A, Vigano M, Magrini U, Ippoliti G, Morra E, Gloghini A, Rambaldi A, Paulli M, Carbone A, Gaidano G. Molecular histogenesis of posttransplantation lymphoproliferative disorders. Blood. 2003;102:3775–3785. doi: 10.1182/blood-2003-05-1683. [DOI] [PubMed] [Google Scholar]
- 33.Arber DA, Braziel RM, Bagg A, Bijwaard KE. Evaluation of T cell receptor testing in lymphoid neoplasms: results of a multicenter study of 29 extracted DNA and paraffin-embedded samples. J Mol Diagn. 2001;3:133–140. doi: 10.1016/S1525-1578(10)60664-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krafft AE, Taubenberger JK, Sheng ZM, Bijwaard KE, Abbondanzo SL, Aguilera NS, Lichy JH. Enhanced sensitivity with a novel TCRgamma PCR assay for clonality studies in 569 formalin-fixed, paraffin-embedded (FFPE) cases. Mol Diagn. 1999;4:119–133. doi: 10.1016/s1084-8592(99)80036-8. [DOI] [PubMed] [Google Scholar]
- 35.Liu H, Bench AJ, Bacon CM, Payne K, Huang Y, Scott MA, Erber WN, Grant JW, Du MQ. A practical strategy for the routine use of BIOMED-2 PCR assays for detection of B- and T-cell clonality in diagnostic haematopathology. Br J Haematol. 2007;138:31–43. doi: 10.1111/j.1365-2141.2007.06618.x. [DOI] [PubMed] [Google Scholar]
- 36.Sandberg Y, van Gastel-Mol EJ, Verhaaf B, Lam KH, van Dongen JJ, Langerak AW. BIOMED-2 multiplex immunoglobulin/T-cell receptor polymerase chain reaction protocols can reliably replace southern blot analysis in routine clonality diagnostics. J Mol Diagn. 2005;7:495–503. doi: 10.1016/S1525-1578(10)60580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szczepański T, Willemse MJ, Brinkhof B, van Wering ER, van der Burg M, van Dongen JJ. Comparative analysis of Ig and TCR gene rearrangements at diagnosis and at relapse of childhood precursor-B-ALL provides improved strategies for selection of stable PCR targets for monitoring of minimal residual disease. Blood. 2002;99:2315–2323. doi: 10.1182/blood.v99.7.2315. [DOI] [PubMed] [Google Scholar]
- 38.Szczepański T, Beishuizen A, Pongers-Willemse MJ, Hahlen K, Van Wering ER, Wijkhuijs AJ, Tibbe GJ, De Bruijn MA, Van Dongen JJ. Cross-lineage T cell receptor gene rearrangements occur in more than ninety percent of childhood precursor-B acute lymphoblastic leukemias: alternative PCR targets for detection of minimal residual disease. Leukemia. 1999;13:196–205. doi: 10.1038/sj.leu.2401277. [DOI] [PubMed] [Google Scholar]
- 39.Lawnicki LC, Rubocki RJ, Chan WC, Lytle DM, Greiner TC. The distribution of gene segments in T-cell receptor gamma gene rearrangements demonstrates the need for multiple primer sets. J Mol Diagn. 2003;5:82–87. doi: 10.1016/s1525-1578(10)60456-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


