Abstract
Small protein B (SmpB) is a requisite component of the transfer messenger RNA (tmRNA)-mediated bacterial translational quality control system known as trans-translation. The initial binding of tmRNA and its subsequent accommodation into the ribosomal A-site are activities intimately linked to SmpB protein function. From a mechanistic perspective, two key unanswered questions that require further investigation are: 1) what constitutes a stalled ribosome recognition complex and 2) does SmpB pre-bind ribosomes to recruit tmRNA. We have assessed, both in vivo and in vitro, the nature and stability of free SmpB interactions with stalled ribosomes and examined whether these interactions are functionally relevant. We present evidence to demonstrate that interaction of free SmpB with ribosomes is salt sensitive and significantly more labile than interaction of the SmpB-tmRNA complex with ribosomes. Upon dissociation of 70S ribosomes SmpB partitions primarily with tmRNA rather than ribosomal subunits. This finding is consistent with biochemical and structural data demonstrating that tmRNA is the high-affinity binding partner of SmpB. Moreover, we show that under normal physiological conditions roughly similar numbers of SmpB and tmRNA molecules are present in cells. Our investigations also reveal that upon induction of a nonstop mRNA, SmpB is enriched in stalled ribosome fractions only in the presence of tmRNA. Based on these findings, we conclude that SmpB does not pre-bind stalled ribosome and that functional SmpB-stalled ribosome interactions require tmRNA. We propose that a 1:1:1 complex of SmpB•tmRNA•EF-Tu (GTP) recognizes and binds a stalled ribosome to initiate trans-translation.
INTRODUCTION
Trans-translation is a bacteria-specific translational quality control mechanism that rescues ribosomes stalled on defective mRNAs, directs the degradation of aberrant protein products, and facilitates the decay of incomplete or damaged mRNAs (1-10). The two unique components of this system are tmRNA (also known as SsrA RNA, or 10Sa RNA) and its required protein partner SmpB. tmRNA is a bi-functional stable RNA molecule with both tRNA- and mRNA-like features and activities. SmpB is a small, basic RNA binding protein that is essential for all known physiological activities of tmRNA (4,11-13). Briefly, trans-translation consists of stalled ribosome recognition and assembly of an SmpB•tmRNA•EF-Tu (GTP)•stalled ribosome complex. The SmpB•tmRNA•EF-Tu (GTP) quaternary complex then behaves in a manner analogous to a tRNA•EF-Tu (GTP) ternary complex. The tRNA-like domain (TLD) of tmRNA enters the ribosomal A-site where the growing peptide is transferred onto it. The ribosomal reading frame then shifts to translate the mRNA sequence contained in the mRNA-like domain of tmRNA. This leads to the addition of a C-terminal proteolysis tag to the newly synthesized polypeptide, which facilitates its degradation by cellular proteases (2,5,14,15). In addition, the SmpB•tmRNA system facilitates the decay of the causative non-stop mRNA (6,8,16). The specific roles of SmpB protein in the trans-translation process remain a matter of some debate. It is clear that SmpB is required for promoting association of tmRNA with stalled ribosomes (4,11,17-22). It has been demonstrated that the unstructured C-terminal tail of SmpB plays a key role in promoting the initial peptide bond formation event in trans-translation (20,23).
SmpB has been shown by numerous studies to bind specifically to tmRNA, however the stoichiometry of this complex is unclear (11,17-22,24). Optical biosensor and melting curve analysis combined with mutational studies suggests that one copy of SmpB binds to a single binding site on the D-arm of tmRNA in Thermus thermophilus (25). These results are mirrored by two co-crystal structure models of SmpB in complex with the TLD of tmRNA from Thermus thermophilus and Aquifex aeolicus (26,27). These models depict a single SmpB binding site on the D-arm of the tmRNA TLD. In contrast, footprinting studies predict multiple SmpB binding sites on tmRNA (28,29), with up to three SmpB molecules binding per tmRNA (29). Additionally, two molecules of free SmpB are suggested to bind 70S ribosomes (both, normal and stalled) (18). Two differing cryo-EM models of the pre-accommodated complex also complicate the issue of the stoichiometry of the SmpB•tmRNA•ribosome complex. The first cryo-EM model depicts a single SmpB molecule bound to the D-loop region of the tmRNA TLD (30). A second model depicts two SmpB molecules in the same pre-accommodated complex (31).
An additional point of uncertainty lies in the order of events in the formation of the SmpB•tmRNA•stalled ribosome complex. One model predicts that a pre-formed SmpB•tmRNA complex recognizes and binds stalled ribosomes to initiate trans-translation. This model is supported by numerous studies that suggest that tmRNA is the specific high affinity RNA binding partner of tmRNA (11,17-22,24). However, based on in vitro SmpB/ribosome binding studies and on in vivo interactions in the absence of tmRNA, it has been proposed that free SmpB might pre-bind the ribosome to recruit tmRNA to stalled ribosomes (32).
To gain deeper insight into SmpB interactions, we set out to study the assembly of the SmpB•tmRNA•stalled-ribosome complex in vivo, under normal log-phase growth conditions, and with canonical SmpB and tmRNA expression levels. We found that under certain non-physiological conditions free SmpB can interact with ribosomes, however this interaction is labile and salt sensitive. We show that ribosome-bound SmpB co-localizes with tmRNA upon dissociation of ribosomal subunits. In addition, we report that SmpB is enriched in stalled ribosomes only in the presence of tmRNA. Taken together, our investigations lead us to conclude that stalled ribosomes are recognized and bound by a pre-formed SmpB•tmRNA complex (along with EF-Tu and GTP) to initiate trans-translation.
MATERIALS AND METHODS
Buffers, Strains, and Plasmids
Buffer A contained 50mM Tris (pH 7.5), 70mM NH4Cl, 30mM KCl, 10mM MgCl2,and 20mM DTT. Buffer B contained 50mM Tris (pH 7.5), 10mM MgCl2, 20mM DTT, and either 100, 200, or 300mM NH4Cl. Buffer C contained 40mM Tris (pH 7.5), 10mM MgCl2, 2mM β-mercaptoethanol, 10% sucrose, and either 100, 200 or 300mM NH4Cl. Buffer D contained 40mM Tris (pH 7.5), 1mM MgCl2, 2mM β-mercaptoethanol, and either 100 or 300mM NH4Cl. Buffer E contained 25mM Tris (pH7.5), 100mM NH4Cl, 30mM KCl, 8mM MgCl2 and 1mM DTT. Buffer F contained 50mM Tris (pH 7.5), 20mM MgCl2 , 2mM β-mercaptoethanol, 10mM Imidazol, and either 100 or 300mM NH4Cl. Buffer G contained 50mM Tris (pH 7.5), 100mM NH4Cl, 10mM MgCl2, 2mM β-mercaptoethanol, and 8M Urea.
The strain W3110 ssrA::CAT has been described previously (20). pλ-cI-N-4-AGG-Flag has been described previously and is referred to in the text as pλ- cI-N AGG (8). Pλ-cI-NS was prepared by sequential rounds of PCR mutagenesis to convert the rare arginine codons (AGG) to abundant codons (CGG) and to introduce a trpA-operon transcriptional terminator near the 3’ end. The plasmid pET28BA was described previously (20). The plasmid ptrnfM for expression of initiator tRNA was a kind gift from Dr. Uttam RajBhandary. The plasmid pETrpS12 for expression of ribosomal protein S12 was a kind gift from Dr. Harry Noller.
Protein and RNA Purification
Protein translation factors IF-1, IF-2, IF-3, EF-Tu, EF-Ts, AlaRS, MetRS, and MTF as well as SmpB were purified using techniques similar to those described for SmpB purification in Sundermeier et al (20). Proteins were purified by batch affinity chromatography using Ni2+-NTA agarose (Qiagen, Valencia,CA) followed by FPLC ion exchange using either a MonoS (HR 10/10) column or a MonoQ (HR 10/10) column (GE Healthcare, Piscataway, NJ) eluted with a linear salt gradient. Ribosomal protein S12 was purified as described (33).
tmRNA was purified from cells as follows. 6L of BL21(DE3)/pLyseS/pET28BA cells were grown to OD600 around 0.5-0.7. SmpB and tmRNA were simultaneously induced for 2 hours with 10μM IPTG. The SmpB•tmRNA complex was purified by affinity chromatography using Ni2+-NTA agarose resin (Qiagen, Valencia,CA). tmRNA was separated from SmpB via RNA extraction using TriReagent LS (MRC, Cincinnatti,OH). tmRNA was further purified via FPLC anion exchange using a MonoQ (HR 10/10) column (GE Healthcare, Piscataway, NJ).
A tRNAfMet enriched tRNA pool was purified by isolating total tRNA from cells expressing tRNAfMet from the plasmid ptrnfM. tRNA purification was performed as described (34). Preparative purification of charged fMet-tRNAfMet was performed as follows. Charging and formylation reactions were performed in buffer E. Reactions (10mL) included 20μM tRNAfMet, 200μM L-methionine, 150μg N10-formyl-tetrahydrofolate, 2μM Met-RS, 5μM MTF, and 3mM ATP, incubated at 37°C for 20min. RNA was precipitated with isopropanol, washed with ethanol, and extracted with TriReagent LS. The charged and formylated fraction was separated from uncharged tRNA via FPLC hydrophobic interactions chromatography using a Hi-Trap Phenyl Sepharose HP column (GE Healthcare, Piscataway, NJ). This product was further purified by FPLC anion exchange using a MonoQ (HR 10/10) column (GE Healthcare, Piscataway, NJ).
Ribosome Association Assays
The in vivo ribosome association assay procedure has been described previously, with the exception that different concentrations of NH4Cl were used as described in the text (20). For separation of ribosomal subunits, 70S ribosomes were purified as described (17,20). Ribosomes were then resuspended in buffer D and loaded onto a 10-40% linear sucrose gradient in buffer D. Gradients were subjected to centrifugation at 25,000 RPM for 16hr in SW28 rotor (Beckman Coulter, Fullerton,CA). Fractions were subjected to western blotting using a previously described SmpB polyclonal antibody (17,20), and Northern blot analysis using a full length tmRNA biotinylated dsDNA probe.
For in vitro SmpB ribosome binding assays, we first assembled fM pre-trans-translation complexes in buffer A. The reaction mix contained 5μM fMet-tRNAfMet, 3μM 70S ribosomes, 3μM IF-1, 3μM IF-2, 3μM IF-3, 1mM GTP and 8μM fM mRNA. Reactions were incubated at 37°C for 45 minutes, and then used directly for ribosome binding assays. tmRNA charging and SmpB ribosome binding reactions were performed in one step. This reaction was performed in buffer B and contained 750nM pre-trans-translation complexes, 750nM SmpB, 750nM EF-Tu, 750nM EF-Ts, 1μM Ala-RS, 2mM ATP, 250μM GTP, 1mM L-alanine, 750nM tmRNA, and 75μM total E. coli tRNA. These reactions (200μL) were incubated at 37°C for 10 minutes. To separate ribosomes from free reaction components, ribosomes were pelleted through 500 μL of a 10% sucrose cushion in buffer C (41,000RPM for 16hr at 4°C in a TLA100.3 rotor (Beckman Coulter, Fullerton, CA)). Ribosomes were resuspended in buffer B, normalized based on A260, and run on 15% tris-tricine PAGE. Westerns were developed using a polyclonal antibody directed against SmpB.
SmpB Intracellular Concentration Measurements
Wild type W3110 cells were grown to mid-log phase, harvested, resuspended in buffer G, and lysed by sonication. Insoluble material was removed by centrifugation. The S30 supernatant fraction, along with a titration of either purified SmpB or ribosomal protein S12, was subjected to western blot analysis using a polyclonal antibody directed against either SmpB or S12. The relationship between Western Blot signal and protein concentration was fit to a linear regression using Microsoft Excel software, and the concentrations of SmpB and S12 were solved based on the equation of the linear fit.
Stalled Ribosome Enrichment
Reporter mRNAs were overexpressed in either W3110 or W3110 ssrA::CAT cells from either pλ-cI-N-AGG or pλ-cI-NS constructs by growing cells to OD600=1.0 and inducing with 1mM IPTG for 45min. Cells were harvested and resuspended in buffer F. Cells were lysed by French Press and ribosomes were purified by pelletting through a 32% sucrose cushion in buffer F as described (20). Ribosomes translating the reporter mRNAs were purified from the total ribosome pool by Ni2+-NTA chromatography.
RESULTS
Interaction of free SmpB with ribosomes is salt sensitive in vivo
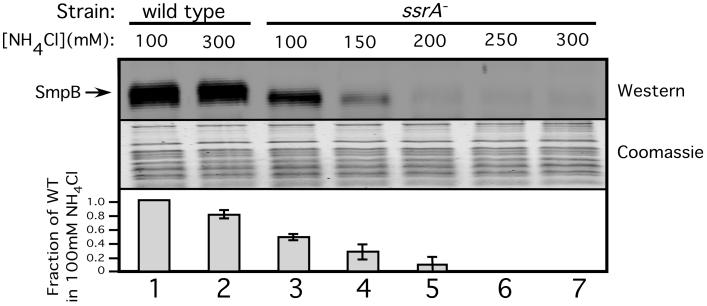
We were interested in gaining detailed mechanistic insights into interactions of SmpB protein with tmRNA and stalled ribosomes. It has been recently proposed that free SmpB, not associated with tmRNA, recognizes stalled ribosomes to recruit tmRNA. However, the evidence for interaction of free SmpB with ribosomes is based mainly on low stringency in vitro experiments or in vivo studies involving SmpB and/or tmRNA overexpression. We, therefore, set out to determine whether free SmpB interacts with ribosomes in vivo, under normal physiological concentrations of all the interacting partners. To this end, we utilized a previously reported high stringency ribosome isolation protocol (20) followed by Western blot analysis (using an SmpB specific antibody) to determine the amount of SmpB protein that co-purifies with ribosomes in vivo. To our surprise, SmpB interaction with ribosomes was only observed in wild type cells (Fig. 1, lanes 2, 7). Interaction of SmpB with ribosomes under these conditions required the presence of tmRNA in cells, as this interaction was completely absent in ssrA- cells. This result disagreed with previous reports regarding SmpB•ribosome interactions (18,29,32,35,36). We surmised that the discrepancy might be due to the different stringency conditions used for ribosome isolation in these studies as compared to ours. Thus, we repeated the experiment using lower stringency conditions (reducing the salt concentration from 300mM to 100mM NH4Cl). Under these low stringency conditions, interaction of free SmpB with ribosomes was observed (Fig. 1, lanes 1, 3), albeit the number of SmpB molecules per ribosome was only around 40% of that value in the otherwise isogenic wild type cells. The decrease in SmpB levels in this assay might be indicative of a decrease in stability of SmpB in the absence of tmRNA, but this phenomenon would not explain the increased salt sensitivity of the interaction of SmpB with ribosomes in the absence of tmRNA. These results demonstrate that interaction of SmpB with ribosomes in the absence of tmRNA is incomplete, labile and salt sensitive, observed only under low stringency conditions.
Figure 1. SmpB ribosome binding is salt sensitive.
Quantitative Western Blot analysis of the co-purification of SmpB with ribosomes performed in the presence of various salt concentrations in the purification buffers. Western blots using an SmpB specific antibody show the presence of SmpB in purified ribosome preparations. Coomassie staining was used as a loading control. Graph represents mean +/- standard deviation of the abundance of SmpB signal relative to lane 1.
Next, we further characterized the observed salt sensitivity of this interaction. We purified ribosomes from ssrA- and wild type cells using a range of salt concentrations (100, 150, 200, 250, and 300mM NH4Cl). With ribosomes isolated from ssrA- cells, association of free SmpB with ribosomes began to decrease at 150mM NH4Cl and was barely detectable at NH4Cl concentrations of 200mM or higher (Fig. 1, lanes 3-7). In contrast, in the presence of tmRNA, we observed little difference in SmpB ribosome association between 100 and 300mM salt concentrations (Fig. 1, lanes 1, 2). Therefore, the salt sensitivity of free SmpB-ribosome interaction is so severe that SmpB binding is disrupted at moderate ionic strength conditions (Less than 200mM salt), even below physiological ionic strength (37,38). These findings argue against the possibility that free SmpB could bind ribosomes in vivo. It’s likely, then, that the observed free SmpB ribosome binding under low stringency conditions is an artifact of the low ionic strength of the purification buffer.
Free SmpB•ribosome interactions are salt sensitive in vitro
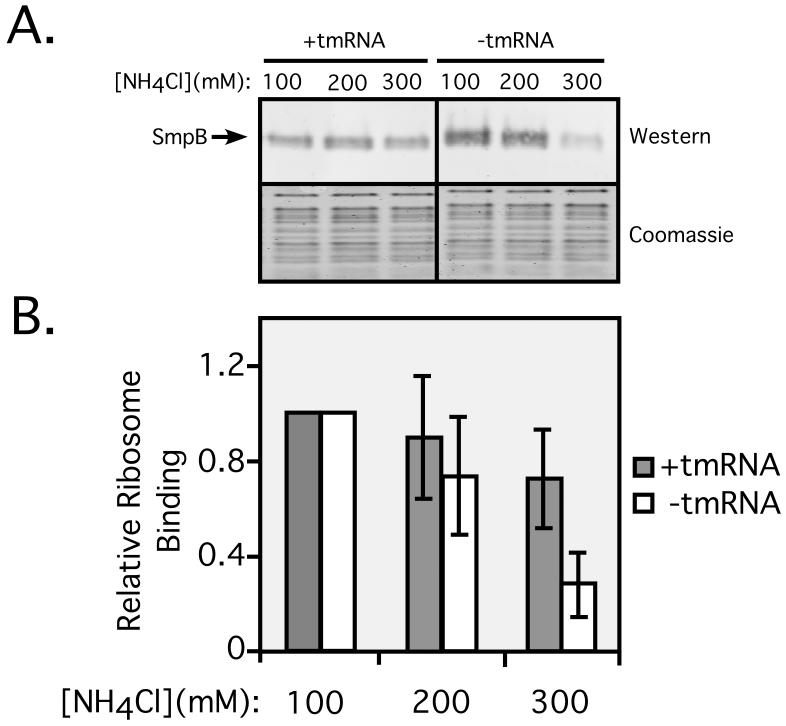
The results of the in vivo ribosome binding assays described above suggest that interactions of SmpB with ribosomes in the absence of tmRNA might be non-specific and physiologically irrelevant. However, those data reflect the binding capacity of free SmpB for the total cellular ribosome pool, only a fraction of which is stalled, or a substrate for SmpB•tmRNA mediated tagging. Hence, we examined the salt sensitivity of free SmpB interactions with stalled ribosomes in vitro. To this end, we programmed ribosomes on a nonstop mRNA (fM-mRNA, containing a 5-UTR, a ribosome binding site, and an initiation codon). These ribosomes contain the AUG initiation codon bound to fMet-tRNAfMet in the ribosomal P-site and an empty A-site, devoid of both tRNA and mRNA. The absence of an A-site mRNA codon renders these in vitro stalled ribosome complexes an ideal substrate for trans-translation. We incubated these stalled ribosomes with SmpB, EF-Tu, GTP, ATP, and Alanyl-tRNA Synthetase (Ala-RS) with or without tmRNA, and added a 100 fold molar excess of total E. coli tRNA as a non-specific competitor. To evaluate the stability of these ribosome complexes, we performed the binding reactions in the presence of either 100, 200, or 300mM NH4Cl. Ribosome complexes formed in these reactions were separated from free components by pelleting through a 10% sucrose cushion, and the amount of SmpB that co-purified with stalled ribosomes was measured by Western blot analysis using an SmpB specific antibody. The results are summarized in figure 3.
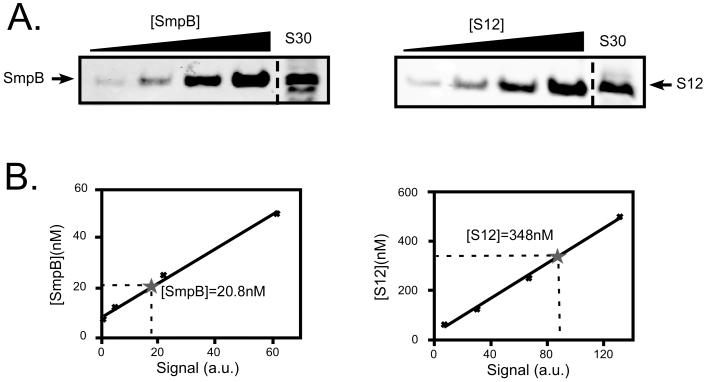
Figure 3. SmpB:ribosome ratio measurements.
Panel A depicts quantitative western blots, measuring the concentrations of SmpB and ribosomal protein S12 in S30 extracts from W3110 cells. S30 extracts, along with titrations of either purified SmpB or purified ribosomal protein S12 were subjected to Western blot analysis with antibodies directed against either SmpB or S12. Panel B shows curve fit of the Western blot signals (in arbitrary units) versus concentration of either SmpB or S12 standards. Asterisks indicate the concentrations of SmpB or S12 in S30. The summary of data from six independent experiments gave an [SmpB]:[S12] ratio of 1:14.2+/-2.4.
In the presence of tmRNA, we observed a small, stepwise decrease in SmpB ribosome binding activity from 100 to 300mM salt. 300mM NH4Cl resulted in only a small (15-20%) reduction in SmpB binding to stalled ribosomes (Fig. 2), suggesting that the vast majority of SmpB-tmRNA-stalled-ribosome complexes generated under these conditions are stable and resistant to the effects of high salt. Increasing salt concentration had a much more dramatic effect on the ribosome binding capacity of free SmpB. 300mM salt reduced the amount of bound SmpB by 80-85% (Fig. 2). Thus, the results of in vitro SmpB-stalled ribosome binding measurements mirror those of in vivo SmpB-total ribosome studies. Both methods show the interaction of SmpB with ribosomes in the absence of tmRNA to be far more labile than the interaction in the presence of tmRNA. The salt sensitivity of the interaction of free SmpB with ribosomes is somewhat less pronounced in vitro, as compared to the observed in vivo salt sensitivity (Fig. 1 vs. Fig. 2). This is most likely due to the myriad of potential non-specific nucleic acid targets present in vivo, which are able to substantially diminish any potential nonspecific interactions of SmpB protein with ribosomes. Therefore, one would expect that non-specific interactions would be less prevalent in vivo than in vitro.
Figure 2. SmpB ribosome binding is salt sensitive in vitro.
Quantitative Western blot analysis of the binding of SmpB to stalled ribosomes in vitro. Western blots using an SmpB specific antibody show SmpB-ribosome co-purification. Coomassie staining was used as a loading control. Graph represents mean +/- standard deviation of the abundance of SmpB signal relative to lane 1.
SmpB and tmRNA are present at a 1:1 ratio in cells
The results of both in vivo and in vitro ribosome binding studies suggested that interaction of free SmpB with ribosome is non-specific. However, it remained formally possible that this interaction is functionally relevant, but too labile and short-lived to survive ribosome purification techniques. In order for SmpB to pre-bind the ribosome it must either: a) possess greater affinity for the ribosome than it does for tmRNA, or b) be present in cells in a molar excess over tmRNA. Moore and Sauer recently measured the ratio of the number of copies of tmRNA to the number of ribosomes, based on comparison to 5S ribosomal RNA (39). They found a ratio of one tmRNA molecule per approximately 15 to 20 ribosomes. To gain deeper insights into the molecular mechanism of SmpB protein function, we performed quantitative western blot analysis to determine the ratio of SmpB to ribosomes, based on comparison to ribosomal protein S12 (Fig. 3). Briefly, we performed western blot analysis with an SmpB specific antibody from S30 extract (total cellular soluble material) of wild type cells along with a titration of purified SmpB. The same S30 extract was also analyzed along with a titration of purified ribosomal protein S12. Analysis of the intensities of the SmpB and S12 bands in the S30 extract, as compared to the intensities of the bands in the purified protein lanes, yielded the intracellular concentrations of SmpB and S12 in the S30 samples. This approach enabled us to calculate the ratio of SmpB to S12. The results of six independent experiments gave an [SmpB]:[S12] ratio of 1:14.2(+/-2.4). These data suggest a roughly 1:1 ratio of SmpB to tmRNA in E. coli. Similarly, a 1:1 ratio of SmpB to tmRNA has been reported in other bacterial species (7,40).
SmpB partitions with tmRNA
Since a similar number of copies of SmpB and tmRNA are present in cells, in order for SmpB to pre-bind the ribosome it must exhibit greater affinity for ribosomes than for tmRNA.
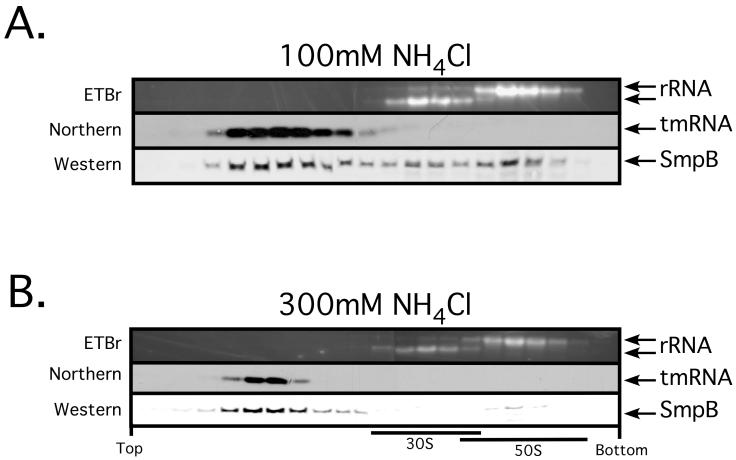
We next set out to determine the relative affinity of SmpB for tmRNA and ribosomes. 70S ribosomes can be dissociated in vitro into 50S and 30S subunits by incubation in low (1 mM) Mg2+. When SmpB•tmRNA•ribosome complexes are dissociated in low Mg2+, tmRNA is released. If ribosomal subunits are then separated by density gradient centrifugation, free tmRNA fails to co-migrate with either subunit and remains in the topmost gradient fractions. We decided to utilize this phenomenon as an indicator of the relative affinities of SmpB for tmRNA and ribosomes. We purified ribosomes from wild type cells, dissociated the subunits in vitro and then separated the subunits using a 10-40% sucrose gradient. If SmpB prefers to bind to tmRNA rather than ribosomes, we would expect it to co-migrate with tmRNA in the top gradient fractions. If, in contrast, the ribosome is the preferred binding target of SmpB, then it should migrate with the ribosomal subunit on which its primary binding site resides. When we performed this separation under low stringency (100mM NH4Cl) conditions, we found that the majority of SmpB co-migrated with tmRNA in the early gradient fractions with a small amount of SmpB bound to either ribosomal subunit (Fig. 4, top panel).
Figure 4. Ribosomal subunit dissociation assay demonstrates the relative affinity of SmpB for tmRNA and ribosomes.
Ethidium bromide staining reveals the positions of 23S and 16S rRNA in the gradient. Western and Northern blots show the migration positions of SmpB and tmRNA, respectively.
Under high stringency conditions (300mM NH4Cl), the partitioning of SmpB protein with tmRNA was even more dramatic. In this case, nearly all of the SmpB co-migrated with tmRNA in the topmost fractions, with only traces of SmpB co-migrating with the 50S subunit (Fig. 4, bottom panel). These results demonstrate that tmRNA is the primary binding partner of SmpB. These results are also in agreement with previously published data regarding this phenomenon (18,35). Since similar numbers to tmRNA and SmpB molecules are present in cells and SmpB partitions primarily with tmRNA, it is unlikely that SmpB can pre-bind the ribosome to recruit tmRNA. We, therefore, propose that a preformed SmpB-tmRNA complex recognizes stalled ribosomes, and that reported pre-binding of free SmpB to ribosome and its subunits is the result of non-specific interactions of a basic RNA binding protein with a large cellular ribonucleoprotein complex under artificial conditions.
SmpB is enriched in stalled ribosomes in vivo, only in the presence of tmRNA
In order to test the hypothesis that SmpB binds stalled ribosomes only in complex with tmRNA, we set out to purify a pool of stalled ribosomes and assess whether SmpB is enriched in these ribosomes over the total ribosome pool. We expected to observe SmpB enrichment in stalled ribosomes purified from wild type cells, but not in stalled ribosomes purified from ssrA- cells. However, if SmpB pre-binds the ribosome in the absence of tmRNA, then SmpB should be enriched in stalled ribosomes purified from either wild type or ssrA- cells. To accomplish this, we expressed a reporter mRNA (λ-CI-N-4-AGG) which codes for a variant of the λ repressor N-terminal domain with an N-terminal 6His epitope and a string of 4 rare arginine codons (AGG), making it a substrate for tmRNA- mediated tagging (8). Briefly, ribosomes stall on the rare codon-containing segment of this construct, while displaying the already translated N-terminal 6His epitope. These ribosomes might then be captured on Ni2+-NTA beads. It should be noted that the product of this purification is not expected to contain only stalled ribosomes, as ribosomes might be captured at any stage beyond translation of the N-terminal 6His epitope. However, as the stalled state is expected to be the most kinetically long-lived state between 6His epitope translation and peptide release, this pool of ribosomes should be greatly enriched for stalled ribosomes. Hereafter, we will refer to this stalled ribosome-enriched pool as stalled ribosomes.
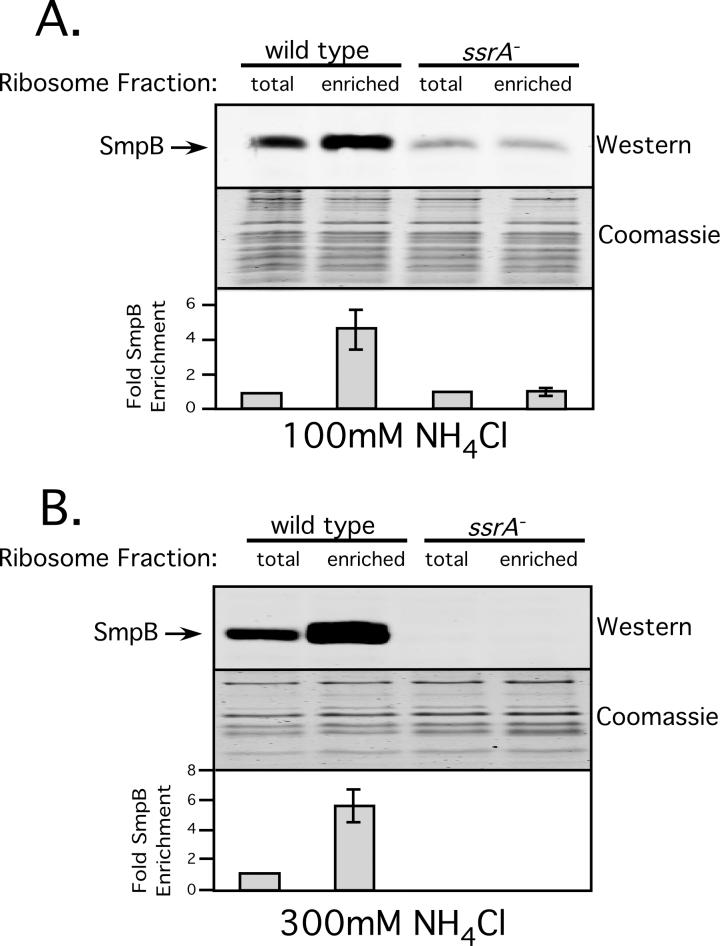
We purified these stalled ribosomes from both wild type cells and ssrA- cells under low (100mM NH4Cl) and high (300mM NH4Cl) stringency conditions. Figure 5 depicts the results of this assay. As expected, in wild type cells, regardless of stringency, we observed around a 5-fold enrichment of SmpB in the stalled ribosome fraction versus total ribosomes. In contrast, no enrichment of SmpB was observed in ssrA- cells under low stringency conditions (Fig. 5A). In ssrA- cells under high stringency conditions, we did not observe SmpB-ribosome binding in either the total or stalled ribosome fractions, consistent with the results of in vivo ribosome association assays described above (Fig. 5B). These results lend further support to the conclusion that functional SmpB•stalled ribosome interactions require the presence of tmRNA.
Figure 5. Enrichment of SmpB in stalled ribosomes requires tmRNA.
Quantitative Western Blot analysis of the relative abundance of SmpB in stalled vs. total ribosome fractions. Graph represents mean +/- standard deviation of the relative amount of SmpB bound per ribosome in the stalled ribosome fraction vs. total ribosomes.
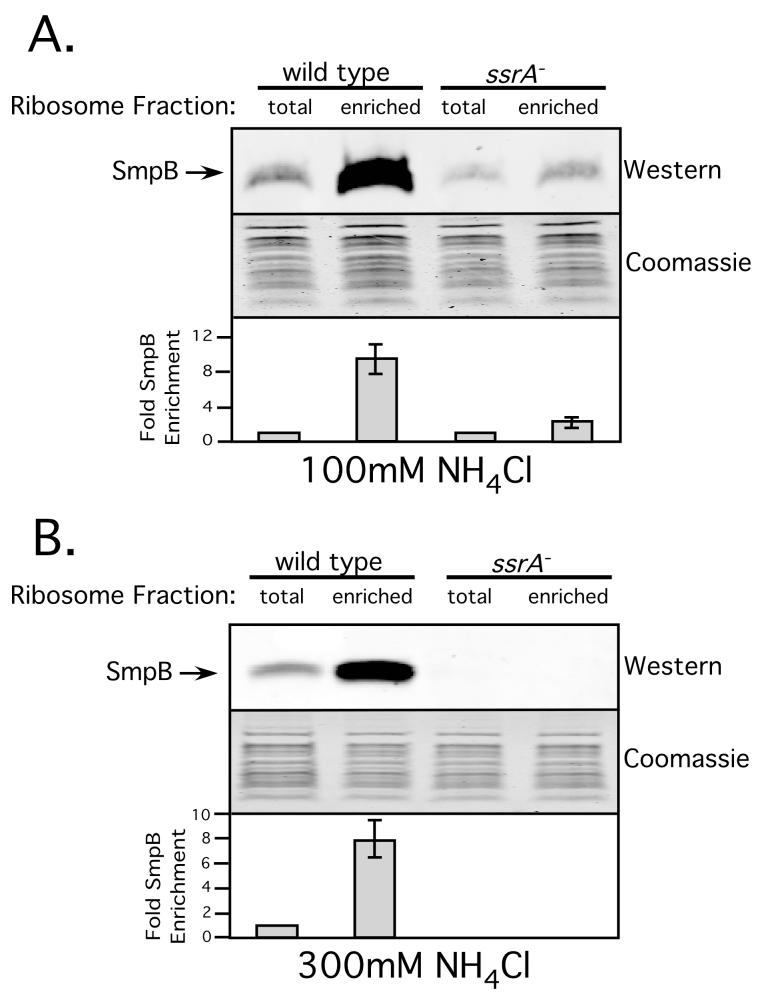
We next repeated this assay using a different trans-translation substrate, a non-stop mRNA reporter. We replaced the rare (AGG) arginine codons with abundant (CGG) codons and inserted a strong trpA-operon transcriptional terminator at the 3’-end. This new reporter (λ-cI-NS) mRNA still codes for the N-terminal 6His epitope but lacks an inframe termination codon. The results of stalled ribosome enrichment experiments with this construct were quite similar to those with the rare codon-containing construct. We observed an approximately 8-fold enrichment of SmpB in stalled ribosomes from wild type cells (Fig. 6). In ssrA- cells, we saw only an insignificant enrichment under low stringency, and no SmpB-ribosome association in either total ribosomes, or the enriched ribosome pool under high stringency conditions (Fig. 6).
Figure 6. Enrichment of SmpB in ribosomes stalled on a non-stop message.
Quantitative Western Blot analysis of the relative abundance of SmpB in stalled vs. total ribosome fractions. Graph represents mean +/- standard deviation of the relative amount of SmpB bound per ribosome in the stalled ribosome fraction vs. total ribosomes.
We have, thus, demonstrated that interaction of free SmpB protein with ribosomes is salt sensitive in vivo and in vitro. Further, we’ve shown that SmpB is present at a similar intracellular concentration relative to tmRNA, and that SmpB partitions with tmRNA upon ribosomal subunit dissociation. Finally, we have demonstrated that SmpB is enriched in stalled ribosome fractions only in the presence of tmRNA. Taken together, these results suggest that tmRNA is required for specific and functional interactions of SmpB with stalled ribosomes. Based on these findings, we propose that SmpB does not prebind the ribosome in vivo, and that SmpB•tmRNA binding occurs prior to stalled ribosome recognition to initiate trans-translation.
DISCUSSION
SmpB is required for tmRNA-mediated recognition and rescue of stalled ribosomes. Despite extensive biochemical and structural studies, the mechanism by which SmpB facilitates the productive engagement of tmRNA with stalled ribosomes has remained a matter of debate. Two competing models have emerged in recent years. The first model suggests that a preformed complex of SmpB•tmRNA•EFTu(GTP) recognizes stalled ribosomes to initiate trans-translation. This model is supported by numerous genetic, biochemical and structural studies that convincingly demonstrate that tmRNA is the specific high-affinity binding partner of SmpB (1,4,12,17,20,25-27,41). This SmpB•tmRNA interaction is illustrated in two recently solved co-crystal structural models. Amino acid residues required for high affinity tmRNA binding are clustered on a unique surface of the protein. Mutations that alter these amino acids disrupt SmpB-tmRNA interactions and prevent ribosome rescue and peptide tagging (17). Moreover, it has been shown that SmpB can bind both the alanine-charged and uncharged forms of tmRNA and that binding of SmpB enhances the aminoacylation of tmRNA by ala-RS (11,21,22). SmpB and EFTu (GTP) can bind simultaneously to the tRNA-like domain of tmRNA and protect the labile ester linkage from hydrolysis (21).
Studies in E. coli and C. crescentus have shown that the intracellular levels of tmRNA and SmpB are reduced when one of the binding partners is missing (7,32,40,42, and this study). Most strikingly, in C. crescentus, the levels and stability of SmpB and tmRNA are under cell cycle control (40). SmpB is reported to protect tmRNA from degradation by RNase R. This study also demonstrates that SmpB binds with high affinity to tmRNA and that this direct binding is responsible for its protection from degradation by RNase R (26,27). Taken together, these finding are consistent with a model wherein the complex of SmpB-tmRNA forms first, is aminoacylated by AlaRS, bound by EF-Tu (GTP) and subsequently delivered to stalled ribosomes. Direct binding of SmpB to tmRNA and its subsequent aminoacylation and delivery to ribosomes would thus protect a large fraction of tmRNA from cellular nucleases, and a large fraction of SmpB protein from proteases. By contrast, if SmpB binds stalled ribosomes first, as proposed by the pre-binding model described below, then its intracellular concentration should not be affected by the presence or absence of tmRNA, as the bulk of SmpB would be bound by ribosomes and inaccessible to cellular proteases.
The second model suggests that SmpB might pre-bind stalled ribosome to recruit tmRNA and initiate trans-translation (29,32,35,36). Two observations led to this conclusion. The first observation was that SmpB co-purifies with 70S ribosomes or the dissociated 50S and 30S subunits in an ssrA-strain (32). Careful examination of the data presented in this report, however, reveals that the SmpB-ribosome preparations were generated under low salt conditions (60-100mM NH4Cl), and as such agrees with our data depicted in figure 1. The question, then, is whether this free SmpB-ribosome interaction, observed only under low stringency conditions, is specific and functionally relevant. We reason that this interaction is likely due to the non-specific binding of a highly basic RNA binding protein, in the absence of its canonical RNA binding partner, to a highly abundant large ribonucleoprotein complex, the ribosome. The labile nature and salt sensitivity of this interaction (Figs. 1, 2, and 3) is consistent with the pre-binding interaction being of a nonspecific nature. In agreement with this notion, two related studies (18,35), aimed at assessing the interactions of free SmpB with ribosomes, demonstrated that when ribosomal subunits are dissociated, the vast majority of SmpB co-localizes with tmRNA and not with ribosomal subunits, even under low stringency conditions. This result, although not interpreted as such, agrees with our finding that tmRNA, and not the ribosome, is the preferred binding partner of tmRNA. Furthermore, if SmpB can pre-bind stalled ribosomes then it should be enriched in stalled ribosome fractions, irrespective of the presence or absence of tmRNA. Our experiments addressing this issue suggest that this is not the case. SmpB is enriched in stalled ribosome fractions only when tmRNA is present, and not in its absence (Figs. 5 and 6).
The second observation that led to the SmpB-ribosome pre-binding hypothesis was that an in vitro generated SmpB ribosome complex could recruit tmRNA to drive peptide bond formation in vitro (32). For this experiment, the authors measured the extent of peptide bond formation at 10 second, 5 minute, and 15 minute time points. Peptide bond formation occurs on a time scale of milliseconds. Therefore, it is entirely conceivable that SmpB could recycle from ribosome to tmRNA, in the time scale of this experiment, to generate the SmpB•tmRNA•EF-Tu (GTP) quaternary complex required for participation in the transpeptidation reaction. A further point of uncertainty associated with the pre-binding model is the suggestion that a pre-formed SmpB•tmRNA•EF-Tu (GTP) complex might still be needed to deliver tmRNA to the pre-bound SmpB-ribosome complex (31,32). This phenomenon is presented as evidence for the requirement of two SmpB molecules to elicit trans-translation. The fact that SmpB partitions primarily with tmRNA and that there are equal numbers of SmpB and tmRNA molecules present per cell is inconsistent with this proposal and can’t be reconciled with the multiple SmpB requirement of this model (see below).
Finally, a cryo-EM reconstruction of a free SmpB-ribosome complex has recently been modeled (35). Low stringency conditions and a large molar excess of SmpB over ribosomes were used to generate these complexes. The density assigned to SmpB was located near the decoding center of the 30S subunit. This positioning of SmpB is provocative, and might represent one of the binding sites of SmpB in the SmpB•tmRNA•EF-Tu (GTP) complex. Interestingly, this SmpB binding site on a stalled ribosome was identified by comparing the differences between cryo-EM maps generated from SmpB-ribosome complexes containing either a nonstop mRNA or a longer mRNA. Therefore, the 30S-decoding center is the only site on the ribosome where a difference in density would be expected.
The results presented in our study are consistent with the first model of stalled ribosome recognition and argue against SmpB ribosome pre-binding as a physiologically relevant phenomenon. We have shown that interaction of free SmpB with ribosomes occurs only under artificially low stringency conditions. In general, several previous studies have looked at interaction of free SmpB with ribosomes but failed to identify its labile and non-specific nature. This highlights the importance of choosing appropriate stringency conditions and the use of competitors to disrupt non-specific interactions when studying binding in vitro. This consideration is particularly critical when looking at interaction of a basic protein with RNA, where charge interactions with the RNA backbone can drive binding to essentially any nucleic acid.
The results of this study also shed new light on the related issue of the stoichiometry of the SmpB•tmRNA•stalled-ribosome complex. We have shown that free SmpB does not specifically bind the ribosome. Further, we have demonstrated that the intracellular concentrations of SmpB and tmRNA are similar. This suggests that the stoichiometry of the SmpB•tmRNA complex is 1:1. The primary SmpB binding site on tmRNA has been conclusively identified (1,4,12,17,20,25-27,41). Since the intracellular concentrations of SmpB and tmRNA are similar, the 1:1 complex, with SmpB bound to the primary high-affinity binding site, would be favored at equilibrium. This conclusion is consistent with recent analyses of the interaction between tmRNA and SmpB, from Thermus thermophilus and Aquifex aeolicus (25-27), which found only one SmpB contacting the D-loop equivalent of tmRNA. Furthermore two co-crystal structure models of SmpB in complex with the tmRNA TLD exhibited a 1:1 SmpB:tmRNA stoichiometry (26,27). Initial cryo-EM derived models of the complex formed by SmpB-tmRNA-EF-Tu-GDP-Kirromycin and 70S ribosomes (30) suggested the presence of a single molecule of SmpB protein in the pre-accommodated complex. Similarly, analysis of affinity purified tmRNA•ribosome complexes also suggested the presence of only one SmpB molecule at a late stage of the trans-translation process (43).
Another cryo-EM reconstructed model of SmpB-tmRNA-EF-Tu-GDP-Kirromycin in complex with a stalled ribosome suggested the presence two molecules of SmpB, one in the decoding center and the other in the A-site compartment of 50S subunit (31). As a consequence of the inclusion of the second SmpB molecule, the tRNA-like domain of tmRNA is now oriented towards the tmRNA ORF (31). It is difficult to visualize how tmRNA could participate in peptide bond formation with its tRNA-like domain facing the ORF rather than the peptidyl-transferase center. This second SmpB molecule will undoubtedly interfere with peptide bond formation. Equally puzzling is the suggestion that both molecules of SmpB interact with two dissimilar parts of the tRNA-like domain of tmRNA, presumably using the same RNA binding surface. It is not clear from these studies how the same SmpB RNA binding residues could interact specifically with two distinct tmRNA sequence and structure elements.
Under normal physiological conditions, stalled ribosomes constitute only a small fraction of the total cellular pool of ribosomes (15,000-20,000 per cell). Normal translating ribosomes are present in vast excess (10 to 20-fold) over SmpB and tmRNA (39). We reason that pre-binding of SmpB to ribosomes is unlikely, as most SmpB would be sequestered and unavailable to support tmRNA recruitment. In contrast, if pre-formed quaternary complexes of SmpB•tmRNA•EFTu (GTP) were responsible for recognizing stalled ribosomes then this surveillance system would be ideally primed to engage and rescue stalled ribosomes. In conclusion, we have demonstrated that specific and functional SmpB-ribosome binding requires tmRNA. Our results rule out the possibility of SmpB pre-binding of ribosomes to recruit tmRNA and initiate trans-translation. Our results also suggest that the stoichiometry of the SmpB•tmRNA•stalled ribosome complex is 1:1:1. We, therefore, propose that ribosome rescue is initiated by a preformed SmpB•tmRNA•EF-Tu (GTP) complex that recognizes and binds the A-site of stalled ribosomes to commence trans-translation.
ACKNOWLEDGEMENTS
We thank Dr. Robert Sauer, Ge Zhiyun, and Dr. Jamie Richards for insightful comments on the manuscript. We also thank members of the Karzai lab for helpful discussions and suggestions. We are grateful to Dr. Jorge L. Benach and members of The Center for Infectious Diseases for their continued support. This work was supported in part by Grants (to AWK) from The National Institute of Health (GM65319, and AI055621), The Northeast Biodefense Center, and The Pew Scholars Program.
REFERENCES
- 1.Dulebohn D, Choy J, Sundermeier T, Okan N, Karzai AW. Biochemistry. 2007;46(16):4681–4693. doi: 10.1021/bi6026055. [DOI] [PubMed] [Google Scholar]
- 2.Gottesman S, Roche E, Zhou Y, Sauer RT. Genes Dev. 1998;12(9):1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haebel PW, Gutmann S, Ban N. Current opinion in structural biology. 2004;14(1):58–65. doi: 10.1016/j.sbi.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Karzai AW, Susskind MM, Sauer RT. EMBO J. 1999;18(13):3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keiler KC, Waller PR, Sauer RT. Science. 1996;271(5251):990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 6.Mehta P, Richards J, Karzai AW. RNA. 2006;12(12):2187–2198. doi: 10.1261/rna.247706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore SD, Sauer RT. Annu Rev Biochem. 2007 doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 8.Richards J, Mehta P, Karzai AW. Molecular microbiology. 2006;62(62):1700–1712. doi: 10.1111/j.1365-2958.2006.05472.x. [DOI] [PubMed] [Google Scholar]
- 9.Tu GF, Reid GE, Zhang JG, Moritz RL, Simpson RJ. J Biol Chem. 1995;270(16):9322–9326. doi: 10.1074/jbc.270.16.9322. [DOI] [PubMed] [Google Scholar]
- 10.Withey JH, Friedman DI. Annu Rev Microbiol. 2003;57:101–123. doi: 10.1146/annurev.micro.57.030502.090945. [DOI] [PubMed] [Google Scholar]
- 11.Hanawa-Suetsugu K, Takagi M, Inokuchi H, Himeno H, Muto A. Nucleic acids research. 2002;30(7):1620–1629. doi: 10.1093/nar/30.7.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karzai AW, Roche ED, Sauer RT. Nature structural biology. 2000;7(6):449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 13.Okan NA, Bliska JB, Karzai AW.PLoS Pathog 200621, e6; 50–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choy JS, Aung LL, Karzai AW. J Bacteriol. 2007 doi: 10.1128/JB.00860-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman C, Thevenet D, Bouloc P, Walker GC, D’Ari R. Genes Dev. 1998;12(9):1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto Y, Sunohara T, Jojima K, Inada T, Aiba H. RNA. 2003;9(4):408–418. doi: 10.1261/rna.2174803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dulebohn DP, Cho HJ, Karzai AW. J Biol Chem. 2006;281(39):28536–28545. doi: 10.1074/jbc.M605137200. [DOI] [PubMed] [Google Scholar]
- 18.Hallier M, Desreac J, Felden B. Nucleic acids research. 2006;34(6):1935–1943. doi: 10.1093/nar/gkl097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizu Y, Ueda T. FEBS Lett. 2002;514(1):74–77. doi: 10.1016/s0014-5793(02)02333-5. [DOI] [PubMed] [Google Scholar]
- 20.Sundermeier TR, Dulebohn DP, Cho HJ, Karzai AW. Proc Natl Acad Sci USA. 2005;102(7):2316–2321. doi: 10.1073/pnas.0409694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barends S, Karzai AW, Sauer RT, Wower J, Kraal B. J Mol Biol. 2001;314(1):9–21. doi: 10.1006/jmbi.2001.5114. [DOI] [PubMed] [Google Scholar]
- 22.Barends S, Wower J, Kraal B. Biochemistry. 2000;39(10):2652–2658. doi: 10.1021/bi992439d. [DOI] [PubMed] [Google Scholar]
- 23.Jacob Y, Sharkady SM, Bhardwaj K, Sanda A, Williams KP. J Biol Chem. 2005;280(7):5503–5509. doi: 10.1074/jbc.M409277200. [DOI] [PubMed] [Google Scholar]
- 24.Barends S, Bjork K, Gultyaev AP, de Smit MH, Pleij CW, Kraal B. FEBS Lett. 2002;514(1):78–83. doi: 10.1016/s0014-5793(02)02306-2. [DOI] [PubMed] [Google Scholar]
- 25.Nameki N, Someya T, Okano S, Suemasa R, Kimoto M, Hanawa-Suetsugu K, Terada T, Shirouzu M, Hirao I, Takaku H, Himeno H, Muto A, Kuramitsu S, Yokoyama S, Kawai G. J Biochem (Tokyo) 2005;138(6):729–739. doi: 10.1093/jb/mvi180. [DOI] [PubMed] [Google Scholar]
- 26.Bessho Y, Shibata R, Sekine S, Murayama K, Higashijima K, Hori-Takemoto C, Shirouzu M, Kuramitsu S, Yokoyama S. Proc Natl Acad Sci USA. 2007;104(20):8293–8298. doi: 10.1073/pnas.0700402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutmann S, Haebel PW, Metzinger L, Sutter M, Felden B, Ban N. Nature. 2003;424(6949):699–703. doi: 10.1038/nature01831. [DOI] [PubMed] [Google Scholar]
- 28.Wower J, Zwieb CW, Hoffman DW, Wower IK. Biochemistry. 2002;41(28):8826–8836. doi: 10.1021/bi0201365. [DOI] [PubMed] [Google Scholar]
- 29.Metzinger L, Hallier M, Felden B. Nucleic acids research. 2005;33(8):2384–2394. doi: 10.1093/nar/gki534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valle M, Gillet R, Kaur S, Henne A, Ramakrishnan V, Frank J. Science. 2003;300(5616):127–130. doi: 10.1126/science.1081798. [DOI] [PubMed] [Google Scholar]
- 31.Kaur S, Gillet R, Li W, Gursky R, Frank J. Proc Natl Acad Sci USA. 2006;103(44):16484–16489. doi: 10.1073/pnas.0607438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hallier M, Ivanova N, Rametti A, Pavlov M, Ehrenberg M, Felden B. J Biol Chem. 2004;279(25):25978–25985. doi: 10.1074/jbc.M314086200. [DOI] [PubMed] [Google Scholar]
- 33.Culver GM, Noller HF. Rna. 1999;5(6):832–843. doi: 10.1017/s1355838299990714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borel F, Hartlein M, Leberman R. FEBS Lett. 1993;324(2):162–166. doi: 10.1016/0014-5793(93)81385-d. [DOI] [PubMed] [Google Scholar]
- 35.Gillet R, Kaur S, Li W, Hallier M, Felden B, Frank J. J Biol Chem. 2007;282(9):6356–6363. doi: 10.1074/jbc.M609658200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanova N, Pavlov MY, Bouakaz E, Ehrenberg M, Schiavone LH. Nucleic acids research. 2005;33(11):3529–3539. doi: 10.1093/nar/gki666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Learn B, Karzai AW, McMacken R. Cold Spring Harb Symp Quant Biol. 1993;58:389–402. doi: 10.1101/sqb.1993.058.01.046. [DOI] [PubMed] [Google Scholar]
- 38.Richey B, Cayley DS, Mossing MC, Kolka C, Anderson CF, Farrar TC, Record MT., Jr. J Biol Chem. 1987;262(15):7157–7164. [PubMed] [Google Scholar]
- 39.Moore SD, Sauer RT. Molecular microbiology. 2005;58(2):456–466. doi: 10.1111/j.1365-2958.2005.04832.x. [DOI] [PubMed] [Google Scholar]
- 40.Hong S-J, Tran Q-A, Keiler KC. Molecular microbiology. 2005;57(2):565–575. doi: 10.1111/j.1365-2958.2005.04709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wower IK, Zwieb C, Wower J. J Biol Chem. 2004;279(52):54202–54209. doi: 10.1074/jbc.M410488200. [DOI] [PubMed] [Google Scholar]
- 42.Keiler KC, Shapiro L. J Bacteriol. 2003;185(6):1825–1830. doi: 10.1128/JB.185.6.1825-1830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shpanchenko OV, Zvereva MI, Ivanov PV, Bugaeva EY, Rozov AS, Bogdanov AA, Kalkum M, Isaksson LA, Nierhaus KH, Dontsova OA. J Biol Chem. 2005;280(18):18368–18374. doi: 10.1074/jbc.M409094200. [DOI] [PubMed] [Google Scholar]