Abstract
Nrf2 is the key transcription factor regulating the antioxidant response. When exposed to oxidative stress, Nrf2 translocates to cell nucleus and forms heterodimer with small Maf proteins (sMaf). Nrf2/sMaf heterodimer binds specifically to a cis-acting enhancer called antioxidant response element and initiates transcription of a battery of antioxidant and detoxification genes. Nrf2 possesses a NESzip motif (nuclear export signal co-localized with the leucine zipper (ZIP) domain). Heterodimerization with MafG via ZIP-ZIP binding enhanced Nrf2 nuclear retention, which could be abrogated by the deletion of the ZIP domain or site-directed mutations targeting at the ZIP domain. In addition, dimerization with MafG precluded Nrf2zip/CRM1 binding, suggesting that Nrf2/MafG heterodimerization may simultaneously mask the NESzip motif. MafG-mediated nuclear retention may enable Nrf2 proteins to evade cytosolic proteasomal degradation and consequently stabilize Nrf2 signaling. For the first time, we show that at the physiological condition, the NESzip motif can be switched-off by heterodimerization.
Keywords: Nrf2, MafG, ZIP, CRM1, FRET
1. Introduction
To adapt to their aerobic life style, mammalian cells have developed elaborate yet highly efficient cytoprotective machinery. When exposed to oxidative stress, these cells can respond with a rapid and coordinated expression of a battery of gene products, including phase II detoxification enzymes/antioxidants and phase III efflux transporters [1–3]. As a consequence, these cells can effectively neutralize and remove excess oxidants to quickly restore redox homeostasis. The antioxidant response is exquisitely regulated. Four components, namely, Nrf2 (NF-E2 related factor 2) [4], Keap1 (Kelch-like ECH-associated protein 1) [5], a group of small musculoaponeurotic fibrosarcoma (Maf) proteins [6] and a cis-acting enhancer called antioxidant response element (ARE) or electrophile responsive element (EpRE) [7–9], are found essential for the regulation of the antioxidant response [10].
Pivotal to the antioxidant response is Nrf2 [4]. Nrf2 is a basic leucine zipper (bZIP) transcription factor featuring a Cap “N” Collar (CNC) structure [4]. Like many other transcription factors, Nrf2 signaling is regulated by compartmental segregation. At unstressed condition, Nrf2 is found mainly sequestered in the cytoplasm by its cytosolic repressor Keap1 [1]. Keap1 is also a Cullin 3-dependent substrate adaptor protein for ubiquitin ligase E3 complex [11–14]. So Nrf2 molecules may not only be sequestered by Keap1 but also subjected to constant degradation in the cytoplasm. When challenged by oxidative stress derived from accumulation of reactive oxygen species (ROS) [15–17] or reactive nitrogen species (RNS) [18, 19], the Keap1-mediated Nrf2 ubiquitination and degradation is impeded in a redox-sensitive manner [20]. In contrast, Nrf2 protein translation is enhanced [21]. The relative abundance of Nrf2 proteins may surpass the Keap1 sequestering capacity. As a consequence, the pool of unbound Nrf2 proteins expands. Since unbound Nrf2 exhibits a graded nuclear translocation correlated with the intensity of oxidation [22], certain amount of Nrf2 proteins translocate into the nucleus and form heterodimer with small Maf proteins. Small Maf (sMaf) proteins, composed of MafF, G and K, are a group of bZIP bi-directional transcription regulators [6, 23]. The sMaf proteins per se lack the transactivation domain, so the sMaf/sMaf homodimers function as transcription repressors [24]. Whereas Nrf2 cannot form homodimer [25, 26], the Nrf2/sMaf heterodimer exhibits high recognition specificity and binding affinity to ARE/EpRE [25] located in the promoter of diverse phase II/III detoxification genes [6, 27]. The binding of Nrf2/sMaf heterodimer to ARE/EpRE thus triggers the transcription of these cytoprotective genes.
Recently, the mechanisms governing the subcellular localization of unbound Nrf2 have been elucidated. One bipartite nuclear localization signal (NLS) is identified in the basic region of Nrf2 [28, 29], called bNLS. One nuclear export signal (NES) is characterized in the ZIP domain of Nrf2 [28, 29], called NESzip. In addition, another NES motif is characterized in the transactivation (TA) domain of Nrf2 [22, 30], called NESTA. The existence of multiple NLS/NES motifs in Nrf2 implies that the subcellular localization of Nrf2 is determined by the collective activities of these motifs. The driving force of individual NLS/NES motif has been analyzed [22]. The combined nuclear exporting activities exerted by both the NESzip and NESTA motifs appear to be able to counteract the nuclear importing activity mediated by the bNLS motif [22]. Disabling of either the NESzip or the NESTA motif by mutations results in Nrf2 nuclear localization [22]. These results naturally raise the question of whether these NLS/NES motifs can be turned on/off under normal physiological conditions and consequently alter the subcellular localization of Nrf2.
Previous studies found that at oxidized condition, the NESTA motif could be disabled, probably by sulfhydryl modification at cysteine 183 (C183) residue embedded in the NESTA motif [22]. The sulfhydryl modification on C183 residue may generate steric hindrance for the binding of nuclear exporting protein chromosome region maintenance 1 (CRM1) [22]. So the NESTA motif appears to be a conditional NES motif that can be turned off by oxidants.
The position of the NESzip motif is overlapped with the ZIP domain [28]. In the present study, we find that heterodimerization with sMaf proteins can simultaneously mask the NESzip motif and preclude NESzip/CRM1 binding. For the first time we show that the NESzip motif can also be turned off at the physiological condition.
2. Materials and Methods
2.1. Cell culture, chemicals and antibodies
Human cervical squamous cancerous HeLa cells and human embryonic kidney (HEK) cells were obtained from ATCC (Manassas, VA). HeLa and HEK cells were cultured as monolayer using minimum essential medium (MEM) supplemented with 10% fetal bovine serum, 2.2 mg/ml sodium bicarbonate, 100 units/ml penicillin and 100 µg/ml streptomycin. Rabbit anti-MafG/K (H-100), anti-Nrf2 (H-300), anti-CRM1 (H-300), anti-Lamin A (H-102), anti-GAPDH (FL-335), anti-HO-1 (H-105) and anti-NQO1 (H-90), mouse anti-GST (B-14) probes were all purchased from St. Cruz Biotech (St. Cruz, CA). Mouse anti-Myc (9B11) probe was purchased from Cell Signaling (Danvers, MA).
2.2. Plasmid construction, site-directed mutagenesis and RT-PCR
The construction of the EGFP-Nrf2zip [28] and pHM6-Nrf2 [31] plasmids have been described before. Human MafG cDNA is a generous gift from Dr. Volker Blank [32]. MafG was PCR amplified and subcloned into pDsRed-Monomer vector (ClonTech, Mountain View, CA). The deletion mutants of MafG, MafGzip (72–162 a.a.) and MafGΔzip (1–71 a.a.), were generated by PCR amplification and inserted into pDsRed-Monomer (mDsRed) vector. For FRET studies, Nrf2zip and MafG were PCR amplified and subcloned into pECFP-C1 and pEYFP-C1 vector (ClonTech), respectively. Alanine substitute mutations were performed using QuikChange XL site-directed mutagenesis kit (Stratagene,La Jolla, CA) according to the manufacturer’s instruction. Briefly, both sense and antisense mutagenic oligonucleotide primers were designed to mutate leucine to alanine. The primers were synthesized and PAGE/HPLC-purified by Integrated DNA Technologies, Inc (Coralville, IA). Mutagenesis reactions were performed in 50 µl reaction solution containing 100 ng template DNA, 125 ng sense and antisense mutagenic primers, 1X reaction buffer with dNTP supplement, 3 µl QuikSolution, 2.5 U Pfu Turbo DNA polymerase and double distilled water. Mutagenesis reaction was performed at the condition of denaturing at 95°C for 1 min, followed by 18 cycles of thermal cycling reaction (95°C for 50 seconds, 60°C for 50 seconds and 68°C for 7 min) and concluded by 7 min extension at 68°C. The parental methylated dsDNA plasmids were subsequently digested by Dpn I at 37°C for 3 h. Afterwards, the thermal cycling products were transformed into ultra-competent XL10-Gold cells (Strategene). The mutant plasmids were extracted and verified by DNA sequencing. We also constructed a pcDNA3.1-Myc-MafG to add a Myc tag (EQKLISEEDL) [33] to the N-terminus of MafG. We also constructed a pcDNA3.1-Nrf2-V5 to add a V5 tag (GKPIPNPLLGLDST) [34] to the C-terminus of Nrf2. To analyze the transcription of phase II genes, 3 µg pcDNA3.1-Nrf2-V5 plasmid was expressed alone or co-expressed with 1 µg pcDNA3.1-Myc-MafG or pcDNA3.1-Myc-MafG2p mutant in HeLa or HEK cells. RNA was extracted using RNeasy mini kit (Qiagen, Valencia, CA) according to manufacturer’s instruction and reverse transcribed (RT) using Superscript First-Strand Synthesis System III kit ( Life Technologies, Rockville, MD). The RT products were further analyzed by PCR reaction. The PCR primers for HO-1, NQO1 [35] and UGT1A1 [36] have been described before. The PCR reaction was denaturing at 95°C for 5 min, followed by 40 cycles of thermal cycling reaction (95°C for 1 min., 55°C for 30 seconds and 68°C for 1 min) and concluded by 10 min extension at 72°C. The RT-PCR products were resolved in 1% agarose gel supplemented with ethidium bromide and visualized in UV light.
2.3. GST pull-down, competitive binding assay and Western blotting
The expression and purification of (His)6-CRM1 proteins has been described before [28]. (His)6-MafG protein was prepared using the similar protocol. Briefly, human MafG was PCR amplified and subcloned into the pQE30 vector (Qiagen). The pQE30-MafG plasmid was transformed into Escherichia coli M15 cells (Qiagen). Expression of (His)6-MafG proteins was induced by 0.5 mM of isopropyl β-D-thiogalactoside (IPTG) for 4 h at 30°C and purified by Ni-NTA slurry (Qiagen). To put a GST tag on Nrf2zip, Nrf2zip was PCR amplified and subcloned into the pGEX-2T vector (GE Healthcare, Piscataway, NJ). The pGEX-Nrf2zip plasmid was transformed into DH5α Escherichia coli and induced by 0.8 mM IPTG at 30°C overnight. GST-Nrf2zip proteins were purified by glutathione (GSH) conjugated beads (Novagen) and eluted with 10 mM GSH in 50 mM Tris buffer. Subsequently, GSH was eliminated in a solution exchange experiment using MicroCon YM-30 spin column (Millipore, Billerica, MA). GST-Nrf2zip protein was preserved in incubation buffer (phosphate-buffered saline (PBS), 0.1% TX-100, pH7.3) supplemented with 1 mM dithiothreitol (DTT) to avoid auto-oxidation. In MafG/CRM1 competitive binding experiment, 1 µg GST-Nrf2zip proteins were first mixed with GSH-conjugated beads in incubation buffer supplemented with fresh 1mM 2-mercaptoethanol (ME) and tumbled at 4°C for 30 min., subsequently, 2 µg (His)6-CRM1 together with 0, 1, 5 µg (His)6-MafG were added into GST-Nrf2zip solution and tumbled at 4°C for 2 hours. The pellets were extensively washed and dissolved in 50 µl gel loading buffer supplemented with 2-ME. The samples were heated at 95°C for 5 min and subjected to Western blotting examination. For western blotting—cell lysates containing 20 µg proteins were resolved by 4–15% linear gradient SDS-polyacrylamide gel (BioRAD, Hercules, CA) electrophoresis and transferred to polyvinylidene difluoride membrane using a semi-dry transfer system (Fisher). The membrane was blocked with 5% nonfat milk in Tris-buffered saline with Tween-20 (TBST) containing 20 mM Tris-HCl, 8 mg/ml NaCl, and 0.2% Tween-20 (pH 7.6) at room temperature for 1 h. The membrane was probed with polyclonal rabbit anti-Nrf2 (1:500), anti-CRM1 (1:500), anti-MafG/K (1:500), anti-GAPDH (1: 10000), anti-Lamin A (1:500), anti-HO-1 (1:500), anti-NQO1 (1:500) and monoclonal anti-GST (1:10,000) and anti-Myc (1:500) in 3% nonfat milk TBST at 4°C overnight. After washing three times with TBST, the membrane was blotted with peroxidase-conjugated secondary antibody (1:5000 dilution) at room temperature for 1 h. Proteins were visualized using the ECL mixture from BioRAD.
2.4. Transient Transfection and Reporter Gene Activity Assays
Transactivation activity assay has been described in detail before [37]. Briefly, HeLa cells were plated in six-well plates at ~4.0 × 105 cells/well. Twenty four hours after plating, cells were transfected using the Lipofectamine method according to manufacturer’s instructions. For each well, 500 ng pARE-TI-Luc reporter containing a single copy of murine GST Ya ARE, 1 µg pHM6-Nrf2 were co-expressed with 0, 1, 10, 25 and 100 ng plasmids expressing wild type or 2 point mutant MafG. Lipofectamine 2000 (Life Technologies) was added into another tube of 125 µl OPTI-MEM in a 1:2.5 ratio to the amount of plasmids and incubated at room temperature for 5 min. The plasmid solution was then mixed with lipofectamine solution with vigorous agitation and incubated at room temperature for 30 min. Cells were incubated with transfection complexes for 3 h, changed to fresh MEM medium and cultured for 16 h. Cells were washed twice with PBS, scraped, and incubated in reporter lysis buffer (Promega, Madison, WI) on ice for 30 min. After centrifugation, 10 µl lysate was mixed with luciferase substrate (Promega) and the ARE-luciferase activity was measured using a Sirius luminometer (Berthold Detection System). Protein concentration was measured using the Bradford method. Luciferase activity was normalized by protein concentration.
2.5. Cell fractionation
The protocol to extract nuclear and cytoplasmic proteins has been described before [38] with minor modification. Briefly, HeLa cells were cultured in 60 mm Petri dishes and transfected with 3 µg pcDNA3.1-Nrf2-V5 alone or with 1 µg pcDNA3.1-Myc-MafG or MafG2p mutant using the Lipo-fectamine method (Life Technologies). After 24 hours, cells were rinsed with ice-cold PBS and harvested with cell lysis buffer A (50 mM Tris-HCl, 10 mM NaCl, 5 mM MgCl2, 0.5% NP-40, pH8.0). After incubation on ice for 10 min, the samples were centrifuged at 12,000×g for 15 min. Supernatants (cytosolic extract) were collected. Nuclear pellets were washed twice with cell lysis buffer A, and then re-suspended in high salt buffer B (20 mM HEPES, 0.5 M NaCl, 1 mM EDTA, 1 mM DTT, pH7.9), vortexed, and centrifuged. Supernatants (nuclear extract) were collected. The protein concentration of each sample was measured. To generate homogenous electrophoretic pattern, cytosolic proteins were diluted in buffer B. 20 µg nuclear proteins and 10 µg cytosolic proteins were loaded for immunoblot analysis.
2.6. Epifluorescent microscopy
The expression and subcellular distribution of EGFP-Nrf2zip at the presence of mDsRed-MafG and its mutants were examined using a Nikon Eclipse E600 epifluorescent microscope and a Nikon C-SHG1 UV light source purchased from Micron-Optics (Cedar Knolls, NJ). HeLa and HEK cells were cultured on ethanol-sterilized glass coverslips and transfected with 1 µg of EGFP-Nrf2zip together with 0.2 µg mDsRed tagged MafG, MafGzip or MafGΔzip using the Lipofectamine method (Life Technologies) and further cultured in MEM for 24 h. The EGFP signals were examined using a FITC filter. The mDsRed signals were examined using a Texas-Red filter. The epifluorescent images were digitized using a Nikon DXM1200 camera and a Nikon ACT-1 software (version 2). Images were superimposed by Adobe Photoshop CS software.
2.7. Confocal microscopy and fluorescence resonance energy transfer (FRET) assay
For FRET assay, HeLa cells were transfected with plasmids expressing ECFP-Nrf2zip (2 µg), and EYFP-MafG or EYFP-MafG mutants (1 µg) in glass bottom dishes (MatTek, Ashland, MA). Twenty four hours after transfection, cells were examined using a Zeiss LSM510 laser scanning confocal microscope (Zeiss, Thornwood, NY) with a 63× water-immersion objective. We used a sensitized emission method for the FRET assay [39, 40]. Three filter sets were used to detect the donor (ECFP), acceptor (EYFP) and FRET signals. The FRET signal is corrected for spectral bleed through and contamination of donor and acceptor fluorescence according to Youvan’s formula (1) [40]:
| (1) |
(Abbreviation: Fc = FRET concentration, bg = background intensity, cf = correction factor, fret = FRET signal, don = Donor signal, acc = Acceptor signal)
The FRET concentration was normalized to donor and acceptor concentrations according to the following formula (2):
| (2) |
For data acquisition, the donor (ECFP) channel was excited with an Argon laser line at 457 nm and the emission was detected using a band pass filter of 475–525 nm. The acceptor (EYFP) channel was excited at 543 nm and its emission was detected at 545–600 nm. The FRET channel was excited at 457 nm and the emission was detected at 545–600 nm. For data analysis, we used the LSM510 SP2 software (version 3.2) to subtract donor and acceptor bleed through and normalize against acceptor (EYFP) and donor (ECFP) intensity.
3. Results
3.1. The NESzip motif co-localizes with the ZIP dimerization domain
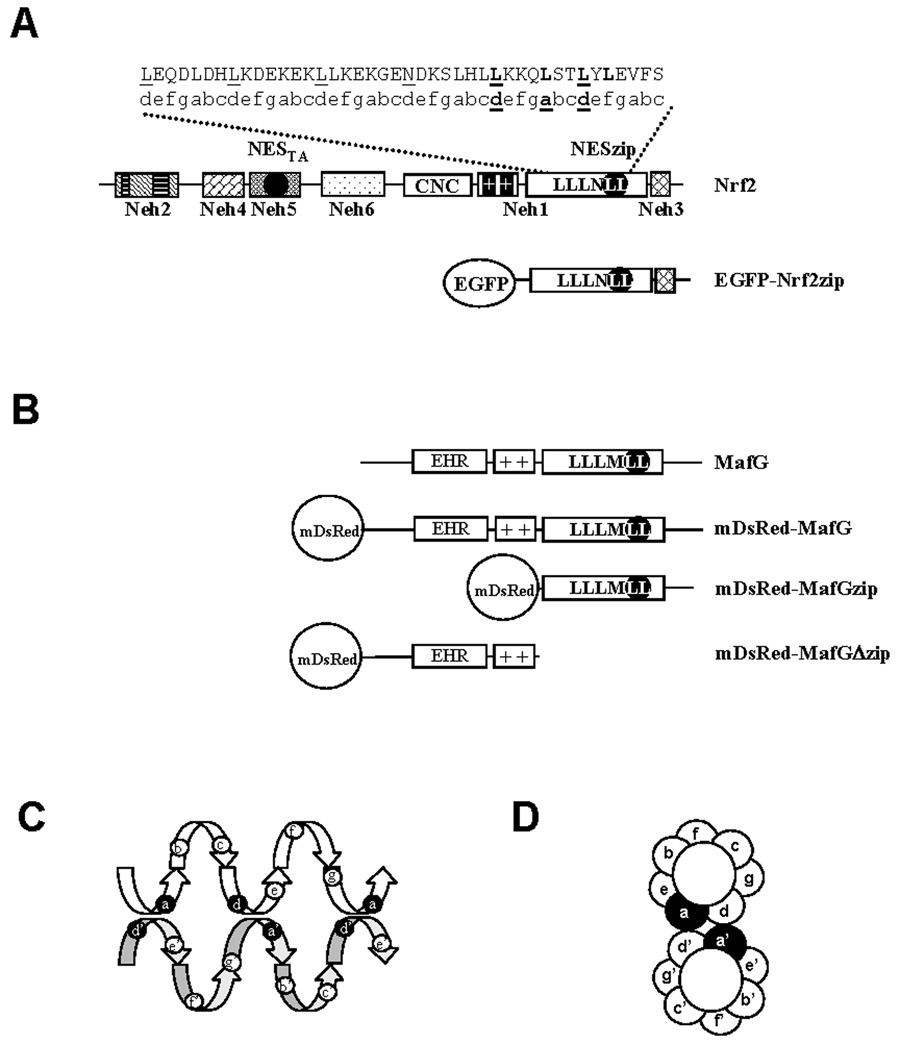
One salient feature of the molecular structure of Nrf2 is the overlapping positioning of functional motifs. The bNLS motif is co-localized with the basic DNA binding domain (Fig. 1A). The NESTA motif is co-localized with the Neh5 transactivation domain (Fig. 1A). The NESzip motif is co-localized with the ZIP dimerization domain [28] (Fig. 1A). Consensus ZIP dimerization domain forms a parallel coiled coil [41] that consists of 4–6 heptads interspersed regularly by leucine residues. Therefore the ZIP domain are also called leucine zipper and formulated as L1L2L3L4L5L6. For Nrf2, the key position of the fourth heptad is a polar asparagine (N) residue, which may preclude the formation of Nrf2/Nrf2 homodimer [25, 26]. So the ZIP domain of Nrf2 can also be formulated as L1L2L3N4L5L6 (Fig.1A). For MafG, the key position of the fourth heptad is a hydrophobic residue methionine, so the ZIP domain of MafG can be represented as L1L2L3M4L5L6 (Fig.1B).
Fig. 1.
Molecular structure of Nrf2 and MafG. (A) Schematic illustration of Nrf2 molecule and plasmid construct. Nrf2 has some highly conserved domains called Nrf2-ECH homology (Neh) domains. The Neh1 contains the ZIP domain (LLLNLL), the basic region (++) and the CNC domain. The Neh2 domain mediates Keap1 binding. The Neh3 domain plays a permissive role of Nrf2 transactivation. The tandem of Neh4 and Neh5 domains mediates cooperative transactivation activity of Nrf2. The Neh6 domain locates in the intervening region. Nrf2 possesses a bipartite NLS (double bars) in the basic region and two NES motifs (black circles). The comprising residues of the ZIP domain of Nrf2 (top panel) are listed according to their position in heptad structure (bottom panel). The demarcation leucines are underlined. The composing leucine residues of the NESzip motif are in bold fonts. (B) Schematic illustration of MafG molecule and plasmid constructs. Typical for small Maf molecules, MafG lacks transactivation domain. MafG has an extensive homology region (EHR) domain, a basic domain (++) and a ZIP domain (LLLMLL). (C) Side view and (D) end view of coiled coil helix of the ZIP motif. When forming dimer, the “a” and “d” residues of one monomer bind with “d’” and “a’” residue of its partner monomer, respectively.
The ZIP domain can also be formulated as (abcdefg)4–6, with each composing residue in every heptad is represented by letter “a” to “g”, respectively. To achieve ZIP-ZIP dimerization, the position “a” and “d” need to be hydrophobic residues. In the process of dimerization, the “a” and “d” residue in one monomer interact with the complementary “d’” and “a’” residue in the opposite monomer, respectively [26] (Fig. 1C–D). The interaction forms a hydrophobic core essential for dimer stability [42].
Canonical NES motif can be formulated as Φ1-(X-X)2–3-Φ2-(X-X)2–3-Φ3-X-Φ4. Φ represents hydrophobic amino acids residues such as leucine, isoleucine, valine, methionine and phenylalanine, and X can be any amino acids [43–45]. In the NESzip motif of Nrf2 (in the 589 a.a. frame), the Φ1 (L537) and Φ3 (L544) residues are located at the “d” position in the fifth and sixth heptad of ZIP domain, respectively. The Φ2 (L541) residue is located at the “a” position in the sixth heptad (Fig.1A). In other words, this NESzip motif occupies three key positions in the dimerization domain. The overlap between the NESzip and the ZIP motif implies that when Nrf2 forms heterodimer via leucine zipper with its obligatory binding partner small Maf proteins, the NESzip motif may be simultaneously masked.
3.2. Dimerization with MafG enhance nuclear retention of Nrf2
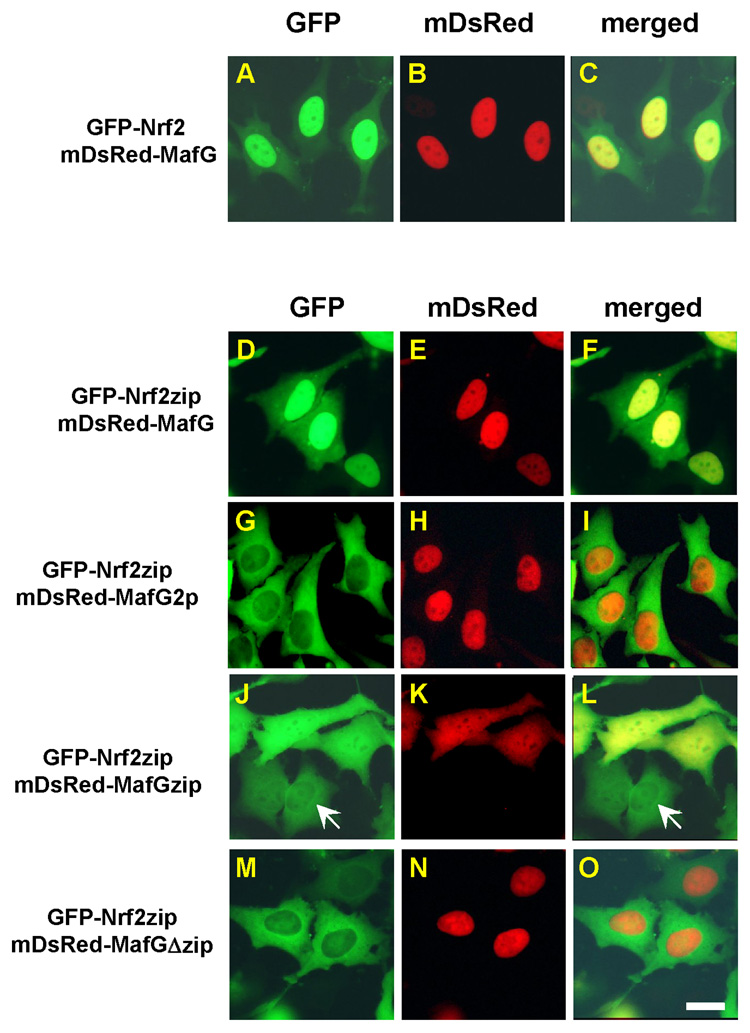
To examine this possibility, we co-expressed an enhanced green fluorescent protein tagged Nrf2 (EGFP-Nrf2) with a monomer Discosoma sp. red fluorescent protein tagged MafG (mDsRed-MafG) in HeLa cells. When expressed alone, EGFP-Nrf2 exhibited a mainly whole cell distribution [22] and mDsRed-MafG exhibited a nuclear distribution (data not shown). When EGFP-Nrf2 was co-expressed with mDsRed-MafG, we observed that mDsRed-MafG could concentrate EGFP-Nrf2 proteins in the nucleus (Fig. 2A–C). This nuclear retention effect appeared to be specific for sMaf proteins, since MafK could also cause accumulation of Nrf2 in cell nucleus (data not shown). In contrast, mDsRed per se failed to alter Nrf2 subcellular distribution (data not shown). These data are also consistent with previous observation that MafK was able to accumulate CNC-bZIP transcription repressor Bach 2 in the nucleus [46].
Fig. 2.
MafG enhances Nrf2 nuclear retention via ZIP-ZIP dimerization in Hela cells. Epifluorescent microscopic examination showed that mDsRed-MafG could arrest EGFP-Nrf2 (A–C) and EGFP-Nrf2zip (D–F) in cell nucleus. In contrast, mDsRed-MafG2p failed to arrest EGFP-Nrf2zip in the nucleus (G–I). The mDsRed-MafGzip showed co-localization with EGFP-Nrf2zip (J–L). In the absence of mDsRed-MafGzip, EGFP-Nrf2zip maintained a cytosolic distribution (arrow, J, L). In contrast, mDsRed-MafGΔzip failed to change EGFP-Nrf2zip distribution (M–O). The left, middle and right column shows EGFP, mDsRed and superimposed images, respectively. Scale bar: 10 µm.
To verify that Nrf2/MafG interaction is mediated by the ZIP domain, we co-expressed EGFP tagged Nrf2zip, a Nrf2 segment that only contains the ZIP domain of Nrf2 (Fig. 1A), with mDsRed-MafG. Whereas EGFP-Nrf2zip mainly exhibited a cytosolic distribution when expressed alone [28], mDsRed-MafG converted it into a nuclear distribution pattern (Fig. 2D–F), suggesting that the Nrf2/MafG interaction is mediated by the ZIP domain. In contrast, when EGFP tagged Nrf2Δzip, a Nrf2 segment with the ZIP domain truncated, was co-expressed with mDsRed-MafG, mDsRed-MafG failed to change the distribution of EGFP-Nrf2Δzip (data not shown). In addition, when the L108 and L115 residues of the ZIP domain of MafG were mutated to alanines, the co-expression of this double point mutant of MafG (MafG2p) failed to alter the cytosolic distribution pattern of Nrf2zip (Fig. 2G–I). We also generated MafG deletion mutants. We truncated the amino-terminus of MafG, including the extensive homology region (EHR) and the basic DNA binding domain, but kept the ZIP domain of MafG intact. The resultant mutant was called MafGzip (Fig. 1B). When mDsRed-MafGzip was expressed alone, it showed a whole cell distribution pattern (data not shown), probably due to the deletion of the NLS motif located in the basic DNA binding region of MafG. When mDsRed-MafGzip was co-expressed with EGFP-Nrf2zip, mDsRed-MafGzip converted the EGFP-Nrf2zip into a whole cell distribution (Fig. 2 J–L). In contrast, in the absence of mDsRed-MafGzip, EGFP-Nrf2zip exhibited a cytosolic distribution (arrow, Fig. 2J, L). The co-localization of MafGzip with Nrf2zip suggested that MafGzip/Nrf2zip binding was very likely mediated by the ZIP domain. We also generated MafG truncation mutant that lacks the ZIP domain (MafGΔzip) (Fig. 1B). In the absence of ZIP domain of MafG, EGFP-Nrf2zip exhibited a cytosolic distribution even at high concentration of mDsRed-MafGΔzip (Fig. 2J–L). In combine, these data suggested that it is the ZIP domain that mediates Nrf2/MafG heterodimerization. Similar results were also observed in HEK cells (Supple. Fig. 1).
3.3. Site mutations disrupting dimerization negated MafG-mediated nuclear retention of Nrf2
To collect more specific evidence that MafG-mediated Nrf2 nuclear retention is mediated by the ZIP-ZIP interaction, we selectively ablated the key leucine residues located in the ZIP domain. In a GST tagged Nrf2zip fusion protein (GST-Nrf2zip), we generated single point (1p) mutant (L537A or L544A), double point (2p) mutant (L537AL544A) and four point (4p) mutant (L537AL541AL544AL546A). In addition, in MafG protein, we also made single point mutant (L108A or L115A) and double point mutant (L108AL115A). The MafG (L108A), MafG (L115A) and MafG2p mutant can also be designated as L5A, L6A, and L5AL6A mutant, respectively.
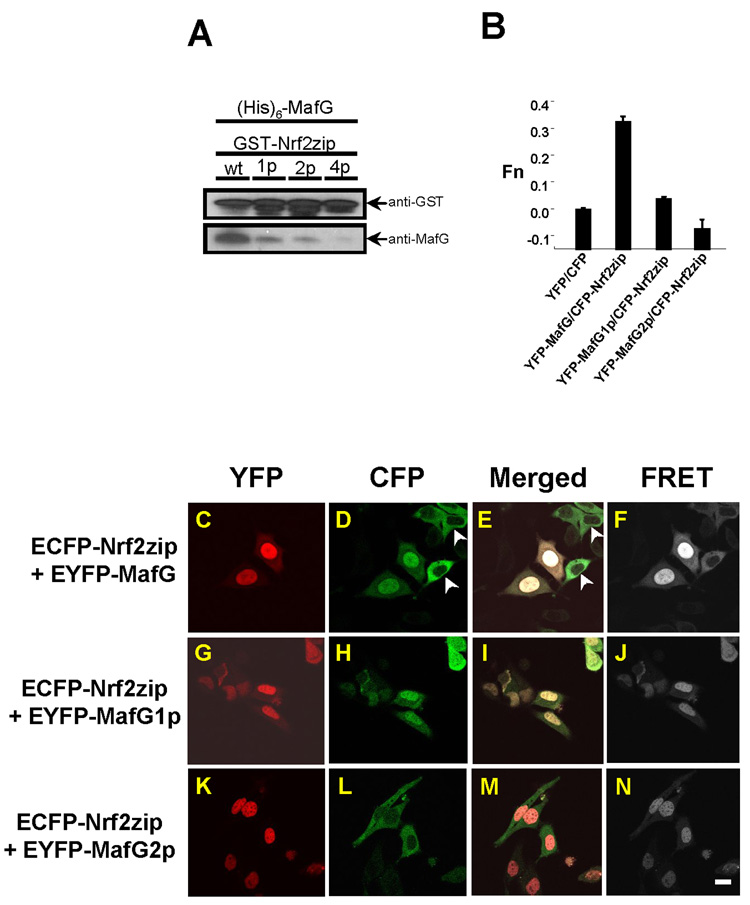
In an in vitro GST pull-down assay, we observed that the single point mutation in the ZIP domain of Nrf2zip attenuated Nrf2zip1p/MafG binding (Fig. 3A). In comparison, the double point mutations could severely reduce Nrf2zip2p/MafG binding (Fig. 3A). The four point mutations completely abolished Nrf2zip4p/MafG binding (Fig. 3A).
Fig. 3.
Dimerization with MafG enhances nuclear retention of Nrf2. (A) GST pull down study showed that wild type Nrf2zip exhibited the strongest binding to MafG. Single point (1p) mutation in Nrf2zip attenuated MafG binding. Two point (2p) mutation further decreased MafG binding. Four point (4p) mutation completely abolished MafG binding. (B) Calculated FRET value showed strong interaction between Nrf2zip/MafG. FRET value was attenuated in Nrf2zip/MafG1p and completely negated in Nrf2zip/MafG2p. As a negative control, CFP/YFP failed to elicit FRET signal. (C–N) Confocal microscopy and FRET assay of MafG/Nrf2zip binding. ECFP-Nrf2zip showed co-localization with EYFP-MafG and EYFP-MafG1p in the nucleus. In the absence of EYFP-MafG, ECFP-Nrf2zip exhibited a cytosolic distribution (arrowheads) (D–E). ECFP-Nrf2zip however, showed a discrete cytosolic distribution, un-overlapped with the nuclear location of EYFP-MafG2p. To enhance visual effect, the EYFP, ECFP and FRET signals are artificially represented with red, green and white color, respectively. Scale bar: 10 µm.
To prove that what we observed in vitro also occur in vivo, we performed the fluorescence resonance emission transfer (FRET) assay. FRET assay has the advantage to discern whether co-localized molecules bind directly to each other [47]. We used a pair of fluorophores enhanced cyan fluorescent protein (ECFP) and enhanced yellow fluorescent protein (EYFP) as FRET donor and acceptor, respectively. We added an ECFP tag to Nrf2zip (ECFP-Nrf2zip) and an EYFP tag to MafG (EYFP-MafG). When expressed alone, EYFP tagged MafG, MafG1p and MafG2p mutants all exhibited a nuclear distribution pattern (data not shown). Like EGFP-Nrf2zip, ECFP-Nrf2zip exhibited a cytosolic distribution when expressed alone (data not shown). These subcellular distribution patterns were consistent with the epifluorescent microscopic observation of mDsRed-MafG, mDsRed-MafG2p and EGFP-Nrf2zip, suggesting that the addition of fluorescent tag did not alter the subcellular distribution of MafG and Nrf2zip.
At the presence of EYFP-MafG (Fig. 3C), condensed nuclear accumulation of ECFP-Nrf2zip was observed (Fig. 3D–E). Strong FRET signals was also detected (Fig. 3B, F), indicating that there was direct binding between Nrf2zip and MafG proteins. In contrast, in the absence of MafG, ECFP-Nrf2zip exhibited a cytosolic distribution (arrowheads, Fig. 3D–E). Single point mutation in MafG (MafG1p) attenuated the FRET signal (Fig. 3B, J) but failed to abolish Nrf2zip nuclear retention (Fig. 3H–I). In contrast, double point mutation in MafG (MafG2p) completely diminished FRET signal (Fig. 3B, N) and nullified Nrf2zip nuclear retention (Fig. 3L–M). In fact, cytosolic distribution of ECFP-Nrf2zip (Fig. 3L) and nuclear distribution of EYFP-MafG2p (Fig. 3K) did not appear to overlap at all (Fig. 3M). Our observation that double point mutation (L5AL6A) in the ZIP domain of MafG disrupted MafG2p/Nrf2zip binding is consistent with the previous reports that double point mutation (L2PM4P) in the ZIP domain of MafK can negate MafK/p45 NF-E2 [48] and MafK/Bach2 [46] heterodimerization.
The same effect was also observed when the ZIP domain of Nrf2 was mutated. Whereas single point mutation in the NESzip motif only attenuated FRET signal, four point mutation could completely diminish the FRET signal and abolish Nrf2zip4p nuclear localization (Supple. Fig. 2).
Therefore mutations disrupting dimerization appeared to concomitantly negate Nrf2 nuclear accumulation.
3.4. Dimerization with MafG precluded CRM1/Nrf2zip binding
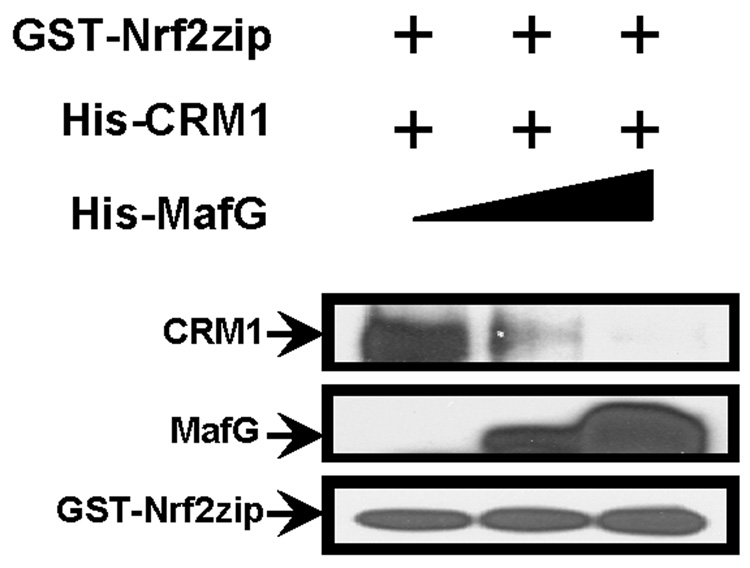
Previous studies showed that nuclear exporting activity mediated by NESzip is CRM1-dependent. In an immunoprecipitation study, CRM1 was found to bind with GFP-Nrf2zip but not GFP-Nrf2zip4p mutant [28]. If Nrf2zip/MafG dimer formation indeed masked the NESzip motif, NESzip/CRM1 binding should be compromised. To examine this possibility, we did a GST pull-down assay. In the absence of MafG protein, GST-Nrf2zip was found to bind with (His)6-CRM1 (Fig. 4). At the presence of increased amount of (His)6-MafG however, Nrf2zip/CRM1 binding was attenuated and eventually disappeared (Fig. 4). Therefore MafG proteins appeared to be able to inhibit Nrf2zip/CRM1 binding in a dose dependent way.
Fig. 4.
Dimerization with MafG precludes Nrf2zip/CRM1 binding. 1 µg of GST-Nrf2zip proteins and 2 µg (His)6-CRM1 proteins were incubated with different amount of (His)6-MafG proteins (0, 1 and 5 µg). GST pull-down results showed that MafG inhibited Nrf2zip/CRM1 binding in a dose-dependent manner.
3.5. MafG-mediated nuclear retention could stabilize Nrf2 proteins
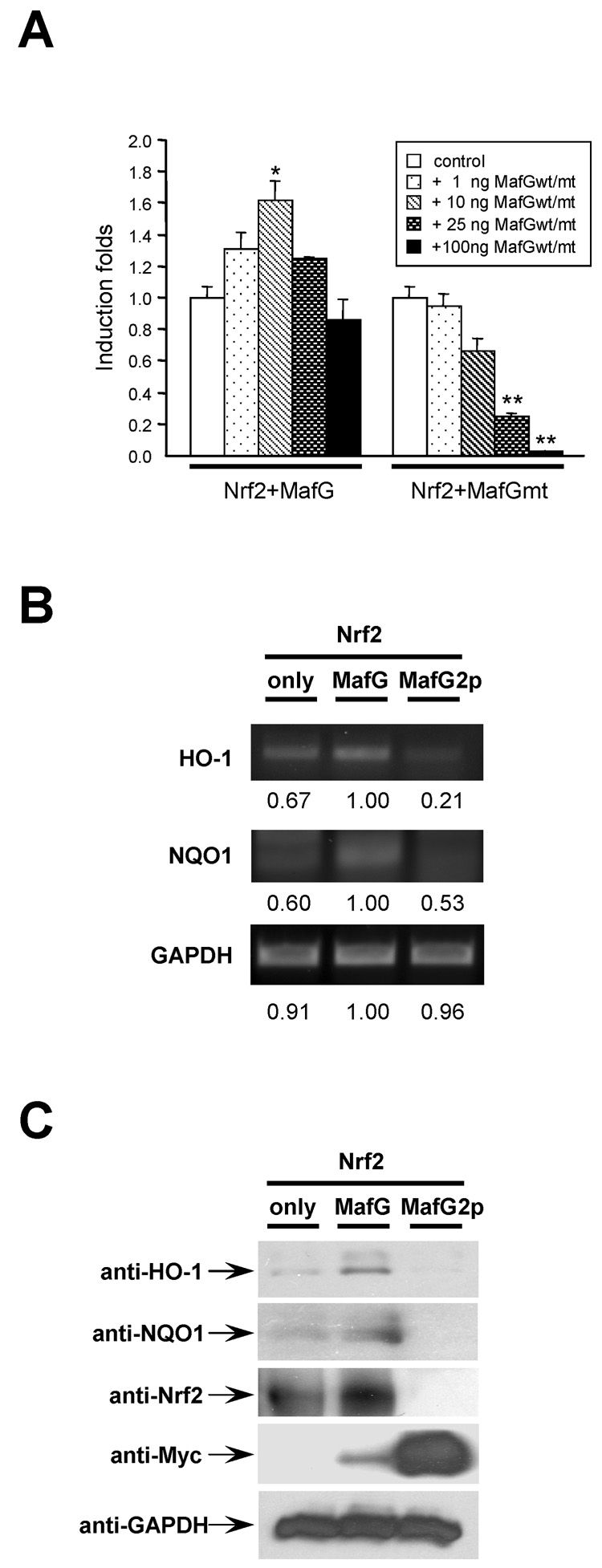
When MafG was co-expressed with Nrf2 in HeLa cells, MafG exerted a bi-directional transcription regulatory effect. At low concentrations, MafG amplified Nrf2 induced ARE-Luciferase activities in a dose-dependent manner (Fig. 5A). At higher concentrations, the amplification effect of MafG was attenuated and even reversed (Fig. 5A). This observation is consistent with previous reports [48, 49], probably due to the reason that overexpressing MafG may favor the formation of MafG/MafG homodimer. Since the MafG/MafG homodimer functions as transcription repressor, they may compete with MafG/Nrf2 heterodimer and alleviate the up-regulatory effect exerted by the MafG/Nrf2 heterodimer. Co-expressing dimerization-deficient mutant of MafG, MafG2p, inhibited Nrf2 signaling in a dose-dependent manner (Fig. 5A). In fact, MafG2p mutant appeared to function as a dominant-negative inhibitor of Nrf2. In agreement with our reporter gene activity assays, our RT-PCR assay showed that the transcription of some phase II genes, including heme oxygenase 1 (HO-1) and NAD(P)H:quinone oxidoreductase 1 (NQO1), was significantly attenuated when Nrf2 was co-expressed with MafG2p mutants (Fig. 5B). At protein level, co-expression of Myc-MafG could remarkably intensify the induction of HO-1 and NQO1 elicited by Nrf2. In contrast, Myc-MafG2p inhibited the induction of HO-1 and NQO1 elicited by Nrf2 (Fig. 5C). The inhibitory effect of Myc-MafG2p was also observed in HEK cells (Supple. Fig. 3). It is noteworthy that the immunoreactivities of Myc-MafG2p were much stronger than that of Myc-MafG (Fig. 5C and Supple. Fig. 3). We also observed more intense expression of MafG2p than MafG with EYFP and mDsRed tags (data not shown). Since both MafG and MafG2p were constructed in an identical expressing vector, their in vivo transcription and translation should be the same. The observed difference of MafG and MafG2p immunoreactivities may be derived from difference in degradation. It suggests that the wild type MafG but not the MafG2p may be controlled by an unraveled negative feedback regulation to avoid hyperactivity of sMaf/Nrf2 signaling.
Fig. 5.
MafG regulates Nrf2 signaling. (A) Reporter gene activity assay. Co-expressing wild type MafG regulated Nrf2 induced ARE-luciferase activities in a bi-directional way. In contrast, co-expressing MafG2p mutant markedly inhibited Nrf2 induced ARE-luciferase activities in a dose dependent way. Hela cells were transfected with 1 µg pHM6-Nrf2, 0.5 µg plasmid expressing ARE-Luc together with 0, 1, 10, 25 and 100 ng plasmids expressing wild type (wt) or 2p mutant (mt) MafG. Twenty four hours after transfection, cells were harvested. Luciferase activity was measured and normalized to protein concentration. Single and double asterisks indicate statistical significance (t-test) of p<0.05 and p<0.01, respectively. (B) RT-PCR analysis of the transcription of phase II genes. 3 µg pcDNA3.1-Nrf2-V5 plasmids were expressed alone or co-expressed with 1 µg pcDNA3.1-Myc-MafG or pcDNA3.1-Myc-MafG2p in HeLa cells. Total RNAs were extracted using RNeasy method and reversed transcribed (RT). Same amount of RT samples were amplified by poly chain reaction (PCR) for 40 cycles and resolved in 1% agarose gel and visualized by ethidium bromide incorporation exited by UV light. The densitometric values of RT-PCR products were labeled underneath. (C) Western blotting results showed that MafG and MafG2p could enhance and attenuate Nrf2-induced HO-1 and NQO1 expression, respectively.
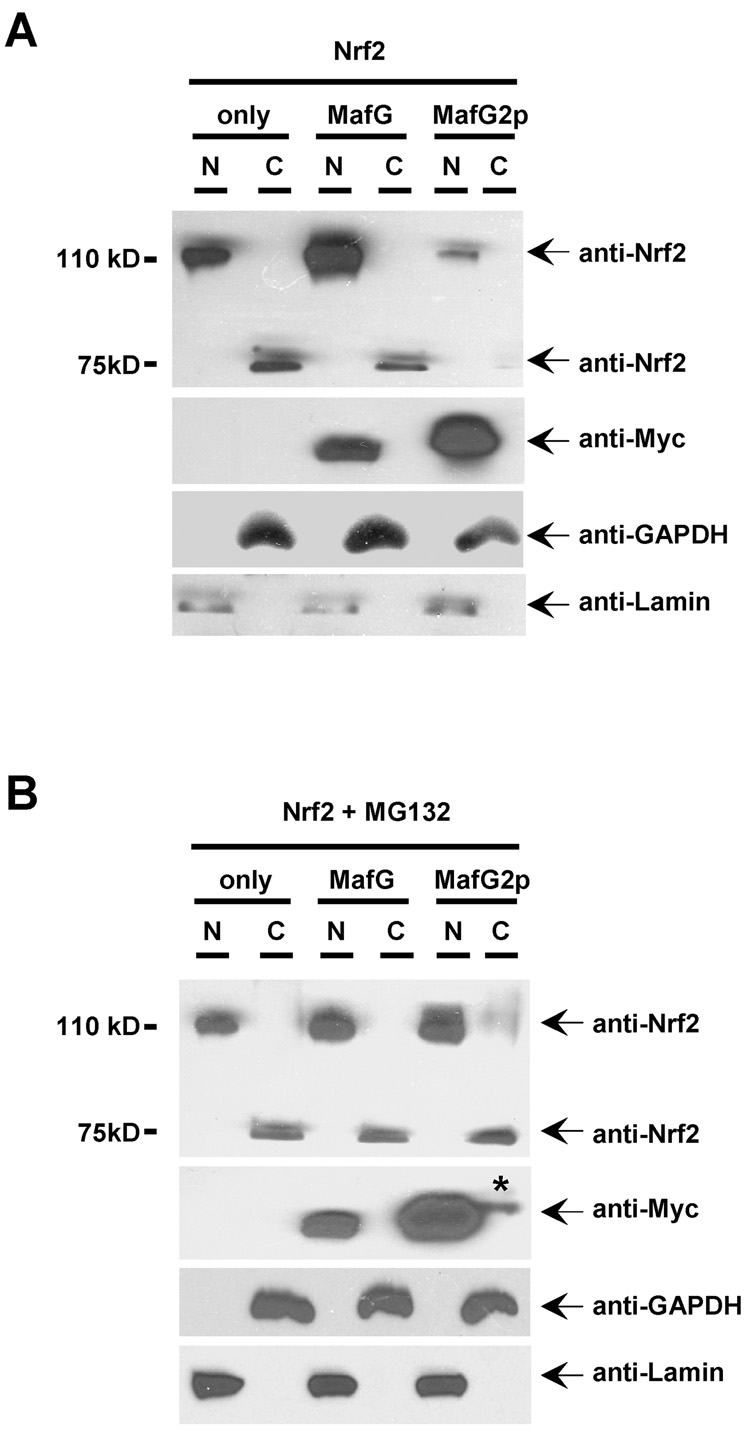
When Nrf2 was expressed alone in Hela cells, Nrf2 immunoreactivities could be detected in the nuclear fraction. Co-expression with Myc-MafG increased Nrf2 immunoreactivities in the nucleus. In contrast, at the presence of Myc-MafG2p, Nrf2 immunoreactivities were significantly attenuated (Fig. 6A). These data suggested that if Nrf2 protein failed to form a heterodimer with MafG and thus be retained in cell nucleus, its exit to the cytoplasm might expose it to proteasomal degradation. In the cytosolic fraction, virtually no Nrf2 immunoreactivities could be detected at ~110kD of full length Nrf2. However, we did observe Nrf2 immunoreactivities at ~75 kD (Fig. 6A). It is unknown whether these ~75 kD products were degraded form of Nrf2. For Nrf1, there is a 65 kD isoform functioning as a dominant negative inhibitor [50]. Further studies are needed to examine this possibility.
Fig. 6.
Nrf2/MafG dimerization stabilizes Nrf2 proteins. (A) Cell fractionation studies showed that, at unstressed condition, Nrf2 immunoreactivities observed in nuclear fraction were enhanced and attenuated when co-expressed with Myc-MafG and Myc-MafG2p, respectively. (B) After overnight MG132 (10 µM) treatment, similar amount of Nrf2 immunoreactivities were observed in cells expessing Nrf2 alone or co-expressing Nrf2 with MafG and MafG2p mutant. Lamin A and GAPDH were used as controls for endogenous nuclear and cytosolic proteins, respectively. The asterisk indicates weak Myc-MafG2p immunoreactivity observed in the cytosolic fraction.
When Hela cells were treated with proteasomal degradation inhibitor MG132 (10 µM) overnight, similar amount of Nrf2 immunoreactivities were detected in nuclear fractions expressing Nrf2 alone and in nuclear fractions co-expressing Nrf2 with Myc-MafG or Myc-MafG2p (Fig. 6B). The validity of MG132 effect was also observed in the increased amount of Myc-MafG2p proteins. Even weak Myc-MafG2p immunoreactivities could be detected in the cytosolic fraction (asterisk, Fig. 6B). Intriguingly, we also detected the ~75 kD bands in cytosolic fractions of MG132 treated samples (Fig. 6B). In contrast, only weak ~110 kD immunoreactivities were detected in the cytosolic fraction (Fig. 6B).
Collectively, these data suggested that MafG-mediated Nrf2 nuclear retention could stabilize Nrf2 protein by preventing its cytosolic degradation.
4. Discussion
4.1. The ZIP domain is necessary and sufficient for Nrf2/MafG heterodimerization
In the present study, we find that heterodimerization with MafG can enhance Nrf2 nuclear retention. Nrf2/MafG dimerization can effectively mask the NESzip motif of Nrf2, as illustrated by the diminished Nrf2zip/CRM1 binding at the presence of MafG. MafG-mediated Nrf2 nuclear accumulation appears to be able to stabilize Nrf2 proteins. For the first time, we delineate that the NESzip activity can be switched off at normal physiological condition.
Our deletion studies show that the ZIP domain is indispensable for Nrf2/MafG binding (Fig. 2). Previously it is reported that DNA binding can facilitate dimerization among bZIP proteins [51, 52]. Since dimerization could be formed between Nrf2zip and MafGzip that lack the basic region (Fig. 1A), our data show that the ZIP domain per se is competent to mediate dimer formation. Furthermore, the absence of the basic region also rules out the possibility that the observed nuclear retention of Nrf2 is actually resulted from the exposure of a hidden bNLS motif. The specificity of ZIP-ZIP interaction is further corroborated by our observation that site-directed mutagenesis ablating key leucine residues in the ZIP domain can negate Nrf2/MafG dimerization and Nrf2 nuclear retention (Fig. 3). Collectively, these data show that the ZIP domain per se is necessary and sufficient for the formation of Nrf2/MafG heterodimer.
4.2. Nrf2/sMafG heterodimerization masks the NESzip motif
Since three composing leucine residues of NESzip are located in the dimerization interface (Fig. 1A), in the process of Nrf2/MafG heterodimerization, these leucine residues are very likely buried in the hydrophobic core and consequently become inaccessible to CRM1 binding. Our in vitro competitive binding assay supported this hypothesis. In a concentration dependent manner, MafG inhibited Nrf2zip binding to CRM1 (Fig. 4). To further verify whether dimerization can mask the NESzip motif, selective labeling and detecting of comprising leucines of the NESzip motif may provide definitive evidence. The analytic methods of hydrogen-deuterium exchange and nuclear magnetic resonance analysis may envision unequivocally whether these leucine residues are masked or not. Unfortunately, these expertise are beyond our capability.
The NESzip motif of Nrf2 is highly conserved across-species, with the only exception of zebra fish [28]. In contrast, this NESzip motif is not conserved in Nrf1 and Nrf3 at all [28]. The high cross-species conservation of Nrf2 NESzip motif implies that the mechanism to switch-off NESzip motif via heterodimerization may be widely employed in various Nrf2 proteins and demand further examination.
Oligomerization-regulated NES/NLS activities have been reported in diverse transcription regulators. A NES motif is characterized in the tetramerization domain of tumor suppressor factor p53. Tetramerization of p53 can occlude this NES and cause p53 nuclear accumulation [53]. The NES motifs in RXRα [54], Survivin [55], CRKL [56] can be masked by homodimerization. The NES motif in breast cancer associated protein BARD1 can be masked by heterodimerization with BRCA1 [57]. In addition, oligomerization can mask the NLS motif and lead to cytosolic accumulation of NF-AT4 [58]. Heterodimerizing with 14-3-3 protein can disable the adjacent NES and NLS motif in hTERT [59] and cdc25 [60], respectively. Therefore oligomerization-mediated switch on/off of NES/NLS activities may be extensively employed as a general regulatory mechanism in cell signaling.
4.3. MafG mediated nuclear retention may potentiate Nrf2 signaling
Nrf2 is a labile protein, with very fast turnover rate [61, 62]. Our present study shows that MafG-mediated Nrf2 nuclear retention can stabilize Nrf2. Cytoplasmic exclusion of Nrf2 proteins may enable Nrf2 proteins to evade proteasomal degradation (Fig. 5C), as corroborated by our MG132 study (Fig. 6B). Stabilized Nrf2 may intensify and prolong antioxidant response (Fig. 5B–C). Previously, small Maf proteins are only portrayed as obligatory binding partners, escorting Nrf2 to recognize and bind to ARE/EpRE [6, 23]. The present study however implies that small Maf proteins may not only initiate but also amplify Nrf2 signaling. Previously, Nioi et al published an elegant chromosomal immunoprecipitation (ChIP) study. They observed that the ratio of Nrf2/MafK binding to the NQO1 ARE could increase more than 10 folds when Hepa-1c1c7 cells were challenged with oxidative stress [9]. Masking of the NESzip motif, in combine with inactivation of the NESTA motif [22], may account for effect recruitment of Nrf2 by sMaf proteins.
Unlike MafG, the heterodimerization-deficient mutant MafG2p failed to stabilize Nrf2 proteins (Fig. 5C and 6A). We were quite surprised to observe the dominant negative inhibitory effect exerted by MafG2p. One prominent observation was that the immunoreactivities of MafG2p were remarkably higher than that of MafG, both in Hela cells (Fig. 5C and Fig. 6) and in HEK cells (Supple. Fig. 3). The presence of robust MafG2p immunoreactivities suggests that MafG2p expression may not be regulated like MafG. How high abundance of MafG2p inhibits Nrf2 signaling in a dominant negative manner? In addition to the likelihood that MafG2p fails to exclude Nrf2 from cytosolic degradation, it is also possible that there is residual binding between MafG2p/sMaf. If the potency of MafG2p/sMaf dimer formation is only partially compromised, since the sMaf/sMaf homodimer function as trans-repressor, the accumulated MafG2p may intensify trans-repression. In future study, we may use in vitro binding assay to measure the binding affinity among Myc and V5 tagged wind type and mutant MafG. Furthermore, we can use FRET assay to examine whether the affinity of MafG/MafG2p binding is different from MafG/MafG binding.
Since both the NESTA motif and NESzip motif can be switched off, it naturally raises a question about their relevant importance in the activation of Nrf2 signaling. Under the homeostatic condition, the NESTA motif may remain active. So the switch on/off of the NESzip motif may be more important to affect Nrf2 subcellular distribution and constitutive induction of phase II genes. If Nrf2 passively enters into the nucleus, Nrf2 can be arrested by sMaf proteins in the nucleus via the masking of the NESzip motif. This may partially explain the observation that the majority cells expressing GFP-Nrf2 exhibited a whole cell or nuclear distribution pattern [22]. Under the oxidative condition however, the switch off of the NESTA motif may play the key role in eliciting Nrf2 nuclear translocation, the masking of NESzip motif may play a subsequent but indispensable role to reinforce Nrf2 nuclear accumulation and amplify Nrf2 signaling. In other words, NESTA and NESzip may be implied in different stages of a sequential Nrf2 activation process.
There is an ARE enhancer in the promoter region of MafG gene [63]. Therefore, the activation of Nrf2 signaling may elicit a positive feedback. An oxidative stimulus may not only induce the transcription of phase II/III genes but also elevate MafG expression. Due to their nuclear localization, small Maf proteins may be safe from proteasomal degradation. Higher MafG activities may sensitize the cell, priming the cell to respond to another oxidative stress more effectively. Further investigations are necessary to examine whether the antioxidant response has a context dependent adaptive nature.
The characterization of dimerization-mediated switch off of NESzip activity may also provide clue to deepen our understanding whether other mechanism(s) also regulates Nrf2 signaling. Can any other factor(s) also modify the formation of Nrf2/sMafG dimer? Recently, it was reported that sumoylation at a consensus SUMO site (13VKRE16) in MafG can attenuate MafG/p45 NF-E2 transcription efficiency [64]. This sumoylation-mediated active repression is sensitive to HDAC inhibition [64], implying intricate interplay with other transcription co-factors. In fact, there is a consensus SUMO site (515LKDE518) located in the ZIP domain of Nrf2. It requires further examination whether this site can be sumoylated. It is very tempting to speculate that sumoylation at this site may inhibit Nrf2/MafG dimerization. Recently, a tyrosine phosphorylation site (Y560) [65] and a consensus MAPK site (S561) has been identified in the vicinity of Nrf2 ZIP domain. Since phosphorylation may have an impact on an adjacent SUMO site [66], it remains to be examined whether phosphorylation at Y560 and/or S561 can have impact on Nrf2/sMaf dimerization.
In conclusion, we found that the NESzip motif functions as a conditional NES. The switch on/off of the NESzip motif may have important functional significance. At unstressed condition, the constant exposure of the NESzip and NESTA motifs maintains nuclear exclusion of unbound Nrf2 protein and subjects it to proteasomal degradation. When Nrf2 signaling is activated by oxidative stress, occlusion of NESzip via dimerization with sMaf proteins switches Nrf2 to a stable nuclear accumulation condition. When the oxidative stress is eased up, the occlusion of NESzip motif may be gradually removed in parallel to the alleviation of stress response. In combine, these data delineate that Nrf2 signaling is delicately orchestrated.
Supplementary Material
Supplementary Fig. 1: MafG enhances Nrf2 nuclear retention via ZIP-ZIP dimerization in HEK cells. Epifluorescent microscopic examination showed that mDsRed-MafG could arrest EGFP-Nrf2 (A–C) and EGFP-Nrf2zip (D–F) in cell nucleus. In contrast, mDsRed-MafG2p failed to arrest EGFP-Nrf2zip in the nucleus (G–I). The mDsRed-MafGzip showed colocalization with EGFP-Nrf2zip (J–L). In contrast, mDsRed-MafGΔzip failed to change EGFP-Nrf2zip distribution (M–O). The left, middle and right column shows EGFP, mDsRed and superimposed images, respectively. Scale bar: 10 µm.
Supplementary Fig. 2: FRET analysis of direct binding among wild type and various mutants of Nrf2zip and MafG.
Supplementary Fig. 3: Western blotting results showed that MafG2p could attenuate Nrf2-induced HO-1 and NQO1 expression in HEK cells.
Acknowledgments
We thank Drs. Jefferson Chan and Yuet W. Kan for providing valuable reagents; and all the members of Dr. Kong’s laboratory for their assistance and critical reading of this manuscript. This work was supported by National Institute of Health grant R01 CA94828 to A.-N.T. K
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Klaassen CD, Slitt AL. Regulation of hepatic transporters by xenobiotic receptors. Curr Drug Metab. 2005;6:309–328. doi: 10.2174/1389200054633826. [DOI] [PubMed] [Google Scholar]
- 3.Mandlekar S, Hong JL, Kong AN. Modulation of metabolic enzymes by dietary phytochemicals: a review of mechanisms underlying beneficial versus unfavorable effects. Curr Drug Metab. 2006;7:661–675. doi: 10.2174/138920006778017795. [DOI] [PubMed] [Google Scholar]
- 4.Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes & development. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motohashi H, O'Connor T, Katsuoka F, Engel JD, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 7.Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. The Journal of biological chemistry. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 9.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. The Biochemical journal. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinkova-Kostova AT, Holtzclaw WD, Kensler TW. The role of Keap1 in cellular protective responses. Chem Res Toxicol. 2005;18:1779–1791. doi: 10.1021/tx050217c. [DOI] [PubMed] [Google Scholar]
- 11.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Molecular and cellular biology. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Molecular and cellular biology. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Molecular and cellular biology. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Molecular and cellular biology. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osburn WO, Wakabayashi N, Misra V, Nilles T, Biswal S, Trush MA, Kensler TW. Nrf2 regulates an adaptive response protecting against oxidative damage following diquat-mediated formation of superoxide anion. Arch Biochem Biophys. 2006;454:7–15. doi: 10.1016/j.abb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson LA, Gemin A, Espiritu R, Singh G. ets-1 is transcriptionally up-regulated by H2O2 via an antioxidant response element. Faseb J. 2005;19:2085–2087. doi: 10.1096/fj.05-4401fje. [DOI] [PubMed] [Google Scholar]
- 17.Itoh K, Tong KI, Yamamoto M. Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic Biol Med. 2004;36:1208–1213. doi: 10.1016/j.freeradbiomed.2004.02.075. [DOI] [PubMed] [Google Scholar]
- 18.Dhakshinamoorthy S, Porter AG. Nitric oxide-induced transcriptional up-regulation of protective genes by Nrf2 via the antioxidant response element counteracts apoptosis of neuroblastoma cells. The Journal of biological chemistry. 2004;279:20096–20107. doi: 10.1074/jbc.M312492200. [DOI] [PubMed] [Google Scholar]
- 19.Park EY, Kim SG. NO signaling in ARE-mediated gene expression. Methods Enzymol. 2005;396:341–349. doi: 10.1016/S0076-6879(05)96028-X. [DOI] [PubMed] [Google Scholar]
- 20.Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biological chemistry. 2006;387:1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- 21.Purdom-Dickinson SE, Sheveleva EV, Sun H, Chen QM. Translational control of nrf2 protein in activation of antioxidant response by oxidants. Molecular pharmacology. 2007;72:1074–1081. doi: 10.1124/mol.107.035360. [DOI] [PubMed] [Google Scholar]
- 22.Li W, Yu SW, Kong AN. Nrf2 possesses a redox-sensitive nuclear exporting signal in the Neh5 transactivation domain. The Journal of biological chemistry. 2006;281:27251–27263. doi: 10.1074/jbc.M602746200. [DOI] [PubMed] [Google Scholar]
- 23.Blank V, Andrews NC. The Maf transcription factors: regulators of differentiation. Trends in biochemical sciences. 1997;22:437–441. doi: 10.1016/s0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- 24.Toki T, Itoh J, Kitazawa J, Arai K, Hatakeyama K, Akasaka J, Igarashi K, Nomura N, Yokoyama M, Yamamoto M, Ito E. Human small Maf proteins form heterodimers with CNC family transcription factors and recognize the NF-E2 motif. Oncogene. 1997;14:1901–1910. doi: 10.1038/sj.onc.1201024. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto T, Kyo M, Kamiya T, Tanaka T, Engel JD, Motohashi H, Yamamoto M. Predictive base substitution rules that determine the binding and transcriptional specificity of Maf recognition elements. Genes Cells. 2006;11:575–591. doi: 10.1111/j.1365-2443.2006.00965.x. [DOI] [PubMed] [Google Scholar]
- 26.Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol. 2002;22:6321–6335. doi: 10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 28.Li W, Jain MR, Chen C, Yue X, Hebbar V, Zhou R, Kong AN. Nrf2 Possesses a redox-insensitive nuclear export signal overlapping with the leucine zipper motif. The Journal of biological chemistry. 2005;280:28430–28438. doi: 10.1074/jbc.M410601200. [DOI] [PubMed] [Google Scholar]
- 29.Jain AK, Bloom DA, Jaiswal AK. Nuclear import and export signals in control of Nrf2. The Journal of biological chemistry. 2005;280:29158–29168. doi: 10.1074/jbc.M502083200. [DOI] [PubMed] [Google Scholar]
- 30.Sun Z, Zhang S, Chan JY, Zhang DD. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Molecular and cellular biology. 2007;27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu R, Chen C, Mo YY, Hebbar V, Owuor ED, Tan TH, Kong AN. Activation of mitogen-activated protein kinase pathways induces antioxidant response element-mediated gene expression via a Nrf2-dependent mechanism. The Journal of biological chemistry. 2000;275:39907–39913. doi: 10.1074/jbc.M004037200. [DOI] [PubMed] [Google Scholar]
- 32.Blank V, Kim MJ, Andrews NC. Human MafG is a functional partner for p45 NF-E2 in activating globin gene expression. Blood. 1997;89:3925–3935. [PubMed] [Google Scholar]
- 33.Munro S, Pelham HR. Use of peptide tagging to detect proteins expressed from cloned genes: deletion mapping functional domains of Drosophila hsp 70. The EMBO journal. 1984;3:3087–3093. doi: 10.1002/j.1460-2075.1984.tb02263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Southern JA, Young DF, Heaney F, Baumgartner WK, Randall RE. Identification of an epitope on the P and V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. The Journal of general virology. 1991;72(Pt 7):1551–1557. doi: 10.1099/0022-1317-72-7-1551. [DOI] [PubMed] [Google Scholar]
- 35.Keum YS, Han YH, Liew C, Kim JH, Xu C, Yuan X, Shakarjian MP, Chong S, Kong AN. Induction of heme oxygenase-1 (HO-1) and NAD[P]H: quinone oxidoreductase 1 (NQO1) by a phenolic antioxidant, butylated hydroxyanisole (BHA) and its metabolite, tert-butylhydroquinone (tBHQ) in primary-cultured human and rat hepatocytes. Pharmaceutical research. 2006;23:2586–2594. doi: 10.1007/s11095-006-9094-2. [DOI] [PubMed] [Google Scholar]
- 36.Wang M, Li YQ, Zhong N, Chen J, Xu XQ, Yuan MB. [Induction of uridine 5'-diphosphate-glucuronosyltransferase gene expression by sulforaphane and its mechanism: experimental study in human colon cancel cells] Zhonghua yi xue za zhi. 2005;85:819–824. [PubMed] [Google Scholar]
- 37.Shen G, Hebbar V, Nair S, Xu C, Li W, Lin W, Keum YS, Han J, Gallo MA, Kong AN. Regulation of Nrf2 transactivation domain activity. The differential effects of mitogen-activated protein kinase cascades and synergistic stimulatory effect of Raf and CREB-binding protein. J Biol Chem. 2004;279:23052–23060. doi: 10.1074/jbc.M401368200. [DOI] [PubMed] [Google Scholar]
- 38.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia Z, Liu Y. Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys J. 2001;81:2395–2402. doi: 10.1016/S0006-3495(01)75886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Youvan DC, Marrs BL. Molecular genetics and the light reactions of photosynthesis. Cell. 1984;39:1–3. doi: 10.1016/0092-8674(84)90185-5. [DOI] [PubMed] [Google Scholar]
- 41.McLachlan AD, Stewart M. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. J Mol Biol. 1975;98:293–304. doi: 10.1016/s0022-2836(75)80119-7. [DOI] [PubMed] [Google Scholar]
- 42.Thompson KS, Vinson CR, Freire E. Thermodynamic characterization of the structural stability of the coiled-coil region of the bZIP transcription factor GCN4. Biochemistry. 1993;32:5491–5496. doi: 10.1021/bi00072a001. [DOI] [PubMed] [Google Scholar]
- 43.Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 44.Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 45.Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katsuoka F, Motohashi H, Tamagawa Y, Kure S, Igarashi K, Engel JD, Yamamoto M. Small Maf compound mutants display central nervous system neuronal degeneration, aberrant transcription, and Bach protein mislocalization coincident with myoclonus and abnormal startle response. Molecular and cellular biology. 2003;23:1163–1174. doi: 10.1128/MCB.23.4.1163-1174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallrabe H, Periasamy A. Imaging protein molecules using FRET and FLIM microscopy. Current opinion in biotechnology. 2005;16:19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 48.Motohashi H, Katsuoka F, Shavit JA, Engel JD, Yamamoto M. Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small Maf proteins. Cell. 2000;103:865–875. doi: 10.1016/s0092-8674(00)00190-2. [DOI] [PubMed] [Google Scholar]
- 49.Marini MG, Chan K, Casula L, Kan YW, Cao A, Moi P. hMAF, a small human transcription factor that heterodimerizes specifically with Nrf1 and Nrf2. The Journal of biological chemistry. 1997;272:16490–16497. doi: 10.1074/jbc.272.26.16490. [DOI] [PubMed] [Google Scholar]
- 50.Wang W, Kwok AM, Chan JY. The p65 isoform of Nrf1 is a dominant negative inhibitor of ARE-mediated transcription. The Journal of biological chemistry. 2007;282:24670–24678. doi: 10.1074/jbc.M700159200. [DOI] [PubMed] [Google Scholar]
- 51.O'Neil KT, Hoess RH, DeGrado WF. Design of DNA-binding peptides based on the leucine zipper motif. Science (New York, N.Y. 1990;249:774–778. doi: 10.1126/science.2389143. [DOI] [PubMed] [Google Scholar]
- 52.Shuman JD, Vinson CR, McKnight SL. Evidence of changes in protease sensitivity and subunit exchange rate on DNA binding by C/EBP. Science (New York, N.Y. 1990;249:771–774. doi: 10.1126/science.2202050. [DOI] [PubMed] [Google Scholar]
- 53.Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. The EMBO journal. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao X, Liu W, Lin F, Li H, Kolluri SK, Lin B, Han YH, Dawson MI, Zhang XK. Retinoid X receptor regulates Nur77/TR3-dependent apoptosis [corrected] by modulating its nuclear export and mitochondrial targeting. Molecular and cellular biology. 2004;24:9705–9725. doi: 10.1128/MCB.24.22.9705-9725.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engelsma D, Rodriguez JA, Fish A, Giaccone G, Fornerod M. Homodimerization antagonizes nuclear export of survivin. Traffic (Copenhagen, Denmark) 2007;8:1495–1502. doi: 10.1111/j.1600-0854.2007.00629.x. [DOI] [PubMed] [Google Scholar]
- 56.Harkiolaki M, Gilbert RJ, Jones EY, Feller SM. The C-terminal SH3 domain of CRKL as a dynamic dimerization module transiently exposing a nuclear export signal. Structure. 2006;14:1741–1753. doi: 10.1016/j.str.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 57.Rodriguez A, Schuchner S, Au WW, Fabbro M, Henderson BR. Nuclear-cytoplasmic shuttling of BARD1 contributes to its proapoptotic activity and is regulated by dimerization with BRCA1. Oncogene. 2004;23:1809–1820. doi: 10.1038/sj.onc.1207302. [DOI] [PubMed] [Google Scholar]
- 58.Zhu J, Shibasaki F, Price R, Guillemot JC, Yano T, Dotsch V, Wagner G, Ferrara P, McKeon F. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell. 1998;93:851–861. doi: 10.1016/s0092-8674(00)81445-2. [DOI] [PubMed] [Google Scholar]
- 59.Seimiya H, Sawada H, Muramatsu Y, Shimizu M, Ohko K, Yamane K, Tsuruo T. Involvement of 14-3-3 proteins in nuclear localization of telomerase. The EMBO journal. 2000;19:2652–2661. doi: 10.1093/emboj/19.11.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumagai A, Dunphy WG. Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes & development. 1999;13:1067–1072. doi: 10.1101/gad.13.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. The Journal of biological chemistry. 2004;279:31556–31567. doi: 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- 62.Stewart D, Killeen E, Naquin R, Alam S, Alam J. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. The Journal of biological chemistry. 2003;278:2396–2402. doi: 10.1074/jbc.M209195200. [DOI] [PubMed] [Google Scholar]
- 63.Katsuoka F, Motohashi H, Engel JD, Yamamoto M. Nrf2 transcriptionally activates the mafG gene through an antioxidant response element. The Journal of biological chemistry. 2005;280:4483–4490. doi: 10.1074/jbc.M411451200. [DOI] [PubMed] [Google Scholar]
- 64.Motohashi H, Katsuoka F, Miyoshi C, Uchimura Y, Saitoh H, Francastel C, Engel JD, Yamamoto M. MafG sumoylation is required for active transcriptional repression. Molecular and cellular biology. 2006;26:4652–4663. doi: 10.1128/MCB.02193-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jain AK, Jaiswal AK. Phosphorylation of tyrosine 568 controls nuclear export of Nrf2. The Journal of biological chemistry. 2006;281:12132–12142. doi: 10.1074/jbc.M511198200. [DOI] [PubMed] [Google Scholar]
- 66.Yang XJ, Gregoire S. A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Molecular cell. 2006;23:779–786. doi: 10.1016/j.molcel.2006.08.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1: MafG enhances Nrf2 nuclear retention via ZIP-ZIP dimerization in HEK cells. Epifluorescent microscopic examination showed that mDsRed-MafG could arrest EGFP-Nrf2 (A–C) and EGFP-Nrf2zip (D–F) in cell nucleus. In contrast, mDsRed-MafG2p failed to arrest EGFP-Nrf2zip in the nucleus (G–I). The mDsRed-MafGzip showed colocalization with EGFP-Nrf2zip (J–L). In contrast, mDsRed-MafGΔzip failed to change EGFP-Nrf2zip distribution (M–O). The left, middle and right column shows EGFP, mDsRed and superimposed images, respectively. Scale bar: 10 µm.
Supplementary Fig. 2: FRET analysis of direct binding among wild type and various mutants of Nrf2zip and MafG.
Supplementary Fig. 3: Western blotting results showed that MafG2p could attenuate Nrf2-induced HO-1 and NQO1 expression in HEK cells.