Abstract
Efforts to elucidate the nature of the bimolecular interaction of parathyroid hormone (PTH) with its cognate receptor, the PTH receptor type 1 (PTHR1), have relied heavily on benzoylphenylalanine- (Bpa-) based photoaffinity cross-linking. However, given the flexibility, size, and shape of Bpa, the resolution at the PTH–PTHR1 interface appears to be reaching the limit of this technique. Here we employ a disulfide-trapping approach developed by others primarily for use in screening compound libraries to identify novel ligands. In this method, cysteine substitutions are introduced into a specific site within the ligand and a region in the receptor predicted to interact with each other. Upon ligand binding, if these cysteines are in close proximity, they form a disulfide bond. Since the geometry governing disulfide bond formation is more constrained than Bpa cross-linking, this novel approach can be employed to generate a more refined molecular model of the PTH–PTHR1 complex. Using a PTH analogue containing a cysteine at position 1, we probed 24 sites and identified 4 in PTHR1 to which cross-linking occurred. Importantly, previous photoaffinity cross-linking studies using a PTH analogue with Bpa at position 1 only identified a single interaction site. The new sites identified by the disulfide-trapping procedure were used as constraints in molecular dynamics simulations to generate an updated model of the PTH–PTHR1 complex. Mapping by disulfide trapping extends and complements photoaffinity cross-linking. It is applicable to other peptide–receptor interfaces and should yield insights about yet unknown sites of ligand–receptor interactions, allowing for generation of more refined models.
Class II G protein-coupled receptors (GPCRs)1 interact with physiologically important peptides. There is intense study of how these peptides associate with their cognate receptors since elucidation of these interactions should provide important insights for the rational design of ligands with enhanced pharmacological properties for use in treating an array of diseases (1, 2). Over the past decade, photoaffinity cross-linking has been employed to study the interaction of a number of peptide–GPCR interactions (3). This technique involves systematically probing the receptor for regions of interaction using a peptide that incorporates a photoreactive moiety that irreversibly cross-links to the receptor. We and others have used this approach extensively to study the interaction of parathyroid hormone (PTH) with its cognate receptor, the PTH receptor type 1 (PTHR1) (4-12). This hormone–receptor system plays an integral role in calcium metabolism and bone biology.
Our research is focused on studying the bimolecular interface of the PTH–PTHR1 complex in order to gain insights that will aid the design of ligands of PTHR1 for the treatment of osteoporosis and other disorders. The extreme N-terminus of PTH is essential for its activity (13). Therefore, elucidating the interaction between this region of the hormone with the receptor should provide valuable information for the future design of novel PTHR1 agonists. Using benzoylphenylalanine- (Bpa-) mediated photoaffinity cross-linking, we previously reported that an analogue of PTH incorporating a Bpa moiety at position 1 (Bpa1-PTH) cross-links to M425 of the receptor, close to the top of transmembrane (TM) 6 (6). However, based on recent findings that Bpa exhibits a strong cross-linking preference for methionine, the precision of this contact site is not well-defined (14-19). Given the methionine preference, coupled with the rotational freedom of the Bpa moiety (encompassing a large radius for cross-linking), further refinement of this PTH/PTHR1 region of interaction using a Bpa photoaffinity cross-linking approach is precluded.
In order to obtain a more detailed and accurate molecular model of the PTH–PTHR1 complex, we used a recently developed approach for cross-linking peptides to receptor in a novel manner. The disulfide bond-mediated approach for cross-linking small molecules and peptides to GPCRs has been used successfully (20, 21), primarily as a means for screening thiol-containing compound libraries in order to identify novel ligands. We now utilize this technique for studying the interactions of PTH and PTHR1 and mapping the interface of their biomolecular complex. After the original submission of this paper, a report of a closely similar approach applied to the C5a receptor appeared (22). This technique monitors disulfide bond formation between a peptide analogue containing a cysteine at a defined position and mutant receptors that have individual cysteines introduced at sites predicted to be close to the natural binding site for the region of the peptide being studied. The rationale is that if the cysteine of the ligand comes into close proximity to an introduced cysteine in the receptor during ligand binding, a disulfide bond will form. Using the disulfide-trapping approach, we probed a total of 24 positions within TM5 and TM6 of PTHR1 for interaction with Cys1. The positions selected for study were based on the previous finding that M425 in TM6 is the contact point for position 1 in PTH (6). Also, the TM5 domain was included in the study based on recent related research from our laboratory (unpublished data) indicating interactions between TM5 and TM6 upon ligand binding. A portion of extracellular loop 3 (EC3), namely, positions 427–429, was included. But the lack of cross-linking confirmed our interest in focusing on TM5 and TM6. Although EC3 is involved in ligand binding, it likely interacts with positions of PTH other than position 1. Application of the disulfide approach enabled us to identify four residues of PTHR1, situated toward the extracellular portions of TM5 and TM6, which form part of the binding pocket for the extreme N-terminus of PTH. Importantly, although 24 positions were probed, only 4 cross-linked. This contrasts to the observation of many cross-linking sites detected due to the “methionine magnet effect” (14 -19). The novel sites of interaction identified by this study generated a molecular model that exhibits important differences from our previous model based on Bpa photoaffinity cross-linking data.
In this report, we demonstrate that the disulfide-trapping approach, when guided by a model based on the widely employed Bpa-mediated photoaffinity cross-linking methodology, can provide higher resolving power than Bpa photo-cross-linking alone. This method should have important applications in mapping ligand–receptor interfaces and in the further refining of existing models, not just for the PTH–PTHR1 system but also for class II GPCRs, in general.
EXPERIMENTAL PROCEDURES
Materials
Iodine-125 was purchased from Amersham Biosciences, and Iodo-Gen precoated tubes were obtained from Pierce (Rockford, IL). DMEM, Opti-MEM serum-free medium, penicillin, streptomycin, fetal bovine serum, and phosphate-buffered saline (PBS) were obtained from Invitrogen (Carlsbad, CA). FuGENE6 was from Roche Diagnostics (Indianapolis, IN). The Dual-Glo luciferase assay system was from Promega (Madison, WI). Plasmid maxi- and mini- prep kits were from Qiagen (Valencia, CA). All of the 5′-phosphorylated oligonucleotides were purchased from Sigma-Genosys (St. Louis, MO). Goat polyclonal anti-PTH/PTHrP-R antibody (E-17) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA), and donkey anti-goat phycoerythrin- (PE-) labeled secondary antibody was obtained from Abcam (Cambridge, MA). Polyacrylamide solution (30%) and molecular weight standards (Precision Plus protein standards, dual color) were from Bio-Rad (Richmond, CA). The GraphPad Prism software was from GraphPad software (San Diego, CA). All other reagents and plastic ware were obtained from Fisher Scientific (Pittsburgh, PA).
Peptide Synthesis and Radioiodination
The Cys1-PTH peptide ([Cys1,Nle8,18, Arg13,26,27,2-Nal23,Tyr34]bPTH-(1–34)-NH2) was synthesized using Fmoc chemistry in our laboratory using a Symphony peptide synthesizer (Protein Technologies, Tucson, AZ) and purified using methods described previously (12). Cys was introduced as Fmoc-Cys (Trt) for protection during synthesis. Deprotection at the end of synthesis was achieved by exposure to 81.5% trifluoroacetic acid, 5% thioanisole, 5% phenol, 5% water, 2.5% dithiothreitol, and 1% trisisopropylsilane. The molecular mass of the purified peptide was confirmed by mass spectroscopy. Radioiodination was performed using a two-step (indirect) procedure based on precoated iodination tubes according to the manufacturer's protocol (Pierce). Preactivation of 125I was performed to prevent the oxidation of the thiol group of Cys1-PTH. Initially, 70 μL of NaH2PO4 and 20 μL of 125I were transferred to an Iodo-Gen precoated tube. After 6 min, the activated iodine solution was transferred to a microcentrifuge tube containing the peptide in 10 mM AcOH (110 μg/11 μL). This was done to prevent direct contact of the peptide with the free oxidizing agent. Dithiothreitol, as a reducing agent, was added (20 μL, 5 mM) following iodination to both quench the reaction and ensure that the peptide is reduced in 130 μL of water. Then the radiolabeled ligand was purified from the excess iodine and reducing agent using HPLC (35–42% acetonitrile gradient over 30 min). The most radioactive samples (counted on γ counter) were analyzed by SDS–PAGE. A band corresponding to PTH monomer was observed even after undergoing oxidation/iodination. Continued protection was afforded by keeping the peptide in a frozen solution until use.
Site-Directed Mutagenesis
wtPTHR1, previously cloned into pcDNA3.1 (23), served as the template for the introduction of cysteines by site-directed mutagenesis using the QuickChange II kit (Stratagene, La Jolla, CA); all reactions were carried out according to the manufacturer's instructions. Cloning was confirmed by DNA sequence analysis (Tufts University Core Facility, Boston, MA).
Cell Culture
Cos-7 cells were used since they do not express endogenous PTHR1. The cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS and 1× penicillin and streptomycin in a humidified 5% CO2 atmosphere at 37 °C.
Adenylyl Cyclase Activity
Cos-7 cells were seeded onto 24-well collagen-coated plates (BD Biocoat cellware) at a density of 60000 cells/well. The cells were transiently transfected the next day with 100 ng/well PTHR1 DNA (wild type or specific mutant receptors), 100 ng/well Cre-luciferase DNA, and 10 ng/well Renilla luciferase DNA (internal control for transfection efficiency). Posttransfection (18–24 h), the media were replaced with DMEM containing 5% FBS. The luciferase assay for measuring hormone-stimulated adenylyl cyclase activity was carried out as described (12). The EC50 was calculated by nonlinear regression analysis using the GraphPad Prism software. The ECmax values are expressed as “fold” increase in activity observed with 1 μM PTH over basal activity.
Flow Cytometry
Cos-7 cells were seeded onto 100 mm plates at a density of 1 × 106 cells/plate and transiently transfected the following day with 1.7 μg of PTHR1 DNA (wild type or specific mutant receptors). Posttransfection (18–24 h), 1 × 106 cells were transferred to 5 mL polypropylene round-bottom tubes and centrifuged at 250g for 5 min. Cells were washed with PBS and subsequently incubated on ice for 1 h with 1 μg of anti-PTHR E-17 antibody. Cells were washed three times with PBS prior to addition of 1 mL of PE-labeled anti-goat secondary antibody (1:200 dilution in PBS). Samples were incubated on ice in the dark for 1 h and subsequently washed three times with PBS. Cells were resuspended in 0.5% formaldehyde and stored at 4 °C. Flow cytometry was performed at the Tufts University Flow Cytometry Core Facility using a MoFlo high-performance cell sorter (Dako, Carpinteria, CA), counting 20000 cells per sample, in duplicate. Nontransfected Cos-7 cells served as a negative control for PTHR1 expression.
Disulfide Trapping
Cos-7 cells were seeded onto 24-well collagen-coated plates (BD Biocoat cellware) at a density of 60000 cells/well. The cells were transiently transfected the next day with 200 ng/well wtPTHR1 or mutant receptor DNA as described above. Posttransfection (18–24 h), the media were replaced with DMEM containing 5% FBS, and 125I-Cys1-PTH was added at 100000 cpm/well. Plates were incubated for 2 h at room temperature, followed by a PBS washing step. SDS–PAGE buffer [62.5 mM Tris (pH 6.8), 10% glycerol, 2% SDS, 0.1% bromophenol blue], with or without 5% β-mercaptoethanol, was added to each well. Following 45 min incubation, cells were scraped from the wells and separated through a 7.5% SDS–polyacrylamide gel. The gels were dried and exposed to film for 1–2 days at −80 °C prior to developing.
Molecular Modeling
The results from the disulfide-trapping experiments were utilized as distance restraints in a molecular dynamics simulation, using the NAMD simulation package, following published procedures (14). In these simulations, a restraint of 6.0 Å was placed between the Cβ atoms of the Ser1 of PTH and the appropriate residue of the receptor: L368, Y421, F424, and M425. As with our previous simulations (employing Bpa cross-linking data), the wild-type ligand and receptor were utilized in order to define as closely as possible the binding mode of the natural ligand. The Cβ–Cβ distance about a Cys-Cys disulfide bond ranges from 3.8 to 4.6 Å. We employed a slightly larger distance restraint during the MD simulations to account for the utilization of the wild-type sequences.
RESULTS
Activity of Cys1-PTH and PTHR1 Cysteine Mutants
We generated a PTH analogue by substituting position 1 (a serine in the native peptide) with a cysteine (Cys1-PTH). Modification of position 1 of PTH can produce profound effects on hormonal bioactivity (13). Therefore, assessing the bioactivity of the Cys1-PTH ligand was prerequisite to initiating the disulfide-trapping strategy. This modified ligand retained the ability to activate wtPTHR1, as judged by a ligand-induced luciferase-reporter assay which monitors the cAMP signaling pathway (Figure 1). Cysteines were introduced individually into 13 positions in the upper region of TM6 of PTHR1 by site-directed mutagenesis. An identical bioassay, using unmodified PTH, confirmed that each of the introduced cysteines had no major detrimental effect on the ability of the mutant receptors to bind hormone and elicit a signal (Table 1). Analysis of cell surface expression of each of the mutant receptors showed all of them to be present on the cell surface (Figure 2).
Figure 1.
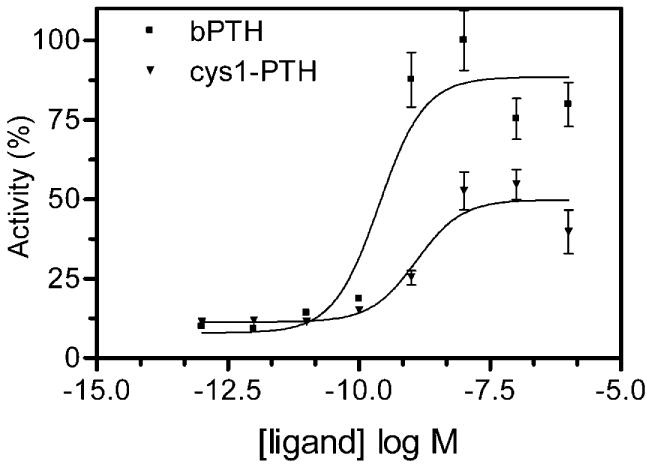
Activation of PTHR1 by Cys1-PTH. Cos-7 cells were transiently transfected with wtPTHR1 DNA and assayed for Cys1-PTH-stimulated adenylyl cyclase activity. The EC50 value, calculated from the concentration curve using GraphPad Prism, was 3.2 nM. Bars show SEM of three or more replicates.
Table 1.
PTH-Stimulated Adenylyl Cyclase Activity of PTHR1 Mutants Containing a Cysteine in TM6a
| PTHR1 mutant | EC50, nM | ECmax fold increase over basal |
|---|---|---|
| wtPTHR1 | 0.2 | 32.0 |
| F417C | 246 | 10.3 |
| G418C | 4.1 | 10.6 |
| V419C | 10.5 | 24.6 |
| H420C | 6.1 | 3.1 |
| Y421C | 0.5 | 7.2 |
| I422C | 2.3 | 1.2 |
| V423C | 0.1 | 5.0 |
| F424C | 1.2 | 8.2 |
| M425C | 0.2 | 5.4 |
| A426C | 0.2 | 5.1 |
| T427C | 0.3 | 2.3 |
| P428C | 5.2 | 11.9 |
| Y429C | 0.9 | 3.2 |
Cos-7 cells were transiently transfected with constructs encoding wtPTHR1 or mutant receptor and assayed for PTH-stimulated adenylyl cyclase activity. All constructs were assayed in triplicate. The EC50 values were calculated from concentration curves using GraphPad Prism. ECmax values are expressed as the fold increase in activity observed with 1 μM PTH over basal activity.
Figure 2.
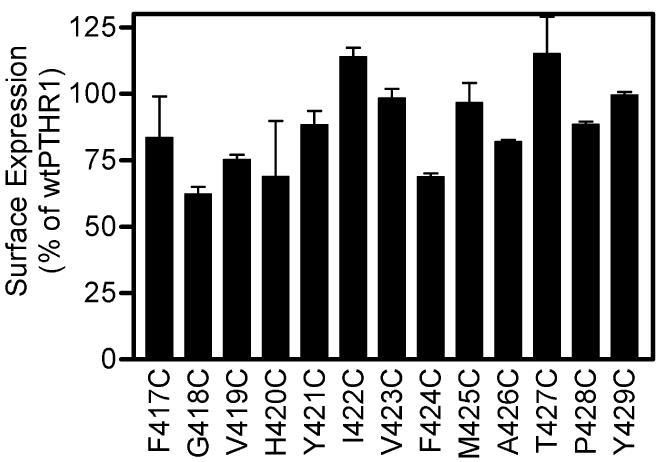
Cell surface expression of PTHR1 mutants with cysteines introduced into TM6. Cos-7 cells were transiently transfected with wtPTHR1 or mutant receptor DNA and analyzed by flow cytometry using an antibody that recognizes the N-terminal extracellular domain of PTHR1 in conjunction with a PE-labeled secondary antibody. The percentage expression levels for wtPTHR1 were set to 100%, and the relative expression levels of all mutant receptors were compared to this. Bars show SEM of two replicates.
Identification of Residues in TM6 in Close Proximity to Position 1 of PTH
When cells transiently expressing mutant PTHR1 constructs were incubated with 125I-labeled Cys1-PTH, lysed, and analyzed by nonreducing SDS–PAGE, three PTHR1 mutants were identified that cross-linked the labeled peptide (Figure 3, upper panel). The cross-linking of the peptide to each of these mutant receptors was abrogated when the samples were analyzed by SDS–PAGE performed under reducing conditions, confirming that the observed cross-linked product results from the formation of a disulfide bond (Figure 3, lower panel). Specificity of the binding was confirmed by performing the experiments in the presence of competing unmodified PTH: no cross-linking of Cys1-PTH to the mutant receptors was observed in the presence on 1 μM PTH (Figure 4).
Figure 3.
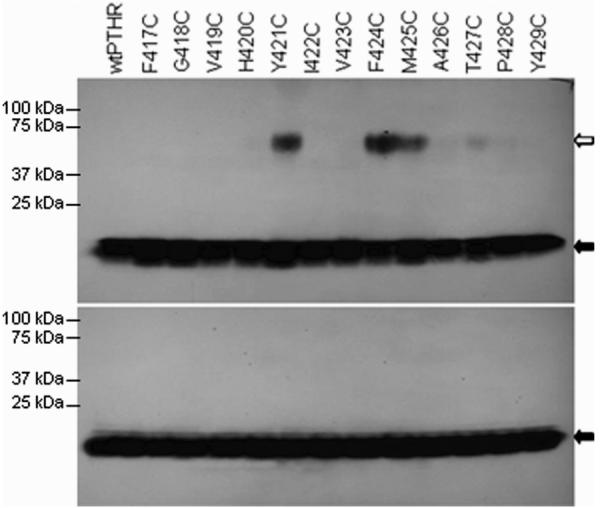
Disulfide trapping of Cys1-PTH to PTHR1 mutants with introduced cysteines in TM6. Cells expressing the various mutant receptors were incubated with 125I-labeled Cys1-PTH and subsequently analyzed by SDS–PAGE in the absence (upper panel) or presence (lower panel) of reducing agent (β-mercaptoethanol). The white arrow indicates Cys1-PTH cross-linked to receptors, and the black arrow indicates the un-cross-linked peptide.
Figure 4.
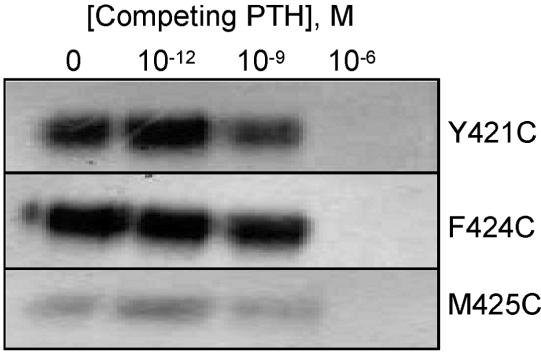
Specificity of interaction of Cys1-PTH with mutant receptors. The mutant PTHR1 forms that cross-linked Cys1-PTH were exposed to 125I-labeled Cys1-PTH in the presence of increasing concentrations of unmodified PTH, and the samples were then analyzed by SDS–PAGE and autoradiography as detailed in Experimental Procedures. Only the region of the gels corresponding to the cross-linked product is shown. In the presence of the highest concentration of PTH used (1 μM), the cross-linking of Cys1-PTH to each receptor mutant was totally abrogated.
Identification of Residues in TM5 in Close Proximity to Position 1 of PTH
We initially focused our attention on residues in the upper region of TM6 since our previous finding, using Bpa1-PTH, showed that M425 appeared to be a contact point for position 1 of the peptide (6). While investigating this region of TM6, ongoing work in our laboratory suggested that, upon activation of the receptor, the upper regions of TM5 and TM6 move closer together (unpublished data). From our finding that the N-terminal residue of PTH is positioned close to the top of TM6, it was presumed that it also comes close to the top of TM5. We tested this hypothesis by introducing cysteines individually into 11 positions in the upper regions of TM5 of PTHR1 (Table 2 and Figure 5) and repeating the disulfide-trapping procedure with Cys1-PTH. Analysis of the data obtained reveals that only a single mutant, L368C, cross-links to the peptide (Figure 6, upper panel). Once again, this cross-linking was abrogated when the sample was analyzed by SDS–PAGE performed under reducing conditions (Figure 6, lower panel).
Table 2.
PTH-Stimulated Adenylyl Cyclase Activity of PTHR1 Mutants Containing a Cysteine in TM5a
| PTHR1 mutant | EC50, nM | ECmax fold increase over basal |
|---|---|---|
| V365C | 11.9 | 16.6 |
| P366C | 1.9 | 10.6 |
| I367C | 6.9 | 26.4 |
| L368C | 12.4 | 31.3 |
| A369C | 13.6 | 31.7 |
| S370C | 13.1 | 56.0 |
| I371C | 13.6 | 41.6 |
| V372C | 7.5 | 72.6 |
| L373C | 17.4 | 52.7 |
| N374C | 24.2 | 50.2 |
| F375C | 22.9 | 62.6 |
Cos-7 cells were transiently transfected with constructs encoding wtPTHR1 or mutant receptor and assayed for PTH-stimulated adenylyl cyclase activity. All constructs were assayed in triplicate. The EC50 values were calculated from concentration curves using GraphPad Prism. ECmax values are expressed as the fold increase in activity observed with 1 μM PTH over basal activity.
Figure 5.
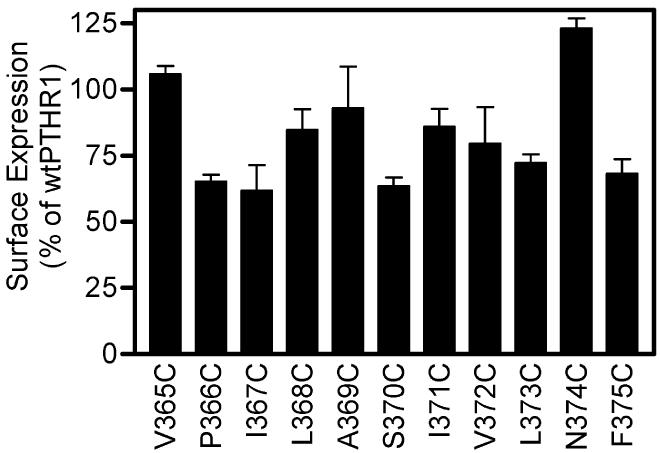
Cell surface expression of PTHR1 mutants with cysteines introduced into TM5. Analysis was performed exactly as described for Figure 2. The percentage expression levels for wtPTHR1 were set to 100%, and the relative expression levels of all mutant receptors were compared to this. Bars show SEM of two replicates.
Figure 6.
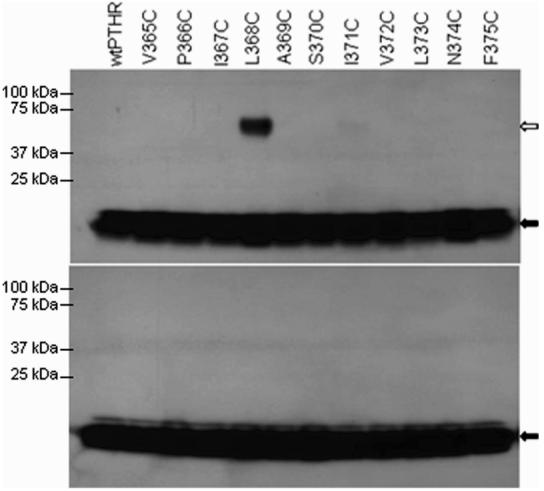
Disulfide trapping of Cys1-PTH to PTHR1 mutants with introduced cysteines in TM5. Cells expressing the various mutant receptors were incubated with 125I-labeled Cys1-PTH and subsequently analyzed by SDS–PAGE in the absence (upper panel) or presence (lower panel) of reducing agent (β-mercaptoethanol). The white arrow indicates Cys1-PTH cross-linked to receptors, and the black arrow indicates the un-cross-linked peptide.
Molecular Modeling of the Site of Interaction of Position 1 of PTH with PTHR1
The new interaction sites identified by disulfide trapping were introduced as distance restraints in molecular dynamics (MD) simulations. All of the simulations were carried out using the wild-type receptor and ligand. The distance restraints were placed onto the Cβ atoms of Ser1 (the native residue at position 1 of PTH) and the appropriate residue of PTHR1 (L368, Y421, F424, or M425). Importantly, each of these MD simulations resulted in essentially identical binding modes, indicating that all of the interactions identified by disulfide trapping are consistent. The resulting model from the F424 simulation is illustrated in Figure 7. The resulting Cβ–Cβ distances between Ser1 and L368, Y421, and F424 are 6.1, 6.3, and 4.1 Å, respectively, for this simulation. The result from the M425 simulation led to a very similar binding mode, but with the N-terminus of the ligand less displaced toward the cytosol, producing a longer distance to L368 (ca. 7.3 Å). In comparison with our previous PTH/PTHR1 model, we observe that the far N-terminus of PTH (Ser1-Val2) shifts more downward (more cytosolic in orientation), with no change in the interactions of the remainder of the ligand. This movement of the PTH N-terminus toward the cytosolic portion of the receptor is led by the interaction with L368 and Y421. The Ser1 residue used in the simulations adopts a position equidistant between the extracellular portion of TM5 and TM6, consistent with the general mode of GPCR activation by ligand.
Figure 7.
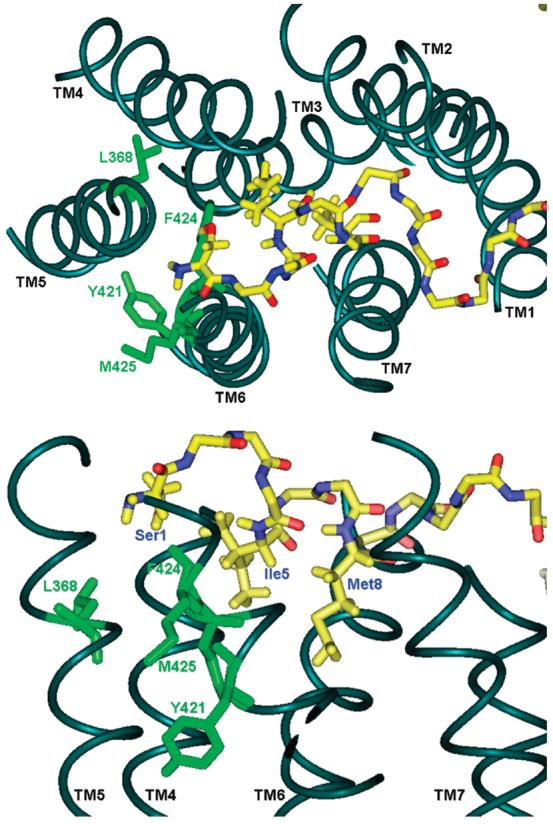
Model of the interaction of position 1 of PTH with PTHR1 obtained by molecular dynamics simulations. The four sites in PTHR1 that formed disulfide bonds with Cys1-PTH are shown in green; the backbone of the ligand is shown in yellow (nitrogen = blue; oxygen = red). The side chains of Ile5 and Met8 of the peptide are displayed for orientation. The top panel depicts a view from above the receptor; the lower panel depicts a side view.
DISCUSSION
Photoaffinity cross-linking has traditionally been used to investigate sites of interaction between peptide hormones and GPCRs (reviewed in ref 3). We have used this technique extensively in our study of the interaction of PTH with PTHR1 (4-6, 8, 9, 12). By combining data collected from extensive photoaffinity cross-linking studies with molecular modeling, a model of the PTH–PTHR1 bimolecular complex has been generated that identifies regions of contact at the interface between the hormone and the receptor. But further iterations of this approach are now yielding diminishing returns. We believe that we are reaching the limit of the resolving power of this technique, making the generation of newer, more detailed models of the PTH–PTHR1 interaction increasingly difficult. Also, we have recently explored an important caveat associated with the commonly employed photoreactive Bpa moiety: we delineated the “methionine magnet effect” over a range of at least 11 amino acid residues in the PTHR1 (14, 19). Although highly useful for surveying a hormone–receptor in order to derive a first-generation map of the bindecular interface, the properties of Bpa have important implications that compromise its utility in photoaffinity cross-linking to delineate contact sites for use in creating high-resolution models of the PTH–PTHR1 interaction and peptide–receptor interactions in general.
A novel cross-linking strategy, mediated through thiol-reactive groups, has been used to investigate the interactions of small molecules and short peptides with GPCRs (20, 21). This disulfide-trapping approach previously utilized principally for screening of compound libraries has now been successfully applied by us to mapping the PTH–PTHR1 bimolecular interface. After the original submission of this paper, a closely similar approach applied to the C5a receptor was reported (22). Unlike Bpa-mediated photoaffinity cross-linking, the disulfide-trapping procedure allows for the identification of numerous residues within the receptor that are in close proximity to a defined position in the peptide being investigated: in this case, position 1 of PTH. Our previous work with Bpa1-PTH identified only a single residue, M425, of TM6 in PTHR1 that was proximal to the N-terminus of the peptide (6). With the Bpa methodology, a single ligand can be used to ask only a single experimental question. In contrast, the disulfide-trapping methodology amplifies the utility of each ligand probe: each probe can be used in a series of experiments to map the surface of interaction along the receptor (although the method is limited to Cys substitutions in both receptor and ligand that are tolerated in terms of biological function). For example, in the investigations reported herein, we interrogated a total of 24 potential contact sites within TM5 and TM6 of PTHR1 with the Cys1-PTH peptide. The probing yields specific results: 20 of 24 sites displayed no cross-linking. On the basis of this 24 position scan, we are now able to define three additional points of interaction between position 1 of PTH and PTHR1. Further delineation of the binding pocket for the extreme N-terminus of PTH using our Bpa1-PTH analogue would have required at best a labor-intensive approach. More likely, further refinement would not be possible using Bpa methodology.
SDS–PAGE analysis consistently showed that the Y421C, F424C, and L368C mutant receptors cross-link the peptide more effectively than the M425C PTHR1 (corresponding to the original site identified by Bpa photoaffinity cross-linking), as judged by the intensity of the cross-linked bands following autoradiography (Figure 4). This suggests that, in the natural binding state, position 1 of the ligand comes closer to Y421, F424, and L368 (or spends more time in their vicinity) than it does to M425. Our previous report of the predominance of Bpa1-PTH interacting with M425 (6) no doubt reflects the preference of the Bpa moiety for methionine residues. The new findings highlight the advantageous properties of the disulfide-trapping approach over Bpa-mediated photoaffinity cross-linking.
The observation that Bpa1-PTH cross-linked to M425 was viewed as somewhat problematic from the outset. M425 is on the extracellular surface of the receptor, but in all models of PTHR1, M425 projects away from the core of the 7TM helices, interacting with the lipophilic alkyl chains of the membrane. We hypothesize that the covalent bond formed with Bpa1 was the result of the preference for methionine coupled with a slight shifting of the bound ligand, such that the Bpa projected into the membrane environment, between TM5 and TM6. This slight alteration in binding can largely be accommodated by rotation about χ1 of Bpa and therefore does not alter the affinity for PTHR1 (as this is largely driven by the C-terminus of the ligand) or its activation profile.
The results obtained from the disulfide-trapping experiments identify a number of residues within the core of the TM bundle forming the interface between TM5 and TM6. The current mode of GPCR activation postulates a translation and rotation of TM6 relative to TM3 (24). The association of Ser1 (of the native ligand) with L368 (TM5) and Y421/F424 (TM6) of the receptor would be expected to initiate conformational changes of the TMs, leading to signaling. The reduced reactive radius of cysteine compared to Bpa in this disulfide-trapping method provides higher resolution in defining important contacts between PTH and PTHR1.
We envision that further work using PTHR1 mutants with cysteines introduced into other TMs, in conjunction with the Cys1-PTH peptide reported here, Cys2-PTH, or other Cys-containing analogues, should enable complete delineation of the binding pocket for the N-terminus of the ligand. Such structural insight should facilitate the design of peptide analogues with improved pharmacological profiles or, ideally, small molecule agonists of PTHR1 that may have utility as the next generation of osteoporosis therapies. Along similar lines, PTHR1 antagonists have potential therapeutic applications in disorders of calcium metabolism, such as hyperparathyroidism and malignancy-associated hypercalcemia. Indeed, the utility of the information obtained from cross-linking approaches has recently been shown in a report describing a small molecule antagonist of PTHR1 (25). Using knowledge gleaned from previous Bpa-mediated cross-linking of PTH to PTHR1, the authors used molecular modeling to manually dock this small molecule into a pocket between the tops of TMs 3, 4, 5, and 6. This structure-based approach to drug design will be aided by future disulfide-trapping experiments to further resolve the nature of the PTH–PTHR1 bimolecular interface.
This disulfide-trapping cross-linking strategy is readily applicable to mapping the interface of other peptide–receptor bimolecular complexes. It is most effectively applied when a first generation experimentally derived model of the hormone–receptor complex, such as can be derived by photoaffinity cross-linking, is available as a structural platform. Knowledge of key putative contact regions between the hormone and receptor obtained from Bpa-mediated photoaffinity cross-linking can be used to efficiently guide the placement of cysteines across the interface in order to resolve the structure. This approach holds promise for providing novel insights useful not only for the design of pharmacological agents but also for elucidating the general principles of hormone–receptor molecular recognition that operate for class II GPCRs.
ACKNOWLEDGMENT
We thank Stephen Kwok for flow cytometry analysis.
Footnotes
This work was supported in part by Grants DK47490 (to M.R.) and GM54082 (to D.F.M.) from the National Institutes of Health.
Abbreviations: Bpa, benzoylphenylalanine; DMEM, Dulbecco's modified Eagle medium; GPCR, G protein-coupled receptor; PTH, parathyroid hormone; PTHR1, PTH receptor type 1; TM, transmembrane; EC, extracellular loop; DTT, dithiothreitol.
REFERENCES
- 1.Harmar AJ. Family-B G protein-coupled receptors. Genome Biol. 2001;2:3013.1–3013.10. doi: 10.1186/gb-2001-2-12-reviews3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoare SR. Mechanisms of peptide and nonpeptide ligand binding to Class B G-protein-coupled receptors. Drug Discov. Today. 2005;10:417–427. doi: 10.1016/S1359-6446(05)03370-2. [DOI] [PubMed] [Google Scholar]
- 3.Pham VI, Sexton PM. Photoaffinity scanning in the mapping of the peptide receptor interface of class II G protein-coupled receptors. J. Pept. Sci. 2004;10:179–203. doi: 10.1002/psc.541. [DOI] [PubMed] [Google Scholar]
- 4.Zhou AT, Besalle R, Bisello A, Nakamoto C, Rosenblatt M, Suva L, Chorev M. Direct mapping of an agonist-binding domain within the parathyroid hormone/parathyroid hormone-related protein receptor by photoaffinity crosslinking. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3644–3649. doi: 10.1073/pnas.94.8.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams AE, Bisello A, Chorev M, Rosenblatt M, Suva LJ. Arginine 186 in the extracellular N-terminal region of the human parathyroid hormone 1 receptor is essential for the contact with position 13 of the hormone. Mol. Endocrinol. 1998;12:1643–1683. doi: 10.1210/mend.12.11.0198. [DOI] [PubMed] [Google Scholar]
- 6.Bisello A, Adams A, Mierke D, Pellegrini M, Rosenblatt M, Suva L, Chorev M. Parathyroid hormone-receptor interactions identified directly by photocross-linking and molecular modeling studies. J. Biol. Chem. 1998;273:22498–22505. doi: 10.1074/jbc.273.35.22498. [DOI] [PubMed] [Google Scholar]
- 7.Mannstadt M, Luck M, Gardella T, Juppner H. Evidence for a ligand interaction site at the amino-terminus of the parathyroid hormone (PTH) PTH-related protein receptor from cross-linking and mutational studies. J. Biol. Chem. 1998;273:16890–16896. doi: 10.1074/jbc.273.27.16890. [DOI] [PubMed] [Google Scholar]
- 8.Behar V, Bisello A, Rosenblatt M, Chorev M. Photoaffinity cross-linking identifies differences in the interactions of an agonist and an antagonist with the parathyroid hormone/parathyroid hormone-related protein receptor. J. Biol. Chem. 2000;275:9–17. doi: 10.1074/jbc.275.1.9. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg Z, Bisello A, Mierke DF, Rosenblatt M, Chorev M. Mapping the bimolecular interface of the parathyroid hormone (PTH)-PTH1 receptor complex: Spatial proximity between Lys27 (of the hormone principal binding domain) and Leu261 (of the first extracellular loop) of the human PTH1 receptor. Biochemistry. 2000;39:8142–8152. doi: 10.1021/bi000195n. [DOI] [PubMed] [Google Scholar]
- 10.Gensure RC, Gardella TJ, Juppner H. Multiple sites of contact between the carboxyl-terminal binding domain of PTHrP-(1–36)analogs and the amino-terminal extracellular domain of the PTH/PTHrP receptor identified by photoaffinity cross-linking. J. Biol. Chem. 2001;276:28650–28658. doi: 10.1074/jbc.M100717200. [DOI] [PubMed] [Google Scholar]
- 11.Gensure RC, Shimizu N, Tsang J, Gardella TJ. Identification of a contact site for residue 19 of parathyroid hormone (PTH) and PTH-related protein analogs in transmembrane domain two of the type 1 PTH receptor. Mol. Endocrinol. 2003;17:2647–2658. doi: 10.1210/me.2003-0275. [DOI] [PubMed] [Google Scholar]
- 12.Wittelsberger A, Corich M, Thomas BE, Lee BK, Barazza A, Czodrowski P, Mierke DF, Chorev M, Rosenblatt M. The mid-region of parathyroid hormone (1–34) serves as a functional docking domain in receptor activation. Biochemistry. 2006;45:2027–2034. doi: 10.1021/bi051833a. [DOI] [PubMed] [Google Scholar]
- 13.Rosenblatt M. Parathyroid hormone: chemistry and structure-activity relations. In: Ioachim HL, editor. Pathobiology Annual. New York: 1981. pp. 53–86. [PubMed] [Google Scholar]
- 14.Wittelsberger A, Thomas B, Mierke D, Rosenblatt M. Methionine acts as a ”magnet” in photoaffinity crosslinking experiments. FEBS Lett. 2006;580:1872–1876. doi: 10.1016/j.febslet.2006.02.050. [DOI] [PubMed] [Google Scholar]
- 15.Hug G, Bobrowski K, Kozubek H, Marciniak B. Photo-oxidation of methionine-containing peptides by the 4-carboxybenzophenone triplet state in aqueous solution. Competition between intramolecular two-centered three-electron bonded (S.S)+ and (S.N)+ formation. Photochem. Photobiol. 2000;72:1–9. doi: 10.1562/0031-8655(2000)072<0001:poomcp>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 16.Clement M, Martin S, Beaulieu ME, Chamberland C, Lavigne P, Leduc R, Guillemette G, Escher E. Determining the environment of the ligand binding pocket of the human angiotensin II type I (hAT1) receptor using the methionine proximity assay. J. Biol. Chem. 2005;280:27121–27129. doi: 10.1074/jbc.M413653200. [DOI] [PubMed] [Google Scholar]
- 17.Rihakova L, Deraet M, Auger-Messier M, Perodin J, Boucard A, Guillemette G, Leduc R, Lavigne P, Escher E. Methionine proximity assay, a novel method for exploring peptide ligand-receptor interaction. Recept. Signal Transduct. Res. 2002;22:297–313. doi: 10.1081/rrs-120014603. [DOI] [PubMed] [Google Scholar]
- 18.Jossart C, Coupal M, McNicoll N, Fournier A, Wilkes BC, DeLean A. Photolabeling study of the ligand domain of natriuretic peptide receptor A: development of a model. Biochemistry. 2005;44:2397–2408. doi: 10.1021/bi048251y. [DOI] [PubMed] [Google Scholar]
- 19.Wittelsberger A, Mierke DF, Rosenblatt M. Mapping ligand-receptor interfaces: approaching the resolution limit of benzophenone-based photoaffinity scanning. Chem. Biol. Drug Des. 2008 doi: 10.1111/j.1747-0285.2008.00646.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buck E, Bourne H, Wells JA. Site-specific disulfide capture of agonist and antagonist peptides on the C5a receptor. J. Biol. Chem. 2005;280:4009–4012. doi: 10.1074/jbc.C400500200. [DOI] [PubMed] [Google Scholar]
- 21.Buck E, Wells JA. Disulfide trapping to localize small-molecule agonists and antagonists for a G-protein coupled receptor. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2719–2724. doi: 10.1073/pnas.0500016102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagemann IS, Miller DL, Klco JM, Nikiforovich GV, Baranski TJ. Structure of the complement factor 5a (C5a) receptor-ligand complex studied by disulfide trapping and molecular modeling. J. Biol. Chem. 2008 doi: 10.1074/jbc.M709467200. (in press) (Epub ahead of print January 14) [DOI] [PubMed] [Google Scholar]
- 23.Adams AE, Pines M, Nakamoto C, Behar V, Yang QM, Besalle R, Chorev M, Rosenblatt M, Levine MA, Suva LJ. Probing the bimolecular interactions of parathyroid hormone and the human parathyroid hormone/parathyroid hormone-related protein receptor. 2. Cloning, characterization, and photoaffinity labeling of the recombinant human receptor. Biochemistry. 1995;34:10553–10559. doi: 10.1021/bi00033a030. [DOI] [PubMed] [Google Scholar]
- 24.Kobilka BK. G protein coupled receptor structure and activitation. Biochim. Biophys. Acta. 2007;1768:794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter PH, Liu RQ, Foster WR, Tamasi JA, Tebben AJ, Favata M, Staal A, Cvijic ME, French MH, Dell V, et al. Discovery of a small molecule antagonist of the parathyroid hormone receptor by using an N-terminal parathyroid hormone peptide probe. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6846–6851. doi: 10.1073/pnas.0605125104. [DOI] [PMC free article] [PubMed] [Google Scholar]


