Abstract
Photoaffinity crosslinking has yielded important insights in the study of G protein-coupled receptors and the mode of ligand binding. The most widely used photolabile moiety is p-benzoylphenylalanine largely because of its reportedly high site specificity, reduced reactivity to water and light, photokinetics, and ease of incorporation into peptide ligands during synthesis. However, in the course of our studies directed at characterizing the binding of parathyroid hormone to its cognate G protein-coupled receptor, we find that inherent properties of p-benzoylphenylalanine, such as its size and conformational flexibility, limit the resulting resolution of the ligand–receptor structure. Here, we examine and define these limits.
Keywords: molecular modeling, parathyroid hormone receptor, p-benzoylphenylalanine, photoaffinity crosslinking, resolution limit
G protein-coupled receptors represent the largest family of membrane proteins, with over 800 members identified in the human genome. They constitute one of the most important groups of targets for drug discovery, with approximately half of all drugs currently in clinical use targeted at G protein-coupled receptors (GPCRs). Yet, the nature of ligand binding to GPCRs and the mechanism of activation of GPCRs are poorly understood; the hydrophobic nature of the seven membrane-spanning helices and the conformational complexity of GPCRs pose a problem for direct structure determination by crystallography or NMR.
To gain insight into ligand–GPCR interactions, photoaffinity scanning has been commonly applied as a means of mapping the points and zones of contact between hormones and their receptors. Using p-benzoylphenylalanine (Bpa)-based photoaffinity scanning, important advances in our understanding of the ligand–receptor interface have been obtained for a number of ligand–GPCR systems. For example, the 27 amino acid hormone secretin was found to adopt an extended conformation while bound to its receptor, a family B GPCR, with the N-terminal residues of the hormone reaching to the extracellular part of TM6 (1,2). Similarly, photoaffinity crosslinking reveals a binding groove for calcitonin on the calcitonin receptor, also a family B GPCR, extending from residues in the N-ECD to contacts in extracellular loop (ECL) 3, with the N-terminus of the ligand positioned in proximity to TM6 (3). In addition, the crosslinking sites for calcitonin agonists versus antagonists differ due to differential interaction of the analogs with active versus inactive receptor states, suggesting conformational changes involving the N-ECD upon receptor activation (4). The angiotensin–receptor system is an example from GPCR family A. Bpa-based crosslinking shows that the eight amino acid ligand adopts an extended conformation which inserts into the TM bundle with position 8 of the ligand contacting residues in TM3, TM6, and TM7 (5). Finally, for the parathyroid hormone (PTH)–receptor (PTHR1) system, molecular dynamics (MD) simulations using experimentally determined contact points between seven different residues in PTH and the receptor yield a model of the ligand–receptor complex that leads to three conclusions: first, PTH traverses between the top of TM1 and TM2; secondly, PTH has a distinct conformation when bound to receptor; thirdly, a helical receptor region of the N-ECD of PTHR1 folds back toward the helical bundle when ligand is bound (6,7).
Using the PTH ligand–receptor system as an example, we find that the resolution of structural detail now being obtained is approaching the maximum obtainable using Bpa-based photoaffinity crosslinking. We analyze a model system and conclude that loose distance constraints (of at least 10 Å) must be used in the molecular modeling of the ligand–receptor complex when Bpa-derived contact points are used as the experimental database.
Methods
Construction of model system
To demonstrate the structural implications of Bpa flexibility, the NMR-derived structure of PTH-(1–34) with a Bpa incorporated at position 20 was created using INSIGHTII. The chi-1 (χ1) dihedral angle was rotated to the three staggered configurations (−60°, 180°, and 60°) and the distances between the reactive carbonyl of Bpa and the side chain of Asn10 and Lys27 were measured.
Simulation using different distance restraints
The molecular model of the PTH receptor was built as described (8). Briefly, the TM bundle is based on the crystal structure of rhodopsin (9). The structural features of the fragment spanning the ectopic portion of TM1 and the juxtamembrane portion of the N-ECD (PTHR1[168–198]), ECL-1 (PTHR1[241–285]), and ECL-3 (PTHR1[420–450]) were incorporated into the model using the experimentally determined distance restraints (10,11).
The receptor was refined by MDs simulations using the GROMACS program (12). A water/decane/water simulation cell (120 ×100 ×100 Å) was used to mimic the hydrophilic/hydrophobic, biphasic nature of the cytoplasmic membrane in a computationally simple model (13,14). During the simulations, the photoaffinity labeling-derived ligand–receptor interactions were maintained by distance restraints of 8 or 14 Å.
Results and Discussion
The use of Bpa as the preferred photoreactive group for photoaffinity crosslinking experiments is largely attributable to the following properties. Benzophenones are chemically more stable than alternative photoreactive moieties such as diazo esters, aryl azides, or diazirines. The wavelength for activation (350–360 nm) has no deleterious effect on the protein or receptor, and Bpa can be manipulated in ambient light. Furthermore, benzophenones react preferentially with C–H bonds, even in the presence of water and other nucleophiles. Unlike with other photoreactive groups, the excitation (to a diradicaloid triplet state) is reversible and the benzophenone group undergoes many excitation–relaxation cycles until a favorable geometry for covalent modification is achieved. This, combined with an optimal life time of the excited state, results in efficient covalent modifications of macromolecules, usually with high site specificity. Finally, the amino acid analog Bpa is commercially available and can be readily introduced into the peptidic ligand during standard solid-phase peptide synthesis (15).
Photoaffinity labeling studies begin with a crosslinking step followed by digestive mapping (6,7), radiosequencing (3,16), or analysis by mass spectrometry analysis (17) to identify the site of interaction between the photoexcited Bpa and the target protein. The structure of the bimolecular interface is commonly obtained by undertaking molecular modeling based on the experimentally determined contact points between specific receptor residue(s) (sometimes a single amino acid or a small segment of residues) and the Bpa site in the ligand. These contact points can be used to derive distance restraints. The confidence in the model of the ligand–receptor complex is enhanced with a greater number of experimental data points. Knowledge of structural features of the ligand or receptor greatly assists in the refinement of the model of the bimolecular complex, based on triangulation amongst multiple Bpa sites on the ligand and their individual receptor contact sites. We have utilized high-resolution NMR methods to determine the structural features of the ligand and isolated receptor domains (11,14).
In the refinement of the structure of the ligand–receptor complex, photolabeling data are commonly incorporated in two different ways. In the first, the computer simulations (MDs simulations are most commonly employed) are carried out with ligand containing the Bpa at the appropriate position. This approach attempts to mimic the experimental set-up and distance constraints of 7 or 8 Å between the carbonyl group of Bpa and the identified residue in the receptor are typically used. Although this approach may recapitulate the photoaffinity labeling experiments, the conversion back to native ligand can be problematic, because the orientation of the Bpa relative to the remainder of the peptide is undefined. As illustrated in Figures 1 and 2, the simple rotation about the χ1 of Bpa produces an enormous range of distances to specific points in the ligand. This demonstrates that even if the position of the Bpa is defined within the receptor, the mapping of the remainder of the ligand is undefined, and depends largely on the computational methods employed. An additional issue for utilization of Bpa in the simulations is the most accurate manner of reflecting the multiple crosslinking points generated for a specific ligand–receptor system. Certainly a ligand incorporating multiple Bpa would not be a good mimic of the experimental conditions.
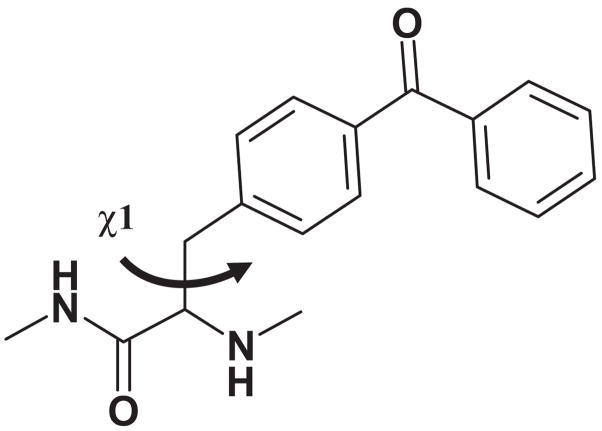
Figure 1.
Structure of p-benzoylphenylalanine with the dihedral angle χ1 denoted.
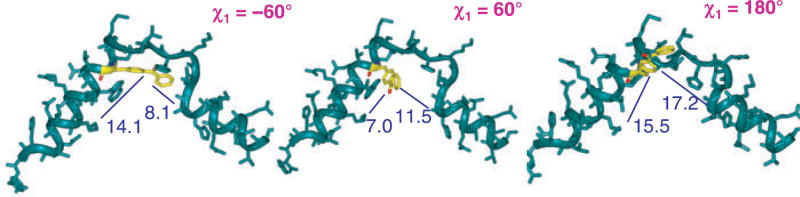
Figure 2.
Model system based on NMR structure of parathyroid hormone (green). Distances (in Å) between the reactive carbonyl group of p-benzoylphenylalanine (yellow) and the side chains of Asn10 and Lys27 are indicated in blue for the three staggered rotamers (χ1 = −60°, 60°, and 180°).
As an alternative simulation approach, we utilize the native peptide ligand in our refinement protocol, not the Bpa-containing analog. Distance restraints are applied to the Cβ atom at the site of Bpa incorporation and to the side-chain of the appropriate residue across the interface in the receptor. To account for use of native ligand and allow for some flexibility of Bpa fitting into the receptor during the crosslinking experiment, we employed a distance of 14 Å (6). This relaxed distance restraint, coupled with the experimentally determined structural features of the ligand and receptor domains, leads to novel insights into the interaction of PTH with its receptor (6,18). However, the question remains, has the resulting resolution now attained reached the limit achievable by these methods?
The reactive radius of the benzophenone group was approximated as a sphere with a radius of 3.1 Å centered on the ketone oxygen (19). In Bpa, rotation around the Cα–Cβ bond defined by angle χ1 allows for significant increases in accessible distances (see Figures 1 and 2). To define the reactive radius of Bpa incorporated into a peptide ligand, we created a model system. We used the solution structure of PTH-(1–34) as determined by NMR (14); Bpa was then introduced at a centrally located position replacing Arg20 (Figure 2). We then rotated the side-chain of Bpa20 from χ1 = −60° to χ1= 180° to χ1 = 60°. For each of these three states, we measured the distances from the reactive carbonyl group to the side-chain of a select residue toward the C-terminus (Lys27) and toward the N-terminus (Asn10) of the hormone. As illustrated in Figure 2, the distances between Bpa20 and K27 range from 7.0 to 15.5 Å; the distances between Bpa20 and Asn10 range from 8.1 to 17.2 Å. Similar conclusions can be drawn for the interactions between Bpa and the receptor. With the same exact ligand-binding mode, depending on the χ1 rotational state of Bpa, a wide range of different amino acids of the receptor would be available for crosslinking; the distance variation from the reactive carbonyl in Bpa is approx. 10 Å. We therefore conclude that it is imperative to use a distance constraint of at least 10 Å for experimentally determined contact points in MD simulations of ligand–receptor complexes. Similar evaluations should be made for benzophenone-derived Lys residues or other photoreactive groups used to experimentally determine contact points when applying distance restraints in computer simulations.
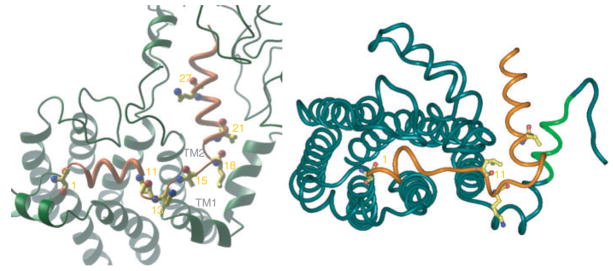
To further examine the effect of tightening of the distance restraint, we carried out a series of MD simulations using a distance of 8 Å for the seven experimentally determined contact points between PTH and its receptor. The simulations followed the standard protocol of building the seven TM helices of PTH1R based on the X-ray structure of rhodopsin (9). The structural features of the proximal N-terminus, ECL1, and ECL3 based on high-resolution NMR studies carried out in a membrane-like environments were incorporated (8). Likewise, the structure of the ligand, consisting of two α-helices as delineated by NMR studies (14), was introduced into the simulations. The structure of the ligand–receptor complex resulting from the simulations is shown in the right panel of Figure 3. When the tight distance constraints are accommodated, the structural features of the ligand are distorted. The N-terminal α-helix, running from Val2 to Gly10, has been reduced to a centrally located loop of helix, attempting to simultaneously fulfill the crosslinking points of Ser1 to M425 (top of TM7) and Leu11 to L174 (proximal N-terminus, located in a second α-helix). In contrast, when using a 14 Å constraint during the simulations (left panel of Figure 3), the helical nature of the N-terminus of PTH is maintained throughout the entire Val2 to Gly10 region.
Figure 3.
Models of the parathyroid hormone (PTH) ligand–receptor complex obtained from molecular dynamic simulations using experimentally determined contact points with distance constraints of 14 Å (left) and 8 Å (right). The ligand is shown in orange. The receptor region [165–176] containing the crosslinking site of Bpa11-PTH is colored light green (right panel).
Conclusions
We conclude that distance restraints based on photoaffinity cross-linking using Bpa must be applied to the molecular modeling of ligand/receptor systems with full knowledge of the limitations and potential pitfalls (20). We suggest that a distance restraint of at least 10 Å is imperative, due to inherent properties of the Bpa group such as its size, physicochemical properties, and conformational flexibility. The Bpa-based technology should be reserved for the first rounds of investigations into a ligand–receptor system to obtain general landmarks and key contact regions.
Acknowledgments
This work was supported by grant DK-47940 (to M.R.) and GM-54082 (to D.F.M.) from the National Institutes of Health.
Abbreviations
- Bpa
p-benzoylphenylalanine
- ECL
extracellular loop
- N-ECD
N-terminal extracellular domain
- PTH
parathyroid hormone
- PTHR1
PTH receptor type 1
- TM
transmembrane helix
References
- 1.Dong M, Li Z, Pinon DI, Lybrand TP, Miller LJ. Spatial approximation between the amino terminus of a peptide agonist and the top of the sixth transmembrane segment of the secretin receptor. J Biol Chem. 2004;279:2894–2903. doi: 10.1074/jbc.M310407200. [DOI] [PubMed] [Google Scholar]
- 2.Dong M, Li Z, Zang M, Pinon DI, Lybrand TP, Miller LJ. Spatial approximation between two residues in the mid-region of secretin and the amino terminus of its receptor. Incorporation of seven sets of such constraints into a three-dimensional model of the agonist-bound secretin receptor. J Biol Chem. 2003;278:48300–48312. doi: 10.1074/jbc.M309166200. [DOI] [PubMed] [Google Scholar]
- 3.Dong M, Pinon DI, Cox RF, Miller LJ. Molecular approximation between a residue in the amino-terminal region of calcitonin and the third extracellular loop of the class B G protein-coupled calcitonin receptor. J Biol Chem. 2004;279:31177–31182. doi: 10.1074/jbc.M404113200. [DOI] [PubMed] [Google Scholar]
- 4.Pham V, Dong M, Wade JD, Miller LJ, Morton CJ, Ng H, Parker MW, Sexton PM. Insights into interactions between the alpha-helical region of the salmon calcitonin antagonists and the human calcitonin receptor using photoaffinity labeling. J Biol Chem. 2005;280:28610–28622. doi: 10.1074/jbc.M503272200. [DOI] [PubMed] [Google Scholar]
- 5.Clement M, Martin SS, Beaulieu M, Chamberland C, Lavigne P, Leduc R, Guillemette G, Escher E. Determining the environment of the ligand binding pocket of the human angiotensin II type I (hAT1) receptor using the methionine proximity assay. J Biol Chem. 2005;280:27121–27129. doi: 10.1074/jbc.M413653200. [DOI] [PubMed] [Google Scholar]
- 6.Wittelsberger A, Corich M, Thomas BE, Lee BK, Barazza A, Czodrowski P, Mierke DF, Chorev M, Rosenblatt M. The mid-region of parathyroid hormone (1–34) serves as a functional docking domain in receptor activation. Biochemistry. 2006;45:2027–2034. doi: 10.1021/bi051833a. [DOI] [PubMed] [Google Scholar]
- 7.Gensure RC, Gardella TJ, Juppner H. Multiple sites of contact between the carboxyl-terminal binding domain of PTHrP-(1–36) analogs and the amino-terminal extracellular domain of the PTH/PTHrP receptor identified by photoaffinity cross-linking. J Biol Chem. 2001;276:28650–28658. doi: 10.1074/jbc.M100717200. [DOI] [PubMed] [Google Scholar]
- 8.Rolz C, Pellegrini M, Mierke DF. Molecular characterization of the receptor-ligand complex for parathyroid hormone. Biochemistry. 1999;38:6397–6405. doi: 10.1021/bi9829276. [DOI] [PubMed] [Google Scholar]
- 9.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 10.Piserchio A, Bisello A, Rosenblatt M, Chorev M, Mierke DF. Characterization of parathyroid hormone/receptor interactions: structure of the first extracellular loop. Biochemistry. 2000;39:8153–8160. doi: 10.1021/bi000196f. [DOI] [PubMed] [Google Scholar]
- 11.Pellegrini M, Bisello A, Rosenblatt M, Chorev M, Mierke DF. Binding domain of human parathyroid hormone receptor: from conformation to function. Biochemistry. 1998;37:12737–12743. doi: 10.1021/bi981265h. [DOI] [PubMed] [Google Scholar]
- 12.Berendson HJC, van der Spoel D, van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91:43–56. [Google Scholar]
- 13.Prado GN, Mierke DF, Pellegrini M, Taylor L, Polgar P. Motif mutation of bradykinin B2 receptor second intracellular loop and proximal C terminus is critical for signal transduction, internalization, and resensitization. J Biol Chem. 1998;273:33548–33555. doi: 10.1074/jbc.273.50.33548. [DOI] [PubMed] [Google Scholar]
- 14.Pellegrini M, Royo M, Rosenblatt M, Chorev M, Mierke DF. Addressing the tertiary structure of human parathyroid hormone-(1–34) J Biol Chem. 1998;273:10420–10427. doi: 10.1074/jbc.273.17.10420. [DOI] [PubMed] [Google Scholar]
- 15.Kauer JC, Erickson-Viitanen S, Wolfe HR, Jr, DeGrado WF. p-Benzoyl-L-phenylalanine, a new photoreactive amino acid. Photolabeling of calmodulin with a synthetic calmodulin-binding peptide. J Biol Chem. 1986;261:10695–10700. [PubMed] [Google Scholar]
- 16.Perodin J, Deraet M, Auger-Messier M, Boucard AA, Rihak-ova L, Beaulieu ME, Lavigne P, Parent JL, Guillemette G, Leduc R, Escher E. Residues 293 and 294 are ligand contact points of the human angiotensin type 1 receptor. Biochemistry. 2002;41:14348–14356. doi: 10.1021/bi0258602. [DOI] [PubMed] [Google Scholar]
- 17.Mills JS, Miettinen HM, Barnidge D, Vlases MJ, Wimer-Mackin S, Dratz EA, Sunner J, Jesaitis AJ. Identification of a ligand binding site in the human neutrophil formyl peptide receptor using a site-specific fluorescent photoaffinity label and mass spectrometry. J Biol Chem. 1998;273:10428–10435. doi: 10.1074/jbc.273.17.10428. [DOI] [PubMed] [Google Scholar]
- 18.Mierke DF, Mao L, Pellegrini M, Piserchio A, Plati J, Tsomaia N. Structural characterization of the parathyroid hormone receptor domains determinant for ligand binding. Biochem Soc Trans. 2007;35:721–723. doi: 10.1042/BST0350721. [DOI] [PubMed] [Google Scholar]
- 19.Dorman G, Prestwich GD. Benzophenone photophores in biochemistry. Biochemistry. 1994;33:5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- 20.Wittelsberger A, Thomas BE, Mierke DF, Rosenblatt M. Methionine acts as a “magnet” in photoaffinity crosslinking experiments. FEBS Lett. 2006;580:1872–1876. doi: 10.1016/j.febslet.2006.02.050. [DOI] [PubMed] [Google Scholar]