Abstract
Cyclotides are disulfide-rich miniproteins with the unique structural features of a circular backbone and knotted arrangement of three conserved disulfide bonds. Cyclotides have been found only in two plant families: in every analyzed species of the violet family (Violaceae) and in few species of the coffee family (Rubiaceae). In this study, we analyzed >200 Rubiaceae species and confirmed the presence of cyclotides in 22 species. Additionally, we analyzed >140 species in related plant families to Rubiaceae and Violaceae and report the occurrence of cyclotides in the Apocynaceae. We further report new cyclotide sequences that provide insights into the mechanistic basis of cyclotide evolution. On the basis of the phylogeny of cyclotide-bearing plants and the analysis of cyclotide precursor gene sequences, we hypothesize that cyclotide evolution occurred independently in various plant families after the divergence of Asterids and Rosids (∼125 million years ago). This is strongly supported by recent findings on the in planta biosynthesis of cyclotides, which involves the serendipitous recruitment of ubiquitous proteolytic enzymes for cyclization. We further predict that the number of cyclotides within the Rubiaceae may exceed tens of thousands, potentially making cyclotides one of the largest protein families in the plant kingdom.
INTRODUCTION
Cyclotides are disulfide-rich peptides recently discovered in plants of the Violaceae and Rubiaceae families (Craik et al., 1999; Colgrave and Craik, 2004). They are ∼30 amino acids in size and have the unique structural features of a head-to-tail cyclized backbone and a knotted arrangement of three-disulfide bonds, referred to as a cyclic cystine knot (CCK) motif (Craik et al., 1999). The compact CCK motif makes cyclotides exceptionally resistant to thermal, chemical, or enzymatic degradation (Colgrave and Craik, 2004). Cyclotides exhibit a range of biological activities, including uterotonic (Gran et al., 2000), hemolytic (Schöpke et al., 1993), antineurotensin (Witherup et al., 1994), anti-HIV (Gustafson et al., 2004), cytotoxic (Lindholm et al., 2002), antibacterial (Tam et al., 1999), and antifouling (Göransson et al., 2004) activities, but their natural function appears to be as plant defense molecules based on their insecticidal (Jennings et al., 2001, 2005; Gruber et al., 2007a; Barbeta et al., 2008) and molluscidal (Plan et al., 2008) properties. Their unique structural framework, range of bioactivities, and sequence diversity make the cyclotides interesting targets for pharmaceutical applications (Craik et al., 2002, 2006a). Figure 1 summarizes the biosynthesis and structure of cyclotides.
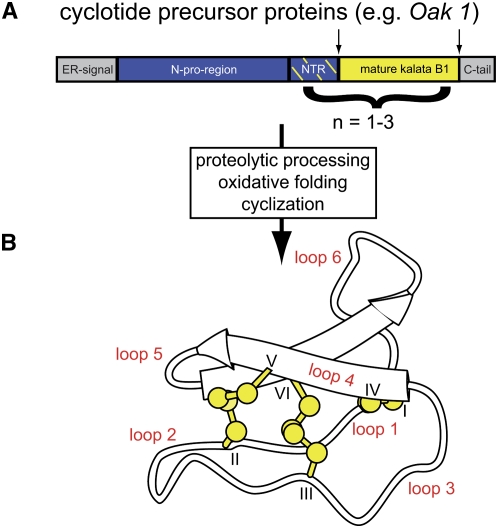
Figure 1.
Biosynthesis and Structure of Cyclotides.
(A) Cyclotides are gene products synthesized on ribosomes and modified to maturation in the secretory pathway (Gruber et al., 2007b). The prototypic cyclotide kalata B1 is synthesized in O. affinis as part of a precursor protein, Oak1 (Oldenlandia affinis kalata B1), comprising an endoplasmic reticulum target signal (gray), N-terminal proregion (blue), an N-terminal repeat (NTR; blue/yellow-dashed), the mature cyclotide domain (yellow), and a C-terminal tail (gray). Other cyclotide precursor proteins contain up to three mature cyclotide domains. Processing of the precursor involves oxidative folding to form three-disulfide bonds, excision of the mature sequence, and head-to-tail cyclization.
(B) Mature cyclotides comprise the typical structural CCK motif, characterized by three disulfide bonds (yellow) in a knotted arrangement. Two disulfide bonds and the adjacent backbone segments form a ring (CI-CIV and CII-CV) that is threaded by the third disulfide bond (CIII-CVI). The Cys residues separate the backbone into six loops, labeled loops 1 to 6. The image shows the backbone structure of kalata B1 with the disulfide bonds indicated in ball-stick representation and the small antiparallel β-sheet indicated with arrows.
Kalata B1, from the Rubiaceae species Oldenlandia affinis, was the first cyclotide discovered (Gran, 1970), although its macrocyclic structure was not elucidated until 1995 (Saether et al., 1995). Aside from cyclotides in the coffee (Rubiaceae) and violet (Violaceae) families, two closely related cyclic knottins have been found in the cucurbit family (Cucurbitaceae) (Chiche et al., 2004). Additionally, two recent studies identified cyclotide-like gene sequences in representatives of the grass family (Poaceae), including important cereal crops, such as wheat (Triticum aestivum), maize (Zea mays), and rice (Oryza sativa) (Basse, 2005; Mulvenna et al., 2006a). So far, the amino acid sequences of >100 cyclotides have been reported, and thousands more have been predicted to be present in the Violaceae. It has been suggested that cyclotides may surpass the well-known plant defensins in number and diversity (Trabi et al., 2004; Simonsen et al., 2005). Although cyclotides appear to occur in every Violaceae species analyzed so far (Simonsen et al., 2005), information about their occurrence, distribution, and evolution in the Rubiaceae and other plant families is very limited.
Rubiaceae is the fourth largest angiosperm family and comprises ∼650 genera and 13,000 species (Delprete, 2004; Govaerts et al., 2006). Recent molecular phylogenetic reconstructions have suggested the division of this family into either two, Cinchonoideae and Rubioideae (Robbrecht and Manen, 2006), or three subfamilies, Cinchonoideae, Ixoroideae, and Rubioideae (Bremer and Jansen, 1991; Bremer et al., 1995, 1999; Bremer, 1996; Rova et al., 2002). The family includes one of the most economically important plants worldwide, namely, coffee. Additionally, it includes timber species, such as bilinga (Nauclea diderrichii), many ornamental plants (e.g., Gardenia spp, Ixora spp, and Mussaenda spp), and important plants for medicinal use, including quinine (Cinchona spp) and ipecac (Carapichea ipecacuanha) (De Wildeman, 1901; Purseglove, 1968). A wide range of other medicinal, ornamental, psychoactive, and aphrodisiac properties of Rubiaceae plants have been reported (Delprete, 2004).
In this study, we analyzed >200 species of Rubiaceae from field collections, living greenhouse collections, and herbarium specimens for the occurrence of cyclotides. We developed a robust and sensitive screening procedure to identify novel cyclotide-producing plants and found 22 novel cyclotide-producing Rubiaceae species. Our results indicate which tribes and genera of Rubiaceae contain cyclotides. This has led to estimates of the number of species likely to contain cyclotides and the number of novel cyclotides to be discovered from those species. Additionally, we analyzed >140 plant species from other plant families related to the Rubiaceae and Violaceae and found 12 novel cyclotide-producing species in the Apocynaceae.
The discovery that circular proteins (cyclotides) are much more numerous than originally anticipated will be valuable in developing strategies for the exploitation of these topologically fascinating proteins for agrochemical and pharmaceutical applications. Based on these findings, together with the analysis of novel cyclotide precursor genes from Rubiaceae and Violaceae and recent advances in Rubiaceae taxonomy, we have derived a mechanistic basis for the evolution of circular proteins within the Rubiaceae in particular and the plant kingdom in general.
RESULTS
Although its circular and knotted nature was not known at the time, the discovery of the first cyclotide, kalata B1, was based on its presence in a tea from the Rubiaceae species O. affinis used in African indigenous medicine to accelerate childbirth (Gran, 1973). It is apparent that many other circular proteins exist, but we lack information about their origin and distribution in plants. To understand the evolution of circular proteins, we screened ∼350 flowering plant species (including >200 Rubiaceae species) for the occurrence of cyclotides and analyzed cyclotide precursor genes from Rubiaceae and Violaceae.
Novel Screening Procedure for Cyclotides
The first aim of this study was to develop an efficient and robust method for cyclotide identification that minimizes the numbers of false-positive and false-negative results. Figure 2 summarizes the three-part (A, B, C) screening procedure that was developed. In a prescreen, plant extracts were prepared and semipurified on a C18 solid phase extraction column. The potential cyclotide-containing fraction was obtained by eluting the column with acetonitrile in water. A decision as to whether a given extract contains cyclotides or not was established in the main screen and confirmed in example cases by a postscreen. In the main screen, all semipure plant extracts were evaluated for their chemical and biophysical properties. The hydrophobicity and mass range of the extract components were analyzed either separately by reverse phase (RP)-HPLC and matrix-assisted laser-desorption ionization time of flight mass spectrometry (MALDI-TOF MS) or in combination by liquid chromatography–mass spectrometry (LC-MS). Based on earlier findings (Craik et al., 1999; Daly et al., 1999), it is clear that cyclotides have very specific chemical and biophysical properties, namely, a hydrophobic surface that accounts for their late elution time on RP chromatography and a mass range between 2500 and 4000 D. Both criteria were critical in the determination of the presence or absence of cyclotides in a species.
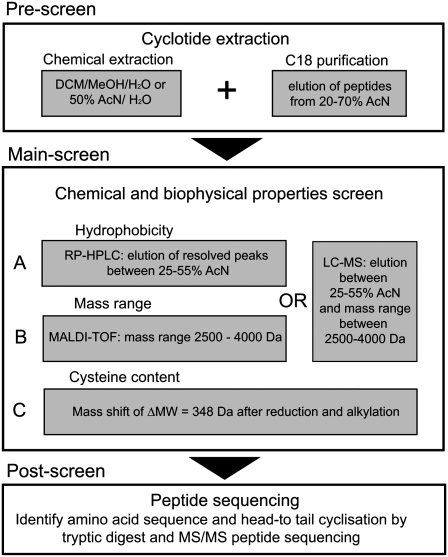
Figure 2.
Flowchart for the Screening of Cyclotides.
A set of criteria was developed to decide whether a species contains cyclotides or not. In a prescreen, all plant material was extracted by either dichloromethane/methanol/water (1:1:1; v/v/v) or acetonitrile/water (1:1; v/v) and prepurified by C18 chromatography. The main screen was developed to analyze the plant compounds for their chemical and biophysical properties and was divided in three screening parts: (A) hydrophobicity, (B) mass range, and (C) Cys content. Each species was scored for the number of peaks (i.e., peptides) that fulfilled the screening criteria in HPLC and MS spectra. If a species scored ≥1 for each of parts A, B, and C, it was classified as cyclotide-containing species. Selected positive hits were confirmed by amino acid sequencing in a postscreen.
Although a significant proportion of plant extracts tested gave an elution profile that passed the chromatographic selection criterion, only some showed a defined set of well-resolved intense HPLC peaks. All species passing the first criterion (hydrophobic elution profile) were further screened by MS for peaks in the required mass range. Only a small proportion of species passed this second criterion. In a third step, we analyzed the chemical nature of the compounds that showed a cyclotide-like HPLC elution profile and mass range. Reduction and alkylation followed by MS analysis were used to analyze the Cys content of the plant compounds. With this method, each Cys residue should mass-shift by 58 D due to the addition of an alkyl group at the side chain of Cys. Hence, a six Cys containing peptide like a cyclotide would shift its mass after reduction and alkylation by 348 D.
In this study, our primary focus was not on the sequencing of novel cyclotides, but on developing a robust screening method that would detect their presence so that we could trace cyclotide occurrence in the plant kingdom. Nevertheless, to illustrate the validity of the screen, we obtained full sequences of two novel cyclotides, CD-1 and PS-1, that were isolated from Chassalia discolor subsp discolor and Psychotria suterella, respectively, as part of the screen. These peptides were initially confirmed as cyclotides based on hydrophobicity profile, mass, and Cys content. Their sequences were determined by a combination of enzymatic digests, tandem MS sequencing, and amino acid analysis. A comparison of these novel peptides with other cyclotide sequences (shown in Figure 3) emphasizes the great diversity of cyclotide sequences as well as the high conservation of key residues within the stable cyclotide framework (i.e., the six Cys residues and a Glu residue in loop 1 of the sequences).
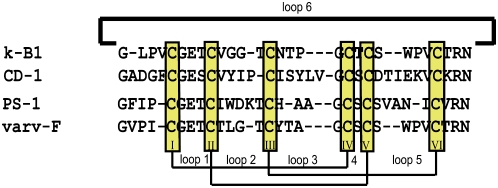
Figure 3.
Sequence Alignment of Novel Cyclotides CD-1 and PS-1.
The novel cyclotides CD-1 and PS-1 from C. discolor and P. suterella, respectively, were aligned to known cyclotide sequences that they are most similar to (kalata B1 and varv F, respectively). The similarity alignment highlights the conserved positions for cyclotides, such as the six highly conserved Cys residues, the Gly and Glu residues in loop 1, the positive residue Arg or Lys in loop 6, and the C-terminal Asn thought to be involved in cyclization of cyclotides. The composition of isobaric residues Leu and Ile of PS-1 were determined by amino acid analysis and their positions differentiated by a chymotrypsin digest. The composition of isobaric residues Leu and Ile of CD-1 were determined by a combination of amino acid analysis and similarity alignment.
In summary, if a species contained peptides showing a cyclotide-like elution profile by RP-HPLC or LC-MS and those peptides had a mass in the range 2500 to 4000 D, and at least one shifted in mass after reduction and alkylation by 348 D, we assumed the presence of cyclotides and their amino acid sequence was confirmed in example cases.
Validation of the Screening Procedure
It was important to cross-validate the screening procedure to minimize the chance of false-positive and false-negative hits. For this purpose, we performed a score-based analysis of parts A, B, and C of the screen for all species studied. Scores were given for the number of peaks that fulfilled the cyclotide-screening criteria (Figure 2). Specifically, any species needed a score ≥1 for each of parts A, B, and C to be classified as cyclotide-producing. Typically, all cyclotide-producing species had significantly higher scores than this threshold, suggesting that the chance of a false-positive hit was low. However, the likelihood of false-negatives may be greater, as it is known that cyclotide expression varies based on environmental conditions such as habitat or season (Trabi et al., 2004); hence, our screening method may be not sensitive enough to account for these variations in some cases.
The following observations provided reassurance that there was a low chance of a false-negative hit: (1) although the expression level of cyclotides may vary, there are no reports of only trace amounts of cyclotides in any given plant. Generally at least one cyclotide is expressed at very high levels when any cyclotides are present (e.g., kalata B1 at ∼1 mg per kg dry plant weight). (2) The data from the analysis of cyclotide occurrence on a genetic level (see below) agree with what is observed on the peptide level (i.e., if a given species contains cyclotide genes, it also contains detectable cyclotides on the peptide level and vice versa). (3) We performed cross-validation analysis on multiple samples of a cyclotide-containing species (P. suterella) and a non-cyclotide-containing species (Psychotria verschuerenii). These species were obtained as herbarium material, glasshouse-grown plants, and field-collected plants from various locations and seasons, and no false-positive or false-negative results were found, confirming the validity of the screening method.
Identification of Novel Cyclotide-Producing Rubiaceae Species
Having developed and validated a cyclotide screening method, the main aim of this study was to collect and analyze Rubiaceae species for the occurrence and distribution of cyclotides. Before this study, only four Rubiaceae plant species had been reported to contain cyclotides, namely, Chassalia parvifolia (Palicoureeae) (Gustafson et al., 1994, 2000), O. affinis (Craik et al., 1999; Plan et al., 2007; Seydel et al., 2007), Psychotria vellosiana (formerly known as Psychotria longipes (Witherup et al., 1994), and Palicourea condensata (Palicoureeae) (Bokesch et al., 2001); another two species, Kadua affinis (known as Hedyotis terminalis, GenBank CB077799) and Kadua centranthoides (Hedyotis centranthoides, GenBank CB084585) (both Spermacoceae) were reported to contain cyclotide precursor sequences.
We tested 208 Rubiaceae species, 60 species belonging to the subfamilies Cinchonoideae/Ixoroideae, and 148 species of the subfamily Rubioideae. From these, we identified 21 novel cyclotide-producing species, all belonging to the subfamily Rubioideae (Table 1), and we confirmed the presence of cyclotides in K. centranthoides by peptide analysis. All 22 species definitively fulfilled the criteria of the prescreens and main screens outlined in Figure 2. Within the Rubioideae, we analyzed plants from all three clades, namely, the basal grade (nine species), the woody clade (48 species), and the herbaceous clade (91 species), including coffee plants (Caffea spp), which do not contain cyclotides. The non-cyclotide-containing Rubiaceae species are listed in Supplemental Table 1 online.
Table 1.
Novel Cyclotide-Containing Rubiaceae Species, All in the Subfamily Rubioideae
| Species | Tribe | Locality of Collection/Native Occurrence | Collector/Reference/Deposition ID |
|---|---|---|---|
| Amphiasma luzuloides | Hedyotideae | Zambia | Dessein et al. 1167 (BR), NBG Belgium (Meise) |
| Amphiasma robijnsii | Hedyotideae | R.D. Congo | Dubois 1196 (BR), NBG Belgium |
| Chassalia discolor subsp discolor | Palicoureeae | Tanzania | Missouri Botanical Garden |
| Geophila repens | Palicoureeae | French Guyana, Brazil, Paraguay | NDa |
| Geophila tenuis | Palicoureeae | French Guyana, Brazil, Venezuela | NYBG, USAb |
| Kadua acuminata | Hedyotideae | Hawaii | NBG Belgium, 19971136-00 |
| Kadua centranthoidesc | Hedyotideae | Hawaii | Wood K.R. 12415 (BISH), Bishop Museum, Hawaii |
| Kadua cordata | Hedyotideae | Hawaii | NTBG Hawaii (Kalaheo), 010067 |
| Kadua lichtlei | Hedyotideae | Marquesas Islands | NTBG Hawaii (Kalaheo), 040036-001 |
| Kadua rapensis | Hedyotideae | French Polynesia, Austral Islands | NTBG Hawaii (Kalaheo), 030142-005 |
| Kadua st.-johnii | Hedyotideae | Hawaii | NTBG Hawaii (Kalaheo), 000030-007 |
| Lasianthus batangensis | Lasiantheae | Cameroon | Sonké B. 1797(BR), NBG Belgium |
| Lasianthus kilimansharicus | Lasiantheae | Kenya | De Block et al. 247(BR), NBG Belgium |
| Palicourea coriacea | Palicoureeae | Bolivia, Brazil | ND |
| Palicourea rigida | Palicoureeae | South America | ND |
| Psychotria brachyceras | Psychotrieae | Brazil | ND |
| Psychotria prunifolia | Psychotrieae | South America | ND |
| Psychotria suterella | Psychotrieae | Brazil | ND |
| Psychotria trichophora | Psychotrieae | Brazil, Guyana | ND |
| Psychotria punctata | Psychotrieae | Africa | NTBG Hawaii (Kalaheo), 980124 |
| Ronabea emetica | Lasiantheae | Central America to Peru | NBG Belgium, 19971040-01 |
| Saldinia axillaris | Lasiantheae | Madagascar | Rabeheritra D. et al. 676 (BR), NBG Belgium |
Documented and identified during field work.
Nonassigned plant sample as gift from New York Botanical Gardens.
Cyclotide precursor gene was identified prior to this study as Hedyotis centranthoides (GenBank CB084585) and has now been confirmed by peptide analysis.
All of the cyclotide-positive species occur in only four of the 19 tribes: Lasiantheae (Lasianthus, Ronabea, and Saldinia), Psychotrieae (Geophila and Psychotria), Palicoureeae (Chassalia and Palicourea), and Spermacoceae (Amphiasma, Kadua, and Oldenlandia; or more specifically within the tribe Hedyotideae sensu) (Delprete et al., 2006), according to the Robbrecht and Manen (2006) classification (see Figure 4).
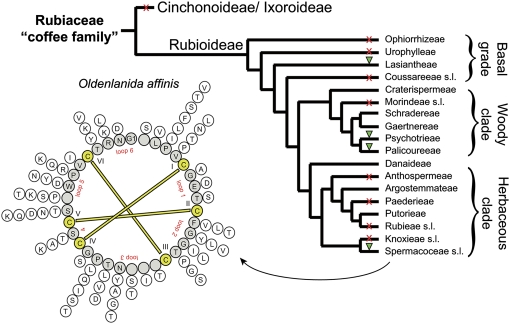
Figure 4.
Distribution of Cyclotides in Rubiaceae.
Summary cladogram of the family Rubiaceae based on Bremer and Manen (2000) showing the relationships between the tribes of the subfamily Rubioideae. The tribal delimitations follow Robbrecht and Manen (2006). The woody and herbaceous clades correspond to the supertribes Psychotriidinae and Rubiidinae, respectively. The presence (green triangle) and absence (red cross) of cyclotides are indicated for all analyzed plant tribes/families. The sequence diversity of cyclotides is indicated for peptides found in the prototypic cyclotide-containing plant O. affinis (Spermacoceae). Variations of amino acids for each position to the cyclotide backbone are indicated in a radial formation on the outside of the backbone circle that was generated by alignment of kalata B1–B18.
Numbers of Unique Cyclotides within One Rubiaceae Species
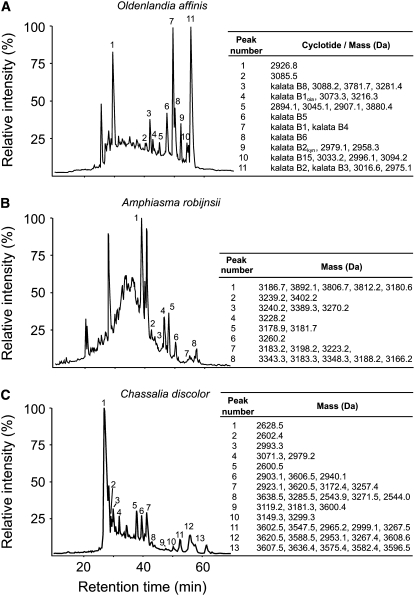
Having established the occurrence and distribution of cyclotides in Rubiaceae species, it was of interest to determine the typical number of unique cyclotides in individual plants. Earlier studies suggested that one species can express 15 to 60 different cyclotides (Trabi et al., 2004; Simonsen et al., 2005). In O. affinis (Rubiaceae), we have so far identified >30 cyclotides (Plan et al., 2007; Seydel et al., 2007), but a recent report suggests this number may increase with varying growth conditions (Seydel et al., 2007). To determine the typical number of individual cyclotides in the cyclotide-producing species identified in this study, we analyzed two sample species, Amphiasma robijnsii and Chassalia discolor subsp discolor (Palicoureeae), by nanospray LC-MS and compared their cyclotide content to O. affinis. As can be seen in Figure 5, A. robijnsii contains at least 22 unique cyclotide-like masses in the range from 3000 to 3900 D eluting between 30 and 55% acetonitrile, and C. discolor contains at least 38 unique cyclotides in the mass range from 2500 to 3700 D eluting between 25 and 60% acetonitrile. Additionally, we identified >18 novel cyclotides from O. affinis in the mass range from 2800 to 3800 D eluting between 30 and 60% acetonitrile. All of these new compounds have masses and retention times different from known cyclotides. Consistent with these results, we identified on average >34 cyclotide masses per cyclotide-producing species using MALDI-TOF analysis (see Supplemental Table 2 online).
Figure 5.
Expression and Numbers of Cyclotides within a Plant Species.
LC-MS profiles of the plant extracts from O. affinis (A), A. robijnsii (B), and C. discolor (C) and are shown to highlight the number of unique cyclotides in each plant species. LC elution profiles are shown from 10 to 70 min at 1% min−1 solvent B (solvent B: 90% acetonitrile in Milli-Q H2O with 0.1% formic acid). Elutions of cyclotides, characterized by mass spectrometry, are indicated with Arabic numbers. Cyclotide masses are given to the right of the graph. Some cyclotides coelute from the reverse phase C18 column; hence, multiple masses are indicated. The representative plant species screened from O. affinis contains at least 18, A. robijnsii at least 22, and C. discolor at least 38 unique and novel cyclotides.
Identification of Novel Cyclotide-Producing Plants Outside the Presently Known Families of Rubiaceae and Violaceae
Based on the distribution of cyclotide-producing plants within the Rubiaceae, and especially their occurrence in basal Rubioideae, we hypothesized that other plant families within the order Gentianales may also test positive for the presence of cyclotides. Hence, we analyzed 66 species from four plant families (Apocynaceae, Gentianaceae, Loganiaceae, and Potaliaceae) within the Gentianales (Table 2; see Supplemental Table 3 online) and found 12 novel species within the Apocynaceae sensu lato (including Asclepiadaceae) to contain cyclotides (Table 2). Additionally, we analyzed many species in several orders of angiosperms, but so far none of these families tested positive for the presence of cyclotides (see Supplemental Table 3 online). After the successful identification at the peptide level of novel cyclotide-producing plants, we extended the study to the nucleic acid level and isolated and characterized cyclotide precursor genes from Rubiaceae and Violaceae species.
Table 2.
Novel Cyclotide-Containing Species in the Family Apocynaceae sensu lato
| Species | Subfamily | Native Source | Deposition IDa |
|---|---|---|---|
| Adenium oleifolium | Apocynoideae | Cultivatedb | 1989-1812 |
| Allamanda neriifolia | Apocynoideae | Cameroon | 1981-0285 |
| Alstonia scholaris | Apocynoideae | India | 1993-1538-75 |
| Caralluma frerei | Asclepiadoideae | Cultivated | 1970-0017 |
| Echidnopsis dammaniana | Asclepiadoideae | Kenya | 1979-0098 |
| Hunteria eburnea | Apocynoideae | Ivory Coast | 1981-0298 |
| Rauvolfia vomitoria | Apocynoideae | Congo | 1987-1146 |
| Stapelianthus decaryi | Asclepiadoideae | Madagascar | 1987-0303 |
| Stephanotis floribunda | Asclepiadoideae | Cultivated | 1965-0283 |
| Strophanthus hispidus | Apocynoideae | Ghana | 1961-5201 |
| Tabernaemontana siphilitica | Apocynoideae | Venezuela | 1964-0544 |
| Tabernanthe iboga | Apocynoideae | Ivory Coast | 1981-0307 |
Accession number from the living plant collection of the National Botanic Garden of Belgium.
Cultivated at the National Botanic Garden of Belgium.
Isolation and Analysis of Cyclotide Precursor Genes
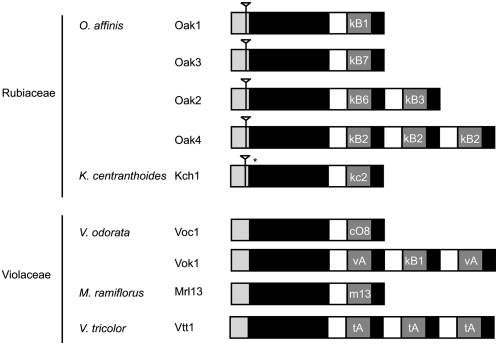
Nine cyclotide precursor genes from O. affinis, K. centranthoides (Rubiaceae), Viola odorata, Viola tricolor, and Melicytus ramiflorus (Violaceae) were isolated from leaf DNA using primers designed to conserved regions of cyclotide precursor transcripts (Figure 6). After characterization of the genes, they were aligned against their respective cDNA sequence, from which the respective primers used to amplify the genes were designed, to identify the gene structure of cyclotides. The organization of cyclotide genes is conserved and similar to that described for cyclotide transcripts (Dutton et al., 2004), where a starting endoplasmic reticulum signal sequence is followed by a propeptide region and one or many copies of cyclotide domains. The main difference between the gene structures isolated from Rubiaceae and Violaceae plants is the presence of a single intron in the signal peptide of genes in Rubiaceae that is not present in the Violaceae genes (shown in Figure 6). The AT content of the introns is high, ranging from 72 to 79%, and is consistent with the AT content of introns from plants and is believed to be involved in the signaling of splicing events (Brown et al., 2002).
Figure 6.
Structure of Cyclotide Precursor Gene.
Cyclotide precursor genes were isolated and characterized from O. affinis (Oak1-4), K. centranthoides (Kch1), Viola odorata (Voc1 and Vok1), Melicytus ramiflorus, (Mrl13), and Viola tricolor (Vtt1). Cyclotide genes have a conserved structure, starting with a signal peptide, followed by a propeptide region and one or more copies of cyclotide coding domains. Introns, indicated by an inverted triangle, are present in the signal region of genes from Rubiaceae plants but are not present in genes from Violaceae plants. The asterisk indicates a premature stop codon.
DISCUSSION
In this multidisciplinary investigation of circular proteins in flowering plants, we analyzed >200 Rubiaceae species, and 22 of them were found to be cyclotide-producing. In this section, we examine the taxonomical relationships between the positive species and compare them to other cyclotide-containing plant families. Based on the distribution of cyclotides, we estimate the number of Rubiaceae species that are likely to contain cyclotides and propose that they form a very large protein family within the plant kingdom, with the number of cyclotides being far larger than earlier anticipated. This proposal is supported by our new discovery reported herein of cyclotides in other plant families of the order Gentianales. The results further suggest that the events that triggered the cyclization of linear CCK proteins to produce cyclotides occurred independently in different plant families. We show that this suggestion is consistent with a recently proposed mechanism for in planta cyclization involving the reverse action of a ubiquitous plant protease.
First, we examined the phylogenetic distribution of the plants that we found to be cyclotide positive. Three different classification schemes are currently available for tribal relationships within the Rubioideae (Bremer and Manen, 2000; Delprete et al., 2006; Robbrecht and Manen, 2006) (see Supplemental Table 5 online). For the tribal placement of the species investigated in this study, we generally followed Robbrecht and Manen (2006), which includes a comprehensive, world-wide survey of the entire family, yet we also took account of the narrower tribal delimitations proposed by Delprete et al. (2006). Our results show that within the Rubiaceae, cyclotides occur in four tribes of the subfamily Rubioideae (Table 1, Figure 4). More broadly, cyclotides occur in the basal grade, the tribe Lasiantheae, and the two main clades, namely, the Psychotrieae/Palicoureeae complex and the tribe Spermacoceae. In the basal grade, cyclotides were detected in three genera (Lasianthus, Ronabea, and Saldinia); therefore, we postulate that cyclotide occurrence had its origin early in the evolution of the Rubioideae.
These findings led us to hypothesize that other families within the order Gentianales are likely to contain cyclotides, too, based on relationships of the families within the order Gentianales (Struwe et al., 1994; Backlund et al., 2000). To test this hypothesis, we analyzed 66 plant species of the Gentianales and found 12 positive cyclotide-producing species within the Apocynaceae (including Asclepiadaceae). This exciting finding further suggests that analyzing other plant families within the order Malpighiales in the future that are closest to Violaceae, (i.e., Passifloraceae and Salicaceae, including Flacourtiaceae) and Turneraceae may be a route to new cyclotide discovery.
In the Rubiaceae, within the subfamily Rubioideae, cyclotides occur in the sister tribes Psychotrieae and Palicoureeae. Within the herbaceous clade, the occurrence is limited to the tribe Spermacoceae or more specifically within the Hedyotideae sensu Delprete (Delprete et al., 2006), and cyclotides seem to be limited to the Hawaiian Kadua species and to some species of the Afro-Madagascan Pentanopsis clade.
Based on the distribution of newly reported cyclotide-containing species in the Rubiaceae, it was of interest to estimate the possible number of species containing cyclotides. The number of species for each of the tribes/genera containing cyclotides is given in Supplemental Table 4 online. Based on these numbers, we estimate that the total number of Rubiaceae species potentially containing cyclotides is ∼3700. Taking into account the incidence of positive cases in our study (30% for Hedyotideae sensu Delprete, 80% for Lasiantheae, and 22% for Psychotrieae/Palicoureeae; Table 3), the total number of likely cyclotide-containing Rubiaceae is ∼980. Earlier studies reported that one plant can express 15 to 60 different cyclotides (Trabi et al., 2004; Simonsen et al., 2005), and consistent with this observation, we found well over 22 different possible cyclotides expressed in a single plant species (Figure 5). Multiplying the numbers of likely cyclotide-containing Rubiaceae species (980) with the numbers of cyclotides within one species (15 to 60) leads to the estimation that there could be between 10,000 and 50,000 unique cyclotides in the Rubiaceae, but one has to note that we have only tested 208 species (1.6% out of 13,000 species in total) and 75 genera (12% out of 650 genera in total; Table 3). However, taking into account the novel cyclotide-producing species within Gentianales (Apocynaceae), it is evident that the number of existing cyclotides may be even higher than our conservative estimate. This potential diversity is consistent with the unique structural features of cyclotides that lend themselves to combinatorial variation. Specifically, cyclotides display remarkable tolerance to variations in the composition and size of their amino acid sequences in the backbone loops of the CCK motif (Craik et al., 2006b; Gruber et al., 2007a) (illustrated in Figure 4). The CCK motif is a highly efficient combinatorial template, and changes in the primary amino acid sequence rarely disrupt the native fold of this template if the conserved Cys residues are not altered.
Table 3.
Statistics of Cyclotide Analysis
| Total No. | No. in Screen | Total (%) | No. Positive | Screen (%) | |
|---|---|---|---|---|---|
| Angiosperms (flowering plants) | >250,000a | 349 | NAb | 34 | 10 |
| Gentianales species (other than Rubiaceae) | 6,650c | 66 | NA | 12 | 18 |
| Rubiaceae species | 13,000 | 208 | 1.6 | 22 | 10 |
| Genera | 650 | 75 | 12.0 | 9 | 12 |
| Hedyotideaed | 734 | 27 | 3.7 | 8 | 30 |
| Lasiantheae | 187 | 5 | 2.7 | 4 | 80 |
| Psychotrieae/Palicoureeae | 2,793 | 46 | 1.6 | 10 | 22 |
Total number of flowering plant species is estimated to be 250,000 to 400,000 (Thorne, 2002; Govaerts, 2003; Scotland and Wortley, 2003).
Numbers are statistically not representative due to small number of species used in this screen.
Estimation based on numbers of species within the families of Apocynaceae (including Asclepiadaceae), Gelsemiaceae (segregated from Loganiaceae), Gentianaceae (including Saccifoliaceae), and Loganiaceae, excluding Rubiaceae according to the APGII classification.
sensu Delprete.
Although some cyclotides are expressed in several species, the occurrence of duplicates has previously been found to be low (<4%, considering we found four sequences multiple times out of 103 cyclotides characterized to date, from 15 species), and this was confirmed in this study as all newly identified cyclotides are novel with respect to mass and hydrophobicity. Previously, we found identical peptides in different plant species in Rubiaceae and Violaceae and within the Violaceae; for example, kalata B1 is found in O. affinis (Rubiaceae) and Viola odorata (Violaceae); varv A has been isolated from Viola tricolor, Viola arvensis, V. odorata, and O. affinis; tricyclon A is found in V. tricolor and V. arvensis; and varv E (=cycloviolacin O12) is found in V. arvensis, V. tricolor, and V. odorata (Craik et al., 1999; Göransson et al., 1999; Mulvenna et al., 2005).
Taken together, the results presented here suggest that cyclotides are much more abundant in the Gentianales than earlier anticipated, and combining this information with that on the occurrence of cyclotides in violet family plants (Simonsen et al., 2005), these structurally unique molecules could form one of the largest distinct protein families in plants. In trying to understand their number and diversity one can ask: how did cyclotides in particular and circular proteins in general evolve in plants? To address this question, we assessed the evidence in terms of the three commonly known mechanisms of evolution that might account for the observed distribution of cyclotides in the plant kingdom: (1) multiple independent gains of the cyclic functionality, (2) lateral gene transfer, and (3) descent from a common ancestor with losses.
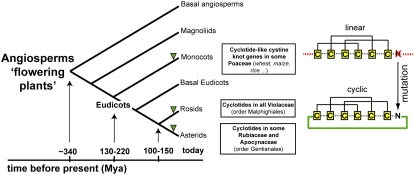
The most parsimonious explanation for the observed distribution is convergent evolution from linear cyclotide-like precursors with at least four independent origins of cyclotides within Rubiaceae. The three families containing cyclotide-bearing plants within the angiosperms (i.e., Rubiaceae/Apocynaceae and Violaceae) belong to Asterids and Rosids (Figure 7), respectively. There is no direct phylogenetic link between Rubiaceae/Apocynaceae and Violaceae, suggesting that cyclotides must have originated independently more than once and in distantly related families. Although lateral gene transfer is a theoretical possibility for the evolution of cyclotides, the differences in the cyclotide precursor gene structure between Rubiaceae and Violaceae, in particular the presence of introns in Rubiaceae but not in Violaceae cyclotide genes (Figure 6), significantly reduces the likelihood that cyclotides evolved by lateral transfer. If lateral gene transfer occurred, cyclotide genes of Rubiaceae species would have had to spontaneously gain intronic sequences, an event usually not linked to lateral gene transfer. Furthermore, the difference in Violaceae and Rubiaceae cyclotide gene structures does not favor the existence of a common cyclotide ancestor between these two plant families, which in turn argues against divergent evolution. As cyclotides have been found in every Violaceae species screened but only found in ∼10% of Rubiaceae species (Table 3), divergent evolution would require loss of cyclotide genes from a significant portion of the Rubiaceae family, which seems unlikely. Thus, overall the information on the distribution of cyclotides throughout the plant kingdom reported here, combined with knowledge of cyclotide gene structures, allows us to suggest that cyclotides most likely arose from convergent evolution in which a cyclizing capability was acquired. Did this capability arise before or after the knotted topology is the next question?
Figure 7.
Distribution and Evolution of Cyclotides in Plants.
Summary cladogram showing the major evolutionary groups of angiosperms and the occurrence of cyclotides within these groups. The timeline of the evolution of flowering plants is indicated in million years ago (Mya). Cyclization of linear proteins may occur by simple mutations of linear cystine knot ancestor genes, which yield appropriately located Asn/Asp residues.
The cystine knot motif of cyclotides is found in linear peptides from animals, plants, fungi, and viruses, although only in cyclotides is it combined with a cyclic peptide backbone. Based on the broad distribution of cystine-knotted peptides in multiple phyla, it seems likely that the evolution of the cystine knot motif occurred first, after which backbone cyclization occurred. The evolution of proteins with a knotted topology has been described based on an analysis of gene structures and protein function, and it has been suggested that convergent evolution is responsible for the structural similarity between animal and plant knotted peptides (Zhu et al., 2003).
The biosynthesis of cyclotides involves oxidative folding, proteolytic processing, and cyclization (summarized in Figure 1A). What evolutionary events triggered the specific processing that leads to cyclization? The recent discovery of linear cyclotide-like sequences in monocots (Poaceae) (Basse, 2005; Mulvenna et al., 2006a) (Figure 7) suggests that a mutation that introduces an Asn/Asp residue at a crucial cyclization point near the ancestral C terminus is the primary driving factor that facilitates the cyclization of linear proteins. This mechanism would explain the independent occurrence of circular proteins in various plant families and is strongly supported by recent reports that a common cellular enzyme (i.e., an asparaginyl-endopeptidase, which is generally involved in the activation and degradation of storage proteins) is implicated in the processing and cyclization of cyclotides (Saska et al., 2007; Gillon et al., 2008), suggesting that the biochemical machinery to make cyclic proteins is ubiquitously present in plants. Likewise, another common protein, protein disulfide isomerase, assists in folding of cyclotides (Gruber et al., 2007b), and so it appears that no special coevolution of a folding and cyclization machinery was necessary to equip a given plant species to produce cyclotides. Cyclotide-producing species simply co-opted existing enzymes to facilitate processing once appropriate mutations were acquired in ancestral linear cystine knot proteins.
Placing this interpretation on an evolutionary timescale, we propose that linear cyclotide-like, cystine knot genes evolved before the divergence of mono- and dicotyledonous plants, which occurred ∼130 to 220 million years ago (Wolfe et al., 1989; Moore et al., 2007). Monocots apparently did not subsequently evolve the necessary genetic mutation that drives cyclization of linear proteins, even though the cyclization enzyme (asparaginyl-endopeptidase) is ubiquitous in plants. The two successful cyclotide-producing groups within the flowering plants, namely, Asterids and Rosids, did however evolve the mutations to introduce Asn/Asp residues at the correct point for cyclization, and we therefore hypothesize that cyclotides evolved independently after the divergence of Asterids and Rosids ∼125 million years ago (Gandolfo et al., 1998; Yang et al., 1999; Bremer et al., 2004). This cyclization mechanism has also successfully evolved independently in cyclotide-related molecules from the cucurbit family (Cucurbitaceae and Rosids) (Hernandez et al., 2000) and trypsin inhibitors from sunflowers (Asteraceae and Asterids) (Korsinczky et al., 2001).
In conclusion, this study demonstrates the value of an interdisciplinary approach for the discovery of novel bioactive molecules from plants. The combination of plant systematics with modern analytical techniques to discover new bioactive molecules has provided an insight into the evolution of a topologically unique family of proteins. It appears that the events that triggered the cyclization of linear proteins to produce cyclotides occurred independently within different plant families by mutation of linear cystine knot genes. Furthermore, circular proteins appear to be much more common than originally anticipated, and we propose that future screening will identify cyclotides in other flowering plant families as the proposed cyclization mechanism may have evolved more ubiquitously. It is likely that cyclotides will ultimately comprise one of the largest protein families within the plant kingdom.
METHODS
Collection and Deposition of Plant Material
Plant material for this study was obtained from the National Botanic Garden of Belgium, the Botanical Gardens in Brisbane and Melbourne (Australia), Auckland, Wellington, and Christchurch (New Zealand), the Botanical Garden of Karl Franzens University Graz (Austria), the National Tropical Botanical Gardens Kalaheo (Hawaii), and the New York Botanical Garden. Field collections were performed in Australia (Queensland and Northern Territory), Sweden (Uppland), Austria (Styria), Hungary (Zala), Tanzania (West Usambara Mountains), and Hawaii (Oahu and Kauai). Analysis was performed on either fresh, dried, or herbarium plants. Plants from Tanzania were identified by Charlotte Taylor or Sigara and have been deposited in the herbarium of Missouri Botanical Gardens or the Tanzanian National Herbarium Lushoto. Hawaiian plants were identified by David Lorence and Ken Wood (National Tropical Botanical Gardens).
Extraction and Purification of Plant Material
Approximately 100 to 800 mg of dried or semidry plant material was homogenized to fine powder with a mortar and pestle in liquid N2 and extracted twice overnight in 50% acetonitrile containing 0.05% trifluoroacetic acid (TFA) or in dichloromethane:methanol (1:1; v/v) as described earlier (Colgrave and Craik, 2004; Trabi et al., 2004; Colgrave et al., 2005). The dried extracts were dissolved in solvent A (Milli-Q H2O with 0.05% TFA) and loaded onto a C18 solid phase extraction column. To separate the hydrophilic noncyclotide compounds from the hydrophobic cyclotide compounds, the column was washed with 20% solvent B (90% acetonitrile in Milli-Q H2O with 0.05% TFA) and eluted with 80% solvent B. The eluates containing cyclotides were analyzed either by LC-MS or MALDI-TOF and RP-HPLC.
Analysis of Peptide Extracts
RP-HPLC, LC-MS, nanospray LC-MS, and MALDI-TOF MS analysis were performed as described earlier (Colgrave and Craik, 2004; Trabi et al., 2004; Colgrave et al., 2005) with minor modifications. For nanospray LC-MS, samples were subjected to a gradient of 90% acetonitrile with 0.1% formic acid over 0.1% formic acid (10 to 70% over 60 min). Molecular mass determinations were done using full-scan mode in the range of 200 to 1800 mass-to-charge ratio using a potential of 900 V applied to the nanospray needle. Reduction and alkylation of peptide extracts was performed as described earlier (Colgrave and Craik, 2004; Colgrave et al., 2005) with minor modifications. Dried peptide extracts were reduced in a buffer containing 0.05 M Tris-HCl, pH 8.3, 4.2 M guanidine-HCl, and 8 mM DTT, and reduced peptides were further alkylated in a buffer containing 0.2 M Tris-HCl, pH 8.3, and 200 mM iodoacetamide. Reduced and alkylated peptide extracts were quenched and analyzed by MS.
Tandem MS Sequencing of Purified Peptides
Novel cyclotides were isolated, purified to homogeneity using RP-HPLC, and prepared for MS/MS sequencing as described earlier (Chen et al., 2005; Ireland et al., 2006). The purified, reduced, and enzymatic digested (trypsin, chymotrypsin, and Endo-GluC) peptides were analyzed by MALDI-TOF MS followed by MALDI-TOF MS/MS sequencing on the Applied Biosystems 4700 MALDI-TOF system. A capillary voltage of 1 kV was applied, and spectra were acquired between mass-to-charge ratios of 60 to 2000 for both TOF spectra and product ion spectra. The MS/MS spectra were examined and sequenced based on N-terminal b-ion fragmentation and C-terminal y-ion fragmentation. Chymotrypsin digests using the same conditions as for trypsin were also conducted to differentiate between the isobaric residues Leu and Ile. Amino acid composition deduced from sequencing was confirmed by high-sensitivity amino acid analysis conducted by the Australian Proteome Analysis Facility. The disulfide connectivity of CysI-IV, CysII-V, and CysIII-VI was assumed based on homology with previously reported cyclotides (Göransson and Craik, 2003).
DNA Extraction and Analysis of Cyclotide Precursor Genes
The protocol used to extract DNA from the leaves of Oldenlandia affinis, Viola odorata, Melicytus ramiflorus, and Viola tricolor was based on a DNA minipreparation method described previously (Chen and Ronald, 1999). In our protocol, we used a modified extraction buffer containing 2% (w/v) CTAB, 1.4 M NaCl, 200 mM Tris, pH 8.0, and 50 mM EDTA. For V. odorata and M. ramiflorus, leaf extracts were viscous and gave poor quality DNA, likely because they contain polysaccharides, polyphenolics, tannins, and secondary metabolites, as is the case for other plant samples (Li et al., 2007). For these plants, 2% (w/v) polyvinylpyrrolidone and 5% (v/v) β-mercaptoethanol were added to the extraction buffer, and 1 M NaCl was used in the isopropanol precipitation step to improve the quality of the DNA extract. DNA from Kadua centranthoides was supplied from the Hawaiian plant DNA library (Morden et al., 1996; Randell and Morden, 1999). Primers to amplify the cyclotide genes were designed against the cDNA sequences of expressed cyclotide transcripts, which can be accessed from the database of circular proteins called CyBase (Mulvenna et al., 2006b; Wang et al., 2008), and are shown in Supplemental Table 6 online. The PCR product was run on a 1.5% agarose gel, and the bands were excised, gel-purified with a QIAquick gel extraction kit (Qiagen), and TOPO cloned into a pCR2.1 vector (Invitrogen) for sequencing by the Australian Genome Research Facility (Brisbane).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FJ211181 (Viola odorata, Vok1), FJ211182 (Melicytus ramiflorus, Mrl13), FJ211183 (Viola tricolor, Vtt1), FJ211184 to FJ211187 (Oldenlandia affinis, Oak1 to -4), FJ211188 (Kadua centranthoides, Kch1), and FJ211189 (Viola odorata, Voc) for cyclotide precursor sequences, as listed in Figure 4.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. MS/MS Sequencing of CD-1.
Supplemental Figure 2. MS/MS Sequencing of PS-1.
Supplemental Table 1. Non-Cyclotide-Containing Rubiaceae Species.
Supplemental Table 2. Number of Novel Cyclotides from Positive Rubiaceae Species.
Supplemental Table 3. Non-Cyclotide-Containing Angiosperm (Non-Rubiaceae and Non-Violaceae) Species.
Supplemental Table 4. Rubioideae Tribes with Potential Cyclotide-Producing Species.
Supplemental Table 5. Comparison of Tribal and Subtribal Classification within the Subfamily Rubioideae.
Supplemental Table 6. Primers Used for Cyclotide Gene Characterization.
Supplementary Material
Acknowledgments
We thank Sigara (Lushoto Herbarium, Tanzania), Charlotte Taylor (Missouri Botanical Gardens), Petra De Block (National Botanic Garden of Belgium), David Lorence, and Ken Wood (National Tropical Botanical Gardens, Hawaii) for help with collection and/or identification of plant material and the New York Botanical Garden for plant specimen and for providing herbarium samples for analysis. We also thank Jason Mulvenna for his help with the isolation of cyclotide precursor genes. This work was supported by grants from the Australian Research Council, the University of Queensland (C.W.G.), and the Swedish Institute (C.W.G.). D.J.C. is an Australian Research Council Professorial Fellow. Part of this research was realized during a fellowship (P.G.D.) for Visiting Scientist from the National Counsel of Technological and Scientific Development of Brazil (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazilian Government) at the Institute of Biological Sciences of the Federal University of Goiás, Goiânia, Goiás, Brazil. Parts of this research have been facilitated by access to the Australian Proteome Analysis Facility established under the Australian Governments Major National Research Facilities Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: David J. Craik (d.craik@imb.uq.edu.au).
Online version contains Web-only data.
References
- Backlund, M., Oxelman, B., and Bremer, B. (2000). Phylogenetic relationships within the Gentianales based on ndhF and rbcL sequences, with particular reference to the Loganiaceae. Am. J. Bot. 87 1029–1043. [PubMed] [Google Scholar]
- Barbeta, B.L., Marshall, A.T., Gillon, A.D., Craik, D.J., and Anderson, M.A. (2008). Plant cyclotides disrupt epithelial cells in the midgut of lepidopteran larvae. Proc. Natl. Acad. Sci. USA 105 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basse, C.W. (2005). Dissecting defense-related and developmental transcriptional responses of maize during Ustilago maydis infection and subsequent tumor formation. Plant Physiol. 138 1774–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokesch, H.R., Pannell, L.K., Cochran, P.K., Sowder, R.C., McKee, T.C., and Boyd, M.R. (2001). A novel anti-HIV macrocyclic peptide from Palicourea condensata. J. Nat. Prod. 64 249–250. [DOI] [PubMed] [Google Scholar]
- Bremer, B. (1996). Phylogenetic studies within Rubiaceae and relationships to other families based on molecular data. Opera Bot. Belg. 7 33–50. [Google Scholar]
- Bremer, B., Andreasen, K., and Olsson, D. (1995). Subfamilial and tribal relationships in the Rubiaceae based on rbcL sequence data. Ann. Mo. Bot. Gard. 82 383–397. [Google Scholar]
- Bremer, B., and Jansen, R.K. (1991). Comparative restriction site mapping of chloroplast DNA implies new phylogenetic relationships within the Rubiaceae. Am. J. Bot. 78 198–213. [Google Scholar]
- Bremer, B., Jansen, R.K., Oxelman, B., Backlund, M., Lantz, H., and Kim, K.-J. (1999). More Characters and more taxa for a robust phylogeny - case study from the coffee family (Rubiaceae). Syst. Biol. 48 413–435. [DOI] [PubMed] [Google Scholar]
- Bremer, B., and Manen, J.-F. (2000). Phylogeny and classification of the subfamily Rubioideae. Plant Syst. Evol. 225 43–72. [Google Scholar]
- Bremer, K., Friis, E.M., and Bremer, B. (2004). Molecular phylogenetic dating of asterid flowering plants shows early Cretaceous diversification. Syst. Biol. 53 496–505. [DOI] [PubMed] [Google Scholar]
- Brown, J.W., Simpson, C.G., Thow, G., Clark, G.P., Jennings, S.N., Medina-Escobar, N., Haupt, S., Chapman, S.C., and Oparka, K.J. (2002). Splicing signals and factors in plant intron removal. Biochem. Soc. Trans. 30 146–149. [DOI] [PubMed] [Google Scholar]
- Chen, B., Colgrave, M.L., Daly, N.L., Rosengren, K.J., Gustafson, K.R., and Craik, D.J. (2005). Isolation and characterization of novel cyclotides from Viola hederaceae: Solution structure and anti-HIV activity of vhl-1, a leaf-specific expressed cyclotide. J. Biol. Chem. 280 22395–22405. [DOI] [PubMed] [Google Scholar]
- Chen, D.H., and Ronald, P.C. (1999). A rapid DNA minipreparation method suitable for AFLP and other PCR applications. Plant Mol. Biol. Rep. 17 53–57. [Google Scholar]
- Chiche, L., Heitz, A., Gelly, J.C., Gracy, J., Chau, P.T., Ha, P.T., Hernandez, J.F., and Le-Nguyen, D. (2004). Squash inhibitors: From structural motifs to macrocyclic knottins. Curr. Protein Pept. Sci. 5 341–349. [DOI] [PubMed] [Google Scholar]
- Colgrave, M.L., and Craik, D.J. (2004). Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: The importance of the cyclic cystine knot. Biochemistry 43 5965–5975. [DOI] [PubMed] [Google Scholar]
- Colgrave, M.L., Jones, A., and Craik, D.J. (2005). Peptide quantification by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry: Investigations of the cyclotide kalata B1 in biological fluids. J. Chromatogr. A. 1091 187–193. [DOI] [PubMed] [Google Scholar]
- Craik, D.J., Cemazar, M., and Daly, N.L. (2006. a). The cyclotides and related macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Devel. 9 251–260. [PubMed] [Google Scholar]
- Craik, D.J., Cemazar, M., Wang, C.K., and Daly, N.L. (2006. b). The cyclotide family of circular mini-proteins: Nature's combinatorial peptide template. Biopolymers 84 250–266. [DOI] [PubMed] [Google Scholar]
- Craik, D.J., Daly, N.L., Bond, T., and Waine, C. (1999). Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J. Mol. Biol. 294 1327–1336. [DOI] [PubMed] [Google Scholar]
- Craik, D.J., Simonsen, S., and Daly, N.L. (2002). The cyclotides: Novel macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Devel. 5 251–260. [PubMed] [Google Scholar]
- Daly, N.L., Love, S., Alewood, P.F., and Craik, D.J. (1999). Chemical synthesis and folding pathways of large cyclic polypeptides: studies of the cystine knot polypeptide kalata B1. Biochemistry 38 10606–10614. [DOI] [PubMed] [Google Scholar]
- Delprete, P.G. (2004). Rubiaceae. In Flowering Plant Families of the American Tropics, N.P. Smith, S.V. Heald, A. Henderson, S.A. Mori, and D.W. Stevenson, eds (New York: Princeton University Press/New York Botanical Garden Press), pp. 328–333.
- Delprete, P.G., Choze, R., Silva, R.A., and Dufrayer, C.R. (2006). Chemotaxonomy and macroclassification of Rubiaceae. Scripta Botanica Belgica. 40 28. [Google Scholar]
- De Wildeman, E. (1901). Notes sur le Ngulu-Maza, bois d'ébénisterie et de construction du Bas-Congo. Rev. Cult. Col. 9 7–10. [Google Scholar]
- Dutton, J.L., Renda, R.F., Waine, C., Clark, R.J., Daly, N.L., Jennings, C.V., Anderson, M.A., and Craik, D.J. (2004). Conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins. J. Biol. Chem. 279 46858–46867. [DOI] [PubMed] [Google Scholar]
- Gandolfo, M.A., Nixon, K.C., and Crepet, W.L. (1998). A new fossil flower from the Turonian of New Jersey: Dressiantha bicarpellata gen. et sp. nov. (Capparales). Am. J. Bot. 85 964–974. [PubMed] [Google Scholar]
- Gillon, A.D., Saska, I., Jennings, C.V., Guarino, R.F., Craik, D.J., and Anderson, M.A. (2008). Biosynthesis of circular proteins in plants. Plant J. 53 505–515. [DOI] [PubMed] [Google Scholar]
- Göransson, U., and Craik, D.J. (2003). Disulfide mapping of the cyclotide kalata B1. Chemical proof of the cyclic cystine knot motif. J. Biol. Chem. 278 48188–48196. [DOI] [PubMed] [Google Scholar]
- Göransson, U., Luijendijk, T., Johansson, S., Bohlin, L., and Claeson, P. (1999). Seven novel macrocyclic polypeptides from Viola arvensis. J. Nat. Prod. 62 283–286. [DOI] [PubMed] [Google Scholar]
- Göransson, U., Sjogren, M., Svangard, E., Claeson, P., and Bohlin, L. (2004). Reversible antifouling effect of the cyclotide cycloviolacin O2 against barnacles. J. Nat. Prod. 67 1287–1290. [DOI] [PubMed] [Google Scholar]
- Govaerts, R. (2003). How many species of seed plants are there? A response. Taxon 52 583–584. [Google Scholar]
- Govaerts, R., Frodin, D.G., Ruhsam, M., Bridson, D.M., and Davis, A.P. (2006). A world checklist of Rubiaceae. Scripta Bot. Belg. 40 35. [Google Scholar]
- Gran, L. (1970). An oxytocic principle found in oldenlandia affinis dc. An indigenous, congolese drug “Kalata-Kalata” used to accelerate delivery. Medd. Nor. Farm. Selsk. 12 173–180. [Google Scholar]
- Gran, L. (1973). On the effect of a polypeptide isolated from “Kalata-Kalata” (Oldenlandia affinis DC) on the oestrogen dominated uterus. Acta Pharmacol. Toxicol. (Copenh.) 33 400–408. [DOI] [PubMed] [Google Scholar]
- Gran, L., Sandberg, F., and Sletten, K. (2000). Oldenlandia affinis (R&S) DC. A plant containing uteroactive peptides used in African traditional medicine. J. Ethnopharmacol. 70 197–203. [DOI] [PubMed] [Google Scholar]
- Gruber, C.W., Cemazar, M., Anderson, M.A., and Craik, D.J. (2007. a). Insecticidal plant cyclotides and related cystine knot toxins. Toxicon 49 561–575. [DOI] [PubMed] [Google Scholar]
- Gruber, C.W., Cemazar, M., Clark, R.J., Horibe, T., Renda, R.F., Anderson, M.A., and Craik, D.J. (2007. b). A novel plant protein disulfide isomerase involved in the oxidative folding of cystine knot defense proteins. J. Biol. Chem. 282 20435–20446. [DOI] [PubMed] [Google Scholar]
- Gustafson, K.R., McKee, T.C., and Bokesch, H.R. (2004). Anti-HIV cyclotides. Curr. Protein Pept. Sci. 5 331–340. [DOI] [PubMed] [Google Scholar]
- Gustafson, K.R., Sowder, R.C., Henderson, L.E.I., Parsons, I.C., Kashman, Y., Cardellina, J.H., McMahon, J.B.I., Buckheit, R.W., Pannell, L.K.J., and Boyd, M.R. (1994). Circulins A and B. Novel human immunodeficiency virus (HIV)-inhibitory macrocyclic peptides from the tropical tree Chassalia parvifolia. J. Am. Chem. Soc. 113 9337–9338. [Google Scholar]
- Gustafson, K.R., Walton, L.K., Sowder, R.C., Jr., Johnson, D.G., Pannell, L.K., Cardellina, J.H., Jr., and Boyd, M.R. (2000). New circulin macrocyclic polypeptides from Chassalia parvifolia. J. Nat. Prod. 63 176–178. [DOI] [PubMed] [Google Scholar]
- Hernandez, J.F., Gagnon, J., Chiche, L., Nguyen, T.M., Andrieu, J.P., Heitz, A., Hong, T.T., Pham, T.T.C., and Nguyen, D.L. (2000). Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry 39 5722–5730. [DOI] [PubMed] [Google Scholar]
- Ireland, D.C., Colgrave, M.L., and Craik, D.J. (2006). A novel suite of cyclotides from Viola odorata: Sequence variation and the implications for structure, function and stability. Biochem. J. 400 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings, C., West, J., Waine, C., Craik, D., and Anderson, M. (2001). Biosynthesis and insecticidal properties of plant cyclotides: The cyclic knotted proteins from Oldenlandia affinis. Proc. Natl. Acad. Sci. USA 98 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings, C.V., Rosengren, K.J., Daly, N.L., Plan, M., Stevens, J., Scanlon, M.J., Waine, C., Norman, D.G., Anderson, M.A., and Craik, D.J. (2005). Isolation, solution structure, and insecticidal activity of kalata B2, a circular protein with a twist: Do Mobius strips exist in nature? Biochemistry 44 851–860. [DOI] [PubMed] [Google Scholar]
- Korsinczky, M.L.J., Schirra, H.J., Rosengren, K.J., West, J., Condie, B.A., Otvos, L., Anderson, M.A., and Craik, D.J. (2001). Solution structures by H-1 NMR of the novel cyclic trypsin inhibitor SFTI-1 from sunflower seeds and an acyclic permutant. J. Mol. Biol. 311 579–591. [DOI] [PubMed] [Google Scholar]
- Li, J.T., Yang, J., Chen, D.C., Zhang, X.L., and Tang, Z.S. (2007). An optimized mini-preparation method to obtain high-quality genomic DNA from mature leaves of sunflower. Genet. Mol. Res. 6 1064–1071. [PubMed] [Google Scholar]
- Lindholm, P., Goransson, U., Johansson, S., Claeson, P., Gulbo, J., Larsson, R., Bohlin, L., and Backlund, A. (2002). Cyclotides: A novel type of cytotoxic agents. Mol. Cancer Ther. 1 365–369. [PubMed] [Google Scholar]
- Moore, M.J., Bell, C.D., Soltis, P.S., and Soltis, D.E. (2007). Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc. Natl. Acad. Sci. USA 104 19363–19368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morden, C.W., Caraway, V.C., and Motley, T.J. (1996). Development of a DNA library for native Hawaiian plants. Pac. Sci. 50 324–335. [Google Scholar]
- Mulvenna, J.P., Mylne, J.S., Bharathi, R., Burton, R.A., Shirley, N.J., Fincher, G.B., Anderson, M.A., and Craik, D.J. (2006. a). Discovery of cyclotide-like protein sequences in graminaceous crop plants: Ancestral precursors of circular proteins? Plant Cell 18 2134–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvenna, J.P., Sando, L., and Craik, D.J. (2005). Processing of a 22 kDa precursor protein to produce the circular protein tricyclon A. Structure 13 691–701. [DOI] [PubMed] [Google Scholar]
- Mulvenna, J.P., Wang, C., and Craik, D.J. (2006. b). CyBase: A database of cyclic protein sequence and structure. Nucleic Acids Res. 34 D192–D194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plan, M.R., Saska, I., Cagauan, A.G., and Craik, D.J. (2008). Backbone cyclised peptides from plants show molluscicidal activity against the rice pest Pomacea canaliculata (golden apple snail). J. Agric. Food Chem. 56 5237–5241. [DOI] [PubMed] [Google Scholar]
- Plan, M.R.R., Göransson, U., Clark, R.J., Daly, N.L., Colgrave, M.L., and Craik, D.J. (2007). The cyclotide fingerprint in Oldenlandia affinis: elucidation of chemically modified, linear and novel macrocyclic peptides. ChemBioChem 8 1001–1011. [DOI] [PubMed] [Google Scholar]
- Purseglove, J.W. (1968). Dicotyledons. (London: Longman).
- Randell, R.A., and Morden, C.W. (1999). Hawaiian Plant DNA library II: Endemic, indigenous, and introduced species. Pac. Sci. 53 401–417. [Google Scholar]
- Robbrecht, E., and Manen, J.F. (2006). The major evolutionary lineages of the coffee family (Rubiaceae, angiosperms). Combined analysis (nDNA and cpDNA) to infer the position of Coptospelta and Luculia, and supertree construction based on rbcL, srp16, trnL-trnF and atpB-rbcL data. A new classification in two subfamilies, Cinchonoideae and Rubioideae. Syst. Geogr. Plants 76 85–146. [Google Scholar]
- Rova, H.E., Delprete, P.G., Andersson, L., and Albert, V.A. (2002). A trnL-F cpDNA sequence study of the Condamineeae-Rondeletieae-Sipaneeae complex with implications on the phylogeny of the Rubiaceae. Am. J. Bot. 89 145–159. [DOI] [PubMed] [Google Scholar]
- Saether, O., Craik, D.J., Campbell, I.D., Sletten, K., Juul, J., and Norman, D.G. (1995). Elucidation of the primary and three-dimensional structure of the uterotonic polypeptide kalata B1. Biochemistry 34 4147–4158. [DOI] [PubMed] [Google Scholar]
- Saska, I., Gillon, A.D., Hatsugai, N., Dietzgen, R.G., Hara-Nishimura, I., Anderson, M.A., and Craik, D.J. (2007). An asparaginyl endopeptidase mediates in vivo protein backbone cyclysation. J. Biol. Chem. 282 29721–29728. [DOI] [PubMed] [Google Scholar]
- Schöpke, T., Hasan Agha, M.I., Kraft, R., Otto, A., and Hiller, K. (1993). Hämolytisch aktive komponenten aus Viola tricolor L. und Viola arvensis Murray. Sci. Pharm. 61 145–153. [Google Scholar]
- Scotland, R.W., and Wortley, A.H. (2003). How many species of seed plants are there? Taxon 52 101–104. [Google Scholar]
- Seydel, P., Gruber, C.W., Craik, D.J., and Dörnenburg, H. (2007). Formation of cyclotides and variations in cyclotide expression in Oldenlandia affinis suspension cultures. Appl. Microbiol. Biotechnol. 77 275–284. [DOI] [PubMed] [Google Scholar]
- Simonsen, S.M., Sando, L., Ireland, D.C., Colgrave, M.L., Bharathi, R., Goransson, U., and Craik, D.J. (2005). A continent of plant defense peptide diversity: Cyclotides in Australian hybanthus (Violaceae). Plant Cell 17 3176–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe, L., Albert, V.A., and Bremer, B. (1994). Cladistics and family level classification of the Gentianales. Cladistics 10 175–205. [Google Scholar]
- Tam, J.P., Lu, Y.A., Yang, J.L., and Chiu, K.W. (1999). An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc. Natl. Acad. Sci. USA 96 8913–8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne, R.F. (2002). How many species of seed plants are there? Taxon 51 511–522. [Google Scholar]
- Trabi, M., Svangard, E., Herrmann, A., Göransson, U., Claeson, P., Craik, D.J., and Bohlin, L. (2004). Variations in cyclotide expression in viola species. J. Nat. Prod. 67 806–810. [DOI] [PubMed] [Google Scholar]
- Wang, C.K., Kaas, Q., Chiche, L., and Craik, D.J. (2008). CyBase: A database of cyclic protein sequences and structures, with applications in protein discovery and engineering. Nucleic Acids Res. 36 D206–D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherup, K.M., Bogusky, M.J., Anderson, P.S., Ramjit, H., Ransom, R.W., Wood, T., and Sardana, M. (1994). Cyclopsychotride A, A biologically active, 31-residue cyclic peptide isolated from Psychotria longipes. J. Nat. Prod. 57 1619–1625. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H., Gouy, M.L., Yang, Y.W., Sharp, P.M., and Li, W.H. (1989). Date of the monocot dicot divergence estimated from chloroplast DNA-sequence data. Proc. Natl. Acad. Sci. USA 86 6201–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.W., Lai, K.N., Tai, P.Y., and Li, W.H. (1999). Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J. Mol. Evol. 48 597–604. [DOI] [PubMed] [Google Scholar]
- Zhu, S.Y., Darbon, H., Dyason, K., Verdonck, F., and Tytgat, J. (2003). Evolutionary origin of inhibitor cystine knot peptides. FASEB J. 17 1765–1767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.