Abstract
Bioactive hormone concentrations are regulated both at the level of hormone synthesis and through controlled inactivation. Based on the ubiquitous presence of 2β-hydroxylated gibberellins (GAs), a major inactivating pathway for the plant hormone GA seems to be via GA 2-oxidation. In this study, we used various approaches to determine the role of C19-GA 2-oxidation in regulating GA concentration and GA-responsive plant growth and development. We show that Arabidopsis thaliana has five C19-GA 2-oxidases, transcripts for one or more of which are present in all organs and at all stages of development examined. Expression of four of the five genes is subject to feed-forward regulation. By knocking out all five Arabidopsis C19-GA 2-oxidases, we show that C19-GA 2-oxidation limits bioactive GA content and regulates plant development at various stages during the plant life cycle: C19-GA 2-oxidases prevent seed germination in the absence of light and cold stimuli, delay the vegetative and floral phase transitions, limit the number of flowers produced per inflorescence, and suppress elongation of the pistil prior to fertilization. Under GA-limited conditions, further roles are revealed, such as limiting elongation of the main stem and side shoots. We conclude that C19-GA 2-oxidation is a major GA inactivation pathway regulating development in Arabidopsis.
INTRODUCTION
For the correct functioning of a chemical communication system, it is essential that there is a mechanism in place to remove active signaling compounds, preventing their progressive accumulation. In addition, removal of the active compounds or their biosynthetic precursors may modulate the amount of the signal being produced. This applies to hormonal communication systems in multicellular organisms, and many hormones are subject to controlled degradation, sequestration, or inactivation. In plants, inactivating pathways are known for all the classical plant hormones, including auxin, cytokinin, abscisic acid, brassinosteroid, and ethylene; they and/or their precursors can be inactivated via specific oxidation and conjugation pathways (Turk et al., 2003; Nambara and Marion-Poll, 2005; Woodward and Bartel, 2005; Sakakibara, 2006). A number of inactivation pathways have similarly been identified for the gibberellin (GA) hormones (Thomas and Hedden, 2006). GAs are a family of hormones regulating many aspects of plant growth and development, including seed germination, stem elongation, and the transition from vegetative growth to flowering (Davies, 2004). They are synthesized from geranylgeranyl diphosphate via a series of conversions involving three classes of enzymes (Hedden and Phillips, 2000). The later conversions involve soluble 2-oxoglutarate–dependent dioxygenases: GA 20-oxidase is involved in the successive oxidation of C-20, leading to its removal and the formation of C19-GAs (GA9 and GA20) containing a C-19,10-lactone that is essential for biological activity. These C19-GAs are converted to the bioactive GAs, GA4 and GA1, by the action of GA 3-oxidase.
A number of mechanisms are known to inactivate GAs. It has long been recognized that GAs can be modified by conjugation, most commonly as ethers and esters of glucose, but a clear physiological role for these processes has not been elucidated (Schneider and Schliemann, 1994). Two novel mechanisms of GA inactivation have recently been reported. First, the ELONGATED UPPERMOST INTERNODE gene of rice (Oryza sativa) encodes a cytochrome P450 monooxygenase that catalyzes epoxidation of the C-16,17 double bond and results in loss of biological activity (Zhu et al., 2006). However, it is not clear to what extent this is a general mechanism that occurs in other plant species. Secondly, methyl transferases specific for GAs have been described in Arabidopsis thaliana, where expression is mainly localized to developing seeds, and analysis of loss-of-function mutants suggests a role in GA inactivation in seed tissues (Varbanova et al., 2007), although accumulation of methylated GAs has not yet been demonstrated.
However, based on the prevalence of 2β-hydroxylated GAs in many plant species (MacMillan, 2002), the most widespread mechanism for GA inactivation seems to be via 2-oxidation, in which the diterpene backbone is hydroxylated at C-2β and, in some cases, further oxidized to a ketone (Sponsel and Macmillan, 1978; Thomas et al., 1999), which undergoes nonenzymatic rearrangement to form a so-called GA catabolite. Biochemical characterization of the enzymes responsible for 2β-hydroxylation showed that, like the GA 20-oxidases and GA 3-oxidases, they belong to the soluble 2-oxoglutarate–dependent dioxygenase class (Griggs et al., 1991). Genes encoding GA 2-oxidases were first identified by screening cDNA expression libraries for 2β-hydroxylase activity (Lester et al., 1999; Martin et al., 1999; Thomas et al., 1999). Five Arabidopsis GA 2-oxidase genes (GA2ox1, -2, -3, -4, and -6; GA2ox5, At3g17203, is a pseudogene) have since been identified based on sequence homology, and their encoded enzyme activities have been confirmed by heterologous expression in Escherichia coli (Thomas et al., 1999; Hedden and Phillips, 2000; Wang et al., 2004; Jasinski et al., 2005), demonstrating that they were active against C19-GAs, such as the bioactive GAs, GA4 and GA1, and their immediate precursors. Evidence that all these enzymes can function in GA inactivation in vivo came from experiments in which overexpression of the genes in Arabidopsis resulted in dwarfing and reductions in bioactive GA levels (S.G. Thomas, A.L. Phillips, and P. Hedden, unpublished data; Wang et al., 2004), and similar results have been obtained in other transgenic plants with related GA 2-oxidases from poplar (Populus spp), rice, and runner bean (Phaseolus coccineus) (Sakamoto et al., 2001; Busov et al., 2003; Appleford et al., 2007). An activation tagging screen for dwarf mutants (Schomburg et al., 2003) identified two further, structurally distinct Arabidopsis genes that encoded 2-oxoglutarate–dependent dioxygenase enzymes with GA 2-oxidase activity. These enzymes, GA2ox7 and GA2ox8, were shown to be active against C20-GAs, such as GA12 and GA53, but are not capable of using C19-GAs as substrates. Therefore, they are not involved in inactivation of bioactive GAs, but rather may be important in regulating GA biosynthesis through the removal of earlier intermediates in the pathway. Phylogenetic analysis has shown that the GA 2-oxidases can be divided into three classes (Lee and Zeevaart, 2005). Members of class III, which includes GA2ox7 and GA2ox8 above, are distinguished by their absolute preference for C20-GAs, but the biochemical and biological significance of the structural division between classes I and II is less clear, although several class I enzymes have been shown to produce GA catabolites, whereas no class II enzymes have been found that have this activity (Serrani et al., 2007).
Despite our knowledge of the various mechanisms of GA inactivation, the importance of GA turnover in the regulation of plant development remains unclear: significantly, there are very few examples of characterized mutations in GA 2-oxidase genes. The pea (Pisum sativum) slender mutant contains a loss-of-function allele of a GA2ox gene, GA2ox1, that is normally active in developing seeds: mature seeds of the mutant have increased levels of GA20, which, after germination, is activated by 3β-hydroxylation, resulting in increased elongation of the seedling (Reid et al., 1992; Ross et al., 1993, 1995; Lester et al., 1999; Martin et al., 1999). Loss of function of the Arabidopsis GA2ox2 gene increases GA4 levels in dark-imbibed seeds and thereby promotes germination in the mutant (Yamauchi et al., 2007). Double mutants in GA2ox7 and GA2ox8 show an increased seed germination rate in the presence of the GA biosynthesis inhibitor ancymidol and have elongated hypocotyls (Schomburg et al., 2003).
In this study, we used a combination of biochemical, molecular, physiological, and reverse-genetic approaches to develop a comprehensive view of the role of C19-GA 2-oxidation in regulating GA levels and GA-responsive plant growth and development in Arabidopsis. We identified loss-of-function mutations in all five C19-GA 2-oxidases. Through analysis of the quintuple C19-GA2ox mutant, we were able to conclude that C19-GA 2-oxidation limits bioactive GA levels and regulates many stages of plant development. In some cases, the phenotypes could be largely attributed to individual GA 2-oxidases with specific roles in key developmental events.
RESULTS
Arabidopsis Contains Five GA 2-Oxidases That Specifically Inactivate C19-GAs
The Arabidopsis genome encodes three GA 2-oxidase (GA2ox) paralogs belonging to class I as defined by Lee and Zeevaart (2005): GA2ox1, GA2ox2, and GA2ox3. All three have been shown to act as C19-GA 2-oxidases (Thomas et al., 1999). Arabidopsis also has two genes belonging to class II: GA2ox4 and GA2ox6. GA2ox4 has been shown to act exclusively as a C19-GA 2-oxidase (Jasinski et al., 2005), while GA2ox6 was similarly reported to have activity against GA4 and GA1 (Wang et al., 2004), although its activity against C20-GAs and other C19-GAs was not tested. Using an in vitro assay with lysates of E. coli expressing GA2ox6 and 14C-labeled GA substrates, we confirmed the C19-GA 2-oxidase activity of GA2ox6 and were unable to find C20-GA 2-oxidase activity against GA12 and GA53 (Table 1). Thus, the Arabidopsis genome encodes five GA 2-oxidases that are active specifically against C19-GAs and are thus capable of inactivating bioactive GAs.
Table 1.
In Vitro Activity of Recombinant GA2ox6
| Substrate | Producta | |
|---|---|---|
| C20-GAs | GA12 | – |
| GA53 | – | |
| C19-GAs | GA9 | GA51 |
| GA20 | GA29 | |
| GA4 | GA34 | |
| GA1 | GA8 | |
| GA7 | – |
Presence of 14C-labeled conversion products was examined by HPLC, and product identity was confirmed by GC-MS (see Methods). –, No product detected.
C19-GA2ox Genes Are Expressed throughout Plant Development
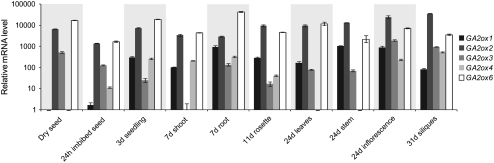
Our results indicate that GA2ox1, -2, -3, -4, and -6 have similar biochemical activities, all being able to catalyze 2β-hydroxylation of C19-GAs. To assess in which organs these enzymes act and to help elucidate the function of each of the C19-GA2ox genes in plant development, we analyzed their developmental expression profiles using real-time quantitative RT-PCR. Relative transcript levels were calibrated using standard curves, allowing for comparison of expression levels between genes. Figure 1 shows that GA2ox transcripts could be detected at all developmental stages analyzed. GA2ox2 and GA2ox6 were the most highly expressed genes in all samples, usually being one or more orders of magnitude higher than the other GA2ox genes. GA2ox6 was dominant early in development, with the highest level found in roots, confirming results obtained with a GA2ox6 promoter-β-glucuronidase reporter line (Wang et al., 2004). GA2ox2 was dominant in the stem, inflorescence, and siliques of mature plants. GA2ox1 transcripts were absent from the dry seed sample and reached highest levels in the root, stem, and inflorescence. GA2ox3 transcripts were more abundant in the inflorescence, silique, and dry seed than in other samples. GA2ox4 transcripts were higher in the 3- to 7-d-old seedling and the inflorescence and silique samples and absent from dry seed, leaf, and stem. These results are consistent with transcript data from a developmental series generated by the AtGenExpress microarray consortium (Schmid et al., 2005). This data set shows highest signals for GA2ox2 and GA2ox6, especially in flowers for GA2ox2, and in roots for GA2ox6. GA2ox1 transcript was found to be more abundant in flowers than in other tissues, GA2ox3 more abundant in young flowers, pollen, and developing seeds, and GA2ox4 more abundant in developing seed and shoot apex.
Figure 1.
Expression Analysis of the Arabidopsis C19-GA2ox Genes.
Developmental expression profile of the C19-GA2ox genes. mRNA levels were determined using real-time quantitative RT-PCR on three biological replicates for each tissue with technical quantitative PCR duplicates for each sample. Standard curves were used to calculate the absolute numbers of GA2ox cDNA molecules in each cDNA sample, and these values were then normalized against three reference genes (see Methods). Results are plotted as the ratio to the lowest detected level (i.e., GA2ox3 in 7 d shoot) ± se. Note that the y axis is on a logarithmic scale.
In conclusion, expression analysis suggests that C19-GA2ox genes function throughout the plant and throughout development. There are no clear indications from expression patterns of highly specific functions for any of the genes, although GA2ox2 and GA2ox6 are the most highly expressed genes at all developmental stages.
Generation of C19-GA2ox Single and Higher-Order Mutants
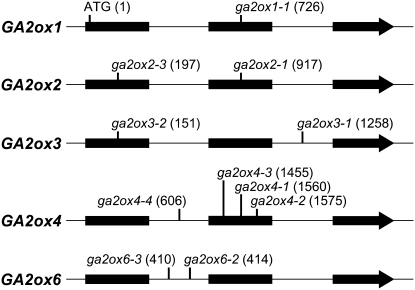
To determine the roles of C19-GA2ox1 through -6 in growth and development of Arabidopsis plants, we searched for insertion alleles for each of these genes in the SALK, GABI-KAT, SLAT, and Wisconsin DsLox collections (Tissier et al., 1999; Alonso et al., 2003; Rosso et al., 2003), which are all in the Columbia (Col) ecotype, and tested for the absence of functional transcripts by RT-PCR. We identified one null line for each of GA2ox1 (ga2ox1-1), GA2ox2 (ga2ox2-1), GA2ox3 (ga2ox3-1), and GA2ox6 (ga2ox6-2) and three lines for GA2ox4 (ga2ox4-1, ga2ox4-2, and ga2ox4-3) (Figure 2; see Supplemental Figure 1 online). In addition, we performed a PCR-based screening of the Wisconsin Knockout Facility Alpha collection (Sussman et al., 2000) and searched the FLAG collection (Samson et al., 2002) for insertions in the Wassilewskija (Ws) ecotype, again confirming the null status by RT-PCR. This resulted in the identification of one null line each for GA2ox2 (ga2ox2-3), GA2ox3 (ga2ox3-2), GA2ox4 (ga2ox4-4), and GA2ox6 (ga2ox6-3) (Figure 2; see Supplemental Figure 1 online). For each line, the exact location of the insertion was determined by sequencing of PCR-generated flanking sequences. Several other potential insertion lines were characterized during this study, but either the presence of the insert could not be confirmed or transcripts were detected in the RT-PCR analyses (see Supplemental Table 1 online).
Figure 2.
Gene Models of GA2ox1, -2, -3, -4, and -6.
The sites of insertions of mutant alleles used in this work are indicated. Numbers between brackets give the positions of the features in nucleotides relative to the start of translation. Thick horizontal lines represent exons, thin horizontal lines between exons represent introns, and vertical lines indicate the position of the feature.
As none of the single loss-of-function mutants displayed any obvious phenotype, we generated higher-order knockouts in the Col ecotype to study the overall role of C19-GA 2-oxidation in Arabidopsis. This included a quintuple mutant that carried loss-of-function alleles of all five C19-GA2ox genes (ga2ox1-1, ga2ox2-1, ga2ox3-1, ga2ox4-1, and ga2ox6-2; further referred to as ga2ox quintuple). Real-time quantitative RT-PCR confirmed that in this background no correctly spliced transcript was produced from the ga2ox3-1 and ga2ox6-2 alleles (see Supplemental Table 2 online).
GA 2-Oxidation and GA-Responsive Gene Expression in the ga2ox quintuple Mutant
Endogenous GA Levels
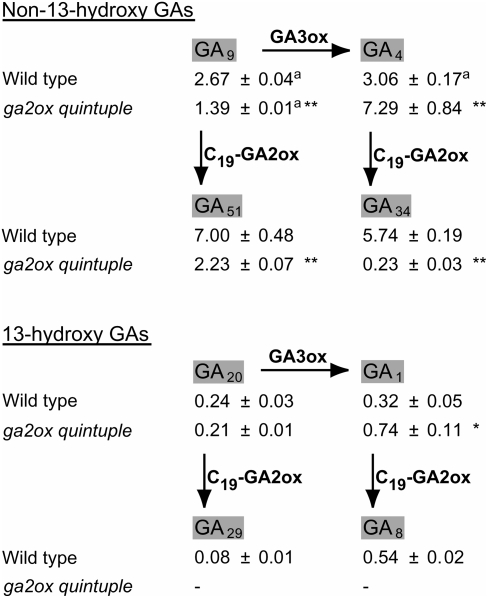
We determined the effect of the combined loss of GA2ox1 through -6 on the endogenous GA content by quantifying the C19-GAs and their 2β-hydroxylated forms in shoots (all aerial parts including the rosette) of wild-type and ga2ox quintuple plants at the bolting stage. Figure 3 shows that the levels of GA4 and GA1 were more than doubled in the ga2ox quintuple mutant, whereas the levels of their 2β-hydroxylated products were severely reduced (by 96% for GA34 and to undetectable levels for GA8). The levels of the 2β-hydroxylated products of GA9 and GA20, GA51, and GA29 were also both decreased in the ga2ox quintuple mutant, in the latter case to an undetectable level (see Discussion for potential routes to production of 2β-hydroxylated GAs in the quintuple mutant). GA9 levels were also reduced to approximately half the amount found in wild-type plants, although GA20 was unchanged.
Figure 3.
Profile of Endogenous GAs in Wild-Type and ga2ox quintuple Plants.
The GA levels (in rosette and bolt of flowering plants) are means of four biological replicates in ng g−1 dry weight ± se, except where indicated. a, Three biological replicates only; *, significantly different from the wild type (t test: P < 0.05); **, significantly different from the wild type (t test: P < 0.01); −, not detected.
GA 2-Oxidation
To determine whether combined knockout of the GA2ox1 through -6 genes resulted in complete absence of C19-GA 2-oxidase activity, we applied 14C-labeled GA9 and GA4 to the apical region of wild-type and ga2ox quintuple plants that had just started to form flower buds and analyzed the products that were formed after 24 h. Whereas in wild-type plants HPLC combined with radiomonitoring showed that 21% of the 14C-GA4 had been converted to 14C-GA34, there was no detectable conversion of 14C-GA4 in the ga2ox quintuple mutant (see Supplemental Figure 2 online), as confirmed by gas chromatography–mass spectrometry (GC-MS) analysis of the products in the appropriate fraction of the HPLC eluate. Similar results were obtained with feeding of 14C-GA9, which was converted to 14C-GA51 in the wild type but not in the ga2ox quintuple. This suggests a complete loss of C19-GA 2-oxidase activity in the ga2ox quintuple mutant.
GA Response at the Molecular Level
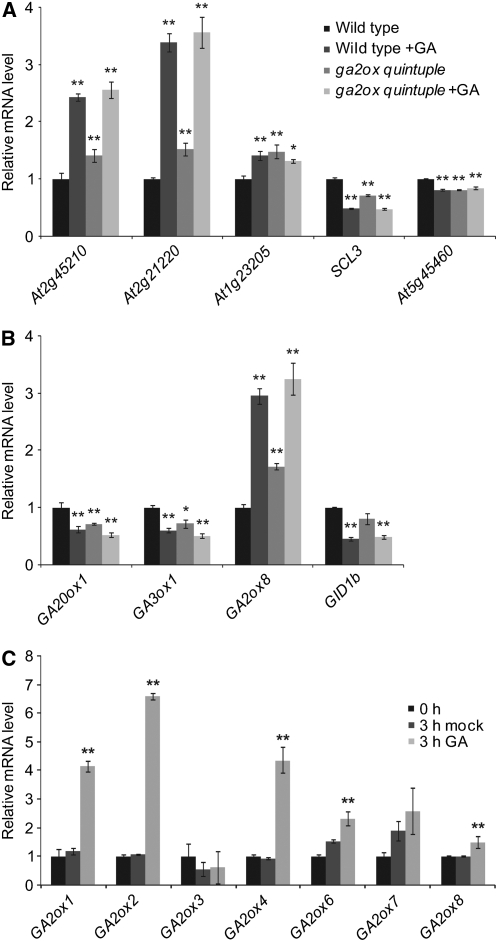
To examine whether the increased levels of bioactive GA in ga2ox quintuple plants resulted in an increased GA response at the molecular level, the expression levels of known GA-regulated genes were compared in shoots of bolting wild-type and ga2ox quintuple plants. A recent analysis of microarray transcript data of hormone-treated Arabidopsis seedlings has generated a list of genes that are induced or repressed by GA (Nemhauser et al., 2006). We tested a number of these genes in bolting shoots of the wild type and the ga2ox quintuple mutant and found their expression to be significantly different in the latter, showing the same trend as in the GA-treated wild type (Figure 4A; see Supplemental Table 2 online). In some cases (At2g45210, At2g12220, and SCARECROW-LIKE3 [SCL3]), further treatment of the ga2ox quintuple mutant with GA3 resulted in an increased response, whereas for others (At1g23205 and At5g45460), the response was already saturated. Many of the genes coding for components of the GA biosynthesis pathway are also regulated by GA (Chiang et al., 1995; Phillips et al., 1995; Thomas et al., 1999; Schomburg et al., 2003; Nemhauser et al., 2006). Figure 4B and Supplemental Table 2 online show that the ga2ox quintuple mutant had significantly reduced expression of GA biosynthetic genes (GA20ox1 and GA3ox1). As with four of the five C19-GA2ox genes, GA2ox3 being the exception, the C20-GA2ox gene GA2ox8 is positively regulated by GA (Figure 4C; see Supplemental Table 3 online). GA2ox8 expression was also increased in the ga2ox quintuple mutant (Figure 4B). These results confirm the presence of increased GA signaling in the ga2ox quintuple mutant and, furthermore, suggest that the loss of C19-GA2ox activity may be partially compensated for by a combination of feedback and feed-forward regulation of various steps in the GA biosynthesis pathway. By contrast, expression of the GA INSENSITIVE DWARF1b (GID1b) GA receptor, which has been reported to be reduced by GA (Griffiths et al., 2006), was not significantly reduced in the ga2ox quintuple mutant (Figure 4B).
Figure 4.
GA-Responsive Gene Expression in Wild-Type and ga2ox quintuple Plants.
(A) and (B) Expression of reported GA-responsive genes (A) and expression of GA20ox1, GA3ox1, GA2ox8, and GID1b (B) in the presence (+GA) and absence of GA. Three biological replicates of each sample type were tested, and reactions were done with two technical (quantitative PCR) replicates. Results are plotted as the ratio to the expression level of each gene in untreated wild-type plants ± se. Key for (B) is as in (A). *, Significantly different from the untreated wild type (P < 0.05); **, significantly different from the untreated wild type (P < 0.01). See Supplemental Table 2 online for mean Ct values and least significant differences (LSDs).
(C) Effect of GA treatment on GA2ox transcript levels. Seedlings were grown on plates containing the GA biosynthesis inhibitor paclobutrazol (1 μM) for 14 d and were sprayed with 100 μM GA3 or an ethanol control solution 3 h before harvesting. Three biological replicates of each sample type were tested, and reactions were done with two technical (quantitative PCR) replicates. Results are plotted as the ratio to the level at 0 h, ± se. **, Significantly different from the 0 h and 3 h mock controls (P < 0.01). See Supplemental Table 8 online for mean Ct values and LSDs.
Developmental Phenotypes of GA2ox Mutants
The ga2ox quintuple mutant was characterized for a number of traits from seed germination, through vegetative development and flowering, to fruit development. In some cases, it was also possible, through analysis of single and double mutants, to identify the one or two C19-GA2ox genes that played the major role in influencing these characters.
Seed Germination
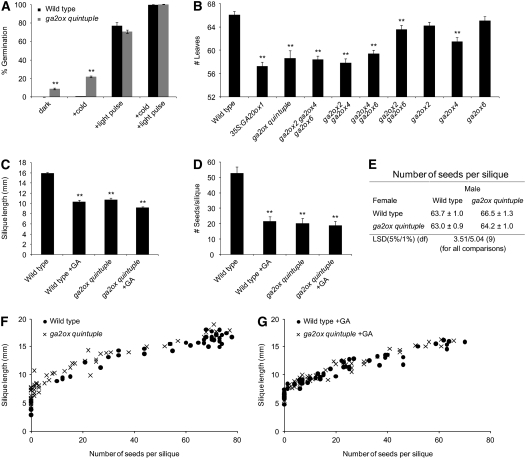
GA acts as a positive regulator of Arabidopsis seed germination, and severely GA-deficient mutant seeds do not germinate because the embryos are not able to emerge through the seed coat (Koornneef and van der Veen, 1980; Debeaujon and Koornneef, 2000; Penfield et al., 2006). GA biosynthesis de novo is a prerequisite for germination because inhibitors of GA biosynthesis applied during imbibition effectively inhibit the process (Nambara et al., 1991; Derkx and Karssen, 1993). Arabidopsis seeds have a very low germination rate in the dark, while both light and cold promote seed germination, at least partially by stimulating GA biosynthesis (Derkx and Karssen, 1994; Toyomasu et al., 1998; Yamaguchi et al., 1998; Yamauchi et al., 2004). Furthermore, light and cold each have been shown to reduce expression of GA2ox2 (Yamauchi et al., 2004; Oh et al., 2006). We tested whether the C19-GA 2-oxidases contribute to the low level of GA in dark-imbibed seed by comparing germination of wild-type and ga2ox quintuple seed incubated in the dark with or without a cold and/or light pretreatment. Compared with wild-type seed, a significantly higher proportion of the ga2ox quintuple seed germinated after cold pretreatment, and a small, but significant, portion of ga2ox quintuple seeds germinated without receiving any pretreatment (Figure 5A; see Supplemental Table 4 online). This increased germination rate in the dark was still present when seeds received a far-red light treatment (see Supplemental Table 5 online), indicating that the different germination rates were not due to a difference in phytochrome activity in the seed batches. However, germination of the ga2ox quintuple seed was completely prevented by application of the GA biosynthesis inhibitor paclobutrazol (PAC) during germination (see Supplemental Table 5 online), implying that the effect of loss of C19-GA2ox activity is on GAs synthesized de novo after imbibition, rather than on the level of bioactive GAs accumulated in the dry seed.
Figure 5.
Developmental Phenotype of GA2ox Mutants.
(A) Germination characteristics of wild-type and ga2ox quintuple seeds. Seeds were germinated in the dark for 4 d, without or with (+) a pretreatment of 4 d of imbibition at 4°C (cold) and/or a 1-min pulse of white light (light pulse). Values represent the average of six independent seed batches ± se. **, Significantly different from the wild type with the same treatment (P < 0.01). See Supplemental Table 9 online for mean values and LSDs.
(B) Number of leaves at the time of floral transition of the ga2ox quintuple and other mutant lines under SD conditions. Values represent the means ± se (n = 15 to 40). **, Significantly different from the wild type (P < 0.01). See Supplemental Table 3 online for mean values and LSDs.
(C) Silique length in the ga2ox quintuple mutant. Values represent the means ± se (n = 36-55). **, Significantly different from the wild type (P < 0.01). See Supplemental Table 10 online for mean values and LSDs.
(D) Seeds per silique in the ga2ox quintuple. Values represent the means ± se (n = 36-55). **, Significantly different from the wild type (P < 0.01). See Supplemental Table 10 online for mean values and LSDs.
(E) Number of seeds per silique after reciprocal hand-pollinations between a wild-type plant and the ga2ox quintuple mutant. Values represent the means ± se (n = 12). See Methods for details on statistical analysis.
(F) and (G) Relationship between silique length and number of seeds in a silique for the wild type and the ga2ox quintuple mutant without (F) and with (G) GA treatment. There is a positive correlation between seed number and silique length, as indicated by high Pearson's correlation coefficients (wild type, r = 0.921; ga2ox quintuple, r = 0.933; wild type + GA, r = 0.946; ga2ox quintuple + GA, r = 0.956).
Hypocotyl and Root Elongation
GA strongly stimulates hypocotyl elongation in the light (Cowling and Harberd, 1999). Compared with the wild type, ga2ox quintuple seedlings had significantly longer hypocotyls, although the increase was less than that caused by GA treatment (Table 2). GA treatment did not significantly increase root length, although roots of the ga2ox quintuple were fractionally longer than those of the wild type (Table 2).
Table 2.
Developmental Phenotype of the ga2ox quintuple Mutant
| Hypocotyl Length (mm) | Root Length (cm) | First Leaf with Trichomes | LD Flowering Time (d) | LD Flowering Time (Leaves) | |
|---|---|---|---|---|---|
| Wild type | 0.9 ± 0.0 (−2.3)a | 5.8 ± 0.1 | 5.1 ± 0.1 | 18.6 ± 0.1 | 15.0 ± 0.3 |
| Wild type + GA | 1.7 ± 0.0 (−1.8)** | 5.9 ± 0.1 | 3.0 ± 0.0** | 15.8 ± 0.1** | 12.7 ± 0.3** |
| ga2ox quintuple | 1.2 ± 0.0 (−2.2)** | 6.0 ± 0.1* | 4.0 ± 0.1** | 17.0 ± 0.2** | 14.2 ± 0.3 |
| ga2ox quintuple + GA | 1.8 ± 0.0 (−1.7)** | 5.9 ± 0.1 | 3.0 ± 0.0** | 15.5 ± 0.1** | 12.8 ± 0.3** |
| LSDwithin GA treatments (5%/1%) (df) | 0.06/0.10a (6) | 0.21/0.28 (67) | 0.45/0.69 (6) | 0.41/0.68 (4) | 0.93/1.55 (4) |
| LSDbetween GA treatments (5%/1%) (df) | 0.09/0.13a (8) | 0.37/0.52 (10) | 0.33/0.48 (8) | 0.28/0.44 (5) | 0.67/1.01 (7) |
| n | 32 | 20–25 | 32 | 18 | 18 |
See Methods for details on design and statistical analysis. The measurements are the means ± se. *, Significantly different from the wild type (P < 0.05); **, significantly different from the wild type (P < 0.01).
Log-transformed values (shown in parentheses) were used for statistical analysis (see Methods), and the LSDs correspond to these values.
Vegetative Phase Change
Abaxial trichomes are a marker for adult leaves and as such may be used as a measure of the vegetative phase change from juvenile to adult, which has been shown to be promoted by GA (Chien and Sussex, 1996; Telfer et al., 1997). In the ga2ox quintuple plants, trichomes were, on average, detected on leaf four, which is one leaf earlier than in wild-type plants (Table 2).
Floral Transition
Arabidopsis plants undergo a second phase change when they move from vegetative to reproductive development (i.e., leaf development is suppressed and lateral meristems give rise to floral rather than vegetative structures). This floral transition is promoted by GA in many species, including Arabidopsis, where flowering is advanced in both long and short days after GA applications (Langridge, 1957; Bagnall, 1992; Wilson et al., 1992). Under long-day conditions (LD), the ga2ox quintuple mutant flowered slightly earlier than the wild type, although this was only significant when measuring the number of days until floral transition, but not when measuring the number of leaves produced on the main stem before floral transition (Table 2). Under short-day (SD) conditions, the ga2ox quintuple mutant flowered significantly earlier than wild-type plants, resembling a 35S:GA20ox1 line, which has increased GA levels due to overproduction of GA 20-oxidase1 (Coles et al., 1999; Figure 5B; see Supplemental Table 6 online). Because GA2ox2, -4, and -6 are all expressed in the region of the shoot apex (Wang et al., 2004; Jasinski et al., 2005; Eriksson, 2006), we hypothesized that these three genes might be responsible for reducing GA levels at the apex and thus suppressing early flowering in wild-type plants in SD conditions. Indeed, the ga2ox2 ga2ox4 ga2ox6 triple mutant flowered as early as the ga2ox quintuple mutant (Figure 5B). Analysis of the various double and single mutant combinations of these three genes indicated that the early flowering phenotype could largely be attributed to loss of GA2ox4, with minor effects of GA2ox2 and GA2ox6. Analysis of all available single mutants further confirmed that GA2ox4 is the main GA2ox gene suppressing early flowering, both in the Col and Ws ecotypes (see Supplemental Table 6 online).
Stem Growth
GA is known to stimulate stem elongation in many plant species. In Arabidopsis, however, GA-promoted elongation of vegetative and inflorescence stem internodes appears to be close to saturation in the wild type, and GA treatment may actually result in a slight reduction in internode length (Rieu et al., 2008; Table 3). However, GA treatment causes an increase in the number of vegetative stem internodes that elongate upon bolting and in the number of flowers (and thus inflorescence internodes) that are formed, resulting in an overall increase in plant height (Rieu et al., 2008). Table 3 shows that loss of function of the five C19-GA2ox genes similarly resulted in taller plants, with an increase of 8% in final plant height. Loss of function of these genes did not significantly affect the number and length of internodes in the vegetative stem under these conditions but did result in an increased number of flowers on the main inflorescence, with slightly reduced internode length, thus giving a phenotype resembling that obtained by treating the wild type with GA. Together, these results indicate that under our standard growth conditions where GA levels are not limiting for vegetative internode elongation, C19-GA2ox genes do not significantly influence internode length, but do affect plant height by negatively regulating the number of flowers produced on the main inflorescence (see Discussion).
Table 3.
Stem Growth Phenotype of the ga2ox quintuple Mutant
| Total Plant Height (cm) | No. of Internodes (Vegetative Stem) | Length of Internodes (Vegetative Stem) (cm) | No. of Flowers/Internodes (Inflorescence) | Length of Internodes (Inflorescence) (cm) | |
|---|---|---|---|---|---|
| Wild type | 62.9 ± 0.7 | 3.6 ± 0.1 | 4.3 ± 0.1 | 59.9 ± 1.0 | 0.80 ± 0.01 |
| Wild type + GA | 67.2 ± 1.0** | 5.8 ± 0.1** | 3.7 ± 0.1** | 72.2 ± 1.2** | 0.63 ± 0.01** |
| ga2ox quintuple | 68.0 ± 0.4** | 4.1 ± 0.2 | 4.2 ± 0.1 | 69.0 ± 0.7** | 0.74 ± 0.01** |
| ga2ox quintuple + GA | 70.5 ± 1.1** | 6.3 ± 0.2** | 3.7 ± 0.1** | 74.6 ± 1.0** | 0.64 ± 0.01** |
| LSDwithin GA treatments (5%/1%) (df) | 1.47/2.44 (4) | 0.76/1.25 (4) | 0.23/0.37 (4) | 3.65/6.05 (4) | 0.022/0.036 (4) |
| LSDbetween GA treatments (5%/1%) (df) | 2.12/3.22 (5) | 0.60/0.88 (7) | 0.27/0.40 (6) | 2.92/4.25 (7) | 0.024/0.035 (7) |
| n | 18 | 18 | 18 | 18 | 18 |
See Methods for details on design and statistical analysis. The measurements are the means ± se. **, Significantly different from the wild type (P < 0.01).
Fruit Development
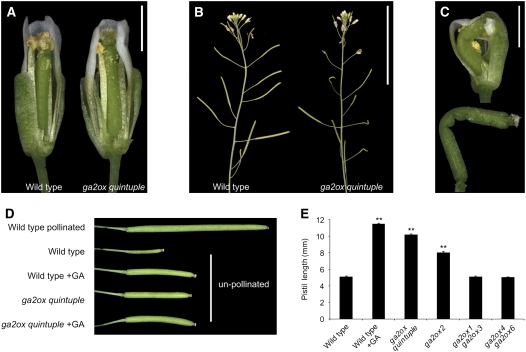
To study whether C19-GA 2-oxidation has a function in fertility and fruit development, we measured the lengths of randomly selected siliques of wild-type and ga2ox quintuple plants and counted the number of seeds that they contained. Both silique length and seed number were greatly reduced in the ga2ox quintuple mutant, to the same extent as in the GA-treated wild type (Figures 5C and 5D; see Supplemental Table 7 online). However, the relationship between seed number and silique length was very similar in the wild type, the ga2ox quintuple mutant, and the GA-treated wild type (Figures 5F and 5G), suggesting that the reduced lengths of the siliques was linked to reduced numbers of seeds per silique. Self and reciprocal hand-pollination of wild-type and ga2ox quintuple flowers showed that male and female fertility potential were unaffected in the mutant (Figure 5E), indicating that the reduction in numbers of seeds per silique was due to reduced efficiency of the transfer of pollen from stamen to pistil. Visual inspection of ga2ox quintuple flowers revealed that the mature anthers were never completely level with the stigma, as they are in wild-type flowers (Figure 6A), resulting in reduced efficiency of pollen transfer to the stigma in mutant flowers.
Figure 6.
Flower Phenotype of GA2ox Mutants.
(A) Flowers of the wild type and the ga2ox quintuple mutant showing unequal elongation of the pistil and stamens in the latter.
(B) Inflorescences of the wild type and the ga2ox quintuple mutant showing short distorted siliques in the latter.
(C) Close-up of bent pistils in ga2ox quintuple flowers.
(D) Silique from a fertilized wild-type plant and pistils from unpollinated wild-type and ga2ox quintuple mutant plants.
(E) Parthenocarpic growth of GA2ox mutants. Values represent the means ± se (n = 24). **, Significantly different from wild-type Col-0 (P < 0.01). See Supplemental Table 4 online for mean values and LSDs.
Bars = 1 mm in (A) and (C), 5 cm in (B), and 10 mm in (D).
We also noticed that a number of the ga2ox quintuple siliques were very short, bent, and seedless, implying that they were actually unfertilized pistils (Figure 6B). Analysis of flower buds at different stages of development showed that in some of the ga2ox quintuple flowers, pistils were deformed within the closed bud (Figure 6C). Interestingly, this phenotype was completely suppressed by GA treatment. Thus, the uncoordinated growth of pistil and sepals or petals was probably caused by unequal increases in GA levels in these organs or differential responsiveness to moderately increased GA levels.
As enhanced GA levels and signaling are known to cause fertilization-independent growth of fruits in Arabidopsis and other species (Schwabe and Mills, 1981; Vivian-Smith and Koltunow, 1999), we hypothesized that elongation of unfertilized pistils would be enhanced in the ga2ox quintuple mutant. To test this, flower buds at stage 12 (according to Smyth et al., 1990) were emasculated, and pistil length was measured after 7 d. Figure 6D shows that unfertilized ga2ox quintuple pistils reached about twice the length of unfertilized wild-type pistils and elongated to the same extent as those of the GA-treated wild type. The siliques derived from unfertilized ga2ox quintuple pistils shattered normally at maturity, as has been described for parthenocarpic siliques generated by GA application (Vivian-Smith and Koltunow, 1999). As GA2ox2 is highly expressed in pistils (Schmid et al., 2005), we tested the parthenocarpic potential of the single ga2ox2-1 and several other GA2ox mutants (Figure 6E). Unfertilized pistils of ga2ox2-1 elongated without fertilization (as did those of the ga2ox2-3 mutant; see Supplemental Table 8 online), but the ga2ox2-1 phenotype was not as pronounced as that of the ga2ox quintuple mutant. Double mutants of the remaining GA2ox genes (ga2ox1-1 ga2ox3-1 and ga2ox4-1 ga2ox6-2) did not show this phenotype, indicating that GA2ox2 is the major GA2ox gene acting in the pistil, together with minor, redundant contributions from one or more of the other GA2ox genes.
Developmental Phenotype of GA2ox Mutants under GA-Limited Conditions
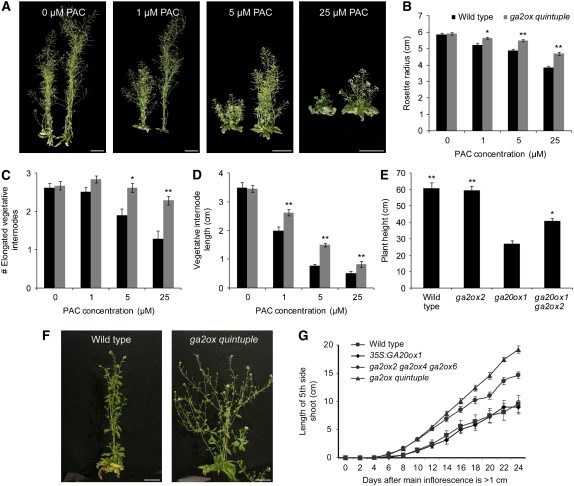
Several developmental processes, such as vegetative internode elongation and rosette leaf elongation, seem to be close to saturation for GA in wild-type Arabidopsis (Rieu et al., 2008); thus, the activity of GA2ox genes in these processes could not be analyzed. We hypothesized that the differences in phenotypes between the wild type and the GA2ox mutants would be enhanced under low GA conditions and compared wild-type and ga2ox quintuple plants grown in the presence of a limiting concentration of the GA biosynthesis inhibitor PAC. Indeed, the ga2ox quintuple knockout partially suppressed the PAC-induced phenotypes (Figure 7A), and several growth characteristics that were not different between the wild type and the ga2ox quintuple without PAC were different in the presence of the inhibitor: rosette radius was significantly larger in the ga2ox quintuple than in wild-type plants (Figure 7B; see Supplemental Table 9 online) and the vegetative stem was longer due to both more and longer vegetative internodes (Figures 7C and 7D). Moreover, the difference in characteristics such as flowering time, total plant height, and number of flowers on the main stem was larger in the presence of PAC than without PAC (see Supplemental Table 9 online).
Figure 7.
Phenotype of the GA2ox Mutants under Low GA Conditions.
(A) to (D) Growth characteristics of wild-type and ga2ox quintuple plants grown on different concentrations of PAC. Whole-plant phenotypes, with each pane showing a wild-type (left) and ga2ox quintuple plant (right) (A), rosette radius (B), number of elongated vegetative internodes (C), and length of elongated vegetative internodes (D). Values represent the means ± se (n = 18). Key for (C) and (D) is as in (B). *, Significantly different from the wild type with the same treatment (P < 0.05); **, significantly different from the wild type with the same treatment (P < 0.01). See Supplemental Table 5 online for mean values and LSDs.
(E) Partial suppression of the ga20ox1-3 stem height phenotype by ga2ox2-1. Values represent the means ± se (n = 3-7). *, Significantly different from the ga20ox1-3 mutant (P < 0.05); **, significantly different from the ga20ox1-3 mutant (P < 0.01). See Supplemental Table 6 online for mean values and LSDs.
(F) and (G) Growth of the side shoots of wild-type and ga2ox quintuple mutant plants in SD conditions. Whole-plant phenotype of 79-d-old plants (F) and graph of fifth side shoot growth over time (G). Values represent the means ± se (n = 15).
Bars = 5 cm in (A) and (F).
As GA2ox2 was the most highly expressed C19-GA2ox gene in the vegetative stem and inflorescence (Figure 1), we tested whether this gene negatively affected their elongation. Because total plant height was not increased in the ga2ox2-1 single mutant (see Supplemental Table 10 online), we used the same rationale as above and reduced the GA level in ga2ox2-1 by introducing it into the partially GA-deficient ga20ox1-3 background (Rieu et al., 2008). Figure 7E shows that ga2ox2-1 partially suppressed the semidwarf phenotype of the ga20ox1-3 line. The double mutant was significantly taller than ga20ox1-3 due to an increase in length of the inflorescence stem (see Supplemental Table 10 online).
When Arabidopsis plants are grown under SD conditions, several GA-regulated processes, such as the floral transition, seem to become more dependent on GA. When the ga2ox quintuple mutant was grown under SD conditions, we noticed a clear difference in plant architecture, compared with the wild type, in that axillary branches were much longer in the mutant (Figure 7F). As a measure of branch growth, we analyzed elongation of the fifth axillary branch. To normalize the growth in relation to the bolting time, we took the point at which the main inflorescence reached 1 cm as time zero. The fifth side shoot of the ga2ox2 ga2ox4 ga2ox6 triple mutant elongated more rapidly than that of the wild type (Figure 7G) and reached a greater final length. This phenotype was further enhanced in the ga2ox quintuple mutant. Under LD conditions, this phenotype was not as apparent; similarly, the 35S:GA20ox1 line did not show increased outgrowth of the side shoots (Figure 7G), implying that the GA2ox genes tightly regulate this process.
Together, these results indicate a role for the GA2ox genes as negative regulators of rosette leaf, vegetative stem and side shoot growth, and, more specifically, for GA2ox2 as one of the GA2ox genes that negatively regulates final inflorescence length.
DISCUSSION
C19-GA 2-Oxidase Activity in Arabidopsis
We and others have shown that Arabidopsis contains five functional genes encoding GA 2-oxidases (GA2ox1, -2, -3, -4, and -6) that are active against C19-GAs, including GA4 and GA1, the main bioactive GAs in Arabidopsis (Thomas et al., 1999; Hedden and Phillips, 2000; Wang et al., 2004; Jasinski et al., 2005; Table 1). Analysis of GA content in plants showed that the levels of the 2β-hydroxylated GAs, GA51, GA34, GA29, and GA8, were much reduced in the ga2ox quintuple mutant, in the case of the 13-hydroxylated GAs to undetectable levels (Figure 3), and feeding experiments with radiolabeled GA demonstrated that floral apices of a ga2ox quintuple mutant were no longer capable of inactivating GA4, by conversion to GA34, or GA9, by conversion to GA51 (see Supplemental Figure 2 online). Thus, it seems likely that no additional C19-GA2ox genes exist in Arabidopsis and that the low amount of GA51 and traces of GA34 still present in the ga2ox quintuple mutant (Figure 3) were generated via 20-oxidation and 3-oxidation of 2β-hydroxylated C20-GAs, such as those produced by GA2ox7 and GA2ox8, as the existence of an early 2-oxidation pathway has been shown previously (Lee and Zeevaart, 2005). The concentration of GA9 was also reduced by ∼50% in the GA2ox quintuple, possibly due to decreased GA 20-oxidase activity as a result of feedback by the elevated levels of bioactive GAs.
Analysis of the GA content of the quintuple mutant also showed increases of approximately twofold in the concentrations of the bioactive GAs, GA4 and GA1, compared with wild-type plants. Although this increase was enough to induce hyperactivation of a number of GA responses, many of these, such as hypocotyl elongation, the number of elongated vegetative internodes (Table 2), and GA-responsive gene expression (Figure 4A), were still not saturated for GA in the mutant and responded further to application of the hormone. The relatively mild increases in the levels of bioactive GAs suggest that other mechanisms limit their accumulation in the ga2ox quintuple mutants in the absence of C19-GA 2-oxidation. Analysis of the transcript levels of GA20ox1 and GA3ox1 in the ga2ox quintuple mutant showed significant downregulation of these genes (Figure 4B), suggesting that feedback regulation of biosynthesis of the bioactive GAs may be one such mechanism. Furthermore, expression of GA2ox8, which encodes a 2-oxidase active against C20-GAs earlier in the biosynthetic pathway, was increased in the ga2ox quintuple, presumably as a result of feed-forward regulation by GA, which might reduce flux through the main part of the pathway by removing C20-GA intermediates.
C19-GA 2-Oxidation Regulates GA-Responsive Growth and Development throughout the Life Cycle
Transcript analysis of the C19-GA2ox gene family (Figure 1) revealed that each gene has a quantitatively different developmental expression profile, but the degree of overlap between expression patterns was such that it was not possible to link specific GA-regulated developmental processes with individual genes. With this degree of redundancy, therefore, it was not surprising that loss-of-function mutants of individual genes did not confer any phenotypes that were immediately apparent. However, detailed quantitative analysis of the ga2ox quintuple mutant enabled identification of a range of phenotypes caused by loss of GA inactivation.
It has been shown recently that GA2ox2 contributes to the suppression of germination in Arabidopsis seeds imbibed in the dark (Yamauchi et al., 2007). Our studies of the ga2ox quintuple mutant provide support for this (Figure 5A). Furthermore, our observation that the effect of loss of GA 2-oxidase activity is on GAs synthesized de novo after imbibition, rather than on the level of bioactive GAs accumulated in the dry seed (see Supplemental Table 5 online), agrees with the finding that similar amounts of GA4 are present in dry seed of the wild type and of the ga2ox2 mutant, whereas seeds of this mutant contain higher levels of GA4 after 48 h of imbibition in the dark (Yamauchi et al., 2007). These results contrast with observations that GA 2β-hydroxylase activity is abundant in seeds of many plants during the later stages of maturation, particularly in legume seeds, which accumulate large amounts of 2β-hydroxylated GAs (Durley et al., 1971; Frydman et al., 1974; Albone et al., 1984). Arabidopsis, which also has high levels of expression of GA biosynthetic genes in the developing seed (Kim et al., 2005; Schmid et al., 2005), must therefore have alternative mechanisms of GA inactivation, such as methylation (Varbanova et al., 2007), in these tissues.
The involvement of GA in the flowering transition is more sharply defined in SDs than in LDs: GAs are absolutely required for flowering in standard (low far-red) SD conditions (Wilson et al., 1992), while they slightly advance flowering of the wild type in LD (Coles et al., 1999). Accordingly, the effects of loss of C19-GA 2-oxidation in the ga2ox quintuple mutant are more pronounced in SD than in LD (cf. Table 2 and Figure 5B). Discrimination of the roles of the different GA2ox genes in suppression of flowering in SD revealed that GA2ox4 plays the major role, with minor contributions from GA2ox2 and GA2ox6. Jasinski et al. (2005) and Eriksson (2006) showed that in both LD and SD shoot expression of GA2ox4 was restricted to the apex with a ring of expression around the meristem, at the base of the young leaf primordia. A similar expression pattern was reported for a putative ortholog of GA2ox4 in rice, GA2ox1. Because in rice GA2ox1 expression below the vegetative meristem was substantially reduced after floral induction, the authors speculated that GA 2-oxidases have a regulatory function in the floral transition (Sakamoto et al., 2001), a hypothesis further elaborated on by King and Evans (2003) to account for the effects of various applied GAs on the floral transition in Lolium temulentum. However, Eriksson et al. (2006) found that GA2ox4 transcript abundance rose as the levels of GA4 and LEAFY mRNA increased in the shoot apex during the floral transition in SDs, consistent with a feed-forward activation of GA2ox4 by the increasing GA levels, suggesting that, in Arabidopsis, C19-GA 2-oxidase activity around the meristem does not play an active role in the floral transition, but acts constantly to prevent low amounts of GAs, possibly originating from the young leaf primordia, from entering the meristem region.
We recently revealed a novel role for GA in Arabidopsis in determining the number of vegetative internodes that elongate to produce the stem and the number of flowers produced (Rieu et al., 2008), these being the major components that contribute to GA-promoted plant height in this species. Under our standard growth conditions, loss of the C19-GA 2-oxidases had no effect on the number of elongated vegetative internodes, but increased overall plant height by increasing the number of flowers produced by the main inflorescence (Table 3). Whether the increase in flower number observed is due to a change in the rate of flower production (plastochron length) or a change in the duration of the flower-producing phase is not immediately apparent. One possibility is that the reduced number of seeds per silique and higher proportion of infertile flowers in the ga2ox quintuple mutant (see below) may have the effect of delaying plant senescence and thus increasing flower number: in Arabidopsis, activity of the inflorescence meristem is suppressed by the presence of developing seeds (Hensel et al., 1994).
Coordinated elongation of the pistil and stamen filament is essential to ensure efficient pollination within the flower. We earlier reported that plants carrying mutations in the biosynthetic genes GA20ox1 and GA20ox2 showed reduced filament elongation, resulting in partial infertility (Rieu et al., 2008). A loss of fertility has also been described for wild-type plants treated with GA (Jacobsen and Olszewski, 1993) and for double mutants in the GA signaling repressors, RGA and GAI (Dill and Sun, 2001). Here, we show that loss of C19-GA 2-oxidation in the ga2ox quintuple mutant similarly results in a partial loss of fertility, which appears to be due to increased length of the pistil relative to the stamen filament and the consequent reduction in the amount of pollen reaching the stigma. Experimental analysis of the cause of this phenomenon in the ga2ox quintuple mutant was not possible because staging of flowers, required for the comparisons, is itself based on the relative length of the floral organs and because a significant proportion of the ga2ox quintuple pistils is malformed (Figures 6B and 6C). However, the ga2ox quintuple shows parthenocarpic pistil elongation (Figure 6D), largely due to the loss of GA2ox2 (Figure 6E), which appears to start before flower opening, as in many cases the pistil impacts against the petals and sepals causing it to bend (Figure 6C). It has recently been shown that the pistils also overelongate prematurely in tomato (Solanum lycopersicum) plants with constitutive GA signaling (Marti et al., 2007).
The relatively minor effects of GA application on many aspects of Arabidopsis development, and similar mild effects of overexpression of GA biosynthetic genes (Coles et al., 1999), suggests that many GA-dependent developmental processes are close to saturation at the hormone levels found in wild-type plants. Therefore, to reveal cryptic phenotypes of GA2ox mutants, we examined growth of the ga2ox quintuple under conditions where GA levels were reduced by application of an inhibitor of GA biosynthesis. Under these conditions, the ga2ox quintuple mutant displayed a more exaggerated phenotype compared with wild-type plants, and some novel phenotypes, such as increased rosette size and increased number and length of vegetative internodes, were observed (Figures 7A to 7D). Similarly, ga2ox quintuple plants grown in SD conditions, which increase the GA dependency of some developmental processes, such as flowering time, also showed new phenotypes, such as increased outgrowth of axillary shoots (Figures 7F and 7G). This suggests that under environmental conditions where GA levels are more limiting, C19-GA 2-oxidases would have a more prominent role in regulating plant development.
In conclusion, a significant number of growth and developmental processes in plants are regulated by the concentrations of bioactive GAs that are the net result of GA biosynthesis, transport, and inactivation within the specific tissues. The main regulatory enzymes identified by previous work have been those involved in the later biosynthetic steps, from GA12 to GA4, which are catalyzed by the 2-oxoglutarate dependent dioxygenases, GA 20-oxidase and GA 3-oxidase (Hedden and Phillips, 2000; Mitchum et al., 2006; Rieu et al., 2008). Both of these enzymes have been shown by overexpression in transgenic plants to be limiting for GA content (Coles et al., 1999; Radi et al., 2006), though which enzyme plays the major regulatory role depends on the species and the tissue. In Arabidopsis, loss of the C20-GA2ox genes GA2ox7 and GA2ox8 yields a limited set of phenotypes related to GA accumulation, including increased hypocotyl elongation similar to that seen in the ga2ox quintuple, indicating that these enzymes are also involved in determining bioactive GA levels. In addition, GA methylation (Varbanova et al., 2007) and epoxidation of the C-16,17 double bond (Zhu et al., 2006) may serve to maintain GA homeostasis, although the two GA-methyl transferases thus far identified in Arabidopsis, GAMT1 and GAMT2, appear to be active only in developing seeds (Varbanova et al., 2007) and 16,17-epoxidation has only been demonstrated in rice (Zhu et al., 2006). It is therefore not known to what extent the operation of these additional GA inactivation mechanisms limit GA accumulation. In this study, we have shown that C19-GA 2-oxidase activity limits GA-responsive growth and development throughout Arabidopsis development and thus constitutes a major GA inactivation pathway. The responsiveness of the C19-GA2ox genes to environmental conditions, including various abiotic stresses such as cold (Achard et al., 2008), drought, salt, and wounding (see Supplemental Figure 3 online), indicates that they may have further roles in the integration of extrinsic signals into the developmental program.
METHODS
Plant Material and Growth Conditions
The wild-type backgrounds used in this study were Col-0, Ws-0, or Ws-4, as specified. The genotype ga20ox1-3 is in the Col-0 background (Rieu et al., 2008); ga2ox1-1 (WiscDsLox_333C08), ga2ox2-1 (SALK_051749), ga2ox3-1 (SALK_042818), ga2ox4-1 (SALK_036923), ga2ox4-2 (SALK_142382), ga2ox4-3 (GABI_171B04), and ga2ox6-2 (SM_3_1859) are in the Col-0 background; ga2ox2-3 (Wisc_2ox2), ga2ox4-4 (Wisc_2ox4), and ga2ox6-3 (Wisc_2ox6) are in the Ws-0 background; and ga2ox3-2 (FLAG_088C02) is in the Ws-4 background. The genotypes ga2ox2-3, ga2ox4-4, and ga2ox6-3 were isolated from the Wisconsin Knockout Facility Alpha T-DNA insertion mutant populations using a PCR-based method. For all lines, homozygous insertion mutants were identified by PCR using allele-specific primers (see Supplemental Table 11 online). The location of the insertions was determined by sequencing the PCR products. Absence of correct transcripts in each of the mutants was confirmed by RT-PCR on total RNA from 7-d-old plants. For ga2ox3-1 and ga2ox6-2, transcripts were also analyzed in the ga2ox quintuple mutant using real-time RT-PCR on total RNA from flowering shoots, as described below for real-time RT-PCR methods. Primers are listed in Supplemental Table 11 online.
Unless stated otherwise, plants were grown on Levington F2 soil in trays, comprising 24 individual 5-cm square pots, under LD conditions (16 h light at 300 μmol m−2 s−1 from Osram HQI-BT 400W/D metal halide lamps, 23°C, 65% relative humidity/8 h darkness, 18°C, 70% relative humidity), watered regularly, and treated with a standard plant nutrient solution once per week. Soil-grown plants treated with GA were sprayed with 100 μM GA3 (Duchefa) three times per week.
Analysis of in Vitro GA 2-Oxidase Activity
The full-length coding sequence of GA2ox6 was amplified by PCR and cloned into pET32a to be expressed as a fusion with thioredoxin in Escherichia coli strain BL21(DE3). Cell lysate was incubated with 14C-labeled GAs and dioxygenase cofactors, and the products were separated by HPLC with online radiomonitoring and identified by GC-MS as described (MacMillan et al., 1997).
Generation of the ga2ox Multiple Mutants
To generate multiple homozygous mutants containing null alleles of up to five GA2ox genes, homozygous lines were crossed, the F1s selfed and homozygous lines identified in the resulting segregating F2 population by PCR-based genotyping using allele-specific primers (see Supplemental Table 11 online). Using this strategy, ga2ox1-1 and ga2ox3-1 were crossed to generate ga2ox1 ga2ox3. ga2ox2-1, ga2ox4-1, and ga2ox6-2 were crossed pairwise to generate the ga2ox2 ga2ox4, the ga2ox2 ga2ox6, and the ga2ox4 ga2ox6 double mutants. ga2ox2 ga2ox4 and ga2ox2 ga2ox6 were crossed to generate the ga2ox2 ga2ox4 ga2ox6 triple mutant. To generate the ga2ox quintuple mutant, ga2ox1-1 was crossed to ga2ox2-1 and ga2ox2-1 was crossed to ga2ox3-1. The resulting ga2ox1 ga2ox2 and ga2ox2 ga2ox3 double mutants were crossed to render ga2ox1 ga2ox2 ga2ox3. This triple mutant was crossed to ga2ox2 ga2ox4 ga2ox6, producing the ga2ox1 ga2ox2 ga2ox3 ga2ox4 ga2ox6 quintuple mutant. During the experiments, we never observed any phenotypes in the mutants that could not be related to GA2ox gene function.
Analysis of Endogenous GA levels
Wild-type and ga2ox quintuple plants were grown in separate trays, which were arranged randomly. Whole shoots were harvested from plants when the first open flower was visible. Here, 23 plants from one tray represented one replicate sample. Four replicate samples (0.5 g dry weight each) of the two genotypes were analyzed by GC-MS as described by Griffiths et al. (2006) with modifications according to Rieu et al. (2008).
Analysis of GA 2-Oxidase Activity in Planta
Wild-type and ga2ox quintuple plants were grown in separate trays, which were arranged randomly. When buds were visible with the naked eye in 50% of the plants, 2000 disintegrations per minute of [17-14C]GA4 (26 pmol) or [17-14C]GA9 (16.5 pmol) was applied to the shoot apex in 2 μL of 10% methanol. Whole shoots were harvested after 24 h and rinsed with water. All 69 shoots from three trays of each genotype were combined as one sample (∼10 g), which was homogenized in 80% methanol water (∼130 mL) using a Polytron PT3100 homogenizer. After stirring overnight at 4°C, the sample was filtered, and the residue re-extracted in methanol (50 mL) for 1 h and filtered, after which the combined filtrates were reduced to dryness in vacuo. A weak acids fraction was then prepared by solvent partitioning, anion exchange (QAE-Sephadex A-25), and C18 SPE as described by Croker et al. (1990), except that the PVP step was omitted. The solvent partition steps were omitted for the experiment with [14C]GA9. The purified samples were analyzed by reverse-phase HPLC with online radiomonitoring as described by MacMillan et al. (1997). The radiolabeled products were identified by GC-MS using a MAT95XP (Rieu et al., 2008) operated in scanning mode, from 650 to 50 atomic mass units at 0.5 s per mass decade. The presence of [12C]GA51 and [14C]GA51 in the relevant HPLC fraction from application of [14C]GA9 to Col-0 was determined by GC-selected ion monitoring of four pairs of ions.
Real-Time Quantitative RT-PCR
Tissue samples for the developmental series have been described previously (Griffiths et al., 2006). To analyze GA responsiveness of the GA2ox genes in the wild type, Col-0 plants were grown in square Petri dishes with 36 plants/dish on 1× Murashige and Skoog salts containing Gamborg B5 vitamins, 1% sucrose, 0.5 g/L MES, pH 5.8, and 0.7% Gelrite for 14 d. Three of the six Petri dishes were then treated by spraying with 100 μM GA3 and three with a mock solution. Petri dishes with different treatments were arranged randomly. Shoots were harvested after 3 h. All plants from one Petri dish represented one replicate. To compare transcript levels between the wild type and the ga2ox quintuple mutant, plants of both genotypes were grown in trays together in a random arrangement with 12 plants/tray. Three of the six trays were treated with GA3 as above. Trays with different treatments were arranged randomly, and whole shoots were harvested when the first flower of the primary inflorescence had opened; six plants of a genotype from one tray represented one replicate sample. Standard curves for the GA2ox genes were obtained from dilution series of known amounts of GA2ox DNA fragments. DNA fragments encompassing at least the full coding sequence of each transcript were first amplified using PCR and cloned into pGEM-T Easy (Promega) using the primers listed in Supplemental Table 11 online. The fragments were then reamplified from these clones using the same primers, purified (QIAquick; Qiagen), quantified (Nanodrop spectrophotometer), and used directly as template for quantitative PCR.
Total RNA extraction, DNase treatment, reverse transcription, and real-time quantitative RT-PCR using SYBR Green have been described previously (Griffiths et al., 2006). In short, total RNA was extracted with on-column DNase treatment (RNeasy; Qiagen), treated again with DNase in solution (Turbo DNA-free kit; Ambion), and used for reverse transcription (SuperScript III Platinum Two-Step qRT-PCR kit with SYBR Green; Invitrogen). The cDNA equivalent of 20 to 40 ng of total RNA was used in a 25-μL PCR reaction on an ABI 7500 real-time PCR system (Applied Biosystems) with Platinum SYBR Green qPCR SuperMix-UDG reagents (Invitrogen). In all experiments, three biological replicates of each sample type were tested and reactions were done with two technical (PCR) replicates. Absence of genomic DNA and primer dimers was confirmed by analysis of RT-minus and water control samples and by examination of dissociation curves. Reactions were assembled using a Beckman Biomek-NX Span-8 liquid handling robot.
To normalize the qPCR data, three reference genes (from Czechowski et al., 2005) were used in each experiment and stability of the reference genes across samples was tested using geNorm software (Vandesompele et al., 2002). In all cases, the sets of reference genes were highly stable (variability V2/3 below the recommended cutoff value of 0.150). Reference genes for the developmental series were as follows: At1g13320, At2g28390, and At4g34270 (V2/3 = 0.118); for the GA response series and for the ga2ox quintuple experiment, At1g13320, At4g26410, and At4g33380 (V2/3 = 0.067 and 0.038, respectively). To calculate the absolute ratios in the developmental series, the number of GA2ox cDNA template molecules per sample was normalized against the corresponding geNorm normalization factor and results from the three biological replicates were averaged. Primers for the following genes have been described before: At1g13320, At2g28390, At4g26410, At4g33380, and At4g34270 (Czechowski et al., 2005), GA20ox1 (Rieu et al., 2008), and GA3ox1 (Griffiths et al., 2006). All other quantitative PCR primers were designed using Primer Express v.2.0 (Applied Biosystems) or GENOPLANTE SPADS software (http://urgi.versailles.inra.fr/tools/spads/) and are listed in Supplemental Table 11 online. Primer pair efficiencies were estimated by analysis of the amplification curves with LinReg software (Ramakers et al., 2003), and the average efficiency of all reactions on a plate (which was always >1.95) was used in calculations (Cook et al., 2004).
Analyses of Mutant Phenotypes
For the germination assay, 100 to 200 seeds were spread on three layers of wet 3MM Whatman filter paper in the dark, and Petri dishes were wrapped in two layers of aluminum foil to prevent exposure to the light. Seed germination was scored by counting all seeds with radical protrusion after 4 d of incubation at 22°C. For cold pretreatment, seeds were incubated at 4°C for 4 d, and for light pretreatment, seeds received 1 min of white light from fluorescent tubes (Phillips Master TL5 54W/840 HO; 200 μmol m−2 s−1) 6 h after the start of incubation at 22°C. There were three replicate Petri dishes of each treatment, arranged randomly.
Wild-type and ga2ox quintuple seedlings used for hypocotyl measurements were grown together and randomly arranged on 1× Murashige and Skoog salts containing Gamborg B5 vitamins, 1% sucrose, 0.5 g/L MES, pH 5.8, and 0.7% Gelrite in vertically standing Petri dishes with 16 seedlings/dish under continuous light at 22°C. Four of the eight dishes contained 100 μM GA3 in the medium. Dishes of different treatments were arranged randomly. After 7 d, they were scanned and measurements were taken using NIH ImageJ software (http://rsb.info.nih.gov/ij/).
Wild-type and ga2ox quintuple plants used to count abaxial leaf trichomes were grown in trays together in a random arrangement with 24 plants/tray. Four of the eight trays were treated with 100 μM GA3. Trays with different treatments were arranged randomly. The presence of trichomes on the abaxial side of leaves of 23-d-old plants was determined using a binocular dissecting microscope.
Wild-type and ga2ox quintuple plants used to determine LD flowering time, internode numbers and lengths, silique lengths, and seeds/silique were grown in trays together in a random arrangement with 12 plants/tray. Three of the six trays were treated with 100 μM GA3. Trays with different treatments were arranged randomly. Flowering time was scored as the total number of leaves produced by the main shoot and also as the number of days until buds could be detected with the naked eye. Internode measurements were done after the plants had stopped flowering. For silique characterization, the length of and number of seeds in randomly selected siliques chosen from siliques number 10 to 20 from the base of the main stem was determined.
Wild-type and ga2ox quintuple plants used to determine LD flowering time, rosette leaf radius, and internode numbers and lengths after paclobutrazol (PAC) treatment were grown in trays together in a random arrangement with 12 plants/tray. Three trays were treated with 0, 1, 5, and 25 μM PAC by soaking the soil (1 dm3) at 5-d intervals, with the initial treatment occurring 7 d after sowing. Rosette leaf radius was determined by taking the average of the two largest leaves when fully elongated. Flowering time and internode measurements were performed as described above.
Plants used to determine the effect of the ga2ox1-1 mutation on stem growth of ga20ox1-1 were segregants identified in the F2 of a cross between homozygous parents. The three wild-type, seven ga2ox2-1, three ga20ox1-1, and three ga20ox1-1 ga2ox2-1 plants were arranged randomly in one tray. Internode measurements were performed after the plants had stopped flowering.
Plants used to determine SD flowering time were grown on a 3:1 mixture of soil and vermiculite in trays comprising 15 individual 5-cm square pots under SD conditions (9 h light at 130 μmol m−2 s−1 from fluorescent lamps, 23°C/15 h darkness, 18°C). From 4 weeks onward, plants were treated with nutrient solution every second week. Genotypes were grown together in trays, with 15 plants of each genotype randomly distributed over a total of 16 trays across three separate experiments. Leaf formation was followed once per week by marking new leaves with a felt pen.
Wild-type and ga2ox quintuple plants used to test the effect of reciprocal hand pollinations were grown in trays together in a random arrangement with 12 plants/tray in four trays. The two pollination types were distributed randomly over each tray.
Plants of eight genotypes used to determine parthenocarpic potential were grown in trays together in a random arrangement with 12 plants/tray in eight trays. Two flowers of stage 11/12 were emasculated from each plant, and pistil lengths were measured with a vernier caliper after 8 d. For GA treatment, wild-type pistils were treated with 1 μL of 100 μM GA3 immediately after emasculation and 1 d later.
Plants used to measure side shoot growth in SD were grown in the same conditions as stated for the SD flowering time determination (see above). The time until the main inflorescence reached 1 cm was scored. The growth of the fifth side shoot, defined as that emerging from the fifth cauline leaf axil counting downwards from the inflorescence, was followed from this day on and for 26 d, taking measurements every second day.
Statistical Analysis
A completely randomized design was used for Petri dishes from which germination measurements were taken. The design for the characterization experiment was a randomized block (providing measurements of SD flowering, side shoot growth, the effect of ga2ox2 on ga20ox1-1, the reciprocal hand pollinations, and parthenocarpic potential) or a split plot (see Gomez and Gomez, 1984) in three blocks (for all other experiments). In the case of the PAC characterization experiment, there were four trays per block, each tray being allocated a concentration of PAC (0, 1, 5, or 25 μM); for the remaining characterization experiments, there were pairs of trays making up each block, with one tray treated with GA and the other kept as control. The trays in both designs were split to contain an equal number of plants of each type, randomized to the cells in the tray.
For statistical analysis of quantitative PCR data, reference-gene-corrected Ct values were used. These were obtained by converting the target gene Ct values to relative quantities, which were corrected with the calculated geometric mean of reference gene relative quantities as normalization factors and then back transformed to Ct values.
Where experiments were balanced for comparison, analysis of variance (ANOVA) was used and otherwise the restricted maximum likelihood (REML) method was applied. To correct for heterogeneity of variance, a logit transformation, namely loge((GE%+0.1)/(100.1-GE%)), where GE% is the percentage germination and an adjustment of 0.1 is used to account for observations of 0 and 100%, was applied to the germination data prior to ANOVA, a loge transformation was applied to hypocotyl length and flowering time (days) on PAC, and a square root transformation was applied to vegetative internode lengths on PAC. Note that, due to a common transformation being applied to all observations, the comparative values between genotypes are not altered and comparisons between them remain valid. A t test was used to assess the significance of differences in germination between far-red light–treated wild-type and ga2ox quintuple seeds. Similarly, t tests were used to test the significance of differences between wild-type and ga2ox quintuple GA levels.
For flowering time under SD conditions, data for the genotypes were derived from three separate experiments under the same conditions. Sources of variation, comprising the design structure of experiments and trays within experiments, were accounted for when using REML.
Following ANOVA or REML analysis, LSDs at 5 and 1% were used to assess significance between particular pairs of genotypes or treatment combinations, on the transformed scale where appropriate. The GenStat statistical system (version 8.2; Lawes Agricultural Trust) was used for all analyses.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At1g78440 (GA2ox1), At1g30040 (GA2ox2), At2g34555 (GA2ox3), At1g47990 (GA2ox4), At1g02400 (GA2ox6), At1g50960 (GA2ox7), At4g21200 (GA2ox8), At4g25420 (GA20ox1), At1g15550 (GA3ox1), At3g63010 (GID1b), and At1g50420 (SCL3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RT-PCR to Confirm Null Status of Insertion Alleles Used in This Study.
Supplemental Figure 2. Metabolism of [17-14C]GA9 and [17-14C]GA4 by the Wild-Type Col-0 and the ga2ox quintuple Mutant.
Supplemental Figure 3. Expression of C19-GA2ox Genes in Response to Abiotic Stress.
Supplemental Table 1. Characterization of Insertion Lines.
Supplemental Table 2. Mean Ct Values and LSDs of Gene Expression Studies Shown in Figures 4A and 4B (GA-Responsive Genes) and of GA2ox3 and GA2ox6.
Supplemental Table 3. Mean Ct Values and LSDs of Gene Expression Studies Shown in Figure 4C.
Supplemental Table 4. Germination Characteristics of Wild-Type and ga2ox quintuple Seeds as Shown in Figure 5A.
Supplemental Table 5. Germination Characteristics of Wild-Type and ga2ox quintuple Seeds after FR and PAC Treatment.
Supplemental Table 6. Flowering Time under Short-Day Conditions as Shown in Figure 5B, Including Single Mutants.
Supplemental Table 7. Silique Phenotypes as Shown in Figures 5C and 5D.
Supplemental Table 8. Parthenocarpic Growth of GA2ox Mutants as Shown in Figure 6E.
Supplemental Table 9. Phenotype of the GA2ox Mutants under Low GA Conditions, Including as Shown in Figures 7B to 7D.
Supplemental Table 10. Suppression of the ga20ox1-3 Phenotype by ga2ox2-1 as Shown in Figure 7E.
Supplemental Table 11. Primers Used for Genotyping, Real-Time Quantitative RT-PCR, and Preparation of Standard Curves.
Supplementary Material
Acknowledgments
We thank Dennis Ward for help with HPLC, Ian Pearman, Anthony Griffiths, and other greenhouse staff for excellent plant material, and Graham Shephard and the Visual Communications Unit for all photographs. We thank G.C. Whitelam (University of Leicester, UK) for help with far-red treatment of seeds. [17-14C]GA4 and [17-14C]GA9 were supplied by L.N. Mander (Australian National University, Canberra). This work was supported by Grant P19317 from the Biotechnology and Biological Sciences Research Council of the UK.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Andrew L. Phillips (andy.phillips@bbsrc.ac.uk).
Online version contains Web-only data.
References
- Achard, P., Gong, F., Cheminant, S., Alioua, M., Hedden, P., and Genschik, P. (2008). The cold-inducible CBF1 factor–dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20 2117–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albone, K.S., Gaskin, P., MacMillan, J., and Sponsel, V.M. (1984). Identification and localization of gibberellins in maturing seeds of the cucurbit Sechium edule, and a comparison between this cucurbit and the legume Phaseolus coccineus. Planta 162 560–565. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide Insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Appleford, N.E.J., Wilkinson, M.D., Ma, Q., Evans, D.J., Stone, M.C., Pearce, S.P., Powers, S.J., Thomas, S.G., Jones, H.D., Phillips, A.L., Hedden, P., and Lenton, J.R. (2007). Decreased shoot stature and grain {alpha}-amylase activity following ectopic expression of a gibberellin 2-oxidase gene in transgenic wheat. J. Exp. Bot. 58 3213–3226. [DOI] [PubMed] [Google Scholar]
- Bagnall, D.J. (1992). Control of flowering in Arabidopsis thaliana by light, vernalization and gibberellins. Aust. J. Plant Physiol. 19 401–409. [Google Scholar]
- Busov, V.B., Meilan, R., Pearce, D.W., Ma, C.P., Rood, S.B., and Strauss, S.H. (2003). Activation tagging of a dominant gibberellin catabolism gene (GA 2-oxidase) from poplar that regulates tree stature. Plant Physiol. 132 1283–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, H.H., Hwang, I., and Goodman, H.M. (1995). Isolation of the Arabidopsis GA4 locus. Plant Cell 7 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, J.C., and Sussex, I.M. (1996). Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 111 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles, J.P., Phillips, A.L., Croker, S.J., Garcia-Lepe, R., Lewis, M.J., and Hedden, P. (1999). Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J. 17 547–556. [DOI] [PubMed] [Google Scholar]
- Cook, P., Fu, C.X., Hickey, M., Han, E.S., and Miller, K.S. (2004). SAS programs for real-time RT PCR having multiple independent samples. Biotechniques 37 990–995. [DOI] [PubMed] [Google Scholar]
- Cowling, R.J., and Harberd, N.P. (1999). Gibberellins control Arabidopsis hypocotyl growth via regulation of cellular elongation. J. Exp. Bot. 50 1351–1357. [Google Scholar]
- Croker, S.J., Hedden, P., Lenton, J.R., and Stoddart, J.L. (1990). Comparison of gibberellins in normal and slender barley seedlings. Plant Physiol. 94 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski, T., Stitt, M., Altmann, T., Udvardi, M.K., and Scheible, W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, P.J. (2004). Plant Hormones: Biosynthesis, Signal Transduction, Action! (Dordrecht, The Netherlands: Kluwer).
- Debeaujon, I., and Koornneef, M. (2000). Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 122 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkx, M.P.M., and Karssen, C.M. (1993). Effects of light and temperature on seed dormancy and gibberellin-stimulated germination in Arabidopsis thaliana - Studies with gibberellin-deficient and gibberellin-insensitive mutants. Physiol. Plant. 89 360–368. [Google Scholar]
- Derkx, M.P.M., and Karssen, C.M. (1994). Are seasonal dormancy patterns in Arabidopsis thaliana regulated by changes in seed sensitivity to light, nitrate and gibberellin? Ann. Bot. (Lond.) 73 129–136. [Google Scholar]
- Dill, A., and Sun, T.P. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durley, R.C., MacMillan, J., and Pryce, R.J. (1971). Investigation of gibberellins and other growth substances in the seeds of Phaseolus multiflorus and of Phaseolus vulgaris by gas chromatography-mass spectrometry. Phytochemistry 10 1891–1908. [Google Scholar]
- Eriksson, S. (2006). The Role of Gibberellins in the Regulation of Arabidopsis Flowering Time. PhD dissertation (Umeå, Sweden: Swedish University of Agricultual Sciences).
- Eriksson, S., Bohlenius, H., Moritz, T., and Nilsson, O. (2006). GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman, V.M., Gaskin, P., and MacMillan, J. (1974). Quantitative and qualitative analysis of gibberellins throughout seed maturation in Pisum sativum cv. Progress No. 9. Planta 118 123–132. [DOI] [PubMed] [Google Scholar]
- Gomez, K.A., and Gomez, A.A. (1984). Statistical Procedures for Agricultural Research, 2nd ed. (Chichester, UK: John Wiley & Sons).
- Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z.L., Powers, S.J., Gong, F., Phillips, A.L., Hedden, P., Sun, T.P., and Thomas, S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs, D.L., Hedden, P., and Lazarus, C.M. (1991). Partial-purification of 2 gibberellin 2β-hydroxylases from cotyledons of Phaseolus vulgaris. Phytochemistry 30 2507–2512. [Google Scholar]
- Hedden, P., and Phillips, A.L. (2000). Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 5 523–530. [DOI] [PubMed] [Google Scholar]
- Hensel, L.L., Nelson, M.A., Richmond, T.A., and Bleecker, A.B. (1994). The fate of inflorescence meristems is controlled by developing fruits in Arabidopsis. Plant Physiol. 106 863–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S., and Olszewski, N. (1993). Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski, S., Piazza, P., Craft, J., Hay, A., Woolley, L., Rieu, L., Phillips, A., Hedden, P., and Tsiantis, M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15 1560–1565. [DOI] [PubMed] [Google Scholar]
- Kim, Y.C., Nakajima, M., Nakayama, A., and Yamaguchi, I. (2005). Contribution of gibberellins to the formation of Arabidopsis seed coat through starch degradation. Plant Cell Physiol. 46 1317–1325. [DOI] [PubMed] [Google Scholar]
- King, R.W., and Evans, L.T. (2003). Gibberellins and flowering of grasses and cereals: Prizing open the lid of the “florigen” black box. Annu. Rev. Plant Biol. 54 307–328. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., and van der Veen, J.H. (1980). Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 58 257–263. [DOI] [PubMed] [Google Scholar]
- Langridge, J. (1957). Effect of day-length and gibberellic acid on the flowering of Arabidopsis. Nature 180 36–37. [Google Scholar]
- Lee, D.J., and Zeevaart, J.A.D. (2005). Molecular cloning of GA 2-oxidase3 from spinach and its ectopic expression in Nicotiana sylvestris. Plant Physiol. 138 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester, D.R., Ross, J.J., Smith, J.J., Elliott, R.C., and Reid, J.B. (1999). Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J. 19 65–73. [DOI] [PubMed] [Google Scholar]
- MacMillan, J. (2002). Occurrence of gibberellins in vascular plants, fungi, and bacteria. J. Plant Growth Regul. 20 387–442. [DOI] [PubMed] [Google Scholar]
- MacMillan, J., Ward, D.A., Phillips, A.L., Sánchez-Beltrán, M.J., Gaskin, P., Lange, T., and Hedden, P. (1997). Gibberellin biosynthesis from gibberellin A12-aldehyde in endosperm and embryos of Marah macrocarpus. Plant Physiol. 113 1369–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti, C., Orzaez, D., Ellul, P., Moreno, V., Carbonell, J., and Granell, A. (2007). Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J. 52 865–876. [DOI] [PubMed] [Google Scholar]
- Martin, D.N., Proebsting, W.M., and Hedden, P. (1999). The SLENDER gene of pea encodes a gibberellin 2-oxidase. Plant Physiol. 121 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum, M.G., Yamaguchi, S., Hanada, A., Kuwahara, A., Yoshioka, Y., Kato, T., Tabata, S., Kamiya, Y., and Sun, T.P. (2006). Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 45 804–818. [DOI] [PubMed] [Google Scholar]
- Nambara, E., Akazawa, T., and McCourt, P. (1991). Effects of the gibberellin biosynthetic inhibitor uniconazol on mutants of Arabidopsis. Plant Physiol. 97 736–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara, E., and Marion-Poll, A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56 165–185. [DOI] [PubMed] [Google Scholar]
- Nemhauser, J.L., Hong, F.X., and Chory, J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 467–475. [DOI] [PubMed] [Google Scholar]
- Oh, E., Yamaguchi, S., Kamiya, Y., Bae, G., Chung, W.I., and Choi, G. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47 124–139. [DOI] [PubMed] [Google Scholar]
- Penfield, S., Gilday, A.D., Halliday, K.J., and Graham, I.A. (2006). DELLA-mediated cotyledon expansion breaks coat-imposed seed dormancy. Curr. Biol. 16 2366–2370. [DOI] [PubMed] [Google Scholar]
- Phillips, A.L., Ward, D.A., Uknes, S., Appleford, N.E.J., Lange, T., Huttly, A.K., Gaskin, P., Graebe, J.E., and Hedden, P. (1995). Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 108 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radi, A., Lange, T., Niki, T., Koshioka, M., and Lange, M.J.P. (2006). Ectopic expression of pumpkin gibberellin oxidases alters gibberellin biosynthesis and development of transgenic Arabidopsis plants. Plant Physiol. 140 528–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers, C., Ruijter, J.M., Deprez, R.H.L., and Moorman, A.F.M. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339 62–66. [DOI] [PubMed] [Google Scholar]
- Reid, J.B., Ross, J.J., and Swain, S.M. (1992). Internode length in Pisum - a new, slender mutant with elevated levels of C19 gibberellins. Planta 188 462–467. [DOI] [PubMed] [Google Scholar]
- Rieu, I., Ruiz-Rivero, O., Fernandez-Garcia, N., Griffiths, J., Powers, S.J., Gong, F., Linhartova, T., Eriksson, S., Nilsson, O., Thomas, S.G., Phillips, A.L., and Hedden, P. (2008). The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 53 488–504. [DOI] [PubMed] [Google Scholar]
- Ross, J.J., Reid, J.B., and Swain, S.M. (1993). Control of stem elongation by gibberellin A1 - Evidence from genetic studies including the SLENDER mutant sln. Aust. J. Plant Physiol. 20 585–599. [Google Scholar]
- Ross, J.J., Reid, J.B., Swain, S.M., Hasan, O., Poole, A.T., Hedden, P., and Willis, C.L. (1995). Genetic regulation of gibberellin deactivation in Pisum. Plant J. 7 513–523. [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Sakakibara, H. (2006). Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57 431–449. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T., Kobayashi, M., Itoh, H., Tagiri, A., Kayano, T., Tanaka, H., Iwahori, S., and Matsuoka, M. (2001). Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 125 1508–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson, F., Brunaud, V., Balzergue, S., Dubreucq, B., Lepiniec, L., Pelletier, G., Caboche, M., and Lecharny, A. (2002). FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Schneider, G., and Schliemann, W. (1994). Gibberellin conjugates - An overview. Plant Growth Regul. 15 247–260. [Google Scholar]
- Schomburg, F.M., Bizzell, C.M., Lee, D.J., Zeevaart, J.A.D., and Amasino, R.M. (2003). Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe, W.W., and Mills, J.J. (1981). Hormones and parthenocarpic fruit set. A literature survey. Hort. Abstr. 51 661–698. [Google Scholar]
- Serrani, J.C., Fos, M., Atares, A., and Garcia-Martinez, J.L. (2007). Effect of gibberellin and auxin on parthenocarpic fruit growth induction in the cv micro-tom of tomato. J. Plant Growth Regul. 26 211–221. [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponsel, V.M., and Macmillan, J. (1978). Metabolism of gibberellin A29 in seeds of Pium sativum cv Progress No9 - Use of H2- and H3-GAs, and identification of a new GA catabolite. Planta 144 69–78. [DOI] [PubMed] [Google Scholar]
- Sussman, M.R., Amasino, R.M., Young, J.C., Krysan, P.J., and Austin-Phillips, S. (2000). The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol. 124 1465–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer, A., Bollman, K.M., and Poethig, R.S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124 645–654. [DOI] [PubMed] [Google Scholar]
- Thomas, S.G., and Hedden, P. (2006). Gibberellin metabolism and signal transduction. In Plant Hormone Signaling, P. Hedden and S.G. Thomas, eds (Oxford, UK: Blackwell), pp. 147–184.
- Thomas, S.G., Phillips, A.L., and Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 96 4698–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier, A.F., Marillonnet, S., Klimyuk, V., Patel, K., Torres, M.A., Murphy, G., and Jones, J.D.G. (1999). Multiple independent defective Suppressor-mutator transposon insertions in Arabidopsis: A tool for functional genomics. Plant Cell 11 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu, T., Kawaide, H., Mitsuhashi, W., Inoue, Y., and Kamiya, Y. (1998). Phytochrome regulates gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiol. 118 1517–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk, E.M., Fujioka, S., Seto, H., Shimada, Y., Takatsuto, S., Yoshida, S., Denzel, M.A., Torres, Q.I., and Neff, M.M. (2003). CYP72B1 inactivates brassinosteroid hormones: An intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol. 133 1643–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., and Speleman, F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3 34.31–34.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varbanova, M., et al. (2007). Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19 32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian-Smith, A., and Koltunow, A.M. (1999). Genetic analysis of growth-regulator-induced parthenocarpy in Arabidopsis. Plant Physiol. 121 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Caruso, L.V., Downie, A.B., and Perry, S.E. (2004). The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell 16 1206–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R.N., Heckman, J.W., and Somerville, C.R. (1992). Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 100 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward, A.W., and Bartel, B. (2005). Auxin: Regulation, action, and interaction. Ann. Bot. (Lond.) 95 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S., Smith, M.W., Brown, R.G.S., Kamiya, Y., and Sun, T.P. (1998). Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi, Y., Ogawa, M., Kuwahara, A., Hanada, A., Kamiya, Y., and Yamaguchi, S. (2004). Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi, Y., Takeda-Kamiya, N., Hanada, A., Ogawa, M., Kuwahara, A., Seo, M., Kamiya, Y., and Yamaguchi, S. (2007). Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol. 48 555–561. [DOI] [PubMed] [Google Scholar]
- Zhu, Y.Y., et al. (2006). ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.