Abstract
Parental imprinting is important for seed development, but few imprinted genes have been identified in plants. The four known imprinted genes in Arabidopsis thaliana encode transcriptional regulators. Here, we describe a novel imprinted gene, MATERNALLY EXPRESSED PAB C-TERMINAL (MPC), which encodes the C-terminal domain of poly(A) binding proteins (PABPs). PABPs play roles in mRNA stability and translation. MPC interacts with proteins that also interact with the C-terminal domain of typical PABPs, suggesting that MPC may regulate translation by modulating PABP activity. In the endosperm, MPC is expressed only from the maternal allele. Reduction of MPC expression affects seed development. In dna methyltransferase1 (met1) mutants, MPC is ectopically expressed, and the paternal allele is active in the endosperm. CGs in the 5′ flanking region and gene body of MPC lose methylation in a met1 background. Both regions are required to confer imprinted reporter expression, suggesting that the gene body contains imprinting control region elements. In Arabidopsis, DEMETER (DME) activates expression of maternal alleles. MPC expression is reduced in flowers and seeds in a dme-4 mutant but only after fertilization in dme-1. We conclude that other factors along with DME promote MPC expression and that DME has indirect effects on imprinted gene expression in endosperm.
INTRODUCTION
In mammals and flowering plants, a subset of genes are expressed only from the maternal or paternal allele, a process termed imprinting. Imprinted genes are often involved in growth control (Wood and Oakey, 2006; Huh et al., 2008). Imprinting in plants has profound consequences for seed development, with dramatic effects on seed size, morphogenesis, and viability (Haig and Westoby, 1991; Gehring et al., 2004), but only a small number of imprinted genes have been identified. The known imprinted genes in Arabidopsis thaliana, FERTILIZATION INDEPENDENT SEED1/MEDEA (FIS1/MEA), FIS2, FLOWERING WAGENINGEN (FWA), and PHERES1 (PHE1), are all involved in transcriptional regulation (Grossniklaus et al., 1998; Vielle-Calzada et al., 1999; Luo et al., 2000; Soppe et al., 2000; Köhler et al., 2003, 2005; Kinoshita et al., 2004; Jullien et al., 2006a). The FWA and PHE1 proteins are both transcription factors (homeodomain and MADS domain, respectively), while FIS1/MEA and FIS2 are members of Polycomb Repressive Complex 2 (PRC2), which inhibits expression of target genes in endosperm, including PHE1.
In this work, we identified a novel type of imprinted gene in Arabidopsis, MATERNALLY EXPRESSED PAB C-TERMINAL (MPC), which encodes a small protein predicted to contain a single conserved domain, the C-terminal region of a poly(A) binding protein (PABP). PABPs, found throughout eukaryotes, bind the poly(A) tails of mRNAs through N-terminal RNA recognition motifs. PABPs also interact with eukaryotic initiation factors associated with the 5′-cap of mRNA, resulting in mRNA circularization, which favors initiation of translation and ribosome recycling. Through their interactions with other factors, PABPs perform a variety of roles, including regulation of mRNA translation and stability (Mangus et al., 2003; Kühn and Wahle, 2004). The Arabidopsis genome contains eight expressed PABPs, constituting the largest known PABP multigene family in any species (Belosotsky, 2003). Most PABPs contain a conserved C-terminal domain (termed the PAB-CT, PABC, or CTC), which binds proteins that can modulate PABP function (Kozlov et al., 2001).
The other maternally expressed imprinted genes in Arabidopsis, FIS1/MEA, FIS2, and FWA, are reported to be regulated by an antagonistic interaction between DNA METHYLTRANSFERASE1 (MET1), which maintains cytosine methylation in CG sequences associated with gene silencing (Finnegan and Dennis, 1993; Kankel et al., 2003), and the DNA glycosylase DEMETER (DME), which excises methylated cytosines (Choi et al., 2002; Xiao et al., 2003; Kinoshita et al., 2004; Gehring et al., 2006; Jullien et al., 2006a; Morales-Ruiz et al., 2006). A current model of imprinting regulation in Arabidopsis (Scott and Spielman, 2004; Huh et al., 2008) presents methylation and silencing as the default state, which is relieved by targeted DME activity in the central cell. This removes cytosine methylation at imprinted loci, allowing expression of the maternal alleles in the central cell and subsequently in the endosperm. However, DME was found not to be expressed in pollen (Choi et al., 2002) and therefore cannot activate the paternal alleles. Maternal FIS1/MEA and FWA behave in accordance with this model, showing no detectable expression in any dme mutant background tested (Choi et al., 2002; Xiao et al., 2003; Kinoshita et al., 2004; Jullien et al., 2006a). Results for FIS2 are less consistent. Jullien et al. (2006a) found FIS2 transcript levels only reduced in flowers of dme-4 mutants, although FWA expression was abolished in the same background. FIS2 reporter expression was likewise diminished but not absent in seeds of presumed null dme-2 as well as weaker dme-4 mutants. Therefore, these authors suggested that other unknown factors as well as DME activate FIS2 expression. More dramatically, Choi et al. (2002) showed no loss of FIS2 expression in dme-1 mutant flowers. In addition, a FIS2 but not a DME reporter was expressed in fem111/agl80 mutant ovules (Portereiko et al., 2006), suggesting that DME is not required for FIS2 expression. Control of paternal expression is also simplified in the model. Paternal FIS1/MEA, FIS2, and FWA are all methylated and silent in pollen. Paternal FIS2 and FWA are ectopically activated in endosperm when contributed by a met1 mutant pollen parent, but hypomethylated paternal FIS1/MEA remains silent in endosperm due to repression by methylation of histone H3 Lys 27 mediated by maternal PRC2 (Kinoshita et al., 2004; Gehring et al., 2006; Jullien et al., 2006a, 2006b).
In this study, we characterized the novel imprinted gene MPC, which is expressed only from the maternally contributed alleles in endosperm. MPC requires MET1 for silencing of the paternal allele in seeds and is regulated by methylation of cytosines in the 5′ flanking region of the MPC locus, as well as the gene body, which lose methylation in a met1 mutant background, causing ectopic gene expression. Unlike the case for FWA or FIS1/MEA, MPC expression is not abolished by mutations in the DME gene; instead, expression is reduced or unchanged, depending on the dme allele and the stage of reproduction. MPC encodes a small protein predicted to contain a protein interaction domain conserved in PABPs but not the domains that interact with RNA. MPC interacts with proteins that also interact with the C-terminal domain of a typical Arabidopsis PABP, suggesting that MPC may affect translation. Reduction of MPC expression using RNA interference resulted in seed abortion, decreased seed size, and embryo and endosperm abnormalities.
RESULTS
At3g19350 (MPC) Is a Novel Imprinted Gene with Maternal-Specific Expression
We set out to identify new imprinted genes in Arabidopsis through transcriptional profiling of seeds in which either parental genomic balance or imprinting regulation were altered. For many species, including Arabidopsis, crossing plants of different ploidies affects seed development, generating reciprocal phenotypes depending on the direction of the cross. One interpretation is that this is due to imbalance in the dosage of active copies of imprinted genes in the seed (Haig and Westoby, 1991; Scott et al., 1998; Spielman et al., 2001). We conducted a microarray experiment (S. Tiwari, R. Schulz, M. Spielman, R.J. Oakey, and R.J. Scott, unpublished data) using RNA extracted from developing siliques produced by balanced crosses (2x × 2x), interploidy crosses (crosses in both directions between 2x and 4x or 2x and 6x plants), and homozygous fis1/mea mutants, which deregulate expression of the imprinted gene PHE1 (Köhler et al., 2003) and therefore potentially deregulate additional imprinted loci. We predicted that interploidy crosses and the fis1/mea mutant would affect expression levels of imprinted genes in patterns distinct from those caused by changes in dosage of biparentally expressed genes. We selected candidate imprinted genes whose expression trends clustered with that of the maternally expressed imprinted gene FWA.
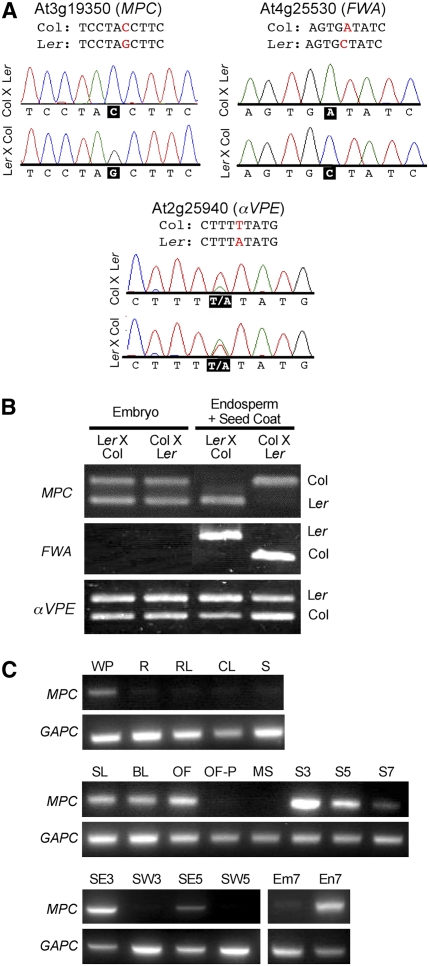
Our set of candidate imprinted genes included the At3g19350 locus, which we subsequently termed MPC (see below). To test parent-specific expression, we identified a polymorphism in the MPC locus between Columbia (Col) and Landsberg erecta (Ler) and sequenced pooled cDNA from siliques at 3 and 5 d after pollination (DAP) generated by reciprocal crosses between these two accessions. In crosses made in either direction, only the maternal allele of MPC is represented in the sequence. We observed the same pattern, as expected, for FWA, while a biallelically expressed gene shows the sequence of both maternal and paternal alleles (Figure 1A). We next separated seeds from the reciprocal crosses into two fractions, embryo and endosperm+seed coat, and performed RT-PCR followed by digestion with an enzyme that could cleave the polymorphic site in Ler but not Col (Figure 1B). This showed maternal-specific expression of MPC in the endosperm+seed coat fraction at 7 DAP and also biallelic expression in embryo at this stage.
Figure 1.
Expression of the MPC Gene.
(A) Allele-specific sequencing of MPC and control cDNAs from Col × Ler and Ler × Col siliques at 5 DAP. Only the maternal alleles of MPC and FWA are represented in the sequence for both crosses (polymorphic nucleotide in black box). The nonimprinted control gene, αVPE, shows a double peak at the polymorphic nucleotide.
(B) RT-PCR and restriction analysis of cDNA from seeds dissected into embryo and endosperm+seed coat fractions at 7 DAP. MPC is expressed only from maternal alleles in endosperm+seed coat, but biallelic in embryo. The imprinted FWA gene is a positive control for the dissection, and αVPE is a nonimprinted control.
(C) RT-PCR analysis of MPC cDNA from vegetative and reproductive organs. MPC is primarily expressed in flower buds, female reproductive organs of open flowers, and young siliques. GAPC, control; WP, whole plant; R, root 5 weeks old; RL, rosette leaf; CL, cauline leaf; S, stem; SL, seedling; BL, late floral buds; OF, open flowers; OF-P, open flowers without pistils; MS, mature stamens; S3, siliques at 3 DAP; S5, siliques 5 DAP; S7, siliques 7 DAP; SE3, seed 3 DAP; SW3, silique wall 3 DAP; SE5, seed 5 DAP; SW5, silique wall 5 DAP; Em7, embryo dissected from 7 DAP seed; En7, endosperm+seed coat dissected from 7 DAP seed.
To assay levels of MPC expression throughout the plant, we performed RT-PCR on RNA extracted from reproductive and vegetative organs (Figure 1C). This showed that MPC is mainly expressed in flower buds and in siliques containing developing seeds, with the highest activity at 3 DAP; there is also low expression in whole plants. The decline in expression between 3 and 7 DAP is correlated with the end of endosperm proliferation and transition to differentiation. We did not detect expression in mature stamens or in flower buds from which pistils had been removed, indicating that MPC is specific to female reproductive development. MPC expression was not detectable in siliques emptied of seeds at 3 and 5 DAP, while high levels of expression were present in the seeds from these siliques. In seeds at 7 DAP, MPC is expressed in endosperm but barely detectable in embryo. Our data are consistent with public microarray data (https://www.genevestigator.ethz.ch/), which shows that At3g19530 is absent in most organs, with highest expression in young siliques.
To further investigate MPC expression in reproductive development, we constructed a reporter gene incorporating the putative promoter-containing region 1.3 kb 5′ of the start codon, plus the MPC gene body (two exons and one intron) excluding the stop codon, translationally fused to nuclear-targeted green fluorescent protein (GFP) (proMPC-MPC-GFP). GFP signal was detected in the central cell before fertilization (Figure 2A) and in the endosperm of seeds from the time of fertilization (Figure 2B) to 4 DAP (Figure 2C). We did not detect signal in silique or seed coat at this or any later stage. This confirms that the maternal-specific expression of MPC observed in whole siliques up to 5 DAP (Figure 1A) and the endosperm+seed coat fraction of dissected seeds at 7 DAP (Figure 1B) could only result from imprinted expression in endosperm, rather than from expression in the maternally derived silique or seed coat. Although RT-PCR showed MPC expression in embryo at 7 DAP, we did not observe embryo expression of the reporter at 7 DAP (data not shown) or at any earlier stage (Figure 2). As embryo expression was clearly visible by RT-PCR only after two rounds of amplification (see Methods), it is likely that endogenous MPC activity in the embryo is very low.
Figure 2.
MPC Expression in the Seed Is Endosperm Specific, and a Translational but Not Transcriptional Reporter Is Imprinted.
(A) to (C), (E), and (F) Nuclear-targeted translational MPC reporter containing the MPC promoter and gene body fused to GFP (proMPC-MPC-GFP).
(A) The reporter is active in the central cell of an unfertilized ovule (arrow and inset).
(B) Reporter expression increases after fertilization; this seed from a self-fertilized proMPC-MPC-GFP plant shows the four nuclei resulting from the first two endosperm divisions. The arrow points to one of the nuclei that lies in a different focal plane to the other three and is therefore fainter.
(C) proMPC-MPC-GFP × proMPC-MPC-GFP.
(D) Wild-type Col (Col × Col).
(E) and (F) The translational reporter is imprinted.
(E) proMPC-MPC-GFP (♀) × Col (♂).
(F) Col (♀) × proMPC-MPC-GFP (♂).
(G) and (H) Nuclear-targeted transcriptional reporter (proMPC-GFP). Confocal images with GFP in green and chlorophyll autofluorescence in red. Some images show green autofluorescence in the seed coat. The transcriptional reporter is expressed when contributed by either parent. All seeds in (C) to (H) are at 4 DAP.
(G) proMPC-GFP (♀) × Col (♂).
(H) Col (♀) × proMPC-GFP (♂).
Bars = 2 μm in (A) and (B) and 100 μm in (C) to (H).
Seeds from crosses between wild-type plants and plants carrying the proMPC-MPC-GFP reporter showed strong GFP signal when the reporter was transmitted by the mother (Figure 2E), but signal was not detectable or barely visible when the reporter was paternally contributed (Figure 2F). By contrast, a reporter in which the putative MPC promoter was transcriptionally fused to nuclear-targeted GFP (proMPC-GFP) showed clear signal when contributed from either parent (Figures 2G and 2H). This suggests that sequences within the gene body as well as the upstream region are required for imprinting of the endogenous MPC locus.
MPC Is Homologous to the C-Terminal Domains of Poly(A) Binding Proteins and Binds PAB-Interacting Proteins in Vitro
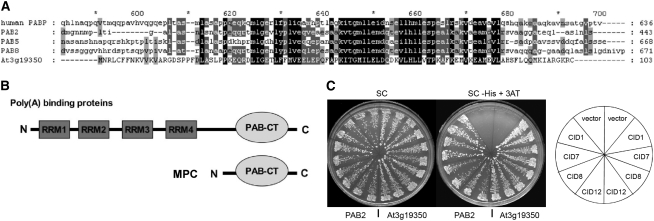
At3g19350 is predicted to encode a small protein of 103 amino acids (www.arabidopsis.org). A BLASTP search (www.ncbi.nlm.nih.gov) identified At3g19350 as containing a single conserved domain, the C-terminal region of a PABP. BLASTP searches show the MPC protein is most closely related to PABPs from Arabidopsis and other plants, including tobacco (Nicotiana tabacum), soybean (Glycine max), and rice (Oryza sativa). Figure 3A presents an alignment of the complete MPC protein with the C-terminal regions of Arabidopsis PAB2 (broadly expressed), PAB5 (reproduction-specific), PAB8 (the Arabidopsis protein with the greatest similarity to MPC), and the major human PABP. Figure 3B shows the basic structure of MPC in relation to typical PABPs.
Figure 3.
The Predicted MPC Protein Has Homology to the C-Terminal Domain of Poly(A) Binding Proteins and Can Bind CID Proteins That Also Interact with a Full-Length Arabidopsis PABP.
(A) Alignment of the complete predicted MPC (At3g19350) protein with the C-terminal domains of the major human PABP and three Arabidopsis PABPs. Conserved amino acid residues are shaded.
(B) The structure of MPC compared with a typical PABP.
(C) A yeast two-hybrid assay indicates that PAM2-containing fragments of CID1, CID7, CID8, or CID12 interact with MPC in a similar manner as with PAB2. The plates show streaks of two representative yeast colonies for each one of the two-hybrid interactions tested. In the left half of the plate are the interaction with PAB2 and in the right half with MPC (At3g19350). Colonies were streaked on synthetic complete (SC) medium to select for both two-hybrid clones and on SC supplemented with 5 mM 3-amino-1,2,4-triazole (SC-His + 3AT) to assess the interaction. The position of the interactions in the plate is indicated on the slices of the diagram to the right of the figure; vector represents the empty cloning vector.
Proteins that interact with the C-terminal domain of PABPs include a PAM2 domain (Khaleghpour et al., 2001b; Roy et al., 2002). In Arabidopsis, 11 CTC-interacting (CID) proteins have been identified that also contain this domain (Bravo et al., 2005). To test whether MPC can function as a PAB-CT, we conducted a two-hybrid interaction assay using the PAM2-containing regions of four CID proteins, CID1/ERD15 (At2g41430), CID7 (At2g26280), CID8 (At1g53650), and CID12/RNA BINDING PROTEIN 37 (At4g10610). All could interact with MPC and also with the PAB2 C-terminal domain (Figure 3C).
Reduced MPC Expression Inhibits Trichome Branching and Seed Growth
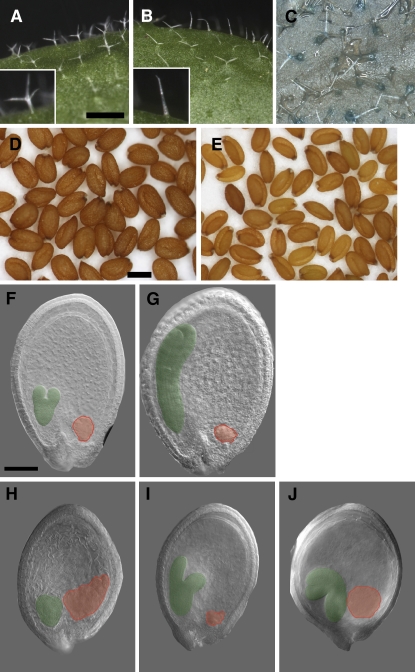
To investigate the broader function of MPC in planta, we generated plants with reduced MPC expression. Since no MPC mutants were publicly available, we sought to abolish MPC function using RNA interference (RNAi). Expression of double-stranded RNA corresponding to a largely unique 133-bp region of MPC (see Supplemental Figure 1 online) was directed by the MPC putative promoter. Plants homozygous for the RNAi transgene had reduced MPC transcript levels in siliques at 3 DAP (see Supplemental Figure 2 online). Some plants had delayed vegetative development and flowering (data not shown), but the most consistently observed vegetative phenotype was reduction of trichome branching (cf. Figures 4A and 4B). All trichomes on rosette leaves of wild-type Col-0 developed three branches (n = 188 trichomes on three plants), while MPC RNAi trichomes had one branch (21.2%), two branches of very unequal length (21.6%), or two symmetrical branches (57.2%) (n = 268 trichomes on three plants sampled at random from 20 homozygotes). A proMPC-MPC-GUS reporter was expressed in trichomes of wild-type plants (Figure 4C), supporting a role for the endogenous MPC gene in promoting trichome branching.
Figure 4.
Reducing MPC Expression Affects Seed and Trichome Development.
(A) Trichomes on rosette leaves of wild-type Col have three branches.
(B) Trichomes on rosette leaves of MPC RNAi plants have one to two branches.
(C) A proMPC-MPC:GUS translational reporter transformed into wild-type Col is expressed in trichomes.
(D) and (E) Mature seeds from wild-type Col (left) and MPC RNAi (right) plant.
(F) to (J) Differential contrast images of developing seeds with embryos pseudocolored green and chalazal endosperm pseudocolored red.
(F) and (G) Wild-type Col at 5 DAP (F) and 7 DAP (G).
(H) to (J) MPC RNAi seeds at 7 DAP showing delayed embryogenesis, abnormal embryo morphology, and enlarged chalazal endosperm ([H] and [J]).
Bars = 1 mm in (A) to (C), 250 μm in (D) and (E), and 100 μm in (F) to (J).
Knockdown of MPC expression disrupted fertilization or seed development and growth. In the wild type, nearly all ovules are fertilized and develop into mature seeds, but siliques from MPC RNAi plants contained unfertilized ovules and aborted seeds. In two independently transformed RNAi lines (which also exhibited the trichome branching phenotype), we found means of 15.27% ± 0.30 (se) unfertilized ovules, 19.64% ± 1.85 aborted seeds, and 7.89% ± 2.40 unfertilized ovules, and 5.12% ± 2.99 aborted seeds, respectively (a minimum of eight siliques scored from each of three plants per line). Seeds that did develop to maturity were smaller than the wild type (cf. Figures 4D and 4E) and had reduced weight. Mean seed weight was 18.7 μg ± 0.07 (se) for wild-type Col-0, and for two RNAi lines, 16.5 μg ± 0.27 and 16.0 μg ± 1.05 (n ≥ 5 plants for each). There were significant differences among the seed weights (Kruskal-Wallis test, P = 0.003). Compared with developing seeds from wild-type siliques, MPC RNAi seeds showed a range of abnormalities, including misshapen embryos (12.5% ± 6.5; three siliques were scored from each of three plants of two lines) and enlarged chalazal endosperm (cf. Figures 4F and 4G with Figures 4H to 4J). We conclude that MPC is important for normal seed growth and development.
MET1 Silences Vegetative and Paternal MPC
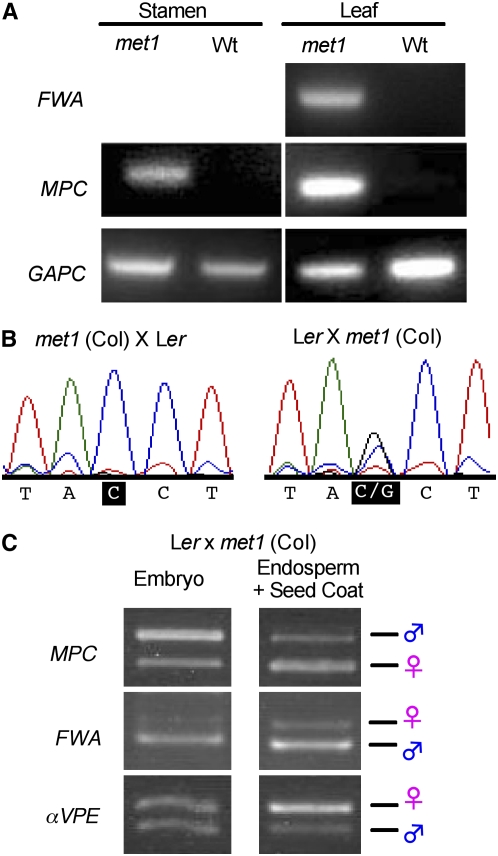
To test whether MPC is silenced by methylation by the methyltransferase MET1, we first examined MPC expression in met1-9 mutants, which harbor a T-DNA insertion in exon 7 (Alonso et al., 2003; http://signal.salk.edu). Plants homozygous for the met1-9 allele display features diagnostic of severe DNA hypomethylation, such as reduced stature and ectopic expression of FWA (Figure 5A; Kinoshita et al., 2004). In homozygous met1-9 plants, MPC was ectopically expressed in leaf and also in stamens containing mature pollen (Figure 5A). Therefore, MET1 is required to silence MPC in vegetative tissue and in male reproduction. We then tested whether the paternal MPC allele is ectopically expressed when transmitted by a hypomethylated pollen parent by comparing expression following reciprocal crosses between met1-9 homozygotes in the Col accession and wild-type Ler plants. Sequencing pooled cDNA of the region of the MPC locus containing a Col/Ler polymorphism showed that at 5 DAP, siliques generated by these crosses contained only maternal MPC transcripts when the pollen parent was wild-type but both maternal and paternal transcripts when the pollen parent was homozygous for met1-9 (Figure 5B). At 7 DAP, seeds from a met1 (Col) × wild type (Ler) cross were dissected into embryo and endosperm+seed coat fractions, and parent-specific expression was assayed using RT-PCR followed by restriction analysis. The paternal MPC allele is expressed in endosperm+seed coat when contributed by a met1-9 pollen parent (Figure 5C; cf Figure 1B). Expression in embryo is biallelic as before. FWA, used as a control for the dissection, shows ectopic paternal expression in endosperm as well as embryo, where this gene is not normally expressed at all (Kinoshita et al., 2004). We also introduced the MPC translational reporter into a met1-9 background and crossed proMPC-MPC-GFP met1-9 plants in both directions with the wild type; we observed ectopic expression when the transgene was contributed by the pollen parent (see Supplemental Figure 3 online). Taken together our data show that MET1 represses the paternal allele of MPC in endosperm.
Figure 5.
Loss of MET1 Function Causes Ectopic MPC Expression.
(A) RT-PCR of MPC RNA from mature stamen and rosette leaf of met1 mutant and wild-type Col plants. Ectopic expression of FWA is a control for hypomethylation in the met1-9 background. GAPC, control.
(B) Allele-specific sequencing of MPC RNA extracted from siliques at 5 DAP. When the pollen parent is wild-type, only the maternal MPC allele is represented in the sequence (left), but when the pollen parent is homozygous for met1-9, both maternal and paternal alleles are detected (right).
(C) RT-PCR and restriction analysis of MPC RNA from embryo and endosperm+seed coat fractions of seeds dissected at 7 DAP shows the paternal MPC allele is ectopically expressed in endosperm when contributed by a met1-9 pollen parent (cf. Figure 1B, which shows the paternal band is absent in endosperm+seed coat when the pollen parent is wild-type). The imprinted FWA gene is a control for the dissection, and αVPE is a nonimprinted control.
Methylated Cytosines in the 5′ Flanking Region and Gene Body of MPC Are Associated with Silencing
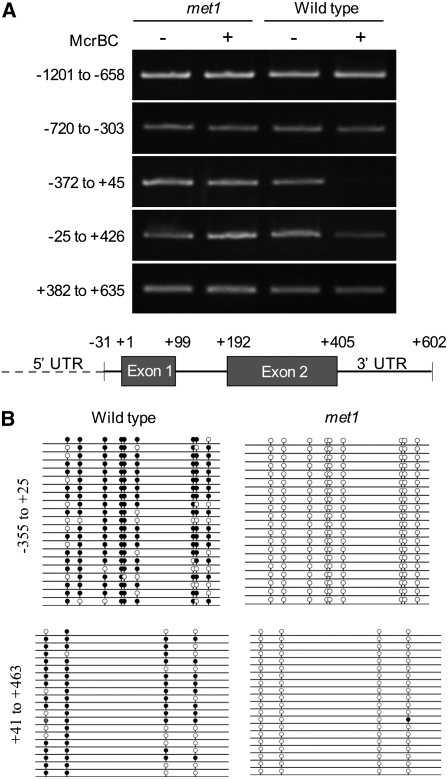
To identify methylated regions of the MPC locus, we used the methylation-specific restriction endonuclease McrBC on DNA extracted from rosette leaves of wild-type Col-0 plants and of plants homozygous for the presumed null met1-6 allele (Xiao et al., 2003). The McrBC enzyme restricts methylated DNA, so methylated regions of McrBC-treated DNA are not amplified by PCR. We amplified overlapping regions of the MPC locus extending from 1.2 kb upstream of the start codon to ∼30 bp downstream of the predicted 3′ untranslated region (UTR) (Figure 6A). The region spanning −372 bp to halfway through exon 1 was amplified from McrBC-treated DNA from met1-6 but not wild-type plants, indicating this region of the locus is methylated in the wild type. There was also reduced amplification of wild-type treated DNA in a region spanning the entire gene body, most of the 5′ UTR and the first 22 bp of the 3′ UTR, suggesting partial methylation of this region.
Figure 6.
MPC Is Methylated in the 5′ Flanking Region and in the Gene Body.
(A) RT-PCR of MPC and surrounding regions treated with the methylation-sensitive restriction endonuclease McrBC (+) or mock treated (–). McrBC digests methylated DNA; the presence of a PCR product indicates a lack of methylation in that genomic fragment. Genomic DNA was extracted from rosette leaf of met1-6 or wild-type Col plants. The start codon is at position +1, and the positions of other gene features are indicated in the schematic below.
(B) CG methylation in multiple clones of the upstream region and gene body of MPC determined by bisulfite sequencing of rosette leaf DNA in met1-9 and wild-type Col backgrounds. A black circle indicates a methylated cytosine in CG context, an open circle is an unmethylated cytosine, and a gray circle is a methylated cytosine in a CN context where N is an unknown base. There are no CG sites between +25 and +45.
We next used bisulfite sequencing of DNA from rosette leaves of wild-type Col-0 and met1-9 homozygous plants to precisely identify the methylated cytosines at the MPC locus that are linked to its expression (Figure 6B; see Supplemental Table 1 online). Consistent with ectopic expression of MPC in met1-9 leaf (Figure 6A), the 5′ flanking region of MPC is depleted in CG methylation in a met1 background. The MPC gene body is also hypomethylated in met1-9 mutants, in accordance with the role of this region in paternal silencing of the transgene (Figure 2). Of nine cytosines in a CG context in the region −355 to +24 from the start codon (region 1), on average 81.5% were methylated in wild-type leaf (n = 21 clones) and none in met1-9 (n = 20). For the gene body, +41 to +483 (region 2), 68.8% of the four cytosines in CG motifs were methylated in the wild type and 1.3% in met1-9 (n = 20 for both). There are no CNG sequences in region 1, but there are nine in region 2, with an average of 18.9% methylation in the wild type, increasing to 50.0% in met1-9. Increased CNG methylation in a met1 background has been observed for other genes (Chan et al., 2005).
Tissue-specific and imprinted expression of FWA are both associated with methylation of promoter direct repeats with sequence related to a SINE retrotransposon (Soppe et al., 2000; Kinoshita et al., 2004, 2007). We searched for transposon-like sequences, direct repeats, and inverted repeats in a region spanning 1.3 kb upstream of the MPC start codon to 1 kb downstream of the coding region (RepeatMasker, http://www.repeatmasker.org; Tandem Repeats Finder [Benson, 1999]). We found no transposon-like sequences or inverted repeats. There are two direct repeats, one of 31 nucleotides at positions −773 and −741 from the start codon with five mismatches, and one of 41 nucleotides at +923 and +963 with three mismatches; however, neither of these contain cytosines in a CG or CNG context.
DME Promotes but Is Not Required for MPC Expression
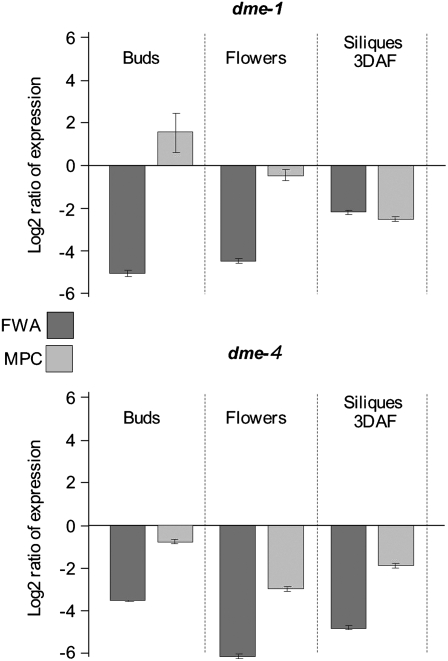
To test whether DME activates maternal MPC, we first performed RT-PCR on floral buds and open flowers from wild-type plants and two homozygous dme mutants, dme-1 (Choi et al., 2002) and dme-4 (Guitton et al., 2004). Although most dme mutant seeds abort, occasionally homozygous mutants can be obtained for these two alleles. RT-PCR showed that MPC expression was unchanged in dme-1 but diminished in the dme-4 background (see Supplemental Figure 4 online). Quantitative real-time PCR (qRT-PCR) performed on floral buds, open flowers, and siliques at 3 d after flowering (DAF) showed that in dme-1 mutants, MPC expression was not reduced in buds or flowers but surprisingly was diminished ∼3.5-fold in siliques (Figure 7; see Supplemental Table 2 online). In dme-4, MPC RNA levels were reduced at all three stages. By contrast, FWA expression was almost undetectable at all stages in both dme mutants, consistent with previous studies (Kinoshita et al., 2004; Jullien et al., 2006a).
Figure 7.
MPC Expression in dme Mutant Backgrounds.
qRT-PCR analysis of MPC and FWA in buds, flowers, and siliques from dme-1 and dme-4. Each bar represents the log2 ratio of expression in the dme mutant to its wild-type background (Ler for dme-1 and C24 for dme-4). Error bars = se.
DISCUSSION
Parental imprinting has significant effects on seed development, growth, and viability (Haig and Westoby, 1991; Gehring et al., 2004). Based on many decades of observation in a wide range of species, imprinting has been associated with control of seed size, hybridization barriers between species, seed abortion following crosses between plants of the same species but different ploidies, and prevention of fully asexual seed development in apomicts. These interpretations are supported by the phenotypes of Arabidopsis plants with mutations in the imprinted FIS1/MEA and FIS2 genes, which include endosperm overproliferation and seed abortion when ovules are fertilized and ectopic division of the central cell in unfertilized ovules. In this work, we identified a new imprinted gene, MPC, which is expressed from maternal but not paternal alleles in endosperm. Reducing MPC transcript levels through RNAi had a variety of effects on seeds, including seed abortion, reduced seed size, and abnormal embryo and endosperm morphology. Therefore, MPC plays a role in normal seed development and growth. The most consistent vegetative phenotype of MPC RNAi plants was reduction of the number of branches on rosette leaf trichomes. Trichome branching is associated with endoreduplication of trichome nuclei (Schwab et al., 2000); one possibility is that loss of MPC function inhibits DNA replication.
MPC encodes a small protein consisting almost entirely of sequence with homology to the conserved C-terminal domain of PABPs. Typical PABPs include RNA recognition motifs that bind the poly(A) tails of mRNAs and a C-terminal domain that interacts with proteins that can modulate PABP activity (Kozlov et al., 2001; Mangus et al., 2003; Kühn and Wahle, 2004). PABP functions include protecting mRNA from degradation and regulating translation. Proteins that bind to the C-terminal domain of typical PABPs have been shown to promote or repress translation (Craig et al., 1998; Khaleghpour et al., 2001a; Roy et al., 2002; Wang and Grumet, 2004; Berlanga et al., 2006). In humans, these interacting proteins include Paip1, Paip2A, and Paip2B, which compete with each other for binding to PABPs. The Paip proteins contain two binding sites for PABPs (PAM1 and PAM2); the latter binds the C-terminal domain (Khaleghpour et al., 2001b; Roy et al., 2002).
In Arabidopsis, two CID proteins were identified that contain the PAM2 motif and bind to the C-terminal domain of the PAB2 protein in a yeast two-hybrid assay; another 11 Arabidopsis proteins containing this motif were subsequently identified with database searches (Bravo et al., 2005). Several of the CID genes have been independently identified by mutant phenotypes: CID1/EARLY RESPONSE TO DEHYDRATION15 (ERD15), induced in response to drought stress (Kiyosue et al., 1994) and proposed to negatively regulate abscisic acid responses (Kariola et al., 2006), and CID5/INCREASED POLYPLOIDY LEVEL IN DARKNESS1, which promotes polyploidy in dark-grown seedlings (Tsumoto et al., 2006). PAM2-containing proteins interacting with PAB-CT have also been identified in cucumber (Cucumis sativus), and one of these, PCI6, was shown to inhibit translation (Wang and Grumet, 2004). We found that the PAM2 domains of CID proteins that interact with the C-terminal domain of a typical Arabidopsis PABP also interact with MPC. As MPC lacks RNA recognition motifs, there is no obvious mechanism for it to bind mRNA directly. We propose that MPC could affect PABP function by titrating proteins that would otherwise interact with the C-terminal domain of a typical PABP that is able to bind mRNA. We believe it is unlikely that MPC would directly affect transcription, like the other known imprinted genes in Arabidopsis. However, we propose that MPC could be a regulatory gene that affects RNA translation.
CG methylation mediated by MET1 plays a role in silencing FIS1/MEA, FIS2, and FWA (Xiao et al., 2003; Kinoshita et al., 2004; Gehring et al., 2006; Jullien et al., 2006a). In a met1 mutant background, we found MPC ectopically expressed in leaf and stamen, and paternal MPC was expressed in endosperm when contributed by a met1 mutant pollen parent. Therefore, MET1 is required to repress expression of MPC in vegetative tissues and paternally derived MPC in seeds. Regulation of MPC expression by MET1 is similar to that observed for FIS2 and FWA—as the paternal alleles of these genes are also ectopically expressed in seeds when donated by a met1 pollen parent—but not FIS1/MEA, which is not paternally activated by hypomethylation. In the upstream region and also the gene body of MPC, we identified cytosines in a CG context that were methylated in wild-type but not met1 mutant leaves. Further work will be needed to test whether these are also methylated on the silent paternal allele of MPC in endosperm. Further work will also determine whether MPC is subject to regulation by histone M3 Lys 27 trimethylation (H3K27me3) mediated by PRC2. However, a whole-genome analysis of H3K27me3 identified FIS1/MEA and PHE1, but not MPC, FWA, or FIS2, as targets of this regulation (Zhang et al., 2007).
A transgene in which 1.3 kb of the MPC upstream region was fused to GFP did not confer imprinted expression on a reporter. However, when the gene body was included in the construct, the reporter was paternally silenced. This suggests that sequences in the gene body of the endogenous MPC locus are required for imprinting. It is possible that these sequences include the methylated cytosines we identified, as methylation in the gene body interferes with transcript elongation in Arabidopsis (Zilberman et al., 2007). For the other four imprinted genes in Arabidopsis, the methylated regions found to be associated with imprinting all lie outside the coding region (Xiao et al., 2003; Kinoshita et al., 2004; Gehring et al., 2006; Jullien et al., 2006a; Makarevich et al., 2008).
Imprinting of FWA is associated with methylation of direct repeats with sequence related to a SINE retrotransposon (Soppe et al., 2000; Kinoshita et al., 2004, 2007), and maternal expression of FIS1/MEA is associated with hypomethylation of direct repeats 3′ of the coding region (Gehring et al., 2006), while paternal expression of PHE1 requires methylation at tandem repeats (Makarevich et al., 2008). However, using bioinformatics analysis we could not identify transposon-like sequences or inverted repeats from 1.3 kb upstream of the MPC start codon to 1 kb downstream of the coding region, and the two direct repeat sequences in this region did not contain any cytosines in a CG or CNG context. Therefore, transposon-like and repetitive sequence elements do not appear to play a role in imprinting of MPC. Similarly, no transposon-like or repetitive sequences were identified near the FIS2 locus; instead, FIS2 imprinting is associated with a 200-bp CG-rich upstream region (Jullien et al., 2006a). These results highlight the diversity of mechanisms involved in imprinting of plant genes.
In the dme-1 background, MPC transcripts were reduced in siliques but not floral buds or flowers, while in dme-4, MPC expression was diminished at all three stages. By contrast, FWA expression was downregulated at all stages in both mutant backgrounds. The dme-1 allele contains a T-DNA insertion in the 5′ UTR, and a low level of transcript predicted to encode a slightly truncated DME protein is detected in dme-1 mutants (Choi et al., 2002). dme-4 contains an early stop codon resulting in a truncated protein that retains the DNA glycosylase domain but not the Cys-rich domain (Guitton et al., 2004). The glycosylase domain of DME is responsible for excising methylated cytosines from DNA, leading to their replacement by unmethylated cytosine (Morales-Ruiz et al., 2006), while the conserved Cys residues of DNA glycosylases provide a link to an iron–sulfur cluster that plays a role in DNA binding (Lukianova and David, 2005). Both dme-1 and dme-4 occasionally produce homozygous mutant seed, supporting the molecular evidence that neither is a null allele. dme-1 mutants could have sufficient DME function to allow full MPC expression, explaining why MPC RNA levels are not reduced in buds and flowers. DME expression diminishes dramatically after fertilization (Choi et al., 2002), and it is therefore thought that reduced expression of DME targets in dme mutant seeds is due to failure to demethylate maternal alleles of imprinted genes in the central cell. In this context, it is surprising that we found reduced MPC RNA levels in dme-1 siliques at 3 DAF. It is possible that DME activity ceases earlier in dme-1 than wild-type seeds; however, if there is sufficient DME activity in the dme-1 mutant central cell to remove methylcytosines from the MPC locus, there would be no methylation for MET1 to maintain after fertilization and, therefore, according to the model of DME-MET1 antagonism, no mechanism for silencing MPC in the dme-1 mutant seed. Therefore, we favor the hypothesis that the dme-1 mutation has an indirect effect on MPC expression after fertilization. The imprinted FIS2 gene was also found to have diminished expression both before and after fertilization in dme-4 (Jullien et al., 2006a) but unchanged expression in dme-1 mutant flowers (Choi et al., 2002). Analysis of FIS2 expression in a dme-1 background after fertilization has not been reported, so it remains to be tested whether there is also an indirect effect of dme-1 on FIS2 during seed development. Taken together, our evidence for MPC, and previous work on FIS2, indicate that DME is not the only positive regulator of the maternal alleles of imprinted genes in Arabidopsis.
METHODS
Plant Stocks
We obtained Col-0, Ler, C24, and met1-9 (Salk_076522) seeds from the Nottingham Arabidopsis Stock Centre; met 1-6 and dme-1 from Robert Fischer (University of California, Berkeley, CA); and dme-4 from Fred Berger (Temasek Lifescience Laboratory, National University of Singapore).
Allele-Specific Sequencing
Reciprocal crosses between Col and Ler were made and siliques harvested at 3 and 5 DAP. Total RNA was extracted from whole siliques using an RNeasy mini kit (Qiagen) following the manufacturer's instructions. First-strand cDNA was synthesized using the ImProm-II reverse transcription system (Promega) using 1 μg of total RNA along with anchored oligo(dT) and random hexamer primers at 47°C for 80 min. Five microliters of cDNA was used in the subsequent PCR with proofreading KOD Hi Fi Polymerase (Toyobo-Novagen) and MPC primers At3g19350_intronspan-F (5′-AAGAACAACGAGATTTGATTGGTG-3′) and At3g19350_ler_capsR (5′-AACATTTCAAAGTTTAGCGGGG-3′). Typical cycling conditions were 95°C for 3 min 30 s followed by 30 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. PCR products were separated on a 2% agarose gel and the 270-bp MPC band was eluted using a gel extraction kit (Qiagen) following the manufacturer's instructions. PCR products were sequenced by Lark Technologies, and the sequence was analyzed using Chromas software (Technelysium). The same protocol was followed for sequencing αVPE and FWA controls. Primer pairs used were αVPE-F (5′-ATCGTCATCAGGCGGATGTTTG-3′) and αVPE-R (5′-ATTTGCCTCATCTGAATCCCTGC-3′), and FWA rt f and FWA dNhe1 as previously described (Kinoshita et al., 2004).
For allele-specific sequencing in a met1 background, siliques were collected at 5 DAP from the crosses met1-9 (Col) × Ler and Ler × met1-9 (Col). RNA extraction and sequencing of MPC were performed as described above.
RT-PCR Analysis
Plant material for expression analysis of endogenous MPC was collected as follows. Col-0 seeds were germinated on Murashige and Skoog agar with 1% sucrose, and seedlings were harvested after 3 weeks. Roots were harvested from 5-week-old plants. Other material harvested was 10 cm of stem from the apex of primary and secondary inflorescences; rosette and cauline leaves; late buds (stages 8 to 12); open flowers (stage 13) (floral stages as in Smyth et al., 1990); open flowers with pistils removed; stamens from stage 14 open flowers; siliques at 3, 5, and 7 DAP; seeds dissected into embryo and endosperm fractions at 7 DAP. For experiments in dme mutant backgrounds, buds were harvested at stages 8 to 12 and open flowers at stage 13.
Total RNA was isolated using an RNeasy plant kit (Qiagen) and subjected to an on-column DNase treatment and a further in-solution DNase treatment. Extracted RNA was further purified and concentrated using an RNeasy MinElute cleanup kit (Qiagen) following manufacturer's instructions. First-strand cDNA was synthesized with Superscript III reverse transcriptase (Invitrogen) using 1 μg of total RNA and oligo(dT) and random primers following the manufacturer's guidelines at 47°C for 60 min. Subsequent PCR was performed as described above but using 22 cycles for GapC, 28 cycles for FWA, and 30 cycles for MPC. Primers for GapC were GApC-F (5′-CACTTGAAGGGTGGTGCCAAG-3′) and GapC-R (5′-CCTGTTGTCGCCAACGAAGTC-3′).
Allele-Specific RT-PCR Analysis of Embryo and Endosperm+Seed Coat Fractions
Reciprocal crosses were performed between the Col and Ler accessions. Seeds were dissected into the embryo and endosperm+seed coat fractions at 7 DAP under a binocular microscope. Total RNA was extracted from each sample using an RNeasy plant mini kit (Qiagen). First-strand cDNA was synthesized from 5 μL of the total RNA with the gene-specific primer MPC-RTr (5′-ACATTTCAAAGTTTAGCGGGGAGCATGGAA-3′) using PrimeScript Reverse Transcriptase (Takara) according to the manufacturer's instructions. The reverse transcribed cDNA was amplified with primers MPC RTf (5′-AAAAGTTGCCCGTGGTGATTCA-3′) and MPC-RTr. The primers MPC RTf2 (5′-TGGAAGAACTTGAGCCACAGT-3′) and MPC-RTr were used for second-round PCR. The amplified PCR fragments were purified and digested with the AluI restriction enzyme. PCR conditions and allele-specific RT-PCR for the αVPE and FWA genes were performed as previously described (Kinoshita et al., 2004).
Quantitative Real-Time PCR
Siliques were collected at 3 DAP from MPC RNAi lines and Col plants. Floral buds (stages 8 to 12), open flowers (stage 13), and siliques at 3 DAF were collected from Ler, homozygous dme-1 (Ler), C24, and homozygous dme-4 (C24) plants. Four biological samples were collected for each dme mutant and wild-type control, and three technical replicates were performed for each sample. Total RNA isolation and cDNA synthesis was similar to that described for RT-PCR. cDNA was diluted 1:10 and 4 μL used for the qRT-PCR. Real-time PCR was done on a DNA Engine mini Opticon 2 (Bio-Rad) using Platinum SYBR green UDG supermix (Invitrogen). Typical cycling conditions were 95°C for 8 min followed by 44 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s; plate read after each cycle; melting curve 38 to 98°C read every 0.5°C; hold for 6 s. Primer pairs used were as follows: At4g25530_Qrt-F (5′-TTATGGACACAGGCAAATGGG-3′) and At4g25530_Qrt-R (5′-TTCTGCTTGAATCTGTTGGAGTG-3′), At3g19350_intronspan-F (5′-AAGAACAACGAGATTTGATTGGTG-3′) and At3g19350_ler_capsR (5′-AACATTTCAAAGTTTAGCGGGG-3′), At3g04120-Qrt-F (5′-GATTCTACAATGGCTGACAAG-3′) and At3g04120-Qrt-R (5′-ACACCAGTAGACTCAACAACG-3′). Calculations were made using the equation 2^-dCT normalized to GapC expression, and then the log2 ratio of expression in dme with relation to wild-type controls was plotted (n was 8 to 12 for each experiment).
Cloning and Transformation
A fragment extending from a 1.3-kb region upstream of the At3g19350 (MPC) start codon to the start codon (5′ flanking region) and a 1.7-kb region including the two exons and intron of MPC but excluding the stop codon (gene body) was amplified using KOD Hi Fi Polymerase (Toyobo-Novagen). For the transcriptional GFP reporter, the 5′ flanking region was fused to NLS-GFP in the vector pBI-GFP(S65T) (gift of Robert Fischer). For the translational GFP reporter, the gene body was placed in-frame with NLS-GFP in the vector containing the 5′ flanking region. For the translational GUS reporter, the 5′ flanking region and gene body were amplified and cloned upstream of the uidA (GUS) reporter gene in the vector BJ60 (gift of Bart Janssen, Horticulture and Food Research Institute, Auckland, New Zealand). The promoter∷MPC-GUS cassette was lifted out of BJ60 with a NotI digest and cloned into the NotI site of the binary vector BJ40 (gift of Bart Janssen).
The binary vectors containing the GUS and GFP expression cassettes were transformed into Agrobacterium tumefaciens strain GV3101. Col plants were transformed following the protocol of Clough and Bent (1998). Transformants were selected on Murashige and Skoog medium containing 50 μg/mL kanamycin.
Primers used for cloning are as follows (linkers containing appropriate restriction sites are in lower case): ProMPC-MPC-GUS, at3g19350_prom-sal1-F (5′-tttgtcgacAATGCTTTTTTCTATTTCTTGC-3′) and at3g19350_prom-trans-kpn1-R (5′-ataggtaccTTTTCCCCTAGCAATTTTC-3′); ProMPC-GFP, at3g19350_prom-sal1-F (5′-tttgtcgacAATGCTTTTTTCTATTTCTTGC-3′) and at3g19350_promdmeGFP-xba1-R (5′-atatctagaGACTTAAGCAATGAAGACGACG-3′); ProMPC-MPC-GFP, at3g19350_prom-sal1-F (5′-tttgtcgacAATGCTTTTTTCTATTTCTTGC-3′) and at3g19350-promdmeGFPtrans-xba-R (5′-atatctagaTTTTCCCCTAGCAATTTTC-3′).
For the RNAi line, a largely unique 133-bp region of MPC was amplified using primers at3g19350-asc1-xba1-F (5′-ttttctagaggcgcgccATGAATCGTTTATGCTTTTTTAAC-3′) and at3g19350-130bp-swa1-bamh1-R (5′-aaaggatccatttaaatCTTCCACCATAAAGAACAAAG-3′) and cloned into pFGC5941 using a two-step cloning method as described on the ChromDB Chromatin Database (http://www.chromdb.org/rnai/vector_info.html). Col plants were transformed as described by Clough and Bent (1998) and transformants selected using 427.5 μL/L BASTA solution (Agro EVO-challenge). Ten independent insertion lines were analyzed.
Confocal Microscopy
Seeds were dissected from siliques, mounted in 25% glycerol, and imaged with an argon ion laser at 488-nm excitation using a Nikon C1 confocal microscope system with a 90i Eclipse microscope and EZ-C1 software. Signal from GFP was collected at 515/530-nm emission and autofluorescence from chlorophyll at ≥650 nm.
Light Microscopy
For GUS staining, whole seedlings 19 d after germination carrying the proMPC-MPC-GUS translational reporter were placed into staining solution (10 mM Tris, pH 7.2, 50 mM NaCl buffer, 1 mg/mL 5-bromo, 4-chloro, 3-indolyl-β-d-glucuronide sodium salt [Melford Laboratories], and 0.1% Triton X-100 [Sigma-Aldrich]), fully submerged, and incubated for 20 h at 37°C. Seedlings were transferred to 70% ethanol for 10 h at room temperature and then 14 h at 4°C. Leaves were mounted on glass slides for observation with a Nikon SMZ1000 microscope. Images were captured with a Nikon Digital Sight DS-U1 camera. To clear seeds for microscopy, seeds were dissected from siliques, mounted in clearing solution (chloral hydrate:water:glycerol, 8w:3v:1v), and imaged using differential contrast optics on a Nikon eclipse 90i microscope and photographed with a Nikon Digital Sight DS-U1 camera.
Two-Hybrid Interaction Assays between MPC and PAM2 Domains
Sequences encoding At3g19350 (MPC) were amplified by PCR from the Salk pUni Clone U84526 using the oligonucleotide primer pairs 5′-GGAATTCAATCGTTTATGCTTTTTTAA-3′ and 5′-GCTGCAGCTAACATCTTTTTCCCCTAG-3′. The product was digested with EcoRI and PstI, purified, and ligated into the pGBKT7 plasmid that carries the coding sequence for the DNA binding domain of Gal4p (Clontech). PAM2-like clones carrying PAM2-like sequences were generated in previously reported work (Bravo et al., 2005). The GAL4-based yeast two-hybrid system was used as described by the supplier (Clontech). The AH109 yeast strain (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2∷ GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3∷ MEL1UAS-MEL1 TATA-lacZ) was cotransformed with the MPC clone and the PAM2-like plasmids. Transformants were selected on SC medium lacking Trp and Leu, purified in the same medium, and then streaked in SC-Trp,-Leu medium and SC-Trp,-Leu,-His medium supplemented with 5 mM 3-amino-1,2,4-triazole; plates were incubated for 3 to 4 d at 30°C.
McrBC Assay
Genomic DNA was extracted from rosette leaves of wild-type Col and met1-6 plants. Genomic DNA (100 ng) was digested with 20 units of McrBC (New England Biolabs) for 12 h at 37°C. Two nanograms of undigested and digested DNA were used as a template in PCR reactions with 34 cycles of amplification. Primer sequences used in PCR reactions are as follows: region −1201 to −658, YI-27(MPC1f) (5′-GTCTAATCTTCTGGTTGATCCTCC-3′) and YI-28(MPC2r) (5′-TCACCAAGTCAACATTCTCTGTATCT-3′); region −720 to −303, YI-29(MPC3f) (5′-TTTGTGCAAAAGATTTCTTACCTTAC-3′) and YI-30(MPC4r) (5′-CGTATGTTGAACAATGCAAAATTT-3′); region −372 to +45, YI-31(MPC5f) (5′-GTTATTTGTAGATATACAAAACAAATGA-3′) and YI-32(MPC6r) (5′-ACGGGCAACTTTTACAACTTTGTT-3′); region −25 to +426, YI-33(MPC7f) (5′-TATCGTCGTCTTCATTGCTTAAGTC-3′) and YI-34(MPC8r) (5′-GAGCATGGAAGGTAGGAACTTG-3′); region +382 to +635, YI-35(MPC9f) (5′-TTGCTAGGGGAAAAAGATGTTAG-3′) and YI-36(MPC10r) (5′-CTAAAACATCAGTTATAACCACTCTAG-3′).
Bisulfite Sequencing
Genomic DNA was extracted from rosette leaves of wild-type Col and met1-9 homozygous plants. Bisulfite sequencing was performed using an Epitect bisulfite kit (Qiagen) according to the manufacturer's instructions. Primer sequences used in PCR reactions were as follows: region 1, −355 to +24: YI-40(MPC-BS1f) (5′-TAAGAGTTAGATAATTTTGTTATTTGTAGATATA-3′) and YI-42(MPC-BS3r) (5′-RRARRTRAATCACCACRRRCAACTTTTACAACTTT-3′); region 2, +41 to 463: YI-45(MPC-BS6f) (5′-ATTGYTTAAGTYATGAATYGTTTATGYTTTTTTAA-3′) and YI-47(MPC-BS8r) (5′-TAAAACTCAACTTAAARCCACATTTTRTTRCTC-3′). The ASA1 gene was used as a positive control for the bisulfite chemical reaction (Jeddeloh et al., 1998). The lollipop diagram of the bisulfite sequencing results was generated using CpG Viewer (Carr et al., 2007).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At3g19350 (MPC), At4g25530 (FWA), At2g25940 (αVPE), At2g41430 (CID1/ERD15), At2g26280 (CID7), At1g53650 (CID8), At4g10610 (CID12), At4g34110 (PAB2), At1g22760 (PAB3), At2g23350 (PAB4), At1g71770 (PAB5), At2g36660 (PAB7), At1g49760 (PAB8), At1g19920 (ASA1), and NP_002559.2 (human PABPC1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of the MPC Coding Sequence with the C-Terminal Regions of Arabidopsis PABs Showing the Sequence Used for the RNAi Construct.
Supplemental Figure 2. qRT-PCR Assay Comparing MPC Expression in Wild-Type Col and MPC RNAi Backgrounds.
Supplemental Figure 3. Paternal proMPC-MPC0-GFP Is Ectopically Expressed When Transmitted by a met1-9 Mutant Pollen Parent.
Supplemental Table 1. Percentage of Methylation at CG, CNG, and Asymmetric Sites in Genomic Regions of MPC Analyzed by Bisulfite Sequencing.
Supplemental Table 2. Expression of MPC and FWA in dme-1 and dme-4 Backgrounds Analyzed by qRT-PCR.
Supplemental Data Set 1. Text File Corresponding to Alignment in Figure 3A.
Supplemental Data Set 2. Text File Corresponding to Alignment in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Nikon UK for supporting the Nikon and University of Bath Imaging Suite; the Biotechnology and Biological Science Research Council (UK) for funding S.T., R.S., L.D., L.M., and M.S.; and Laura Aguilar-Henonin and Nancy Mendoza for technical support.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Rod J. Scott (bssrjs@bath.ac.uk).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Belosotsky, D.A. (2003). Unexpected complexity of poly(A)-binding protein gene families in flowering plants: Three conserved lineages that are at least 200 million years old and possible auto- and cross-regulation. Genetics 163 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanga, J.J., Baass, A., and Sonenberg, N. (2006). Regulation of poly(A) binding protein function in translation: Characterization of the Paip2 homolog, Paip2B. RNA 12 1556–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo, J., Aguilar-Henonin, L., Olmedo, G., and Guzmán, P. (2005). Four distinct classes of proteins as interaction partners of the PABC domain of Arabidopsis thaliana poly(A)-binding proteins. Mol. Genet. Genomics 272 651–665. [DOI] [PubMed] [Google Scholar]
- Carr, I.M., Valleley, E.M.A., Cordery, S.F., Markham, A.F., and Bonthron, D.T. (2007). Sequence analysis and editing for bisulphite genomic sequencing projects. Nucleic Acids Res. 35 e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S.W.L., Henderson, I.R., and Jacobsen, S.E. (2005). Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6 351–360. [DOI] [PubMed] [Google Scholar]
- Choi, Y., Gehring, M., Johnson, L., Hannon, M., Harada, J.J., Goldberg, R.B., Jacobsen, S.E., and Fischer, R.L. (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110 33–42. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Craig, A.W.B., Haghighat, A., Yu, A.T.K., and Sonenberg, N. (1998). Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature 392 520–523. [DOI] [PubMed] [Google Scholar]
- Finnegan, E.J., and Dennis, E. (1993). Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 21 2383–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring, M., Choi, Y., and Fischer, R.L. (2004). Imprinting and seed development. Plant Cell 16 S203–S213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring, M., Huh, J.H., Hsieh, T.-F., Penterman, J., Choi, Y., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (2006). DEMETER DNA glycosylase establishes MEDEA Polycomb gene self-imprinting by allele-specific demethylation. Cell 124 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.-P., Hoeppner, M.A., and Gagliano, W. (1998). Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science 280 446–450. [DOI] [PubMed] [Google Scholar]
- Guitton, A.-E., Page, D.R., Chambrier, P., Lionnet, C., Faure, J.-E., Grossniklaus, U., and Berger, F. (2004). Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in control of seed development in Arabidopsis thaliana. Development 131 2971–2981. [DOI] [PubMed] [Google Scholar]
- Haig, D., and Westoby, M. (1991). Genomic imprinting in endosperm: Its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 333 1–13. [Google Scholar]
- Huh, J.H., Bauer, M.J., Hsieh, T.-F., and Fischer, R.L. (2008). Cellular programming of plant gene imprinting. Cell 132 735–744. [DOI] [PubMed] [Google Scholar]
- Jeddeloh, J.A., Bender, J., and Richard, E.J. (1998). The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev. 12 1714–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien, P.E., Katz, A., Oliva, M., Ohad, N., and Berger, F. (2006. b). Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr. Biol. 16 486–492. [DOI] [PubMed] [Google Scholar]
- Jullien, P.E., Kinoshita, T., Ohad, N., and Berger, F. (2006. a). Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18 1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel, M.W., Ramsey, D.E., Stokes, T.L., Flowers, S.K., Haag, J.R., Jeddeloh, H.A., Riddle, N.C., Verbsky, M.L., and Richard, E.J. (2003). Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163 1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariola, T., Grader, G., Helenius, E., Li, J., Heino, P., and Palva, E.T. (2006). EARLY RESPONSE TO DEHYDRATION 15, a negative regulator of abscisic acid responses in Arabidopsis. Plant Physiol. 42 1559–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour, K., Kahvejian, A., De Crescenzo, G., Roy, G., Svitkin, Y.V., Imataka, H., O'Connor-McCourt, M., and Sonenberg, N. (2001. b). Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol. Cell. Biol. 21 5200–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour, K., Svitkin, Y.V., Craig, A.W., DeMaria, C.T., Deo, R.C., Burley, S.K., and Sonenberg, N. (2001. a). Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell 7 205–216. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Miura, A., Choi, Y., Kinoshita, Y., Cao, X., Jacobsen, S.E., Fischer, R.L., and Kakutani, T. (2004). One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303 521–523. [DOI] [PubMed] [Google Scholar]
- Kinoshita, Y., Saze, H., Kinoshita, T., Miura, A., Soppe, W.J.J., Koornneef, M., and Kakutani, T. (2007). Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J. 49 38–45. [DOI] [PubMed] [Google Scholar]
- Kiyosue, T., Yamaguchi-Shimozaki, K., and Shinozaki, K. (1994). Cloning of cDNAs for genes that are early-responsive to dehydration stress (ERDs) in Arabidopsis thaliana L.: Identification of three ERDs as HSP cognate genes. Plant Mol. Biol. 25 791–798. [DOI] [PubMed] [Google Scholar]
- Köhler, C., Hennig, L., Spillane, C., Pien, S., Gruissem, W., and Grossniklaus, U. (2003). The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler, C., Page, D.R., Gagliardini, V., and Grossniklaus, U. (2005). The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat. Genet. 37 28–30. [DOI] [PubMed] [Google Scholar]
- Kozlov, G., Trempe, J.F., Khaleghpour, K., Kahvejian, A., Ekiel, I., and Gehring, K. (2001). Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl. Acad. Sci. USA 8 4409–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn, U., and Wahle, E. (2004). Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta 1678 67–84. [DOI] [PubMed] [Google Scholar]
- Lukianova, O.A., and David, S.S. (2005). A role for iron–sulfur clusters in DNA repair. Curr. Opin. Chem. Biol. 9 145–151. [DOI] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Dennis, E.S., Peacock, W.J., and Chaudhury, A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevich, G., Villar, C.B.R., Eriova, A., and Kohler, C. (2008). Mechanism of PHERES1 imprinting in Arabidopsis. J. Cell Sci. 121 906–912. [DOI] [PubMed] [Google Scholar]
- Mangus, D.A., Evans, M.C., and Jacobsen, A. (2003). Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Ruiz, T., Ortega-Galisteo, A.P., Ponferrada-Marín, M.I., Martínez-Macías, M.I., Ariza, R.R., and Roldán-Arjona, T. (2006). DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl. Acad. Sci. USA 103 6853–6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portereiko, M.F., Lloyd, A., Steffen, J.G., Punwani, J.A., Otsuga, D., and Drews, G.N. (2006). AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18 1862–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, G., De Crescenzo, G., Khaleghpour, K., Kahvejian, A., O'Connor-McCourt, M., and Sonenberg, N. (2002). Paip1 interacts with Poly(A) binding protein through two independent binding motifs. Mol. Cell. Biol. 22 3769–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, R.J., Spielman, M., Bailey, J., and Dickinson, H.G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125 3329–3341. [DOI] [PubMed] [Google Scholar]
- Scott, R.J., and Spielman, M. (2004). Imprinting in plants and mammals—The same but different? Curr. Biol. 14 R201–R203. [DOI] [PubMed] [Google Scholar]
- Schwab, B., Folkers, U., Ilgenfritz, H., and Hülskamp, M. (2000). Trichome morphogenesis in Arabidopsis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355 879–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe, W.J.J., Jacobsen, S.E., Alonso-Blanco, C., Jackson, J.P., Kakutani, T., Koornneef, M., and Peeters, A.J.M. (2000). The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol. Cell 6 791–802. [DOI] [PubMed] [Google Scholar]
- Spielman, M., Vinkenoog, R., and Scott, R.J. (2001). The epigenetic basis of gender in flowering plants and mammals. Trends Genet. 17 705–711. [DOI] [PubMed] [Google Scholar]
- Tsumoto, Y., Yoshizumi, T., Kuroda, H., Kawashima, M., Ichikawa, T., Nakazawa, M., Yamamoto, N., and Matsui, M. (2006). Light-dependent polyploidy control by a CUE protein variant in Arabidopsis. Plant Mol. Biol. 61 817–828. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada, J.P., Thomas, J., Spillane, C., Coluccio, A., Hoeppner, M.A., and Grossniklaus, U. (1999). Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 13 2971–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., and Grumet, R. (2004). Identification and characterization of proteins that interact with the carboxy terminus of poly(A)-binding protein and inhibit translation in vitro. Plant Mol. Biol. 54 85–98. [DOI] [PubMed] [Google Scholar]
- Wood, A.J., and Oakey, R.J. (2006). Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet. 2 1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, W., Gehring, M., Choi, Y., Margossian, L., Pu, H., Harada, J.J., Goldberg, R.B., Pennell, R.I., and Fischer, R.L. (2003). Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev. Cell 5 891–901. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Clarenz, O., Cokus, S., Bernatavichute, Y.V., Pellegrini, M., Goodrich, J., and Jacobsen, S.E. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5 e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D., Gehring, M., Tran, R.K., Ballinger, T., and Henikoff, S. (2007). Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39 61–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.