Abstract
The rice (Oryza sativa) DELLA protein SLR1 acts as a repressor of gibberellin (GA) signaling. GA perception by GID1 causes SLR1 protein degradation involving the F-box protein GID2; this triggers GA-associated responses such as shoot elongation and seed germination. In GA-insensitive and GA biosynthesis mutants, SLENDER RICE1 (SLR1) accumulates to high levels, and the severity of dwarfism is usually correlated with the level of SLR1 accumulation. An exception is the GA-insensitive F-box mutant gid2, which shows milder dwarfism than mutants such as gid1 and cps even though it accumulates higher levels of SLR1. The level of SLR1 protein in gid2 was decreased by loss of GID1 function or treatment with a GA biosynthesis inhibitor, and dwarfism was enhanced. Conversely, overproduction of GID1 or treatment with GA3 increased the SLR1 level in gid2 and reduced dwarfism. These results indicate that derepression of SLR1 repressive activity can be accomplished by GA and GID1 alone and does not require F-box (GID2) function. Evidence for GA signaling without GID2 was also provided by the expression behavior of GA-regulated genes such as GA-20oxidase1, GID1, and SLR1 in the gid2 mutant. Based on these observations, we propose a model for the release of GA suppression that does not require DELLA protein degradation.
INTRODUCTION
Gibberellins (GAs) are a large family of tetracyclic diterpenoid plant hormones that induce a wide range of plant growth responses, including seed germination, stem elongation, leaf expansion, flowering, and pollen maturation (Richards et al., 2001; Thomas et al., 2005). The recent discovery of three important GA signaling factors through genetic studies using rice (Oryza sativa) and Arabidopsis thaliana mutants has led to advances in our understanding of GA signal transduction. The first factor to be isolated, DELLA protein, acts as a repressor of GA signaling (Peng et al., 1997, 1999; Silverstone et al., 1998; Ikeda et al., 2001; Itoh et al., 2002; Chandler et al., 2002). Rice has one DELLA protein, SLENDER RICE1 (SLR1), whereas Arabidopsis has five (Repressor of ga1-3 [RGA], GA-INSENSITIVE [GAI], RGA-LIKE1 [RGL1], RGL2, and RGL3; Sun et al., 2004). The second factor to be identified, an F-box protein, is a subunit of SCF E3 ubiquitin ligase involved in degradation of DELLA protein: this factor is referred to as GA INSENSITIVE2 (GID2) in rice (Sasaki et al., 2003) and SLEEPY1 (SLY1) in Arabidopsis (McGinnis et al., 2003). The third factor is a GA receptor, GID1, which specifically interacts with active GAs such as GA4 and GA1 in vitro with reasonable affinity; its loss-of-function mutant in rice shows a severe GA-insensitive dwarf phenotype (Ueguchi-Tanaka et al., 2005). While rice possesses only a single GID1 gene, there are three Arabidopsis genes homologous to rice GID1. Nakajima et al. (2006) confirmed that the products of these Arabidopsis genes also interact with active GAs, and Griffiths et al. (2006), Willige et al. (2007), and Iuchi et al. (2007) showed that the Arabidopsis triple mutant gid1a gid1b gid1c results in a severe GA-insensitive dwarf phenotype. Based on the results of functional and biochemical analyses of these three GA signaling factors, the following molecular mechanism of GA perception has been proposed (Ueguchi-Tanaka et al., 2007b; Hirano et al., 2008). In the absence of GA, DELLA protein represses various GA responses in planta. When GA is present, the GA receptor GID1 binds to GA, triggering an interaction between GID1 and DELLA protein. DELLA protein is then degraded through the 26S-proteasome pathway, with the aid of the SCFGID2/SLY1 complex, resulting in various GA-dependent responses.
In this GA perception model, DELLA protein degradation by GA in collaboration with GID1 and F-box protein is considered to be a key event in GA signaling. This idea is supported by observations in rice and Arabidopsis that DELLA protein accumulates to abnormally high levels in GA-deficient and GA-insensitive mutants (Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006; Willige et al., 2007; Iuchi et al., 2007). In addition, there is a positive correlation between the accumulation of the rice DELLA protein SLR1 and the severity of dwarfism in rice GA-related mutants. This suggests that SLR1 negatively regulates GA responses in a quantitative manner. However, there is an exception in which dwarf severity and SLR1 level are not correlated. The level of SLR1 protein in the gid2 mutants is much higher than that in the most severe rice GA mutants, such as gid1 and cps (a loss-of-function mutant in ent-copalyl diphosphate synthase; Sakamoto et al., 2004), although dwarfism of gid2 is much milder than these mutants. Griffiths et al. (2006) also reported that the accumulation level of Arabidopsis DELLA protein RGA is higher in the sly1 mutant than in the gid1a gid1b gid1c triple mutant or in ga1-3, the most severe GA-deficient mutant in Arabidopsis. To explain this, Griffiths et al. (2006) proposed that there might be a basal level of RGA degradation mediated by SCFSLY1 but independent of GA and GID1, leading to higher accumulation of RGA in sly1 than in ga1-3. However, this hypothesis cannot explain why sly1 shows a milder dwarf phenotype than ga1-3 even though DELLA protein accumulates to higher levels in sly1. Consequently, the molecular mechanism of high-level accumulation of DELLA protein in F-box mutants with mild phenotypes (e.g., rice gid2 and Arabidopsis sly1) is one of the remaining questions in GA signaling.
In this study, we revealed that both the high-level accumulation of SLR1 and its decreased repressive function in gid2 depend on the presence of GA and GID1. Taking into account the previous observation that SLR1 protein interacts with GID1 in a GA-dependent manner (Nakajima et al., 2006; Ueguchi-Tanaka et al., 2007a), we propose that SLR1 loses its repressive function when it forms a complex with GA and GID1.
RESULTS
GA Is Essential for Abnormal Accumulation of SLR1 Protein in gid2
We first investigated the correlation between the amount of SLR1 and the severity of dwarfism among rice GA-related mutants (Figure 1; see Supplemental Table 1 online). The GA-deficient and GA-insensitive mutants we examined show varying severity in dwarfism. The amounts of phosphorylated (SLR1-P) and nonphosphorylated (SLR1) SLR1 proteins (Itoh et al., 2005) were usually correlated with the severity of dwarfism, indicating that GA action is negatively regulated by SLR1 in a quantitative manner in these mutants. There was one exception to this rule: the gid2 mutant alleles gid2-1, -2, and -5 accumulated the highest level of both phosphorylated and nonphosphorylated forms of SLR1 of all the mutants examined, whereas the severities of dwarfism were milder than those of mutants such as cps1-1 and gid1-3. This strongly suggests that the suppressive activity of SLR1 accumulating in gid2 is weaker than in other GA-related mutants.
Figure 1.
Endogenous SLR1 Protein Levels Correlate with the Severity of Dwarfism in GA-Deficient and GA-Insensitive Mutants, with the Exception of gid2.
(A) Gross morphology of GA-deficient mutants (left panel) and various alleles of gid1 and gid2 mutants (right panel) grown for 2 weeks. The wild type is shown as a control. Closed and open arrowheads represent the uppermost positions of the 2nd and 3rd leaf sheaths, respectively. Bars = 5 cm.
(B) Top panel: SLR1 protein in the plant lines from (A) detected by protein gel blot analysis. SLR1-P, phosphorylated SLR1; SLR1, nonphosphorylated SLR1. Bottom panel: Coomassie blue (CBB) staining to show that approximately equal amounts of total protein (10 μg) were loaded in each lane.
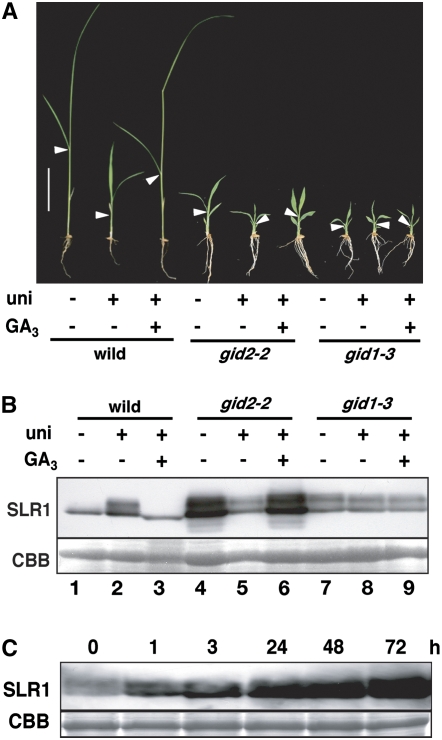
We next investigated the effect of GA on dwarfism and on the level of SLR1 protein. Since gid1 and gid2 mutants accumulate endogenous GA (Sasaki et al., 2003; Ueguchi-Tanaka et al., 2005), we pretreated plants with uniconazole (an inhibitor of GA biosynthesis) to reduce the endogenous GA level and then treated them with GA3 (Figure 2; see Supplemental Table 2 online). In the wild type, the amount of SLR1 protein was increased by uniconazole treatment; this increase was reversed by subsequent treatment with GA3 (Figure 2B, lanes 1 to 3). This confirms previous observations that a decrease in the endogenous GA level induces accumulation of the SLR1 protein and, conversely, that presence of GA leads to SLR1 degradation (Itoh et al., 2001). In untreated gid2, the level of SLR1 was much higher than in the wild type (Figure 2B, compare lanes 4 and 1). However, the level of SLR1 was decreased following uniconazole treatment and restored by later addition of GA3 (Figure 2B, lanes 4 to 6). In gid1-3, these treatments did not significantly change the amount of SLR1 (Figure 2B, lanes 7 to 9). It is noteworthy that the amount of SLR1 protein in gid2-2 plants treated with uniconazole (Figure 2B, lane 5) was almost the same as in uniconazole-treated wild type (Figure 2B, lane 2) and the same as in gid1-3 under any conditions (lanes 7 to 9). This indicates that the SLR1 level in the absence of GA in gid2 is similar to the level in plants that cannot perceive GA (e.g., wild-type plants under GA-deficient condition or gid1 mutant plants).
Figure 2.
Effects of GA on Dwarfism and Levels of SLR1 Protein in the Wild Type, gid2, and gid1.
(A) Gross morphology of the wild type, gid2-2, and gid1-3 under GA-rich or GA-deficient conditions. The seedlings were grown with (uni +, GA3 −) or without (uni −, GA3 −) 10−6 M uniconazole (an inhibitor of GA synthesis) for 3 weeks, or the seedlings were grown with 10−6 M uniconazole for 2 weeks and then treated with GA3 for 1 more week (uni +, GA3 +). Closed arrowheads represent the uppermost positions of the 4th leaf sheath. Bar = 5 cm.
(B) Top panel: protein gel blot showing the level of SLR1 protein in seedlings grown under the conditions presented in (A). Bottom panel: Coomassie blue (CBB) staining to show approximately equal amounts of total protein (10 μg) were loaded in each lane.
(C) Time course of GA-dependent accumulation of SLR1 in gid2-2. The seedlings were pretreated with 10−6 M uniconazole for 2 weeks and then treated with GA3 for the indicated period.
To estimate the repressive activity of these SLR1 proteins, we compared the heights of several rice genotypes under various GA levels. When gid2-2 was treated with uniconazole, plant height was reduced; subsequent treatment with GA3 restored plant height (Figure 2A; see Supplemental Table 2 online). On the other hand, the plant height of gid1-3 was not affected by either treatment. By comparing the degree of dwarfism to the level of SLR1, we concluded that the suppressive activity of SLR1 in uniconazole-treated gid2-2 (i.e., under GA-deficient conditions) is much higher than that of untreated or uniconazole/GA3 treated gid2-2 and similar to that in gid1. We then examined the time course of GA-dependent accumulation of SLR1 in gid2-2 (Figure 2C). For this experiment, gid2-2 plants were first pretreated with uniconazole for 2 weeks, treated with GA3, and then sampled at each indicated time point. The amount of SLR1 protein gradually increased during the 3 d following treatment with GA3. This indicates that SLR1 proteins accumulate over time in a GA-dependent manner in the absence of the GID2-mediated proteolysis, while the degradation of SLR1 protein proceeds within 30 min after treatment with GAs (Ueguchi-Tanaka et al., 2005).
The necessity of GA for high-level SLR1 accumulation in gid2 was genetically confirmed by crossing gid2-2 with a GA-deficient mutant, cps1-1 (Figure 3). CPS encodes ent-copalyl diphosphate synthase, which catalyzes the first step of GA biosynthesis. As previously mentioned, accumulation of SLR1 in cps1-1 was much lower than in gid2-2, although cps1-1 shows more severe dwarfism than gid2-2 (Figure 3; see Supplemental Table 3 online). The gid2-2 cps1-1 double mutant was similar to the cps1-1 single mutant in terms of both height and SLR1 protein level. This observation confirms that GA is essential for higher accumulation of SLR1 in the gid2 mutant but that the accumulated SLR1 in gid2 does not effectively function as a repressor to GA signaling.
Figure 3.
Dwarfism and Accumulation of SLR1 Protein in gid2, cps1, and gid2 cps1.
(A) Gross morphology of the wild type, gid2-2, cps1-1, and the gid2-2 cps1-1 double mutant grown for 4 weeks. Closed and open arrowheads represent the uppermost positions of the 4th and 6th leaf sheath, respectively. Bar = 5 cm. Inset: close-up of the three mutants (bar = 1 cm).
(B) Top panel: protein gel blot showing the level of SLR1 protein accumulated in the seedlings shown in (A). SLR1-P, phosphorylated SLR1; SLR1, nonphosphorylated SLR1. Bottom panel, Coomassie blue (CBB) staining to show that approximately equal amounts of total protein (10 μg) were loaded in each lane.
GID1 Is Essential for Abnormal Accumulation of SLR1 Protein in gid2-2
The above results suggest that GA perception by GID1 is important for high-level accumulation of SLR1 and its lower repression activity in gid2. To examine this hypothesis, we produced double mutants of gid1-3 and gid2-2 by crossing plants heterozygous for each of the single mutants. The genotypes of GID1 and GID2 loci in the F2 segregated seedlings were determined by PCR analysis. As we expected, the dwarf severity of the gid1-3 gid2-2 double mutant was similar to the gid1-3 single mutant and much more severe than that of gid2-2 single mutant (Figure 4A; see Supplemental Table 4 online). We noticed that among the homozygous gid2 plants, there were three different phenotypes: plants showing exactly the same dwarfism as gid2, those showing more severe dwarfing than gid2 but milder than gid1, and same as gid1 (gid1-3 gid2-2 double mutant). Genotyping of the GID1 locus of the two former plants revealed that plants carrying heterozygous alleles of GID1 (+/−) show more severe dwarfing than plants homozygous for the wild-type allele of GID1 (+/+). This suggests that the amount of GID1 protein may be related to the repressive activity of SLR1.
Figure 4.
Dwarfism and Accumulation of SLR1 Protein in Plants with GID1 and/or GID2 Mutant Alleles.
(A) Gross morphology of 4-week-old plants carrying wild-type or mutant alleles of GID1 and GID2. Closed and open arrowheads represent the uppermost positions of the 4th and 6th leaf sheaths, respectively. Bar = 10 cm in the left panel and 5 cm in the right panel.
(B) Top panel: Protein gel blot showing the level of SLR1 protein accumulated in the seedlings shown in (A). SLR1-P, phosphorylated SLR1; SLR1, nonphosphorylated SLR1. Bottom panel, Coomassie blue (CBB) staining to show that approximately equal amounts of total protein (10 μg) were loaded in each lane.
We also examined the level of SLR1 protein in these mutants. The amount of SLR1 in the gid1-3 gid2-2 double mutant (Figure 4B, lane 4) was the same as in the gid1-3 single mutant (lane 6) and much less than in gid2-2 (lane 2). Plants containing homozygous mutant alleles of gid2 (−/−) and heterozygous alleles of gid1 (+/−) (lane 3) accumulated a much higher level of SLR1 protein than did gid1-3 gid2-2 double and gid1-3 single mutants (lanes 4 and 6, respectively), but this level was lower than that of the gid2-2 single mutant (lane 2). These observations support the hypothesis that the GID1-mediated GA perception system is essential for high-level accumulation of SLR1 in gid2, and the repressive activity of SLR1 becomes much milder in gid2 when both GA and GID1 are present. Furthermore, these results suggest that these phenomena are dose dependent on the level of GID1.
According to this hypothesis, overproducing GID1 protein in gid2 mutant plants should further increase the amount of SLR1 protein and decrease its suppressive activity. To test this, we introduced a GID1-overproducing transgene into a gid2-2 mutant background. We distinguished the transcript of introduced GID1 cDNA (labeled “trans” in Figure 5A) from that of the endogenous GID1 (labeled “endo”) by the difference in their molecular sizes. As expected, overproduction of GID1 in gid2-2 alleviated the severity of dwarfism of gid2-2 (Figure 5B) and enhanced the accumulation of SLR1 protein relative to the gid2-2 control (Figure 5C).
Figure 5.
Enhanced Accumulation of SLR1 Protein in gid2 Plants Overproducing GID1.
(A) Top panel: RNA gel blot analysis of the endogenous and transformed GID1 transcripts. vec WT, wild type transformed with the empty pActNos vector; GID1-over gid2-2, gid2-2 plant transformed with the GID1 overproducing construct; vec gid2-2, gid2-2 plant transformed with the empty vector; endo, the GID1 transcript derived from the endogenous GID1 gene; trans, the GID1 transcript derived from the introduced GID1 gene. Bottom panel: Ethidium bromide (EtBr) staining to show equal loading.
(B) Gross morphology of transgenic seedlings grown for 2 months after regeneration. Bar = 10 cm.
(C) Protein gel blot showing the level of SLR1 protein in the seedlings shown in (B). Approximately 10 μg (top panel) and 2 μg (middle panel) of total protein was loaded in each lane. SLR1-P, phosphorylated SLR1; SLR1, non-phosphorylated SLR1. Bottom panel, Coomassie blue (CBB) staining of the blot from the top panel to show that approximately equal amounts of total protein (10 μg) were loaded in each lane.
Next, we examined the effect of GA on GID1-overproducing gid2 plants. The gid2 plants overproducing GID1 were treated with or without 10−4M GA3 for 3 weeks. The dwarfism of the control gid2 plants (transformed with the empty vector) was slightly restored by GA3 treatment, while gid2 plants overproducing GID1 responded to GA3 and attained a plant height similar to the wild type (Figure 6). This result indicates that the repressive activity of SLR1 is almost completely suppressed in gid2 under the conditions of excess GA and GID1.
Figure 6.
Rescue of the Dwarf Phenotype of gid2 by Excess GA and GID1.
Gross morphology of transgenic plants grown for 6 weeks with or without GA treatment. Three-week-old regenerated transgenic plants were transferred to soil containing 10−4 M GA3 (+) or solvent (−) and grown for further 3 weeks. The symbols for each plant are identical to those in Figure 5. GA3 − and + of vec gid2-2 were from same transformant line, and GA3 − and + of GID1-ov gid2-2 were also from same transformant line. The same results were obtained from another three independent vec gid2-2 and GID1-ov gid2-2 lines. Bar = 10 cm.
GA Signaling Occurs in the gid2 Mutant
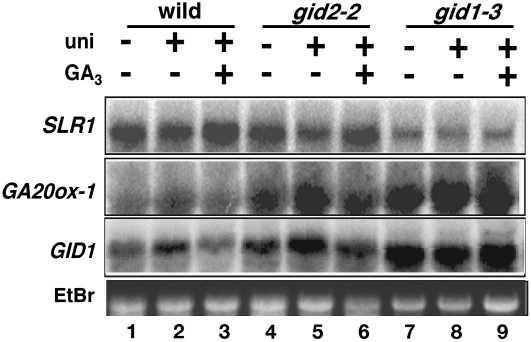
The facts that (1) uniconazole treatment of gid2 enhanced dwarfism and (2) applying GA alleviated this dwarfism suggest that GA signaling works even in gid2. To investigate the possibility of GA signaling in gid2, we next examined the expression of three genes, SLR1, GA20ox-1, and GID1, whose expression is regulated by GA signaling. Expression of GA20ox-1, which encodes an enzyme catalyzing the later steps of the GA synthetic pathway, and GID1 are downregulated by GA via feedback through the GA signaling pathway (Olszewski et al., 2002; Griffiths et al., 2006), whereas SLR1 expression is upregulated by GA (Itoh et al., 2001). Although the default levels of GA20ox-1 and GID1 transcripts were higher in gid2-2 than in the wild type, in these plants, the mRNA levels were increased by uniconazole and diminished by application of GA (lanes 1 to 3 for the wild type and lanes 4 to 6 for gid2-2 in Figure 7). On the other hand, the level of SLR1 mRNA was slightly decreased by uniconazole, and this decrease was reversed by GA application in both the wild type and in gid2-2. These results demonstrate that GA signaling works similarly in wild-type and gid2 plants. In gid1-3, the GA20ox-1 and GID1 mRNAs were maintained at high levels, and the SLR1 mRNA was maintained at a low level under any conditions tested (lanes 7 to 9), indicating that the feedback regulation mediated by GA signaling does not work in this mutant due to the lack of functional GID1 receptor.
Figure 7.
GA Signaling Works Partially in the gid2 Mutant.
Expression of SLR1, GA20ox-1, and GID1 Genes in wild-type, gid2-2, and gid1-3 plants under GA-rich or GA-deficient conditions. Growth conditions of the wild type, gid2-2, and gid1-3 are the same as in Figure 2A. Total RNA (5, 12.5, and 7.5 μg) was loaded to detect SLR1, GID20ox-1, and GID1 transcripts, respectively. EtBr; EtBr control corresponding to the 5-μg samples.
Increased Transcription of SLR1 Is Essential for Its High-Level Accumulation in gid2
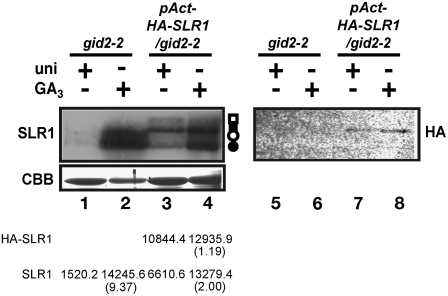
The RNA gel blot anaylsis of SLR1 in gid2 (Figure 7) lead us to hypothesize that the abnormal accumulation of SLR1 protein in gid2 is due to the result of increased transcription of SLR1 mRNA. To confirm this possibility, we generated a transgenic gid2-2 mutant by introduction of HA-SLR1 cDNA under the control of the actin promoter, which produces SLR1 independently from GA or GA signaling, and compared the level of SLR1 protein derived from the introduced and endogenous genes (Figure 8). Besides endogenous SLR1 protein bands with or without phosphorylation (open and closed circles in lanes 1 to 4, respectively), the SLR1 antibody detected two new bands with lower mobility in the transgenic lines (closed and open squares in lanes 3 and 4). The HA antibody recognized the lower band of the two (lanes 7 and 8), indicating that this band was derived from the transgene and probably corresponds to the HA-SLR1 without phosphorylation (closed square in lanes 3 and 4). On the other hand, the new upper band, which was detected by SLR1 antibody, may correspond to HA-SLR1 with phosphorylation (open square in lanes 3 and 4). Though it was not detected by the HA antibody, it was probably because of its lower sensitivity than SLR1 antibody. As previously mentioned, the SLR1 proteins with or without phosphorylation accumulated with GA treatment in both nontransgenic and transgenic gid2-2 mutants (lanes 1 to 4). On the other hand, the band intensity of nonphosphorylated and phosphorylated HA-SLR1 proteins, detected by the SLR1 antibody, was almost the same under the GA-rich and -deficient conditions (closed and open squares in lanes 3 and 4). These results indicate that the increased transcription of SLR1 by its own promoter is essential for GA-dependent accumulation of SLR1 protein in the gid2-2 mutant.
Figure 8.
No Change in the SLR1 Protein Level When SLR1 Is Expressed under the Control of a Constitutive Promoter in gid2-2.
HA-SLR1cDNA was expressed by the actin promoter under GA-rich or -deficient conditions in gid2-2. Left panel: SLR1 protein was detected by protein gel blot analysis with anti-SLR1 antibody. Open circle, phosphorylated SLR1; closed circle, nonphosphorylated SLR1; open square, phosphorylated HA-SLR1; closed square, nonphosphorylated HA-SLR1. Bottom panel: Coomassie blue (CBB) staining to show that approximately equal amounts of total protein (5 μg) were loaded in each lane. Right panel: HA-SLR1 protein was detected with anti-HA antibody. Total amount of protein in each lane was same as left panel. Numbers represent the band intensities (arbitrary units) of SLR1 and HA-SLR1 of each lane. Numbers in parentheses indicate the ratio of the band intensities between uni − GA3 + and uni + GA3 −.
DISCUSSION
A Model for Reversal of DELLA Repression Activity in the Absence of GID2 Function
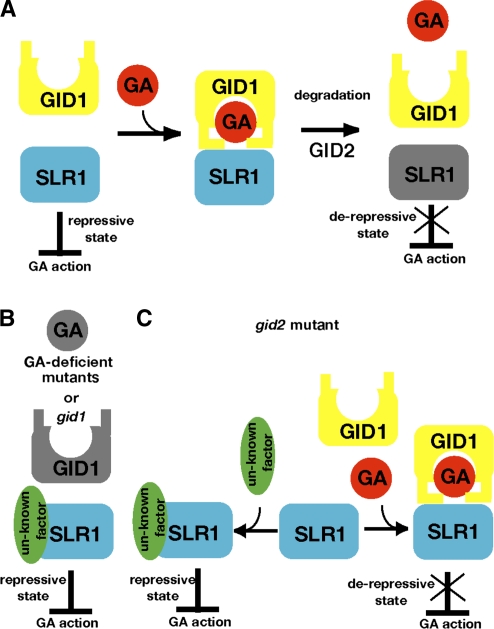
At present, derepression of DELLA protein repressive activity has been considered a key step in GA signaling (Sun and Gubler, 2004; Ueguchi-Tanaka et al., 2007b). According to the current DELLA derepression model, when GA is absent, DELLA protein (SLR1) represses GA actions (repressive state in Figure 9A). When GA is present, the GID1 receptor binds to GA, which allows GID1 to interact with SLR1. SLR1 interacting with GA-GID1 is degraded with the aid of the SCFGID2 complex, through the 26S-proteasome pathway; this degradation releases the repressive state and activates GA actions (derepressed state in Figure 9A). In this model, degradation of DELLA protein mediated by GA, GID1, and GID2 is a critical step for derepressing SLR1's repressive activity. In this study, however, we demonstrated that there is an alternative way to derepress the repressive activity of SLR1 that is mediated only by GA and GID1 and does not depend on the activity of GID2.
Figure 9.
Molecular Models for the Regulation of SLR1 Repressive Activity.
(A) GID2-dependent DELLA derepression in the wild type. When GA is absent, SLR1 represses GA actions (repressive state). When GA is present, the GID1 receptor binds to GA, which allows GID1 to interact with SLR1. The GID1-GA-SLR1 complex is in turn targeted by the SCFGID2 complex, and the SLR1 protein is degraded by the 26S proteosome, which releases the repressed state of GA responses.
(B) In gid1 and GA-deficient mutants, SLR1 is unable to interact with a GA-GID1 complex. SLR1 might interact with one or more unknown proteins to express its suppressive activity.
(C) In gid2, the GA-GID1-SLR1 complex accumulates. The repressive activity of SLR1 in the GA-GID1-SLR1 complex is lower than that of SLR1 interacting with unknown protein(s), leading to a partially derepressed state.
In GA-deficient mutants and gid1, SLR1 cannot interact with a GA-GID1 complex, since GA or active GID1 does not exist (Figure 9B). As a consequence, SLR1 can constitutively maintain strong repressive activity (constitutive repressive state). It is very possible that SLR1 interacts with one or more unknown proteins to function as a repressor in GA signaling because SLR1 does not contain a DNA binding domain even though DELLA proteins have been considered to function as transcription factors (Cao et al., 2006; Zentella et al., 2007). Thus, in GA-deficient mutants and in gid1, SLR1 would constitutively interact with the unknown protein(s) to express its suppressive activity. Actually, Feng et al. (2008) and de Lucas et al. (2008) reported that Arabidopsis DELLA proteins interact with phytochrome-interacting factors (bHLH-type transcription factors) in the absence of GA to prevent the elongation of hypocotyl. This supports the above idea that interaction of SLR1 with the unknown protein(s) is important for expression of the SLR1 function in GA signaling.
On the other hand, gid2 contains some level of GID1 protein and high levels of GA (Sasaki et al., 2003). Under these conditions, SLR1 would be pulled from both sides, that is, by interaction with the postulated unknown protein(s) and by interaction with the GA-GID1 complex (Figure 9C). Consequently, a certain amount of SLR1 would interact with GID1 to form a GA-GID1-SLR1 complex, and this complex would stably exist due to the absence of the SCFGID2-mediated degradation system. All results obtained in this study suggest that the repressive activity of SLR1 in the GA-GID1-SLR1 complex is much lower than that of SLR1 (alone or interacting with unknown protein). Overproduction of GID1 or GA treatment, both of which should pull toward formation of the GA-GID1-SLR1 complex and away from formation of the SLR1 unknown protein complex, induced GA actions such as elongation of leaves (Figures 2A, 5B, and 6), upregulation of SLR1 transcription (Figure 7), and downregulation of GA20ox-1 and GID1 transcription (Figure 7). Conversely, treatment with uniconazole or introduction of gid1 or cps mutations, all of which inhibit the formation of the GA-GID1-SLR1 complex, enhanced the repressive state in gid2, causing more severe dwarfism (Figures 2A, 3A, and 4A), downregulation of SLR1 transcript (Figure 7), and upregulation of GA20ox-1 and GID1 expression (Figure 7). All these results demonstrate that derepression of SLR1 repression activity can be caused by the interaction of SLR1 with GA-GID1, without degradation of SLR1 (Figure 9); therefore, the step of GA-GID1-SLR1 complex formation can be considered as a mechanism for derepression of SLR1 repressive activity. Recently, Ariizumi and Steber (2007) reported that seed germination of the sly1 mutant does not require RGL2 protein degradation in Arabidopsis. Furthermore, GA treatment increased the percentage of germination of all sly1 alleles tested, and the improvement of the germination was dependent on the concentration of GA applied. These phenomena in Arabidopsis are essentially the same as our observations in the rice gid2 mutant described above; therefore, the model proposed in Figure 9 may also explain the derepression of DELLA repressive activity in Arabidopsis (Ariizumi et al., 2008). The biological meaning of the derepression of SLR1 repressive activity that does not accompany the GID2-mediated SLR1 degradation has not been clarified in wild-type plants yet, but it is possible that this mechanism might be important under stress conditions, where GID2 cannot actively function to degrade the GA-GID1-DELLA protein complex (Achard et al., 2006, 2007).
If the interaction of SLR1 with GA-GID1 is sufficient to cause derepression, why have plants developed a specific system to degrade DELLA through the SCFGID2/SLY1 complex? In the proposed model, GA-GID1-SLR1 complex formation competes with SLR1 unknown protein complex formation. In this situation, large amounts of GID1 and GA would be necessary to efficiently reverse the repressive activity of SLR1. In fact, excess GA and overproduced GID1 were needed to completely release the repressive state of GA action by SLR1 in gid2 mutant plants (Figure 6). On the other hand, when SLR1 interacting with GA-GID1 is degraded by the 26S-proteasome pathway, derepression can be easily accomplished even under limited amounts of GA and GID1; in addition, GA and GID1 can be reused for further derepression of SLR1. In this context, derepression via the 26S-proteasome pathway is a much more sophisticated system than one using only GA and GID1. It is interesting to speculate that a derepression system depending merely on GA and GID1 was a precursor to the system involving the 26S-proteasome pathway, a pathway which possibly evolved at a later stage. Our previous study on GID1 and F-box proteins in the lycophyte Selaginella moellendorffii revealed that a GID1 gene from this lower plant completely rescued the dwarf phenotype of the rice gid1 mutant, but the GID2 homologous genes found in this plant could only partially rescue the dwarf phenotype of rice gid2 mutant (Hirano et al., 2007). This suggests that the interaction between Selaginella GID1 and rice DELLA (SLR1) effectively occurs in rice cells, while interaction between Selaginella GID2 proteins and rice SLR1 does not. This supports the idea that a core derepression system of DELLA repression activity, composed of GA and GID1, is well conserved, but its appended system with F-box protein is not as conserved. Further study on the affinity of interaction between DELLA and F-box proteins from various plant species would be necessary to test this hypothesis.
Abnormally High Accumulation of SLR1 in gid2
High-level accumulation of SLR1 protein is one of the unique characteristics of gid2 mutants. Although other GA-related mutants such as GA-deficient mutants and gid1 also accumulate more SLR1 than does the wild type, the accumulation in these mutants is much lower than that in gid2 (Figure 1). A similar phenomenon has been observed in the Arabidopsis F-box protein mutant sly1. Griffiths et al. (2006) reported that sly1 contains a much higher level of RGA, an Arabidopsis DELLA protein, than does Arabidopsis gid1a gid1b gid1c, a triple mutant of three GA receptor genes. They suggested that there might be a basal level of SCFSLY1-mediated RGA degradation independent of GID1. It is known that Arabidopsis F-box protein can interact with RGA and GAI in the absence of At GID1 proteins or GA, at least in yeast cells (Dill et al., 2004). On the other hand, the rice F-box protein GID2 does not interact with SLR1 in yeast cells. Furthermore and more importantly, the SLR1 protein level in the gid1 gid2 double mutant plants was similar to that of the gid1 single mutant (Figure 4B). This result indicates that GA-independent degradation of SLR1 by GID2 is not the main reason for the difference in SLR1 levels between gid1 and gid2.
The results of this study have demonstrated that higher accumulation of SLR1 in gid2 than in gid1 is due to elevated SLR1 mRNA levels in the gid2 mutant. The level of SLR1 mRNA in untreated gid2-2 was higher than in gid1-3 and slightly lower than in the wild type (Figure 7). The reason the SLR1 mRNA level in gid2 is higher than in gid1 may be because GA signaling is at least partly functional in gid2, and gene expression is regulated by GA as expected. In gid2, the SLR1 mRNA level was decreased by uniconazole treatment and increased by GA treatment (Figure 7), demonstrating that the SLR1 expression is positively regulated by GA. gid2 constitutively produces high amounts of active GA, showing that GA signaling is constitutively working to activate SLR1 expression. On the other hand, gid1 does not perceive GA even at high levels, GA signaling does not work at all, and the SLR1 mRNA level is maintained at a low level. The model that elevated SLR1 mRNA levels cause high accumulation of SLR1 in gid2 was also supported by the observation that SLR1 accumulation did not occur when SLR1 expression was driven by a constitutive promoter (Figure 8). Taken together, these observations suggest that even slightly higher amounts of SLR1 mRNA in gid2 than in gid1 directly cause the higher accumulation of SLR1 protein, since the 26S-proteasome pathway via the SCFGID2 complex does not function in gid2 mutant plants.
METHODS
Plant Materials and Growth Conditions
Primer sequences for genotyping are listed in Supplemental Table 5 online. The rice cultivar Oryza sativa cv Taichung 65 (wild type); GA-deficient mutants d35 (mutant of ent-kaurene oxidase; Itoh et al., 2004), Waito-C (mutant of OsGA3ox2; Itoh et al., 2001), and cps1-1 (mutant of ent-copalyl diphosphate synthase; Sakamoto et al., 2004); and GA-insensitive mutants gid1-3, -7, and -8 (mutants of GA receptor GID1; Ueguchi-Tanaka et al., 2005, 2007a) and gid2-1, -2, and -5 (mutants of F-box protein; Sasaki et al., 2003) were used in this study. To generate rice plants containing various combinations of GID1 and GID2 alleles, genetic crosses were performed between plants heterozygous for each gene (i.e., GID1/gid1-3 and GID2/gid2-2). The genotype of each F2 plant was identified by PCR using primers gid1-26U and gid1-26L for the GID1 locus and primers gid2-2U and gid2-2L for the GID2 locus. To generate cps1-1 gid2-2 double mutants, plants heterozygous for each gene were crossed, and the genotype of each F2 plant was identified by PCR using two sets of primers, cpsTos1U and LTR4A, and cpsTos1U and cpsTos1L for the CPS1 locus and gid2-2U and gid2-2L for the GID2 locus. The cps1-1 gid2-2 double mutants could be obtained only at a very low frequency because of the biased production of cps1-1 mutant homozygous plant as described previously (Chhun et al., 2007). Rice seeds were immersed in water for 3 d and grown for 2 or 4 weeks in a greenhouse.
pActNos and pActGID1 (Ueguchi-Tanaka et al., 2005) were introduced into rice gid2-2 mutant (Sasaki et al., 2003) and wild-type plants by Agrobacterium tumefaciens–mediated transformation (Hiei et al., 1994). pActHA-SLR1 (described below) was also introduced into the gid2-2 mutant. The genotype of gid2-2 mutant callus was identified by PCR using primers gid2-2U and gid2-2L.
GA and Uniconazole Treatment of Rice Seedlings
Seeds were immersed in water or 10−6 M uniconazole solution for 3 d. The seeds were then sown onto soil containing same solutions as above and grown in a greenhouse at 30°C (day) and 24°C (night). After 2 weeks, half of the seedlings treated with uniconazole were sprayed with 10−4 M GA3 solution containing 0.02% Tween 20 for 1 week at 1-d intervals. Three-week-old seedlings were harvested and then immediately frozen at −80°C until they were used for protein and RNA gel blot analyses. For time-course experiments, 2-week-old seedlings with 10−6 M uniconazole were sprayed with 10−4 M GA3 solution containing 0.02% Tween 20 and harvested at the time indicated. For GA treatment of GID1-overproducing gid2 plants, each line of 3-week-old transformants was divided into two; one was transferred to the soil containing 10−4 M GA3 with 0.1% ethanol as a solvent and the other was transferred to the soil containing 0.1% ethanol as a control. These plants were grown for 3 more weeks in the greenhouse in the conditions described above. After 3 weeks, the effect of GA on the gross morphology was examined by comparing GA-treated and -untreated transgenic plants of same line.
Plasmid Construction
For construction of HA-SLR1 in the pActNos/Hm2 vector, the HA-SLR1 fragment was amplified using HA-SLR1-U and HA-SLR1-liter primers using pGADT7-SLR1 as a template (Ueguchi-Tanaka et al., 2005) and cloned into the XbaI-SmaI site of the pBluescript II SK+ vector (Stratagene) to generate HA-SLR1/pBS. HA-SLR1/pBS was digested with XbaI/SmaI and cloned into pActNos/Hm2 at the XbaI/SmaI target site to generate pAct HA-SLR1.
Immunoblot Analysis of SLR1 Protein
Crude protein extracts of rice seedlings were prepared by grinding with liquid nitrogen using a mortar and pestle in the presence of sea sand (425 to 850 μm; Wako Pure Chemical). An equal volume of 2× sample buffer was added, and samples were then boiled for 5 min. Protein samples were separated by 7.5% SDS-PAGE and transferred to Hybond enhanced chemiluminescence nitrocellulose membrane (GE Healthcare). For the detection of SLR1 protein, the blots were treated with 5% skim milk in TBST (0.1% Tween 20 in 2 mM Tris-HCl, pH 7.6, and 13.7 mM NaCl) for 1 h and subsequently incubated with anti-Os SLR1 antiserum (1:10,000 dilution) raised in rabbit (Itoh et al., 2002) for 1 h. Blots were washed three times with TBST for 15 min each. The membrane was incubated with goat anti-rabbit IgG horseradish peroxidase–conjugated secondary antibody (1:25,000 dilution) for 45 min, and blots were washed following the same procedure described above. All reactions were conducted at room temperature. Detection of peroxidase activity was performed according to the instruction manual from Pierce. HA-SLR1 protein was also detected with an anti-HA antibody raised in mouse (1:2000 dilution) (H9658; Sigma-Aldrich) and anti-rabbit IgG horseradish peroxidase–conjugated secondary antibody (1:10,000 dilution). The band intensity was quantified using ImageJ (http://rsbweb.nih.gov/ij/index.html).
RNA Isolation and RNA Gel Blot Analysis
Total RNA was isolated from seedlings as described by Chomczynski and Sacchi (1987). Total RNA (5, 7.5, and 12.5 μg) was electrophoresed for RNA gel blot analyses of SLR1, GID1, and GA20ox-1 transcripts, respectively, on a 1% agarose gel and then transferred to Hybond N+ membrane (Amersham Pharmacia Biotech). Hybridization was performed at 65°C in 6× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate), 5× Denhardt's solution (1× Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA), 0.5% SDS, 10% dextran sulfate, and 0.1 mg·mL−1 denatured salmon sperm DNA. Filters were washed twice with 2× SSC and 0.1% SDS at 65°C for 30 min and once with 0.2× SSC and 0.1% SDS at 65°C for 10 min. The 0.8-kb NotI fragment (N-terminal region) of the SLR1 cDNA and the 0.7-kb SmaI fragment of GA20ox-1 cDNA were used as probes. The 1.1-kb full-length cDNA fragment was used for the GID1 probe. DNA probes were made by random-probe labeling of these fragments with [α-32P]dCTP using the BcaBEST labeling kit (Takara) according to the instruction manual.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: GID1 (AK074026), GID2 (AB100246), and SLR1 (BAE96289).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Comparison of the 2nd and 3rd Leaf Sheath Length among Various GA-Deficient and GA-Insensitive Rice Mutants.
Supplemental Table 2. Comparison of the 4th Leaf Sheath Length among the Wild Type, gid2, and gid1, Treated with/without Uniconazole and GA3.
Supplemental Table 3. Comparison of the 4th and 6th Leaf Sheath Length among the Wild Type, gid2, cps1, and gid2 cps1.
Supplemental Table 4. Comparison of the 4th and 6th Leaf Sheath Length among the Wild Type and Plants with Mutant Alleles at the GID1 and/or GID2 Loci.
Supplemental Table 5. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Mayuko Kawamura and Hiroko Ohmiya for expert technical assistance. We also thank Kenji Asano for construction of pActHA-SLR1. This work was supported in part by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (18107001 for M.M. and 19570037 for M.U.-T.), by Target Proteins Research Program (M.M.), and by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Green Technology Project IPG0003 for M.M.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Miyako Ueguchi-Tanaka (mueguchi@agr.nagoya-u.ac.jp) and Makoto Matsuoka (makoto@nuagr1.agr.nagoya-u.ac.jp).
Online version contains Web-only data.
References
- Achard, P., Baghour, M., Chapple, A., Hedden, P., Van der Straeten, D., Genschik, P., Moritz, T., and Harberd, N.P. (2007). The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation ot floral meristem-identity genes. Proc. Natl. Acad. Sci. USA 104 6484–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., Van der Straeten, D., Peng, J.R., and Harberd, N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311 91–94. [DOI] [PubMed] [Google Scholar]
- Ariizumi, T., Murase, K., Sun, T.-P., and Steber, C.M. (2008). Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 20 2447–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi, T., and Steber, C.M. (2007). Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell 19 791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, D., Cheng, H., Wu, W., Soo, H.M., and Peng, J. (2006). Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 142 509–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, P.M., Marion-Poll, A., Ellis, M., and Gubler, F. (2002). Mutants at the Slender1 locus of barley cv. Himalaya: Molecular and physiological characterization. Plant Physiol. 129 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhun, T., Aya, K., Asano, K., Yamamoto, E., Morinaka, Y., Watanabe, M., Kitano, H., Ashikari, M., Matsuoka, M., and Ueguchi-Tanaka, M. (2007). Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell 19 3876–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162 156–159. [DOI] [PubMed] [Google Scholar]
- de Lucas, M., Daviere, J.M., Rodriguez-Falcon, M., Pontin, M., Iglesias-Pedraz, J.M., Lorrain, S., Fankhauser, C., Blazquez, M.A., Titarenko, E., and Prat, S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451 480–484. [DOI] [PubMed] [Google Scholar]
- Dill, A., Thomas, S.G., Hu, J., Steber, C.M., and Sun, T.-P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z.-L., Powers, S.J., Gong, F., Phillips, A.L., Hedden, P., Sun, T.-P., and Thomas, S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Hirano, K., et al. (2007). The GID1-mediated gibberellin perception mechanism is conserved in the lycophyte Selaginella moellendorffii but not in the bryophyte Physcomitrella patens. Plant Cell 19 3058–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, K., Ueguchi-Tanaka, M., and Matsuoka, M. (2008). GID1-mediated gibberellin signaling in plants. Trends Plant Sci. 13 192–199. [DOI] [PubMed] [Google Scholar]
- Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., Matsuoka, M., and Yamaguchi, J. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, H., Sasaki, A., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Hasegawa, Y., Minami, E., Ashikari, M., and Matsuoka, M. (2005). Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1. Plant Cell Physiol. 46 1392–1399. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Tatsumi, T., Sakamoto, T., Otomo, K., Toyomasu, T., Kitano, H., Ashikari, M., Ichihara, S., and Matsuoka, M. (2004). A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol. Biol. 54 533–547. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sentoku, N., Kitano, H., Matsuoka, M., and Kobayashi, M. (2001). Cloning and functional analysis of two gibberellin 3b-hydroxyase genes that are differently expressed during the growth of rice. Proc. Natl. Acad. Sci. USA 98 8909–8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi, S., Suzuki, H., Kim, Y.-C., Iuchi, A., Kuromori, T., Ueguchi-Tanaka, M., Asami, T., Yamaguchi, I., Matsuoka, M., Kobayashi, M., and Nakajima, M. (2007). Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J. 50 958–966. [DOI] [PubMed] [Google Scholar]
- McGinnis, K.M., Thomas, S.G., Soule, F.D., Strader, L.C., Zale, J.M., Sun, T.-p., and Steber, C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, M., et al. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46 880–889. [DOI] [PubMed] [Google Scholar]
- Olszewski, N., Sun, T.P., and Gubler, F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14 (suppl.): S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400 256–261. [DOI] [PubMed] [Google Scholar]
- Richards, D.E., King, K.E., Ait-ali, T., and Harberd, N.P. (2001). How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 67–88. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T., et al. (2004). An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134 1642–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Jeong, D.H., An, G., Kitano, H., Ashikari, M., and Matsuoka, M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299 1896–1898. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L., Ciampaglio, C.N., and Sun, T.-p. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T.-p., and Gubler, F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55 197–223. [DOI] [PubMed] [Google Scholar]
- Thomas, S.G., Rieu, I., and Steber, C.M. (2005). Gibberellin metabolism and signaling. Vitam. Horm. 72 289–338. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T.-Y., Hsing, Y.-i., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Nakajima, M., Katoh, E., Ohmiya, H., Asano, K., Saji, S., Hongyu, X., Ashikari, M., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2007. a). Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1 and gibberellin. Plant Cell 19 2140–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Nakajima, M., Motoyuki, A., and Matsuoka, M. (2007. b). Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 58 183–198. [DOI] [PubMed] [Google Scholar]
- Willige, B.C., Ghosh, S., Nill, C., Zourelidou, M., Dohmann, E.M.N., Maier, A., and Schwechheimer, C. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella, R., Zhang, Z.L., Park, M., Thomas, S.G., Endo, A., Murase, K., Fleet, C.M., Jikumaru, Y., Nambara, E., Kamiya, Y., and Sun, T.P. (2007). Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19 3037–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.