Abstract
Plants are simultaneously exposed to abiotic and biotic hazards. Here, we show that local and systemic acclimation in Arabidopsis thaliana leaves in response to excess excitation energy (EEE) is associated with cell death and is regulated by specific redox changes of the plastoquinone (PQ) pool. These redox changes cause a rapid decrease of stomatal conductance, global induction of ASCORBATE PEROXIDASE2 and PATHOGEN RESISTANCE1, and increased production of reactive oxygen species (ROS) and ethylene that signals through ETHYLENE INSENSITIVE2 (EIN2). We provide evidence that multiple hormonal/ROS signaling pathways regulate the plant's response to EEE and that EEE stimulates systemic acquired resistance and basal defenses to virulent biotrophic bacteria. In the Arabidopsis LESION SIMULATING DISEASE1 (lsd1) null mutant that is deregulated for EEE acclimation responses, propagation of EEE-induced programmed cell death depends on the plant defense regulators ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) and PHYTOALEXIN DEFICIENT4 (PAD4). We find that EDS1 and PAD4 operate upstream of ethylene and ROS production in the EEE response. The data suggest that the balanced activities of LSD1, EDS1, PAD4, and EIN2 regulate signaling of programmed cell death, light acclimation, and holistic defense responses that are initiated, at least in part, by redox changes of the PQ pool.
INTRODUCTION
When low light adapted plants are exposed to a sudden increase in light intensity, they experience a large increase in absorbed light energy. Only a proportion of this energy can be used by photosynthetic metabolism (Asada, 1999; Niyogi, 2000). Excess excitation energy (EEE), the amount of energy exceeding that required for photosynthetic CO2 assimilation, is dissipated in the form of heat via the xanthophyll cycle or by the provision of alternative electron acceptors, such as oxygen, in the water–water cycle and photorespiration (Willekens et al., 1997; Asada, 1999; Niyogi, 2000; Ort and Baker, 2002; Muller-Moule et al., 2003). Reactive oxygen species (ROS), such as singlet oxygen (1O2), superoxide anion (O2.−), and hydrogen peroxide (H2O2), are produced during an increase in excitation energy (Asada, 1999; Karpiński et al., 1999; Niyogi, 1999; Karpińska et al., 2000; Fufezan et al., 2002). The response to EEE stress does not only involve changes in photosynthetic flux but is also accompanied by alterations in leaf water status and temperature and is as a consequence also associated with increased activity of heat shock transcription factors, raised abscisic acid (ABA) levels, changes of the redox state of glutathione, and decreased stomatal conductance (Karpiński et al., 1997, 1999; Panchuk et al., 2002; Fryer et al., 2003; Ball et al., 2004; Chang et al., 2004).
Light energy and photosynthetic flux also play important roles in the plant immune response to pathogens, and certain characteristics of plant defense are shared with its reaction to increased light intensity (Karpiński et al., 2003; Ball et al., 2004; Mateo et al., 2004). These include changes in chlorophyll a fluorescence parameters and in foliar water status as well as triggering of cell death and bursts of ROS associated with local and systemic signal transmission (Karpiński et al., 2003; Ball et al., 2004; Mateo et al., 2004; Bechtold et al., 2005). Acclimation of plant leaves to EEE is controlled, at least in part, by specific changes in redox status of the photosynthetic electron carrier chain, namely the plastoquinone (PQ) pool (Karpiński et al., 1997, 1999; Pfannschmidt et al., 1999; Karpińska et al., 2000; Pfannschmidt, 2003; Kruk and Karpiński, 2006).
During the photoperiod, fine control of redox homeostasis is needed to prevent ROS overload due to excess light or pathogen attack since the largest producers of ROS are the chloroplasts and the peroxisomes (Kozaki and Takeba, 1996; Asada, 1999; Foyer and Noctor, 2003; Mateo et al., 2004). Thus, in conditions where plants are exposed to excess light, they are also at risk of overflowing ROS-dependent signaling systems and metabolic processes. Failure to maintain redox balance results in growth defects or initiation of cell death (Karpiński et al., 1999; Dat et al., 2000; Karpińska et al., 2000; Mateo et al., 2004). H2O2 and O2.− have emerged as the two key ROS that together with the hormones salicylic acid (SA) and ethylene contribute to the propagation of programmed cell death (Mazel and Levine, 2001; Overmyer et al., 2003).
In Arabidopsis thaliana, numerous mutants have been isolated that are defective in regulating programmed cell death. These initiate programmed cell death spontaneously or in response to a stress stimulus, such as pathogen infection or the application of ROS. In wild-type plants and in several of these mutants, programmed cell death is potentiated by light (Seo et al., 2000; Mach et al., 2001; Samuilov et al., 2003; Mateo et al., 2004; Mühlenbock et al., 2007). One of the best-characterized deregulated programmed cell death phenotypes is caused by recessive mutations in the LESION SIMULATING DISEASE RESISTANCE1 (LSD1) gene (Jabs et al., 1996; Dietrich et al., 1997; Rusterucci et al., 2001; Epple et al., 2003; Torres et al., 2005). The lsd1 mutant fails to limit the spread of programmed cell death under long (>16 h) photoperiods or after infection with an avirulent pathogen. LSD1 is also necessary for acclimation to conditions that promote photooxidative stress. The lsd1 plants have reduced stomatal conductance and lower peroxisomal catalase activity under permissive (low light) conditions that would make them vulnerable to ROS overload during EEE stress. Consistent with this idea, photooxidative stress will cause further limitations of stomatal gas exchange in lsd1 that in turn will cause an increase in EEE-induced photorespiratory H2O2 and exacerbated programmed cell death (Mateo et al., 2004). Notably, all defects of Arabidopsis lsd1 plants in restricting programmed cell death depended on the immune regulators ENHANCED DISEASE SENSITIVITY1 (EDS1) and PHYTOALEXIN DEFICIENT4 (PAD4) (Rusterucci et al., 2001; Mateo et al., 2004).
The SA pathway leading to systemic immunity is negatively controlled by MITOGEN PROTEIN KINASE4 (MPK4) (Petersen et al., 2000; Rusterucci et al., 2001; Brodersen et al., 2002, 2006). The SA signaling pathway also regulates light acclimatory processes (Karpiński et al., 2003; Mateo et al., 2004) and is genetically and functionally interconnected with redox changes in the glutathione pool and with changes in hydrogen peroxide levels (Karpińska et al., 2000; Karpiński et al., 2003; Mateo et al., 2006). It has also been suggested that EDS1 and PAD4 amplify ethylene and SA signals by processing ROS that are essential for regulating the expression of cellular immunity against biotrophic pathogens (Rusterucci et al., 2001; Brodersen et al., 2006, Mühlenbock et al., 2007). EDS1 signaling complexes are nucleocytoplasmic, and evidence suggests that dynamic interactions between cell compartments are important for effective stress signal relay (Mateo et al., 2004; Feys et al., 2005).
Recently, two components of Arabidopsis immunity were identified using gene expression microarrays combined with reverse genetics (Bartsch et al., 2006). A flavin-dependent monooxygenase (FMO1) positively regulates the EDS1 pathway, and one member (NUDT7) of a family of cytosolic Nudix hydrolases exerts negative control of EDS1 signaling. A common theme, underlying the functions of counterparts of these proteins in animals and fungi, is in redox stress responses.
LSD1, PAD4, EDS1, and MPK4 affect many aspects of plant defense and acclimation by regulating SA, jasmonate/ethylene, and other as yet unidentified signal intermediates that lead to programmed cell death (Wiermer et al., 2005), systemic immunity, systemic acquired acclimation (SAA) to EEE (Mateo et al., 2004, 2006) as well as to hypoxia-induced, tissue-specific lysigenous aerenchyma formation in Arabidopsis (Mühlenbock et al., 2007). All of these features point to an intrinsic activity of LSD1, EDS1, and PAD4 in processing and transduction of redox-derived signals from different subcellular compartments and different plant organs and tissues during a variety of environmental stresses.
While the importance of ROS/hormonal cellular homeostasis is clear, research in both plants and animals raises more questions than answers. For example, how are different cellular responses elicited from ROS produced from different subcellular compartments? What are the key proteins that perceive changes in ROS/hormone levels and what are their functions under differing cellular redox states? How many genes, proteins, and metabolites are adjusted in response to a specific ROS/hormonal signaling? Which are the DNA cis-regulatory elements in ROS/hormonal-responsive genes that interact with trans-regulatory proteins? These questions apply equally to fungus, animal, and plant cells.
We aimed to unravel processes controlling plant acclimation to EEE and retrograde chloroplast to nucleus signaling. Here, we show that ethylene plays a role during acclimation to EEE of wild-type Arabidopsis plants and that its formation is dependent, in part, on the redox status of photosynthetic electron transport carriers (e.g., the PQ pool). We further establish that ethylene and ROS signaling are necessary for EEE-induced programmed cell death and acclimation in wild-type leaves and runaway cell death in lsd1 leaves (see also Mühlenbock et al., 2007). We also demonstrate that EEE regulates programmed cell death not only in exposed leaves but also in leaves undergoing SAA. Using photosystem I– and photosystem II (PSII)–specific wavelengths of light, photosynthetic electron transport inhibitors, and lsd1 mutants as a test system for deregulated EEE acclimation (Mateo et al., 2004), we find that programmed cell death propagation signals and production of ethylene require EIN2, EDS1, and PAD4 activity. Activation of the SA signaling pathway in response to EEE was observed after an initial induction of ethylene/ROS signaling. We conclude that ethylene and ROS contribute to plant acclimation to EEE downstream of EDS1 and PAD4 in a pathway modulated by LSD1 and induced by redox changes in the photosynthetic electron transport carriers (e.g., PQ pool).
RESULTS
EEE-Induced Local and Systemic Programmed Cell Death Depends on Redox-Regulated Foliar Ethylene and ROS
In Arabidopsis wild-type (Columbia [Col-0]) leaves, excess light (for 1 h) caused programmed cell death that was detected by lactophenol-trypan blue (TB) staining (Figures 1A and 1B). The induction of programmed cell death in adjacent but untreated leaves (leaves undergoing SAA exposed to the ambient growth light) reflects an active process that we refer to as EEE-induced programmed cell death. Acclimation to EEE is signaled by chloroplastic, peroxisomal, cytosolic, and plasma membrane–specific signal transduction pathways (e.g., Karpiński et al., 2003, Mateo et al., 2004; Kleine et al., 2007). The redox state of the PQ pool was demonstrated to be involved in signaling of EEE acclimation and can be selectively manipulated by treatment with light enriched in 680-nm (light-2 [L2]) or 700-nm (light-1 [L1]) wavelength or by use of specific photosynthetic electron transport inhibitors (Escoubas et al., 1995; Karpiński et al., 1999; Pfannschmidt et al., 1999; Kruk and Karpiński, 2006). Treatment of low light–grown plants with DCMU or 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB) elicits similar effects on the redox status of the PQ pool as light enriched with the wavelengths 700 nm (L1) or 680 nm (L2), respectively (Kruk and Karpiński, 2006). DCMU and L1 keep the PQ pool more oxidized, while DBMIB and L2 keep the PQ pool more reduced (Escoubas et al., 1995; Karpiński et al., 1999; Pfannschmidt et al., 1999; Mateo et al., 2004, Kruk and Karpiński 2006). Arabidopsis wild-type leaves that were either exposed to DCMU and held under ambient light for 3 h followed by exposure to excess light (for 1 h) or that received the L1 treatment were associated with strongly reduced programmed cell death (Figure 1).
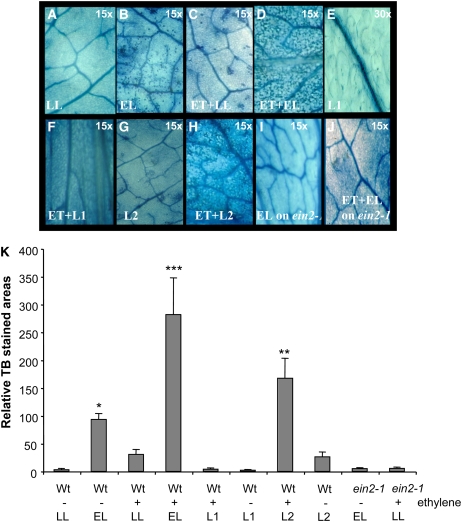
Figure 1.
Plant Programmed Cell Death in Response to Excess Light Is Modulated by Redox Status of the PQ Pool.
(A) to (J) TB staining of Col-0 leaves after low light (LL; 100 ± 20 μmol photons m−2 s−1) (A), 1 h of excess light (EL; 2200 ± 200 μmol m−2 s−1) (B), fumigated with 7.5 ppb ethylene for 24 h in LL (ET+LL) (C), fumigated with ethylene for 24 h and exposed to 1 h of EL (ET+EL) (D), or exposed to 4 h of light-1 (L1; enriched in 700-nm wavelength of energy of 10.24 J s−1 m−2) (E); Col-0 leaves fumigated with ethylene for 24 h and then exposed to L1 (ET+L1) (F); leaves exposed to light-2 (L2; enriched in 680-nm wavelength of energy of 10.24 J s−1 m−2) (G) or fumigated for 24 h with ethylene and exposed to 4 h of L2 (ET+L2) (H); leaves of the ethylene insensitive 2-1 null mutant exposed to 1 h of EL (EL on ein2) (I); and leaves of the ein2-1 null mutant exposed to 24 h of ethylene and then to 1 h of EL (ET+EL on ein2-1) (J). Images are representative of at least 27 leaves per treatment from three independent experiments (n = 27). Magnification is as indicated on each image (×15 or ×30).
(K) Areas of TB-stained foliar tissues in Col-0 and the ein2-1 null mutant cultivated in LL and then exposed to light and ethylene conditions as described in (A) to (J). Images are representative of at least nine leaves per treatment from three independent experiments (n = 3, n = 27 ± sd). Confidence levels were tested by a Student's t test (*, P < 0.05, **, P < 0.01; ***, P < 0.001). Ethylene fumigation (for 24 h) was performed directly before exposure to different light conditions, and fumigated samples are indicated by a plus sign. Samples receiving different light treatments only are indicated by a minus sign. Symbols for light treatment are the same as in (A) to (J). Only sporadic programmed cell death appears in ein1-2 and in L1 and ET+L1 conditions.
Fumigation of plants grown under low light conditions with low concentrations of ethylene (7.5 parts per billion) caused a significant increase in programmed cell death (Figures 1A and 1B) to ∼30% of the level observed in plants treated with excess light. However, ethylene fumigation in combination with excess light (for 1 h) or light-2 (for 4 h) or DBMIB treatments increased the incidence of programmed cell death synergistically (Figures 1A and 1B).
These data suggest that a more reduced PQ pool (programmed cell death induced by EL and L2 and synergistically increased due to additional ethylene and lack or weak programmed cell death in L1 and L1+ethylene of the wild type and in ein2; Figure 1) causes EEE-induced programmed cell death and light acclimatory responses that are regulated by EEE-induced ROS and ethylene homeostasis. Additionally, we quantified the areas of TB staining in leaves exposed to the different light regimens and observed that increases in programmed cell death occurred in wild-type leaves only under excess light stress or 4 h of light-2 (Figure 1B) and that it was strongly reduced in the ethylene insensitive2 (ein2) null mutant (Guzman and Ecker, 1990).
Previously, it was reported that ROS and ethylene regulate photosynthesis (Kays and Pallas, 1980; Asada, 1999; Karpiński et al., 1999) and that ethylene potentiates the oxidative burst (Ge et al., 2000; de Jong et al., 2002; Tuominen et al., 2004). Therefore, we measured levels of hydrogen peroxide (H2O2) and 1-aminocyclopropane-1-carboxylate (ACC), which is the immediate biosynthetic precursor of ethylene and a reliable marker of ethylene levels in the plant (Adams and Yang, 1979). We also measured relative (to the ambient growth condition) stomatal conductance (RSC), since it is known that EEE induces stomata closure and limitation of CO2 uptake and in consequence a photorespiratory burst of H2O2 (Mateo et al., 2004). Exposure of plants to 1 h of excess light caused a significant increase in foliar H2O2 and ACC concentrations in local and systemic leaves (Figures 2A to 2C) and a significant decrease in RSC (Figure 2D). Also, plants exposed to DBMIB treatment under low light conditions or plants exposed to L2 conditions for 4 h had significantly increased foliar levels of H2O2 and ACC and decreased levels of RSC (Figure 2). By contrast, low light–adapted plants that were treated with DCMU and then exposed to excess light or plants exposed to L1 had lower levels of H2O and ACC but not significantly reduced RSC compared with control plants grown under ambient (low light) conditions and exhibited only sporadic incidences of programmed cell death (Figures 1 and 2). Thus, we determined that there is a positive correlation between ethylene/ROS production and the incidence of programmed cell death and a negative correlation between RSC and the incidence of programmed cell death in the treatments that caused a stronger reduction of the PQ pool. Therefore, we concluded that the redox status of the PQ pool regulates cellular ROS/ethylene homeostasis, and consequently programmed cell death, in wild-type leaves exposed to EEE.
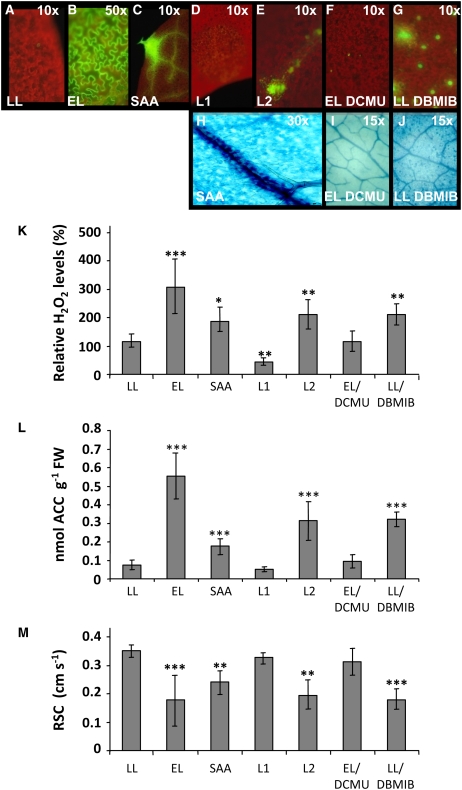
Figure 2.
Plant Cellular ROS/Ethylene Homeostasis, Programmed Cell Death, and Stomatal Conductance Are Modulated by Redox Status of the PQ Pool.
(A) to (J) Detection of hydrogen peroxide in leaves stained with 10 μM 2′,7′-dichlorofluorescin diacetate and visualized with fluorescence microscopy (10-, 30-, and 50-fold magnification). Dark-red color is derived from chlorophyll fluorescence, and green indicates peroxide. Leaves were either exposed to low light ([A]; 100 ± 20 μmol m−2 s−1) or to 1 h of excess light ([B]; 2200 ± 200 μmol m−2 s−1), were developing SAA (C) (Karpiński et al., 1999), were exposed to 4 h of light-1 (L1 enriched in 700-nm wavelength of energy of 10.24 J s−1 m−2) (D) or exposed to 4 h of light-2 (L2 enriched in 680-nm wavelength of energy of 10.24 J s−1 m−2) (F), treated with 8 μM of DCMU under LL conditions for 3 h and then exposed to 1 h EL ([G]; EL DCMU), or treated with 14 μM DBMIB under LL conditions for 4 h (LL DBMIB) (H). DBMIB and DCMU treatments caused an ∼20 and 40% reduction of photosynthetic electron transport, respectively, as indicated by chlorophyll a fluorescence parameters. Additionally, we show TB-stained Col-0 leaves that are developing SAA (I) (Karpiński et al., 1999), leaves treated with 8 μM DCMU under LL conditions for 3 h and then exposed to 1 h of EL ([J]; EL DCMU), and leaves treated with 14 μM DBMIB under LL conditions for 4 h ([K]; LL DBMIB). Images are representative of at least 19 leaves per treatment from three independent experiments.
(K) to (M) Levels of foliar H2O2 (K), foliar ACC (nmol per g of fresh weight; [L]), and RSC (in comparison to control plants cultivated in LL; [M]) in Arabidopsis Col-0 leaves exposed to LL, after 1 h of exposure to EL, or during development of SAA (Karpiński et al., 1999), in leaves of a plant exposed to L1 or to L2, in leaves treated with 8 μM DCMU under LL conditions for 3 h and exposed to EL for 1 h, or in leaves treated with 14 μM DBMIB under LL conditions for 4 h. Data are representative of a triplicate sample of pooled leaves from four independent experiments (n = 4 ± sd). Confidence levels were tested by a Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
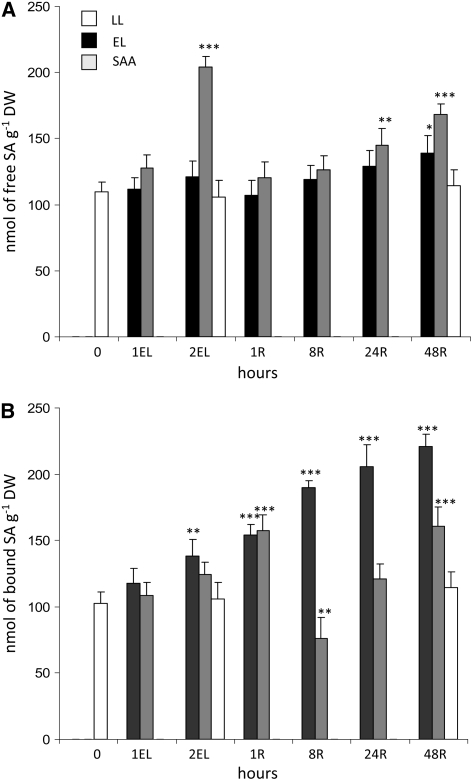
Recently we demonstrated that SA and glutathione signaling pathways are functionally and genetically integrated in the regulation of acclimation to EEE (Mateo et al., 2006). We therefore proceeded to measure the levels of free and bound foliar SA in response to EEE (Figure 3). We observed significant changes in free foliar SA levels after 2 h of excess light exposure, but only in leaves undergoing SAA (Figure 3A). In the case of conjugated foliar SA, we observed a significant increase in levels several hours after EEE stress (Figure 3B). These results indicate that SA signaling is activated in recovery and post-stress acclimation responses, both in leaves directly exposed to excess light and in leaves undergoing SAA. The results also suggest that the initial response to EEE (up to 1 h) is predominately governed by ethylene and ROS signaling (Figures 1 and 2).
Figure 3.
Free and Bound Foliar SA Is Specifically Induced in Post-Stress Recovery and Acclimation Response to Excess Light.
Free (A) and bound (B) foliar SA levels, estimated in dry weight (DW) during 1 and 2 h of excess light (2200 ± 200 μmol photons m−2 s−1) and after 1, 8, 24, and 48 h of recovery in low light (100 ± 20 μmol photons m−2 s−1) measured in leaves directly exposed to EL and in leaves undergoing SAA in LL (systemic response) (Karpiński et al., 1999). Data are representative of pooled leaf samples from four independent experiments (n = 4 ± sd). Confidence levels were tested by a Student's t test (*, P < 0.01; **, P < 0.005; ***, P < 0.001).
Taking into consideration the above results, we also analyzed expression of two robust molecular markers, ASCORBATE PEROXIDASE2 (APX2), a marker of SAA, and PATHOGEN RESISTANCE1 (PR1), a marker of systemic acquired resistance (SAR), in ein2 and salicylic insensitive2 (sid2) null mutants (Wildermuth et al., 2001) (see Supplemental Figure 1 online). We found that EIN2 is not required for appropriate induction of APX2 in leaves directly exposed to excess light (local leaves [LO]). By contrast, in leaves undergoing SAA (systemic leaves [SY]), EIN2 is required for systemic induction of APX2 and SAA. In the case of PR1 regulation, we found that EIN2 is required for delay of PR1 induction in response to excess light (EEE) in both directly exposed leaves (LO) and in leaves undergoing SAA (SY) (see Supplemental Figure 1 online). SID2 delays the induction of APX2 in directly exposed leaves but not in leaves undergoing SAA (SY leaves). Consistent with previous observations that PR1 is regulated by SA (Dangl and Jones, 2001), SID2 is required for appropriate induction of PR1 in response to excess light in both LO and SY leaves (see Supplemental Figure 1 online). These results confirm that SAA and SAR are functionally and genetically integrated in acclimatory responses to EEE.
EEE Causes a Concerted Induction of Acclimation and of Defense-Related Genes
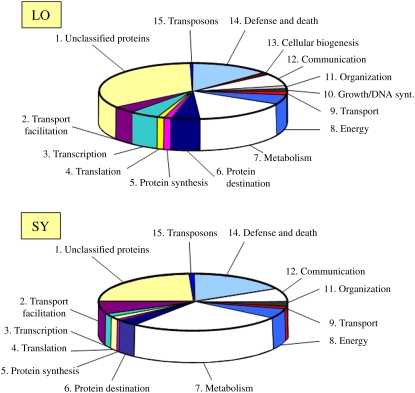
To further investigate the genetic mechanisms controlling responses to EEE and SAA, two separate suppression subtractive hybridization cDNA libraries were constructed and analyzed. In the first library (LO), we identified 730 ESTs representing 278 different genes induced in response to 40 min of excess light exposure (Figure 4; see Supplemental Data Set 1 online). In the second subtractive library constructed from leaves undergoing SAA (SY), we obtained 462 EST clones, representing 148 different genes (Figure 4; see Supplemental Data Set 1 online). We also found groups of genes that were only detected in excess light or SAA libraries (see Supplemental Data Set 1 online). The EST clones identified in both libraries were assigned to 15 different functional groups, including categories and subcategories (Figure 4). Taking the imperfection of the experimental system we used into consideration (Ji et al., 2002), we did not perform a comparative analysis between local and systemic libraries. However, we can conclude that plant global response to EEE involves gene expression reprogramming of different categories of genes that were previously classified either as biotic or abiotic stress-responsive genes.
Figure 4.
Functional Categorization of Subtraction Suppressed Subtractive Hybridization EST Libraries.
mRNA for these libraries was isolated from partially exposed rosettes, from leaves (local [LO]) directly exposed to 40 min of excess light (2200 ± 200 μmol photons m−2 s−1), and from leaves undergoing SAA (systemic [SY]) in low light (100 ± 20 μmol photons m−2 s−1) for 40 min (i.e., LO and SY leaf samples collected at the same time). List of all sequenced ESTs from these libraries is presented in Supplemental Data Set 1 online.
Since redox status of the photosynthetic electron transport carriers plays an important regulatory role in EEE responses (Karpiński et al., 1997, 1999; Karpińska et al., 2000; Kruk and Karpiński, 2006), we tested the EEE-induced ESTs (see Supplemental Data Set 1 online) for opposite regulation by DCMU and DBMIB treatments. We found that 25 ESTs from the libraries (Table 1) that were induced by excess light or SAA were suppressed by DCMU and induced by DBMIB. We will refer to these 25 ESTs as the PQ regulon. In this novel regulon, we identified the presence of several robust markers for light acclimation, pathogen defense, ozone, drought, low-temperature responses, and genes regulated by ethylene, ROS, glutathione, SA, ABA, sugar, and auxin (indole-3-acetic acid [IAA]) signaling (Figure 4, Table 1; Geisler et al., 2006). In the PQ regulon, we also detected a putative regulator of ribulose-1,5-bis-phosphate carboxylase/oxygenase and other nuclear-encoded chloroplast genes that are controlled by sugars and light signaling (Table 1). It is important to note that the set of genes presented in Table 1 does not represent the complete PQ regulon.
Table 1.
Pool of Genes Regulated by Changes in Redox Status of the PQ Pool (PQ Regulon) and Induced by Excess Light and by the SAA Mechanism
| Genes Detected in EL or SAA EST Libraries | DB Number | ELa | SAAa | DCa | DBa |
|---|---|---|---|---|---|
| Heat shock protein 3 (HSC70-3) | AT3G09440 | 6.3 | 4.10 | 0.20 | 5.10 |
| Beta Carotene Hydroxylase | AT5G52570 | 8.7 | 4.50 | 0.20 | 6.20 |
| Plastid-lipid associated protein (PAP) | AT4G04020 | 11.3 | 6.50 | 0.10 | 5.30 |
| Pathogenesis receptor kinase (PR5K) | AT4G36010 | 4.2 | 2.70 | 0.45 | 3.60 |
| Plant defensin-fusion protein (PDF1.2b) | AT2G26020 | 12.1 | 6.50 | 0.30 | 4.70 |
| DNAJ heat shock protein, putative (J3) | AT3G44110 | 4.4 | 2.60 | 0.40 | 2.50 |
| Encodes an auxin-inducible GST | AT2G29450 | 4.4 | 2.55 | 0.60 | 4.70 |
| Glycosyl hydrolase family 17 protein | AT4G16260 | 6.4 | 4.60 | 0.10 | 4.90 |
| Pyruvate decarboxylase | AT5G54960 | 5.9 | 4.70 | 0.10 | 4.80 |
| Dehydrin Xero2 | AT3G50970 | 8.3 | 6.10 | 0.10 | 5.90 |
| Disease resistance (TIR-NBS-LRR) | AT4G16860 | 6.5 | 4.50 | 0.10 | 4.30 |
| Cytochrome P450 81D1 (CYP81D1) | AT5G36220 | 9.6 | 6.50 | 0.20 | 6.50 |
| Phenylalanine ammonia-layse (PAL1) | AT3G53260 | 6.1 | 2.50 | 0.10 | 5,80 |
| Glutathione S-Transferase 6 (GST6) | AT2G47730 | 11.7 | 6.50 | 0.20 | 7.10 |
| Superoxide dismutase (Cu-Zn) (CSD1) | AT1G08830 | 13.4 | 6.40 | 0.10 | 6.30 |
| Monodehydroascorbate reductase (MDR) | AT3G09940 | 9.4 | 5.80 | 0.20 | 6.10 |
| Phospholipase D α 1 (PLD1) | AT3G15730 | 6.4 | 2.50 | 0.30 | 5.60 |
| Phospholipase C (PLC2) | AT3G08510 | 17.6 | 6.50 | 0.10 | 8.10 |
| Rubisco subunit binding protein β subunit | AT3G13470 | 8.3 | 2.40 | 0.40 | 5.60 |
| Aminotransferase, putative | AT2G24850 | 14.8 | 7.60 | 0.10 | 6.20 |
| Xyloglucan endo-1,4-β-D-glucanase, SEN4 | AT4G30270 | 5.5 | 2.20 | 0.30 | 5.10 |
| Pfam domain, PF00069: protein kinase | AT4G23150 | 12.4 | 6.40 | 0.30 | 7.10 |
| Vegetative storage protein (VSP2) | AT5G24770 | 8.4 | 6.80 | 0.20 | 4.10 |
| S-Linalool synthase | AT1G61120 | 4.5 | 2.30 | 0.30 | 3.10 |
Custom gene arrays (see Supplemental Data Set 1 online) were hybridized to P33-labeled cDNAs from Arabidopsis leaves exposed to EL (2200 ± 200 μmol m−2 s−1, 40 min), leaves undergoing SAA, leaves treated with 8 μM DCMU (DC) for 3 h, and leaves treated with 14 μM DBMIB (DB) for 3 h in ambient low light (LL) as described by Karpiński et al. (1999).
Relative transcript level values represent ratios of mean mRNA abundance to mean mRNA abundance in LL after normalization to mean actin mRNA levels. Data are representative for pooled samples of 10 leaves from four independent experiments (n = 4). χ2 analysis based on the values observed in leaves induced and suppressed by DBMIB and DCMU, respectively, and values compared with that of neutral control. All presented differences in relative transcript abundance are significant for 2 degrees of freedom (P < 0.005).
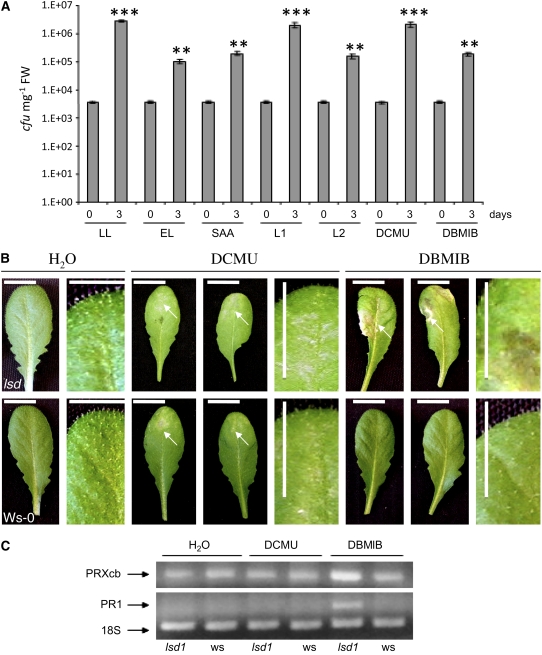
The above results indicate strong similarities between EEE acclimation and pathogen defense responses (SAR). Therefore, we tested whether leaves undergoing local and systemic acclimation to EEE were altered in pathogen defense and found that they limited growth of virulent biotrophic Pseudomonas syringae DC3000 after infection compared with nonacclimated control plants (Figure 5A). Leaves treated with DCMU for 3 h and then exposed to EEE for 1 h or leaves exposed to 4 h of L1 before infection permitted similar growth of P. syringae DC3000 as control plants (Figure 5A). By contrast, treatment of leaves with DBMIB in ambient light conditions or with 4 h of exposure to L2 caused significantly reduced growth of P. syringae DC3000 compared with wild-type plants (Figure 5A). Together, these data show that the redox status of the PQ pool regulates, at least in part, acclimation to EEE, programmed cell death, and basal defense responses to virulent biotrophic bacteria.
Figure 5.
Redox Status of the PQ Pool Affects Defense against Virulent Biotrophic Bacteria Infection in Arabidopsis Leaves.
(A) Growth of P. syringae pathovar DC3000 in infected Col-0 leaves 1 h and 0 d and 72 h and 3 d after infection and estimated in fresh weight (FW). Infections were made on leaves with various light acclimatory conditions and consequently changed the PQ redox status. Leaves before infection were acclimated to low light (control) (100 ± 20 μmol photons m−2 s−1), exposed for 1 h to excess light (2200 ± 200 μmol photons m−2 s−1), were developing SAA (Karpiński et al., 1999), or were exposed to 4 h of light-1 (L1 enriched in 700-nm wavelength of energy of 10.24 J s−1 m−2), exposed to 4 h of light-2 (L2 enriched in 680-nm wavelength of energy of 10.24 J s−1 m−2), treated with 8 μM of DCMU under LL conditions for 3 h and then exposed to 1 h EL, or treated with 14 μM DBMIB under LL conditions for 4 h. DBMIB and DCMU treatments caused an ∼20 and 40% reduction of photosynthetic electron transport, respectively, as indicated by chlorophyll a fluorescence parameters. Results represent three independent experiments and 15 infections per experiment and treatment (n = 45 ± sd). Confidence levels were tested in a Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). cfu, colony-forming units.
(B) Treatment of leaves with 8 μM DCMU in low light conditions (100 ± 20 μmol photons m−2 s−1) caused formation of discrete spots of chlorosis (indicated by arrows) in both Ws-0 and Ws lsd1. By contrast, application of 14 μM DBMIB under low light conditions induced spreading lesions only in Ws lsd1 (indicated by arrows) (n = 5 from two independent experiments, n = 2). Chlorosis in Ws-0 and in lsd1 was observed 24 h after DCMU, and runaway cell death in lsd1 mutants was also observed 24 h after DBMIB treatment. Bars = 1 cm.
(C) Relative transcript levels of PRXcb and PR1 measured by RT-PCR (n = 4). Induction of PR1 transcript was detected 2 h after DBMIB treatment.
Redox Status of the PQ Pool Affects Runaway Cell Death and ROS/Ethylene Signaling in the lsd1 Mutant
Previously, we showed that excitation energy from PSII induces runaway cell death in lsd1 mutant plants and that this is regulated by the same light conditions that control the redox status of the PQ pool (Mateo et al., 2004). We reasoned here and before (Karpiński et al., 1999; Karpińska et al., 2000; Ślesak et al., 2003; Mateo et al., 2004) that EEE passing through PSII likely contributes to excess oxidation of components of the photosynthetic electron transport chain, causing increased production of ROS and changes in PQ redox status. Here, we observed that lsd1 mutants and wild-type plants treated with DBMIB or DCMU in ambient light conditions developed localized chlorosis (DCMU) and runaway cell death (DBMIB) within 48 h (Figure 5B). Additionally, in Figure 1A, we demonstrated that DCMU inhibited EEE-induced programmed cell death in Col-0. However, only DBMIB caused spreading runaway cell death in lsd1 (Figure 5B). This was associated with induction of gene markers for SAR (PR1) and a marker for programmed cell death (PRXcb) (Jabs et al., 1996) (Figure 5C). PR1 and PRXcb induction was detected several hours after DBMIB treatment and preceded the development of runaway cell death. To determine the extent of oxidation or reduction of the PQ pool during DCMU or DBMIB treatments, respectively, we monitored chlorophyll a fluorescence parameters (Karpiński et al., 1999). We concluded that reduction of the PQ pool contributes to the induction of pathogen defense genes and to propagation of runaway cell death in lsd1.
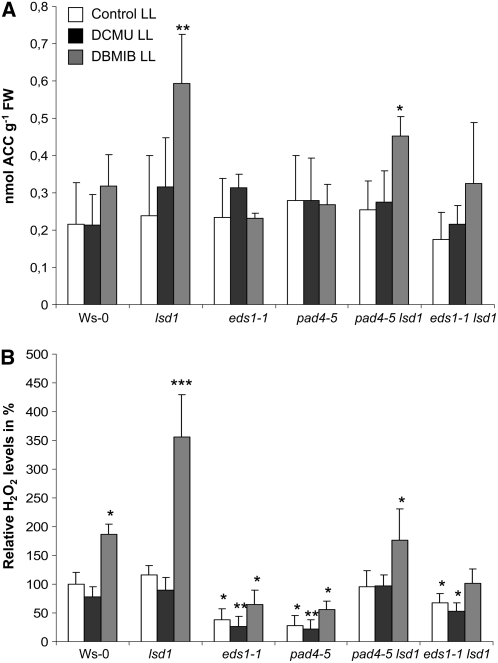
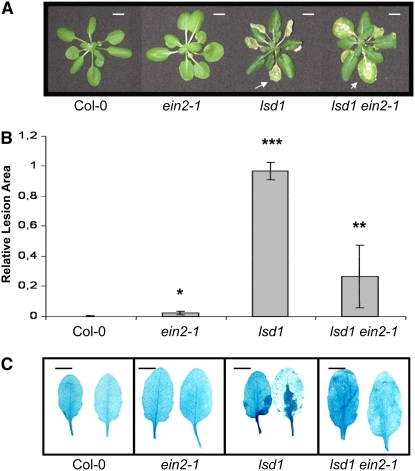
In a further analysis, we demonstrated that the induction of foliar ethylene and H2O2 in response to redox changes of the PQ pool is under negative control of LSD1 and under positive control of EDS1 and PAD4 (Figure 6). Single and double mutant analysis confirmed that LSD1 suppressed EDS1- and PAD4-dependent ROS and ethylene signaling (Figure 6) and that EIN2 acts downstream of EDS1 and PAD4 (Figures 6 and 7). Propagation of programmed cell death was dependent, at least in part, on the ethylene signal transduction encoded by EIN2, since the Col ein2-1 lsd1 double mutant had significantly reduced runaway cell death compared with Col lsd1 after restricting stomatal conductance (restricting gas exchange) (Figure 7). These results confirm that EEE-induced programmed cell death through EIN2 signaling is regulated by LSD1.
Figure 6.
LSD1 and Redox Status of the PQ Pool Regulate Foliar Ethylene and H2O2 Levels.
Foliar ACC (A) and foliar H2O2 (B) levels relative to those observed in control Ws-0 plants. ACC and H2O2 were measured in Ws-0, Ws-lsd1, Ws-eds1-1, and Ws-pad4-5 mutants and in Ws-pad4-5 lsd1 and Ws-eds1-1 lsd1 double mutants 4 h after treatment of leaves with 8 μM DCMU and 14 μM DBMIB under low light conditions (100 ± 20 μmol photons m−2 s−1). ACC and H2O2 were estimated in fresh weight (FW) of leaves. Data are representative of pooled leaf samples from four independent experiments (n = 4 ± sd). Confidence levels were tested by a Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Figure 7.
Dysfunction of EIN2 Partially Reverts Propagation of EEE-Induced Runaway Cell Death in lsd1.
(A) Representative images of rosettes of Col-0, Col-ein2-1, Col-lsd1, and Col-lsd ein2-1 plants 72 h after artificially restricting gas exchange with smear of lanoline apply on adaxial side of a one leaf (R.G). Bars = 1 cm.
(B) Lesion areas were measured relative to the control wild-type Col-0 (taken as zero or a few lesions) in leaves of Col-ein2-1, Col-lsd1, and Col-lsd ein2-1 plants 72 h after R.G. The lesion area (runaway cell death) was significantly reduced (restricted) in the lsd1 ein2-1 double mutant compared with the lsd1 single mutant. Confidence levels were tested by a Student's t test (n = 18 ± sd; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
(C) TB-stained Col-0, Col-ein2-1, Col-lsd1, and Col-lsd ein2 leaves after being exposed to photorespiratory conditions for 24 h (by R.G. with lanoline smear apply on adaxial side of a single leaf). Foliar chlorosis could be observed in the lsd1 ein2-1 double mutant, but they were not stained by TB, indicating that runaway cell death was restricted in the double mutant.
The EEE-Induced Ethylene Response Is Modulated by LSD1
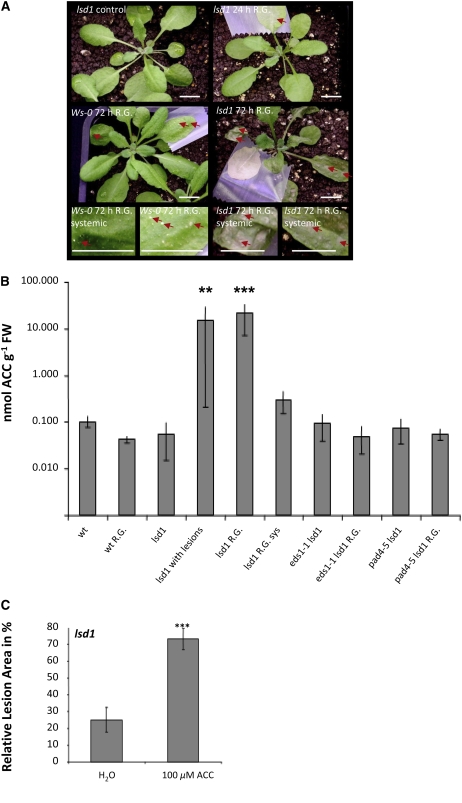
Previously, we showed that EEE acclimatory responses, stomatal conductance, and the photorespiratory burst of ROS require LSD1 activity (Mateo et al., 2004). Therefore, we decided to determine whether LSD1 had a similar regulatory impact on ethylene during the photorespiratory condition, due to EEE. Such a condition can be induced naturally, for example, during heat shock and/or drought stress or artificially, by restricting foliar gas exchange with a smear of lanoline wax applied on the adaxial side of a leaf or with semitransparent scotch tape (Figure 8). It is important to note that the lsd1 mutant plant can be cultivated for 5 to 7 weeks under permissive conditions (LL, short photoperiod, high relative humidity, e.g., 85%) without displaying visual symptoms of runaway cell death (lsd1 without lesions). However, any nonpermissive conditions (pathogen attack, EEE, SA, and glutathione) induce runaway cell death (lsd1 with lesions) after several days. Finally, we presented evidence that runaway cell death depends on photorespiration (Mateo et al., 2004). In that work, we demonstrated that low photorespiratory conditions (higher CO2 and ambient oxygen or ambient CO2 and reduced oxygen) stopped runaway cell death that had already been induced in lsd1 mutants.
Figure 8.
Photorespiratory Conditions in Single Rosette Leaves Induce an Ethylene Burst and Systemic Runaway Cell Death in the Ws-lsd1 Rosette.
(A) R.G., obtained by applying semitransparent Scotch tape to the adaxial surface of a single leaf, induced local runaway cell death followed by systemic runaway cell death in older (red arrows), but not in younger, rosette leaves. In Ws-0 plants exposed to R.G., sporadic programmed cell death was observed in both the treated and in the oldest systemic leaves. The images are representative of 17 independent experiments. Similar effects were observed in Col-0 and in Col-lsd1 plants 24 h after R.G. was achieved by applying a smear of lanoline wax on the adaxial surface of a single leaf (see Supplemental Figure 2 online). Bars = 1 cm.
(B) R.G. by applying a smear of lanoline wax on the adaxial surface of a leaf significantly induced foliar ACC production in Ws-lsd1 but not in wild-type (Ws-0) plants after 24 h of treatment. The eds1 lsd1 and pad4 lsd1 double mutants similarly to Ws-0 did not significantly induce ACC in the same treatments (one-way analysis of variance with Tukey-Kramer procedure; **, P < 0.01; ***, P < 0.001; n = 4 ± sd for each treatment). Similar R.G. treatment induced foliar ethylene emission (see Supplemental Figure 3 online).
(C) Injection of 100 μM ACC solution into leaves induced runaway cell death after 48 h in Ws-lsd1 leaves. Compare with leaves injected with water (Student's t test; ***, P < 0.001; n = 24 ± sd).
Here, we demonstrated that systemic spreading of programmed cell death in wild-type plants (Figure 8, see Wassilewskija [Ws-0] leaves with red arrows) and levels of foliar ACC (Figure 8B) under photorespiratory conditions (after artificially restricted gas exchange [R.G.]) is dependent on LSD1 activity. Restricting gas exchange in a single leaf of a 5-week-old lsd1 rosette growing under ambient conditions (permissive low light conditions) led to runaway cell death of the oldest rosette leaves 72 h after R.G. (Figure 8, in lsd1 leaves 72 h after R.G. indicated by red arrows). Interestingly, younger lsd1 leaves (near the center of the rosette) didn't die under these conditions. In Ws-0 plants exposed to photorespiratory conditions (R.G.), only sporadic programmed cell death was detected 72 h after R.G. (Figure 8, Ws-0 72 h R.G., red arrows). Similar results of runaway cell death induction were obtained with the smear of lanoline used for R.G. and induction of photorespiratory conditions (Mateo et al., 2004) and in Col-lsd1 (see Supplemental Figure 2 online).
Under low light conditions, artificially restricted gas exchange had no significant effect on foliar ACC levels during the first 24 h of R.G. treatment in Ws-0 plants (Figure 8B). By contrast, lsd1 mutants produced high levels of ACC and lsd1 leaves emitted ethylene (∼350-fold higher than control plants within 24 h after R.G.) before exhibiting runaway cell death on systemic leaves (Figures 8A and 8B; see Supplemental Figure 3 online). Increased ACC concentrations in lsd1 mutant leaves exposed to R.G. conditions were paralleled by a simultaneous peak in ethylene emissions from leaves (see Supplemental Figure 3 online). Under such R.G. stress, ACC levels did not increase in pad4-5 lsd1 and eds1-4 lsd1 double mutants (Figure 8B). This is in agreement with the data presented in Figures 6 and 8B. Similar to the above, changes in foliar ACC levels in single lsd1 null mutants and in double eds1 lsd1 and pad4 lsd1 were observed after 1 h exposure to excess light (Mühlenbock et al., 2007). These results showed that LSD1, PAD4, and EDS1 belong to the same signaling system that controls ROS and ethylene levels (Mühlenbock et al., 2007) and concomitant responses in Arabidopsis.
Previously, we demonstrated that foliar H2O2 levels in lsd1 mutants during restricted gas exchange follow similar patterns to those observed for ACC (Mateo et al., 2004). Collectively, these results indicate that in daylight, the hypersensitive disease response, SAR, and SAA are strongly exacerbated by EEE that was induced, for example, by excess light or photorespiratory conditions. It is important to note that restricted foliar gas exchange limits not only CO2 uptake but also aeration of leaf vapors, such as H2O2, methyl jasmonate, methyl salicylate, and ethylene, and that it mimics the effects of elicitors of the hypersensitive disease response (Figure 8A).
Analysis of the single eds1-1 and pad4-5 mutants as well as of the eds1-1 lsd1 and pad4-5 lsd1 double mutants revealed that the increase in ACC under EEE conditions in lsd1 requires the defense regulators EDS1 and PAD4 (Figures 6 and 8). Similar results were observed for foliar H2O2 levels after EEE was induced by restricting gas exchange (Mateo et al., 2004). To investigate the role of ethylene further, we measured the effects of application of ethylene on the propagation of runaway cell death in the lsd1 mutant. When exposed to 100 μM ACC for 48 h, lsd1 plants exhibited significantly larger areas of runaway cell death than the control plants (Figure 8C). Collectively, the data presented in Figures 6 and 8 and data presented in Figure 6 in Mühlenbock et al. (2007) lead us to conclude that EDS1 and PAD4 operate upstream of ethylene/ROS production in EEE stress signaling and result in the propagation of runaway cell death in the lsd1 mutant.
DISCUSSION
Role of Ethylene in EEE-Induced Programmed Cell Death
Exposure of wild-type Col-0 plants to excess light caused rapid increases in foliar ACC, the direct precursor of ethylene, in both leaves directly exposed to excess light and in leaves undergoing SAA (Figure 1). We provide several pieces of evidence that the production of ACC, ROS, and expression of genes regulated by ethylene, ROS, SA, glutathione, ABA, IAA, and sugars under excess light stress are regulated, at least in part, by signaling originating from redox changes in the photosynthetic transport carriers, for example, the PQ pool (Table 1, Figures 1 to 8; see Supplemental Data Set 1 and Supplemental Figures 1 to 3 online). Increase of free and conjugated foliar SA in response to EEE was detected after a significant increase of ethylene and ROS levels. This suggests that ethylene and ROS (Liu and Zhang, 2004, Geisler et al., 2006; Mühlenbock et al., 2007) rather than SA signaling are important for the initial and acute response to EEE. Additionally, we previously reported that SA- and jasmonate-dependent signaling pathways are not required for rapid EEE induction of the robust genetic marker for EEE acclimation, APX2 (Karpiński et al., 1997, 1999; Chang et al., 2004), indicating that yet another signaling system predominates in the early EEE response. After several hours of exposure to EEE, post-stress recovery and acclimatory responses do, however, induce foliar glutathione (Karpiński et al., 1997) and free and conjugated SA in leaves directly exposed and in leaves undergoing SAA (Figure 3). The recent observation that injecting SA or glutathione under the leaf epidermis induces glutathione or SA synthesis, respectively, demonstrates that foliar glutathione and SA levels are interconnected (Mateo et al., 2006). In addition, we have demonstrated that plants with deregulated SA synthesis or deregulated SA signaling are also deregulated in glutathione synthesis or signaling, respectively, and that SID2 function is required for optimal photosynthetic performance of a plant (Mateo et al., 2006). Moreover, higher foliar SA levels than those detected in low light acclimated plants were observed in plants acclimated to EEE (cultivated in high light conditions; Figure 1 in Karpiński et al., 2003). Collectively, these data suggest that ethylene and ROS production and signal relay precede changes in the glutathione pool and accumulation of SA in the early response to EEE in Arabidopsis leaves.
Our results reveal that changes in the redox status of the PQ pool regulate EEE-dependent ethylene/ROS signaling of both acclimatory and defense responses. Significantly, only 1 h of exposure to EEE was sufficient to restrict growth of virulent biotrophic bacteria (Figure 5). The effects of pharmacological (DCMU and DBMIB) or specific wavelength light treatments and restricted gas exchange argue against phytochromes playing a major role in regulation of EEE responses. Also, light-1/DCMU and light-2/DBMIB antagonistically regulate the redox status of the PQ pool (Kruk and Karpiński, 2006). Thus, ethylene and ROS production in response to the redox status of PQ appears to be a necessary component of an acute programmed cell death response to EEE and of the subsequent acclimatory and defense responses (Figures 1, 2, 5, 6, and 8, Table 1). The synergistic effect of ethylene fumigation (at low concentrations) combined with either excess light or light-2 (Figure 1) on programmed cell death supports our reasoning that ethylene and ROS combined (Figure 1, 5, and 6) are the initial signals promoting programmed cell death in plants under EEE stress. The observation that EEE-induced programmed cell death occurs in systemic tissues and that local and systemic tissues exposed to excess light or undergoing SAA are resistance toward biotrophic virulent bacteria infection suggests that the same processes may regulate SAA and SAR (Karpiński et al., 1999). This is supported by several observations. First, leaves directly exposed to EEE, leaves undergoing SAA, or leaves treated with DBMIB or light-2 (but not with DCMU or light-1) displayed significantly higher resistance toward biotrophic virulent pathogen infection than that observed in control plants. Second, EEE induced numerous genes involved in light acclimatory and pathogen defense responses (Figure 4, Table 1; see Supplemental Data Set 1 and Supplemental Figure 1 online). EEE and subsequent changes in redox status of the PQ pool also induced programmed cell death and higher resistance toward biotrophic virulent pathogen infection in unexposed parts of a plant undergoing SAA. Third, EEE induced by photorespiratory conditions (achieved by artificially restricting foliar gas exchange) led to runaway cell death in other leaves not directly challenged with such stress (Figure 8). Together, our results strongly suggest that light acclimatory and biotrophic pathogen defense responses are modulated by crosstalk at the level of ROS and hormone signaling that is regulated by the PQ pool redox status and in general redox changes in the chloroplast. Interestingly, it was demonstrated that chloroplasts indeed participate in regulation of programmed cell death in response to the infection of leaves by some biotrophic virulent pathogens (Krzymowska et al., 2007).
However, it was recently demonstrated that SAA is distinct from pathogen-stimulated SAR and apparently involves a novel signal or combination of signals that specifically preacclimate photosynthetic tissues to upcoming EEE (Rossel et al., 2007). The authors mainly concluded this on the basis of mRNA profiling of wild-type plants and of mutant plants with several deregulated gens, including the Cys2/His2-type zinc finger transcription factor ZAT10. This conclusion was not supported by subsequent tests of virulent biotrophic bacterial growth after infection of leaves that are undergoing or developing SAA in wild-type and mutant plants (Rossel et al., 2007). Our results demonstrate that SAA is rapidly able to induce SAR (within the first hour of EEE treatment) in leaves directly exposed to excess light and in leaves undergoing SAA.
LSD1, PAD4, and EDS1 Regulate EEE Stress-Induced Programmed Cell Death by Modulating Foliar ROS/Hormonal Homeostasis
Previously, LSD1 was identified as a negative regulator of SA-dependent programmed cell death and plant defense responses to pathogens (Dietrich et al., 1997; Torres et al., 2005) and as a regulator of light acclimation processes (Mateo et al., 2004). Deregulated programmed cell death in both of these stresses depends on the plant defense components EDS1 and PAD4 (Rusterucci et al., 2001). Could the antagonistic activities of LSD1 and EDS1/PAD4 be important in processing the SA, ethylene, and ROS pathways? Here, we show that runaway cell death in response to EEE induced by photorespiratory conditions or by exposure to excess light in lsd1 mutants causes significantly increased production of the ethylene precursor ACC within hours of initiation and is promoted by ethylene and ROS signaling (Figures 1, 2, 6, 8B, and 8C). We further demonstrate that propagation of programmed cell death in lsd1 depends, at least in part, on the redox status of the PQ pool. Significantly, the enhanced production of ethylene and ROS in lsd1 under EEE conditions requires EDS1 and PAD4 (Figures 6 and 8B). This finding was unexpected since EDS1 and PAD4 are known to positively regulate the SA pathway in plant defense against pathogens (Wiermer et al., 2005). Also, EDS1 and PAD4 are required to repress jasmonate/ethylene signaling in the Arabidopsis mpk4 mutant that has constitutively active SA signaling through EDS1 and PAD4 (Brodersen et al., 2002, 2006). This apparent alternation of EDS1/PAD4-dependent signal relay may be governed by a particular input stimulus and points to a promiscuity in EDS1/PAD4 activities that was not previously appreciated, although signals other than SA were recently shown to be regulated by EDS1/PAD4 in pathogen-triggered defense and programmed cell death (Bartsch et al., 2006).
We conclude that EDS1 and PAD4 act upstream of ethylene in the EEE response (Figures 6, 8B, and 9). In light stress, EDS1 and PAD4 positively control redox signal relay (Mateo et al., 2004). We propose now that this control is through promotion of ethylene/ROS signaling. However, partial reversion of runaway cell death in the double ein2-1 lsd1 mutant (Figure 7) indicates that ethylene is not the only programmed cell death signal that is suppressed by LSD1. Alternatively, another analysis reveals that a bZIP transcription factor in Arabidopsis, bZIP10, that is held in the cytoplasm by LSD1 contributes to programmed cell death in the oxidative stress response (Kaminaka et al., 2006). Kaminaka et al. (2006) suggested an alternative pathway to cell death that is controlled by bZIP, and this may be supported by our results presented above. Our recent results suggest complicated inter- and intracellular homeostasis between auxins, ABA, ethylene, glutathione, SA, ROS, and plasma membrane electrical action potential. It is also important to note here that the youngest leaves in lsd1 rosettes did not die after induction of runaway cell death (Figure 8). This effect may be due to hormonal sink-source regulation by auxins or cytokinins, and perhaps an increased understanding of bZIP function in Arabidopsis could provide an explanation for this exciting and largely unexplored phenomenon.
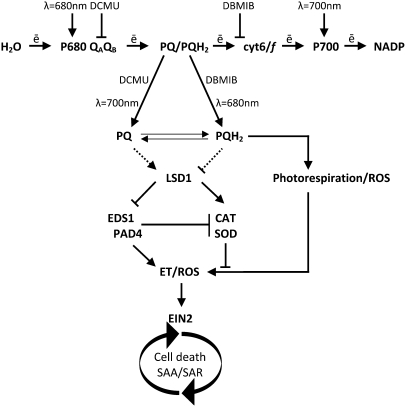
Figure 9.
Model for EEE-Induced Programmed Cell Death Controlled by the Chloroplast Redox Signaling, Photorespiration, and LSD1.
The programmed cell death redox signaling mechanism initiated by redox changes in the PQ pool is regulated by LSD1 that acts to limit the spread of cell death. The regulatory mode of LSD1 suppresses ROS production from photorespiration (Mateo et al., 2004). PAD4- and EDS1-dependent cellular ethylene production, together with EIN2, modulate ethylene (ET)–induced programmed cell death signaling during acclimatory and biotrophic pathogen defense responses. LSD1 positively regulates, either directly or indirectly, superoxide dismutase (SOD) and catalase (CAT) gene expression and activities and thus controls cellular ROS production (Jabs et al., 1996; Kliebenstein et al., 1999; Mateo et al., 2004). We propose that LSD1, EDS1, and PAD4 constitute a ROS/ethylene homeostatic switch, controlling light acclimation (SAA) and pathogen defense (SAR) holistic responses.
Additionally, many ethylene mutants have disrupted ABA signaling, and the ein2-1 mutant was shown to be ABA sensitive, which may have led to additional EEE stress and accounted for a faster induction of APX2. Importantly, however, ethylene signaling appears to act upstream of these effects, and the complicated and unresolved crosstalk between ABA and ET in light stress may have contributed to downstream effects that are outside the scope of this study.
The large increase in ACC observed here in response to EEE has been described in other studies and referred to as stress ethylene, an early response that amplifies programmed cell death signals (Ge et al., 2000; Overmyer et al., 2000; Tuominen et al., 2004). Our results show that LSD1 suppresses an ethylene/ROS-dependent programmed cell death pathway that requires the functions of EDS1 and PAD4 (Figure 9). Our evidence further points to an important role of the chloroplast in modulating defense responses through the redox status of its PQ pool (Figure 9, Table 1). It may be that plants have evolved unique defense mechanisms that depend on EEE and redox signaling originating from the chloroplast. Such response systems are likely to be crucial to plants growing in the natural environment where acclimation to prevailing abiotic stress factors (EEE) and defense responses have to be integrated. Rapid changes in light intensity and quality, in humidity, and temperature make acclimation to the natural environment an ever-present problem. Therefore, any biotic stress response in plants will be specifically adjusted in a concerted manner by a prevailing abiotic stress condition. LSD1, together with EDS1 and PAD4, may be key homeostatic regulators of a plant global acclimatory and defense mechanism. This was recently supported by the observation that LSD1, EDS1, and PAD4 are essential for regulation of lysigenous aerenchyma formation in Arabidopsis during root hypoxia (Mühlenbock et al., 2007). Together, these results indicate that plants' acclimatory and basal defense strategies are orchestrated through crosstalk by a genetic system that acts as a hub between redox signals from the chloroplast and ROS/hormonal signals arising in response to specific stress factors.
METHODS
Plant Materials, Growth, Light Conditions, and Pharmacological Treatments
The following mutants were in Arabidopsis thaliana accession Col-0: lsd1 (Jabs et al., 1996), ein2-1 (Guzman and Ecker, 1990), and the double mutant ein2 lsd1. All other mutants (lsd1, eds1-1, pad4-5, eds1-1 lsd1, and pad4-5 lsd1) were in accession Ws-0. The double mutant lsd1 ein2-1 was produced by crossing Col ein2-1 and Col lsd1. Plants were selected from a segregating F2 population by observing a triple response phenotype (for ein2-1) (Guzman and Ecker, 1990) and sensitivity to restricted gas exchange (lsd1). PCR was used to confirm homozygosity of lsd1 with the primer set forward 5′-GTGTGTGTTTGGATGAAAGTAGCAG-3′ and reverse 5′-GCTAAATGACAACAGCTTAGACGC-3′ and ein2-1 with the primer set forward 5′-CTAGCTCGGTCTATGATTTGC-3′ and reverse 5′-CGGAATGAAGGAGGACCATC-3′ followed by restriction enzyme cleavage with AflII (BspTI) to ensure correct fragment amplification.
Plants were grown in low light chambers at 100 ± 25 μmol photons m−2 s−1 (NIALOX NAV high-pressure sodium vapor lamps for experiments in Figure 1 and HQI ME-lamps 400 and 250 W [Osram] for experiments in Figures 2 and 3), with a relative air humidity of 50 to 60%, a temperature of 20 ± 1°C, and a short 9-h photoperiod. For excess light, Arabidopsis plants were treated with 2200 ± 250 μmol photons m−2 s−1, HMI (halogen metal lamps)1200 W photo-optic lamp (Osram) for up to 2 h. Photoperiod, temperature (±3°C), and humidity (±5%) for all experiments were similar to those in the LL chamber. Treatment with light-1 and light-2 was performed as described by Mateo et al. (2004) with the following modifications. Four-week-old plants cultivated in low light were exposed to light enriched with either the 680- or 700-nm wavelength, and in both cases, the light provided at the leaf surface (∼100 μmol photons m−2 s−1) was adjusted to the same energy levels for both wavelengths (10.24 J s−1 m−2). During 4 h of exposure to such light conditions, plants were also exposed to ethylene gas (7.5 ppm). Plants were also sprayed with or exposed to a 30 to 40 μL drop of DCMU (8 μM) or DBMIB (14 μM) applied to the leaf 2 to 3 h before EL treatment as described (Karpiński et al., 1999). Approximately 20 μL of 100 μM ACC was injected into leaves, and water injection was used as a control (we conducted two replicates of 12 plants/treatment). The area covered by lesions was calculated by automated pixel analysis of grayscale digital photos of the same resolution, using the software Image J (National Institutes of Health; adapted from Chaerle et al., 2004).
TB Staining
Cell necrosis was detected as described by Koch and Slusarenko (1990). Areas of TB-stained leaves were calculated with the help of a raster grid (grid cell area of 1 mm2) and a light microscope (Olympus) at 15-fold magnification. TB staining was estimated per 0.5 cm2 of randomly chosen leaf areas within the grid. In each area we scored TB staining at 25, 50, 75, or 100% of the dead zone aria. Experiments were performed at least three times at different time points and with several repetitions per experiment.
Bacterial Pathogen Infections
In the pathogen proliferation tests, 5-week-old Col-0 plants were exposed to excess light (2000 ± 250 μmol photons m−2 s−1) for up to 1 h or light-1 and light-2 for up to 4 h and then treated with DBMIB for up to 4 h in ambient (low) light or with DCMU for up to 3 h in ambient light and then exposed to excess light as above for 1 h. Two h after such treatments, leaves were infected with the virulent Pseudomonas syringae pathovar DC3000. Bacterial grow was inspected as described (Rusterucci et al., 2001) 1 h (0 time) and 72 h after infection (n = 5 for each treatment).
Stomatal Conductance and Restricting Gas Exchange
Stomatal conductance was measured as previously described (Mateo et al., 2004). Artificial restriction of gas exchange was achieved either by application of a lanolin wax smear or by adhering strips of semitransparent tape to the adaxial surfaces of leaves covering two-thirds of the leaf or the whole leaf blade (Mateo et al., 2004).
ACC, Ethylene, and SA Quantifications
ACC production was determined as previously described (Concepcion et al., 1979; Langebartels et al., 1991). Gas samples were analyzed using gas chromatography (GC-8A equipped with a 2 m Porapak N80/100 mesh, 3-mm i.d. column; Shimadzu). Determination of SA was performed as described by Meuwly and Métraux (1993) from leaves snap-frozen in liquid nitrogen. Ethylene was measured according to Du and Yamamoto (2003). The samples were injected into a gas chromatograph (Hitachi 263-50) with a flame ionization detector and a spiral glass column (0.35 × 200 cm) packed with 60/80 mesh activated alumina. Chromatographic conditions were as follows: column, injector, and detector temperatures were 80, 120, and 120°C, respectively; the carrier gas was He at 35 mL min−1. Ethylene concentration was calculated on the basis of leaf surface area compared with the chromatographic data of a 10.6 mg L−1 nitrogen balanced ethylene standard (Sanso). Experiments were performed at least in triplicate at different time points.
Hydrogen Peroxide Quantification
Hydrogen peroxide was quantified as described previously (Guilbault et al., 1968; Jimenez et al., 2002) with the following modification: 100 mg of fresh Arabidopsis leaf tissue of 5-week-old plants was used per 1 mL of extraction medium. We also visualized hydrogen peroxide by staining Arabidopsis leaves in the dark for 15 min with 10 μM 2′,7′-dichlorofluorescin diacetate and visualizing the stain by fluorescence microscopy (Axiophot; Zeiss).
Subtractive Suppression cDNA Libraries
A standard protocol was used for large-scale total RNA extraction (Karpiński et al., 1997; CTAB extraction buffer, lithium chloride precipitation followed by phenol:chloroform 1:1 [v/v] extraction). Subtractive suppression hybridization was performed using the PCR-Select cDNA subtraction kit (Clontech) according to the manufacturer's instructions. Double-stranded cDNA was prepared from 2 mg of poly(A)+ mRNA of tester and driver populations. cDNA prepared from leaves directly exposed to EEE or from leaves undergoing SAA were used as the testers and that from the control samples (low light–treated plants) as the driver for the forward subtraction to isolate fragments corresponding to genes whose expression level was increased in response to EEE or during SAA. The cDNA was then digested with RsaI. In two separate ligations, tester cDNA was ligated to kit adapters 1 and 2. In the first hybridization, an excess of driver cDNA was hybridized at 68°C for 8 h with tester cDNA ligated to adapter 2 in reaction 2. In the second hybridization, reactions 1 and 2 were hybridized together in the presence of fresh driver cDNA at 68°C overnight. Hybrid sequences were then removed. The subtractive products were amplified by PCR using kit primers that were complementary to adapters 1 and 2. PCR was performed using the following conditions: 75°C for 5 min and 27 cycles at 94°C for 30s, 66°C for 30s, and 72°C for 1.5 min. Then, a nested PCR was performed as follows: 12 cycles at 94°C for 30s, 66°C for 30s, and 72°C for 1.5 min. The final PCR products were identified as upregulated in local and systemic leaves in relation to low light–treated leaves. Consequently, PCR fragments were cloned into a pT-Adv vector (AdvanTAge PCR cloning kit; Clontech). Escherichia coli cells with high efficiency were then transformed (Epicurian Coli electroporation-competent cells) by electroporation. PCR products of SSH were cloned into the T/A cloning vector pGEM-T Easy (Promega) according to the manufacturer's instructions and introduced into TOP10-competent E. coli cells (Invitrogen). Positive clones were then selected on ampicillin plates with IPTG and X-gal selection of recombinants and were later sequenced. Several hundred colonies were obtained from each subtraction experiment. Randomly picked single colonies were grown in 1 mL of liquid Luria-Bertani medium with 100 μg/mL of ampicillin. Plasmid DNA isolated from these cultures was subjected to sequencing. Sequencing and the analysis of NCBI blast function was performed by the Laboratory of DNA Sequencing at Umeå Plant Science Center.
Different cDNA ESTs identified from Arabidopsis custom EL- and SAA-induced subtractive suppression cDNA libraries (see Supplemental Data Set 1 online) selected for their high homology to known and reported Arabidopsis and other plant genes were assembled on nylon membranes. PCR amplification of the selected cDNA clones was performed using the universal M13 pair of primers. The amplification of a unique and specific cDNA fragment was confirmed by electrophoresis of an aliquot of the purified PCR product on agarose gel. Purified PCR product (100 ng) from each cDNA clone was loaded on the gene array. After hybridization with labeled cDNA, made from mRNA isolated from EEE-exposed leaves, from leaves undergoing SAA, and leaves treated with DCMU and DBMIB, membranes were exposed to P33-sensitive screens and scanned using a Typhoon 8600 (Molecular Dynamics, Amersham-Pharmacia Biotech). Quantitative analysis of the gene array images was made using Image-Quant 5.0 (Molecular Dynamics, Amersham-Pharmacia Biotech). Data represent pooled leaf samples from four independent experiments (n = 4). χ2 P values < 0.05 were considered weakly significant and < 0.001 strongly significant.
RT-PCR Analysis
Transcript levels for PR1 and PRXcb (Jabs et al., 1996) and APX2 (Karpiński et al., 1997) were determined by RT-PCR using the following primers: forward 5′-GTCCGCTGCTCAACTCACCCCTACC-3′ and reverse 5′-GAGTGTAGGGTCGGGTAAACCTGTGTTGC 3′; APX2, forward 5′-AAGAAAGCTGTTCAGAGATGC-3′ and reverse 5′-CGGTTGGTAGTTGAAGTC-3′. They were analyzed as described by Karpińska et al. (2001) using the RETROscript first strand synthesis kit (Ambion) and the Ambion Quantum RNA kit using 18sRNA as an internal standard.
Microarray Data Meta-Analyses
Experimental statistics and normalization were performed by the Nottingham Arabidopsis Stock Centre (NASC) using MAS 5.0 scaling or by The Arabidopsis Information Resource using the print-tip-group Loess method. Normalized signal values were used to generate ratios of treated or mutant plants to untreated or wild-type plants in the same experiment. NASC-generated detection call and P values were used to evaluate whether a gene was induced, suppressed, neutral, or not expressed (in both the control and the treatment). χ2 P values < 0.05 were considered weakly significant and < 0.001 were strongly significant.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases and are listed in Table 1 (DB column).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Robust Molecular Markers of SAA and SAR (APX2 and PR1, Respectively) Are Differentially Regulated by SID2 and EIN2 in Response to Excess Light.
Supplemental Figure 2. Photorespiratory Conditions in Single Rosette Leaves Induce Runaway Cell Death in Col-lsd1 and Ws-lsd1 Rosettes.
Supplemental Figure 3. Photorespiratory Conditions Induced Emission of Gaseous Ethylene in Ws-lsd1 Plants.
Supplemental Data Set 1. List of Identified and Sequenced ESTs from Subtractive Suppression cDNA Libraries Generated from Leaves Undergoing Molecular Reprogramming in Arabidopsis Rosettes Partially Exposed to Excess Light.
Acknowledgments
We thank Alfonso Mateo and Dietmar Funck for assistance during data generation for Figure 5 and for editing Supplemental Data Set 1 online, respectively. S.K. is grateful for financial support from the Polish Ministry of Science and Higher Education (Grants N301 075 31 and N301 2550 33). M.S.-H. was supported by the Marie Curie postdoctoral fellowship MEIF-CF-2005-024914. S.K. and B.K. thank the Swedish Research Councils (VR), the Swedish Strategic Foundation, and the Knut and Alice Wallenberg Foundation for funding. J.E.P. is grateful to The Alexander von Humboldt Foundation and a DFG ‘SFB 670’ grant for funding. P.M.M. acknowledges support from the UK Biotechnology and Biological Sciences Research Council.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Per Mühlenbock (pemuh@psb.ugent.be).
Online version contains Web-only data.
References
- Adams, D.O., and Yang, S.F. (1979). Ethylene biosynthesis: Identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl. Acad. Sci. USA 76 170–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada, K. (1999). The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 601–639. [DOI] [PubMed] [Google Scholar]
- Ball, L., Accotto, G.-P., Bechtold, U., Creissen, G., Funck, D., Jimenez, A., Kular, B., Leyland, N., Mejia-Carranza, J., Reynolds, H., Karpiński, S., and Mullineaux, P.M. (2004). Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell 16 2448–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch, M., Gobbato, E., Bednarek, P., Debey, S., Schultze, J.L., Bautor, J., and Parker, J.E. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase NUDT7. Plant Cell 18 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, U., Karpiński, S., and Mullineaux, P.M. (2005). The influence of the light environment and photosynthesis on oxidative signaling responses in plant-biotrophic pathogen interactions. Plant Cell Environ. 28 1046–1055. [Google Scholar]
- Brodersen, P., Petersen, M., Nielsen, H.B., Zhu, S., Newman, M.A., Shokat, K.M., Rietz, S., Parker, J., and Mundy, J. (2006). Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 47 532–546. [DOI] [PubMed] [Google Scholar]
- Brodersen, P., Petersen, M., Pike, H.M., Olszak, B., Skov, S., Odum, N., Jorgensen, L.B., Brown, R.E., and Mundy, J. (2002). Knockout of Arabidopsis ACCELERATED-CELL-DEATH11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 16 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston, V., Earl, S., Harrison, E., Mathas, E., Navabpour, S., Page, T., and Pink, D. (2003). The molecular analysis of leaf senescence - A genomics approach. Plant Biotechnol. J. 1 3–22. [DOI] [PubMed] [Google Scholar]
- Chaerle, L., Hagenbeek, D., De Bruyne, E., Valcke, R., and Van Der Straeten, D. (2004). Thermal and chlorophyll-fluorescence imaging distinguish plant-pathogen interactions at an early stage. Plant Cell Physiol. 45 887–896. [DOI] [PubMed] [Google Scholar]
- Chang, C.C.-C., Ball, L., Fryer, M.J., Baker, N.R., Karpiński, S., and Mullineaux, P.M. (2004). Induction of ASCORBATE PEROXIDASE 2 expression in wounded Arabidopsis leaves does not involve known wound-signaling pathways but is associated with changes in photosynthesis. Plant J. 38 499–511. [DOI] [PubMed] [Google Scholar]
- Concepcion, M., Lizada, C., and Yang, S.F. (1979). A simple and sensitive assay for 1-aminocyclopropane-1-carboxylic acid. Anal. Biochem. 100 140–147. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defense responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- Dat, J., Vandenabeele, S., Vranova, E., Van Montagu, M., Inze, D., and Van Breusegem, F. (2000). Dual action of the active oxygen species during plant stress responses. Cell. Mol. Life Sci. 57 779–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, A., Yakimova, E., Kapchina, V., and Woltering, E. (2002). A critical role for ethylene in hydrogen peroxide release during programmed cell death in tomato suspension cells. Planta 214 537–545. [DOI] [PubMed] [Google Scholar]
- Dietrich, R.A., Richberg, M.H., Schmidt, R., Dean, C., and Dangl, J.L. (1997). A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88 685–694. [DOI] [PubMed] [Google Scholar]
- Du, S., and Yamamoto, F. (2003). Ethylene evolution changes in the stems of Metasequoia glyptostroboides and Aesculus turbinate in relation to gravity-induced reaction wood formation. Trees (Berl.) 17 522–528. [Google Scholar]
- Epple, P., Mack, A.A., Morris, V.R., and Dangl, J.L. (2003). Antagonistic control of oxidative stress-induced cell death in Arabidopsis by two related, plant-specific zinc finger proteins. Proc. Natl. Acad. Sci. USA 100 6831–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoubas, J., Lomas, M., LaRoche, J., and Falkowski, P.G. (1995). Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc. Natl. Acad. Sci. USA 92 10237–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., Wiermer, M., Bhat, R.A., Moisan, L.J., Medina-Escobar, N., Neu, C., Cabral, A., and Parker, J.E. (2005). Arabidopsis SENESCENCE ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer, C.H., and Noctor, G. (2003). Redox sensing and signaling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol. Plant. 119 355–364. [Google Scholar]
- Fryer, M.J., Ball, L., Oxborough, K., Karpiński, S., Mullineaux, P.M., and Baker, N.R. (2003). Control of Ascorbate Peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J. 33 691–705. [DOI] [PubMed] [Google Scholar]
- Fufezan, C., Rutherford, A.W., and Krieger-Liszkay, A. (2002). Singlet oxygen production in herbicide-treated photosystem II. FEBS Lett. 532 407–412. [DOI] [PubMed] [Google Scholar]
- Ge, L., Liu, J.Z., Wong, W.S., Hsiao, W.L.W., Chong, K., Xu, Z.K., Yang, S.F., Kung, S.D., and Li, N. (2000). Identification of a novel multiple environmental factor-responsive 1-aminocyclopropane-1-carboxylate synthase gene, NT-ACS2, from tobacco. Plant Cell Environ. 23 1169–1182. [Google Scholar]
- Geisler, M., Kleczkowski, L.A., and Karpiński, S. (2006). A universal algorithm for genome-wide in silicio identification of biologically significant gene promoter putative cis-regulatory-elements; identification of new elements for reactive oxygen species and sucrose signaling in Arabidopsis. Plant J. 45 384–398. [DOI] [PubMed] [Google Scholar]
- Guilbault, G.G., Brignac, J.P., and Zimmer, M. (1968). Homovanillic acid as a fluorometric substrate for oxidative enzymes. Analytical applications of the peroxidase, glucose oxidase and xanthine oxidase systems. Anal. Chem. 40 190–196. [DOI] [PubMed] [Google Scholar]
- Guzman, P., and Ecker, J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabs, T., Dietrich, R.A., and Dangl, J.L. (1996). Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273 1853–1856. [DOI] [PubMed] [Google Scholar]
- Ji, W., Wright, M.B., Cai, L., Flament, A., and Lindpaintner, K. (2002). Efficacy of SSH PCR in isolating differentially expressed genes. BMC Genomics 3 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez, A., Creissen, G., Kular, B., Firmin, J., Robinson, S., Verhoeyen, M., and Mullineaux, P. (2002). Changes in oxidative processes and components of the antioxidant system during tomato fruit ripening. Planta 214 751–758. [DOI] [PubMed] [Google Scholar]
- Kaminaka, H., Näke, C.H., Epple, P., Dittgen, J., Schütze, K., Chaban, C.H., Holt, B.H., Merkle, T., Schäfer, E., Harter, K., and Dang, J.L. (2006). bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 25 4400–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpińska, B., Karlsson, M., Schinkel, H., Streller, S., Suss, K.H., Melzer, M., and Wingsle, G. (2001). A novel superoxide dismutase with a high isoelectric point in higher plants. Expression, regulation, and protein localization. Plant Physiol. 126 1668–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpińska, B., Wingsle, G., and Karpiński, S. (2000). Antagonistic effects of hydrogen peroxide and glutathione on acclimation to excess excitation energy in Arabidopsis. IUBMB Life 50 21–26. [DOI] [PubMed] [Google Scholar]
- Karpiński, S., Escobar, C., Karpińska, B., Creissen, G., and Mullineaux, P. (1997). Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpiński, S., Gabrys, H., Mateo, A., Karpińska, B., and Mullineaux, P.M. (2003). Light perception in plant disease defence signalling. Curr. Opin. Plant Biol. 6 390–396. [DOI] [PubMed] [Google Scholar]
- Karpiński, S., Reynolds, H., Karpińska, B., Wingsle, G., Creissen, G., and Mullineaux, P. (1999). Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284 654–657. [DOI] [PubMed] [Google Scholar]
- Kays, S.J., and Pallas, J.E. (1980). Inhibition of photosynthesis by ethylene. Nature 285 51–54. [Google Scholar]
- Kleine, T., Kindgren, P., Benedict, C., Hendrickson, L., and Strand, A. (2007). Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 144 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Dietrich, R.A., Martin, A.C., Last, R.L., and Dangl, J.L. (1999). LSD1 regulates salicylic acid induction of copper zinc superoxide dismutase in Arabidopsis thaliana. Mol. Plant Microbe Interact. 12 1022–1026. [DOI] [PubMed] [Google Scholar]
- Koch, E., and Slusarenko, A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki, A., and Takeba, G. (1996). Photorespiration protects C3 plants from photooxidation. Nature 384 557–560. [Google Scholar]
- Kruk, J., and Karpiński, S. (2006). An HPLC-based method of estimation of the total redox state of plastoquinone in chloroplasts, the size of the photochemically active plastoquinone-pool and its redox state in thylakoids of Arabidopsis. Biochim. Biophys. Acta 1757 1669–1675. [DOI] [PubMed] [Google Scholar]
- Krzymowska, M., Konopka-Postupolska, D., Sobczak, M., Macioszek, V., Ellis, B.E., and Hennig, J. (2007). Infection of tobacco with different Pseudomonas syringae pathovars leads to distinct morphotypes of programmed cell death. Plant J. 50 253–264. [DOI] [PubMed] [Google Scholar]
- Langebartels, C., Kerner, K., Leonardi, S., Schraudner, M., Trost, M., Heller, W., and Sandermann, H., Jr. (1991). Biochemical plant responses to ozone: Differential induction of polyamine and ethylene biosynthesis in Tobacco. Plant Physiol. 95 882–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., and Zhang, S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach, J.M., Castillo, A.R., Hoogstraten, R., and Greenberg, J.T. (2001). The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc. Natl. Acad. Sci. USA 98 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo, A., Funck, D., Mühlenbock, P., Kular, B., Mullineaux, P.M., and Karpiński, S. (2006). Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J. Exp. Bot. 57 1795–1807. [DOI] [PubMed] [Google Scholar]
- Mateo, A., Mühlenbock, P., Rusterucci, C., Chang, C.C.-C., Miszalski, Z., Karpińska, B., Parker, J.E., Mullineaux, P.M., and Karpiński, S. (2004). LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 136 2818–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel, A., and Levine, A. (2001). Induction of cell death in Arabidopsis by superoxide in combination with salicylic acid or with protein synthesis inhibitors. Free Radic. Biol. Med. 30 98–106. [DOI] [PubMed] [Google Scholar]
- Meuwly, P., and Métraux, J.P. (1993). Ortho-anisic acid as internal standard for the simultaneous quantitation of salicylic acid and its putative biosynthetic precursors in cucumber leaves. Anal. Biochem. 214 500–505. [DOI] [PubMed] [Google Scholar]
- Mühlenbock, P., Plaszczyca, M., Plaszczyca, M., Mellerowicz, E., and Karpiński, S. (2007). Lysigenous aerenchyma formation in Arabidopsis is controlled by LESION STIMULATING DISEASE1. Plant Cell 19 3819–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Moule, P., Havaux, M., and Niyogi, K.K. (2003). Zeaxanthin deficiency enhances the high light sensitivity of an ascorbate-deficient mutant of Arabidopsis. Plant Physiol. 133 748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi, K. (2000). Safety valves for photosynthesis. Curr. Opin. Plant Biol. 3 455–460. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K. (1999). Photoprotection revisited: genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 333–359. [DOI] [PubMed] [Google Scholar]
- Ort, D.R., and Baker, N.R. (2002). A photoprotective role for O2 as an alternative electron sink in photosynthesis? Curr. Opin. Plant Biol. 5 193–198. [DOI] [PubMed] [Google Scholar]
- Overmyer, K., Brosche, M., and Kangasjarvi, J. (2003). Reactive oxygen species and hormonal control of cell death. Trends Plant Sci. 8 335–339. [DOI] [PubMed] [Google Scholar]
- Overmyer, K., Tuominen, H., Kettunen, R., Betz, C., Langebartels, C., Heinrich Sandermann, J., and Kangasjärvi, J. (2000). Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 12 1849–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchuk, I.I., Volkov, R.A., and Schoffl, F. (2002). Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol. 129 838–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103 1111–1120. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt, T. (2003). Chloroplast redox signals: How photosynthesis controls its own genes. Trends Plant Sci. 8 33–41. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt, T., Nilsson, A., and Allen, J.F. (1999). Photosynthetic control of chloroplast gene expression. Nature 397 625–628. [Google Scholar]
- Rossel, J.B., Wilson, P.B., Hussain, D., Woo, N.S., Gordon, M.J., Mewett, O.P., Howell, K.A., Whelan, J., Kazan, K., and Pogson, B.J. (2007). Systemic and intracellular response to photooxidative stress in Arabidopsis. Plant Cell 19 4091–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusterucci, C., Aviv, D.H., Holt III, B.F., Dangl, J.L., and Parker, J.E. (2001). The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13 2211–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuilov, V.D., Lagunova, E.M., Kiselevsky, D.B., Dzyubinskaya, E.V., Makarova, Y.V., and Gusev, M.V. (2003). Participation of chloroplasts in plant apoptosis. Biosci. Rep. 23 103–117. [DOI] [PubMed] [Google Scholar]
- Seo, S., Okamoto, M., Iwai, T., Iwano, M., Fukui, K., Isogai, A., Nakajima, N., and Ohashi, Y. (2000). Reduced levels of chloroplast FtsH protein in tobacco mosaic virus-infected tobacco leaves accelerate the hypersensitive reaction. Plant Cell 12 917–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ślesak, I., Karpińska, B., Surówka, E., Miszalski, Z., and Karpiński, S. (2003). Redox changes in the chloroplast and hydrogen peroxide are essential for regulation of C3-CAM transition and photooxidative stress responses in the facultative CAM plant Mesembryanthemum crystallinum L. Plant Cell Physol. 44 573–581. [DOI] [PubMed] [Google Scholar]
- Torres, M.A., Jones, J.D.G., and Dangl, J.L. (2005). Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 37 1130–1134. [DOI] [PubMed] [Google Scholar]
- Tuominen, H., Overmyer, K., Keinanen, M., Kollist, H., and Kangasjärvi, J. (2004). Mutual antagonism of ethylene and jasmonic acid regulates ozone-induced spreading cell death in Arabidopsis. Plant J. 39 59–69. [DOI] [PubMed] [Google Scholar]
- Wiermer, M., Feys, B.J., and Parker, J.E. (2005). Plant immunity: The EDS1 regulatory node. Curr. Opin. Plant Biol. 8 383–388. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C., Dewdney, J., Wu, G., and Ausubel, F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 562–565. [DOI] [PubMed] [Google Scholar]
- Willekens, H., Chamnongpol, S., Davey, M., Schraudner, M., Langebartels, C., Van Montagu, M., Inze, D., and Van Camp, W. (1997). Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J. 16 4806–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]