Abstract
This article presents evidence that DELLA repression of gibberellin (GA) signaling is relieved both by proteolysis-dependent and -independent pathways in Arabidopsis thaliana. DELLA proteins are negative regulators of GA responses, including seed germination, stem elongation, and fertility. GA stimulates GA responses by causing DELLA repressor degradation via the ubiquitin-proteasome pathway. DELLA degradation requires GA biosynthesis, three functionally redundant GA receptors GIBBERELLIN INSENSITIVE DWARF1 (GID1a, b, and c), and the SLEEPY1 (SLY1) F-box subunit of an SCF E3 ubiquitin ligase. The sly1 mutants accumulate more DELLA proteins but display less severe dwarf and germination phenotypes than the GA biosynthesis mutant ga1-3 or the gid1abc triple mutant. Interestingly, GID1 overexpression rescued the sly1 dwarf and infertility phenotypes without decreasing the accumulation of the DELLA protein REPRESSOR OF ga1-3. GID1 rescue of sly1 mutants was dependent on the level of GID1 protein, GA, and the presence of a functional DELLA motif. Since DELLA shows increasing interaction with GID1 with increasing GA levels, it appears that GA-bound GID1 can block DELLA repressor activity by direct protein–protein interaction with the DELLA domain. Thus, a SLY1-independent mechanism for GA signaling may function without DELLA degradation.
INTRODUCTION
This article investigates the proteolysis-independent regulation of DELLA proteins, negative regulators of plant growth. Active gibberellins (GAs) are tetracyclic diterpenoid hormones that stimulate many stages in plant development, including seed germination, stem and root elongation, transition to flowering, fruit expansion, and pollen tube elongation (Richards et al., 2001; Swain et al., 2004; Swain and Singh, 2005; Thomas et al., 2005). GA stimulates these processes by targeting the proteins of the DELLA family of negative regulators for destruction by the 26S proteasome. If GA production is blocked as in the GA biosynthesis mutant ga1-3, overaccumulation of DELLA repressors results in serious growth defects, including dwarf stature, decreased germination capacity, delayed flowering, and reduced fertility (Sun and Kamiya, 1994; Cheng et al., 2004; Tyler et al., 2004; Yu et al., 2004). The rescue of these GA-deficient mutants by GA application is associated with the rapid disappearance of DELLA repressors (Sun and Gubler, 2004; Ueguchi-Tanaka et al., 2007a). In addition to GA biosynthesis, the disappearance of DELLA proteins also requires the GA receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1) and the F-box subunit of an E3 ubiquitin ligase, SLEEPY1 (SLY1).
DELLA proteins are a subfamily of the GRAS family of putative transcription factors that act as repressors of GA responses. Recently, chromatin immunoprecipitation was used to document that the DELLA protein REPRESSOR OF ga1-3 (RGA) associates with and appears to activate the expression of promoters of downstream negative regulators of GA signaling (Zentella et al., 2007). DELLA proteins also appear to repress transcription of PHYTOCHROME INTERACTING FACTOR3 (PIF3) and PIF4-activated promoters through direct binding of the PIF transcription factors needed for their expression (de Lucas et al., 2008; Feng et al., 2008). There are five genes in the Arabidopsis thaliana DELLA family with partly overlapping functions. DELLAs RGA and GA-INSENSITIVE (GAI) are the main negative regulators in stem elongation (Peng and Harberd, 1993; Peng et al., 1997; Silverstone et al., 1997; Dill and Sun, 2001; King et al., 2001; Dill et al., 2004; Fu et al., 2004). DELLAs RGA, RGA-LIKE1 (RGL1), and RGL2 function in flower development (Wen and Chang, 2002; Cheng et al., 2004; Swain et al., 2004; Tyler et al., 2004). RGL2 is the main negative regulator of seed germination (Lee et al., 2002; Tyler et al., 2004; Cao et al., 2005; Ariizumi and Steber, 2007). These five proteins share the N-terminal DELLA motif required for GA regulation as well as the C-terminal GRAS functional domain (Sun and Gubler, 2004). Like GA treatment, loss-of-function DELLA mutations rescue the phenotypes of the ga1-3 mutant (Cheng et al., 2004; Tyler et al., 2004; Cao et al., 2005). Deletions within the DELLA motif, including amino acids DELLA and TVHYNP, result in a gain-of-function phenotype similar to that of ga1-3 associated with loss of GA-dependent DELLA degradation (Peng et al., 1999; Dill et al., 2001; Itoh et al., 2002; Wen and Chang, 2002; Willige et al., 2007). These mutants are GA insensitive and cannot be GA rescued.
Failure to degrade DELLA protein in response to GA is also associated with GA-insensitive phenotypes due to recessive mutations in the GA receptor GID1 and F-box gene SLY1. The nuclear-localized GA receptor GID1 was first identified in rice (Oryza sativa) as a protein predicted to have homology to human hormone-sensitive lipase (Ueguchi-Tanaka et al., 2005). Whereas there is a single GID1 gene in rice, there are three orthologs in Arabidopsis named GID1a, GID1b, and GID1c (Nakajima et al., 2006). Disruption of a single GID1 gene does not cause a strong growth defect, double mutants show mild growth defects, and triple gid1a gid1b gid1c mutants display extreme GA-insensitive growth defects similar to the ga1-3 mutant (Griffiths et al., 2006; Iuchi et al., 2007; Willige et al., 2007). Among the three GID1 genes, mutations in GID1a or GID1c seem to have a stronger effect on vegetative development, whereas a single mutant in GID1b causes decreased GA sensitivity in seed germination. Thus, it appears that the three GID1 genes are functionally redundant with partially specialized functions. The GID1 protein acts by GA-dependent binding to the DELLA and TVHYNP motifs of the DELLA protein (Griffiths et al., 2006; Ueguchi-Tanaka et al., 2007b; Willige et al., 2007; Feng et al., 2008). Glutathione S-transferase (GST)-GID1 shows increasing interaction with RGA with increasing GA concentrations, suggesting that GID1-GA-RGA complex formation is GA dependent (Griffiths et al., 2006). Deletion of the 17–amino acid DELLA motif in gai-1 and rga-Δ17 results in failure to interact with GID1 even in the presence of GA (Griffiths et al., 2006; Willige et al., 2007; Feng et al., 2008).
Arabidopsis SLY1 and rice GID2 encode orthologous F-box subunits that determine the substrate specificity of an SCF (for Skp1, Cullin, F-box) E3 ubiquitin ligase complex (Steber et al., 1998; Steber and McCourt, 2001; McGinnis et al., 2003; Sasaki et al., 2003; Dill et al., 2004; Fu et al., 2004; Gomi et al., 2004). It appears that binding of DELLA RGA by GID1-GA stimulates the interaction of RGA with SCFSLY1, and ubiquitination of DELLA by SCFSLY1 targets DELLA for destruction by the 26S proteasome (Griffiths et al., 2006). The recessive sly1 mutants show increasing severity of phenotypes with increasing severity of allele (Ariizumi and Steber, 2007). The sly1-10 allele, which lacks the last eight amino acids, is less severe than the sly1-2 allele, which lacks the last 40 amino acids (Steber et al., 1998; Steber and McCourt, 2001), and the sly1-2 allele is less severe than the sly1-t2 allele, which contains a T-DNA in the F-box motif leading to loss of the last 78 amino acids (Ariizumi and Steber, 2007). The sly1 mutant phenotypes of increased seed dormancy, dwarfism, and reduced fertility are similar to but not as severe as those of the ga1-3 and gid1a gid1b gid1c triple mutants (Steber et al., 1998; McGinnis et al., 2003; Griffiths et al., 2006; Ariizumi and Steber, 2007; Willige et al., 2007). Interestingly, while the sly1 mutant phenotype is less severe, it is associated with higher levels of DELLA protein accumulation. Thus, it appears that GA regulation of seed germination, stem elongation, and fertility depends on factors other than the absolute level of DELLA proteins. Moreover, we previously showed that dormant sly1-2 seeds acquire the ability to germinate upon after-ripening, even though they continue to accumulate high levels of DELLA RGL2 and RGA (Ariizumi and Steber, 2007). After-ripening of sly1-2 seeds also resulted in increased levels of GA-inducible transcripts compared with dormant sly1-2 and ga1-3 seeds. These data suggested that either after-ripening bypassed DELLA repression of seed germination or resulted in inactivation of DELLA protein as a repressor of GA-regulated gene expression (Ariizumi and Steber, 2007).
This article uses the sly1 mutant background to explore the notion that GID1 can block DELLA repression of stem elongation and fertility through a proteolysis-independent mechanism. To address this question, we created Arabidopsis lines in which each of the three GID1 genes is overexpressed from the constitutive cauliflower mosaic virus (CaMV) 35S promoter. We found that ectopic expression of GID1 genes partly rescued the stem elongation and fertility defects of the GA-insensitive sly1 mutants without altering DELLA RGA protein levels. This rescue required the presence of GA and a functional DELLA motif. These results suggest that formation of the GID1-GA-DELLA complex reduces repressor activity of DELLA proteins.
RESULTS
GID1 Overexpression Rescues the Dwarfism of sly1 Mutants
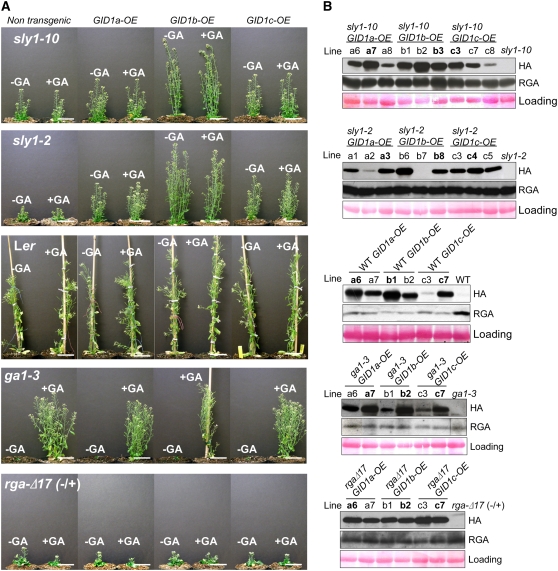
To explore the role of the GID1 genes in Arabidopsis GA signaling, we introduced three chimeric constructs each containing an N-terminal HA fusion of one of three GID1 genes (GID1a, GIDb, or GID1c) under control of the 35S CaMV promoter into several Arabidopsis backgrounds including wild-type Landsberg erecta (Ler), ga1-3, sly1-10, sly1-2, rga-Δ17 (heterozygous; +/−), and gai-1 (homozygous; −/−) (Figure 1; see Supplemental Figure 1 online). Figure 1A shows 36-d-old plants grown with or without a 10 μM GA4 treatment. GID1 gene overexpression suppressed the sly1-10 plant height and fertility defects. GID1b-overexpression (GID1b-OE) had the strongest effect on plant height causing some increase in wild-type Ler height and restoration of the sly1-10 mutant to a final height similar to the wild type (see Supplemental Table 1 online). However, plant growth in sly1-10 GIDb-OE appeared somewhat slower than in the wild type. By contrast, GID1a- and GID1c-overexpression (GID1a-OE and GID1c-OE) resulted in plants slightly taller than the vector-only and untransformed sly1-10 controls (Figure 1A; see Supplemental Figure 2 and Supplemental Table 1 online). Similar results were observed when the GID1 overexpression (GID1-OE) constructs were introduced into the more severe sly1-2 mutant (Figure 1A). Final plant height appeared to be independent of the sly1 allele severity (see Supplemental Table 1 online).
Figure 1.
The Effect of GID1 Overexpression on Plant Growth and RGA Protein Accumulation.
(A) Three chimeric constructs (HA:GID1a, HA:GID1b, and HA:GID1c) were introduced into wild-type Ler, sly1-10, sly1-2, ga1-3, and rga-Δ17(+/−) backgrounds. Heterozygous (+/−) plants were used because homozygous rgaΔ-17 (−/−) plants are infertile. Representative 36-d-old T3 transgenic plants in which HA:GID1a, HA:GID1b, and HA:GID1c are overexpressed are shown. Plants were treated with (+GA) or without (−GA) 10 μM GA4 every 3 d. Bars = 5 cm.
(B) Protein blot analysis of HA:GID1 fusions and RGA protein accumulation in independent T3 lines was performed using HA and RGA antibodies. Total protein (150 μg) from wild-type Ler and 40 μg total protein isolated from all other genotypes in the absence of GA application was loaded. Representative pictures shown in (A) correspond to lanes shown in bold in (B). Equal protein loading was confirmed by Ponceau staining (bottom panels).
GA also stimulates reproductive development, transition to flowering, and fertility in Arabidopsis (Cheng et al., 2004; Tyler et al., 2004). To explore the effect of GID1-OE on fertility, we determined the number of seeds per silique for each line (see Supplemental Table 1 online). The infertility of sly1-2 and sly1-10 was better suppressed by transformation with the GID1c-OE construct than by transformation with GID1a-OE or GID1b-OE (see Supplemental Table 2 online). GA treatment of sly1 transformants had no effect on plant height and fertility (Figure 1A; see Supplemental Tables 1 and 2 online), suggesting that endogenous GA levels were not a limiting factor in determining the degree of sly1 phenotype restoration.
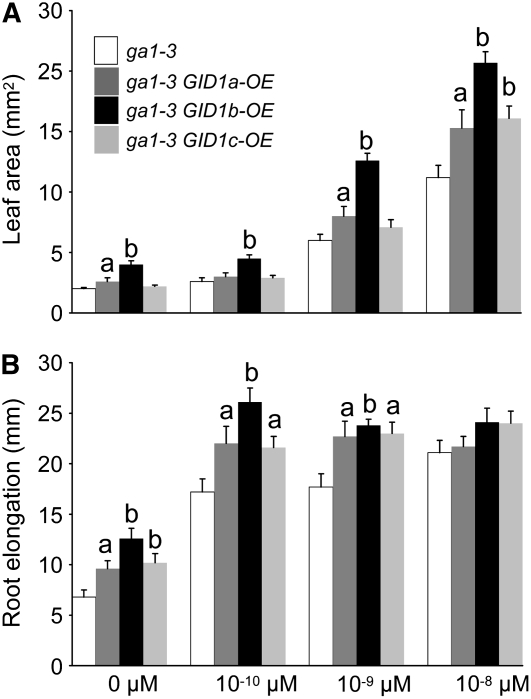
To determine whether GID1-OE can suppress the stem elongation and fertility phenotypes of a GA biosynthesis mutant, the same constructs were introduced into the ga1-3 mutant. None of the GID1-OE constructs rescued plant height or fertility in the absence of GA, suggesting that endogenous GA is necessary (Figure 1A). The GA-treated ga1-3 GID1b-OE lines were the tallest, suggesting that the GID1b-OE lines are more GA sensitive (Figure 1A; see Supplemental Table 1 online). To assess this, we examined the effect of increasing GA concentrations on seedling leaf area and root elongation. Seeds were incubated in GA4 to stimulate seed germination, washed, and transferred to medium containing increasing concentrations of GA4. After 10 d of incubation, seedling leaf area and root length were determined. The GID1b-OE lines appeared to have larger leaf area and longer primary root, suggesting that these lines are more GA sensitive (Figure 2; see Supplemental Figure 3 online). GID1b-OE lines also had larger leaves on media containing no hormone, possibly due to prolonged response to the GA treatment used to stimulate seed germination.
Figure 2.
GID1 Overexpression in ga1-3 Enhanced Vegetative Sensitivity to GA Treatment.
The ga1-3 mutant and the transgenic ga1-3 mutant plants overexpressing each HA:GID1 fusion were grown on MS-agar containing different concentrations of GA4 (0, 10−10, 10−9, and 10−8 M). After 10 d of incubation at 22°C, the leaf area and root elongation of these seedlings was measured. Error bars are se (n = 10). A significant difference from untransformed ga1-3 is indicated: a, P < 0.05; b, P < 0.01, as determined by t test.
GID1 Overexpression Does Not Cause RGA Disappearance in sly1 Mutants
GA stimulates plant growth by triggering the disappearance of RGA protein via the action of GID1, SCFSLY1, and the 26S proteasome (Itoh et al., 2003; Sun and Gubler, 2004; Ueguchi-Tanaka et al., 2007a). Based on this, we hypothesized that GID1 overexpression may stimulate plant growth in the wild-type Ler, sly1, and ga1-3 backgrounds by stimulating DELLA degradation. Protein blot analysis was used to examine the accumulation of DELLA RGA and HA:GID1 protein in the wild type, sly1, and ga1-3 (Figure 1B). GID1-OE caused no change in RGA protein accumulation in the ga1-3 mutant (Figure 1B; see Supplemental Figure 4 online). GID1-OE in wild-type Ler resulted in lower RGA protein levels and increased final plant height compared with the untransformed control (Figure 1A; see Supplemental Table 1 online). This is consistent with previously published results showing that GID1a-OE caused decreased accumulation of RGA (Willige et al., 2007). By contrast, RGA protein destruction was blocked by the sly1-10 and sly1-2 mutations. GID1 overexpression and increased plant height did not correlate with decreased RGA protein accumulation (Figure 1B). Thus, GID1 overexpression in the sly1 mutants restored plant height without altering the RGA protein levels. Whereas the final plant height of sly1 GID1-OE lines was not strongly correlated with the level of HA:GID1 accumulation (Figure 1B; see Supplemental Table 1 online), examination of 10-d-old seedlings revealed that higher levels of HA:GID1b accumulation correlated with more rapid hypocotyl and root elongation (Figure 1B; see Supplemental Figure 5 online). This suggests that GID1 can stimulate GA signaling in sly1 mutants without DELLA proteolysis and that the rate of stem elongation is not determined by the absolute DELLA protein level but by the amount of GID1 accumulation. There are two possible explanations for this. GID1 overexpression may be able to deactivate the DELLA/RGA repression of stem elongation without proteolysis, or it may stimulate stem elongation via a DELLA-independent mechanism. If the latter model is true, we would expect GID1 overexpression to suppress the dwarf phenotype of DELLA motif deletion mutants, gai-1 and rga-Δ17.
GID1-OE Rescue of sly1 Mutants Requires a Functional DELLA Motif
We introduced the three GID1-OE constructs into the gai-1 (−/−) and rga-Δ17 (+/−) backgrounds, two mutants missing the 17–amino acid DELLA motif known to be essential for DELLA protein interaction with the GID1 GA receptor (Peng and Harberd, 1997; Dill et al., 2001; Griffiths et al., 2006; Ueguchi-Tanaka et al., 2007b; Willige et al., 2007; Feng et al., 2008). Transformants carrying the GID1-OE constructs in these backgrounds showed no change in plant height (Figure 1A; see Supplemental Figure 6 online) and no change in DELLA RGA protein levels (Figure 1B). This suggests that GID1 must bind to DELLA protein to stimulate stem elongation, ruling out a DELLA-independent mechanism. Previous work has shown that the gai-1 and rga-Δ17 mutant proteins are resistant to GA-induced degradation and fail to interact with GID1 in the presence of GA, suggesting that the resulting dwarf phenotype is due to inability to undergo GID1/GA-induced protein destruction or deactivation (Dill et al., 2001; Griffiths et al., 2006; Willige et al., 2007; Feng et al., 2008).
Although sly1 mutants accumulate higher levels of DELLA protein than do ga1-3 or the gid1a gid1b gid1c triple mutant, sly1 mutants are taller and show less severe germination and fertility phenotypes (McGinnis et al., 2003; Griffiths et al., 2006; Ariizumi and Steber, 2007; Willige et al., 2007). We previously hypothesized that the intermediate sly1 phenotypes result from DELLA accumulation in a less active form (Ariizumi and Steber, 2007). The GID1 overexpression data suggest that the less active DELLA form may result from interaction with GID1-GA. If so, introduction of the DELLA deletion mutations rga-Δ17 or gai-1 into the sly1 mutant background should block DELLA interaction with GID1 leading to a more severe dwarf phenotype. Indeed, sly1-10 rga-Δ17 (+/−) and sly1-10 gai-1 (−/−) double mutants were completely infertile and showed a more extreme dwarf phenotype than the sly1-10, rga-Δ17 (+/−), and gai-1 (−/−) single mutants (Figures 3A and 3B). This additive effect indicates that the functional DELLA motif is required for the sly1 intermediate fertility and plant height phenotype and suggests that the GID1-GA-DELLA protein complex that accumulates in the sly1 mutant is less effective at repressing stem elongation than is unbound DELLA protein.
Figure 3.
The sly1 Intermediate Phenotype Is Dependent on the Presence of a Functional DELLA Motif in RGA and GAI Proteins.
(A) Shown are 50-d-old sly1-10, sly1-10 rga-Δ17, and rga-Δ17 mutants. Enlarged magnification of the sly1-10 rga-Δ17 double mutant is shown in the right panel.
(B) Shown are 50-d-old sly1-10, gai-1, and sly1-10 gai-1 mutants. Bars = 1 cm.
(C) GAI and RGA protein accumulation in wild-type Ler, ga1-3, sly1-2, sly1-2 gai-t6, gai-1, sly1-10 gai-1, and sly1-10 was determined by protein blot analysis of 40 μg of total protein extracted from 50-d-old rosette leaves. The asterisk denotes nonspecific bands.
Next, the effect of the DELLA motif deletion in gai-1 on protein accumulation in the sly1-10 mutant background was examined. Protein blot analysis of sly1-10, sly1-10 gai-1 (−/−), and gai-1 (−/−) mutants showed that the level of GAI protein accumulation was lower in the sly1-10 gai-1 and gai-1 mutant than in the sly1-10 single mutant (Figure 3C). By contrast, the RGA protein level in sly1-10 gai-1 was equivalent to the sly1-10 mutant. Thus, it appears that GAI regulates its own protein accumulation but not RGA protein accumulation. This suggests that the increased dwarfism and decreased gai-1 protein accumulation in sly1-10 gai-1 compared with the sly1-10 single mutant may be due in part to a failure of the gai-1 protein to interact with GID1.
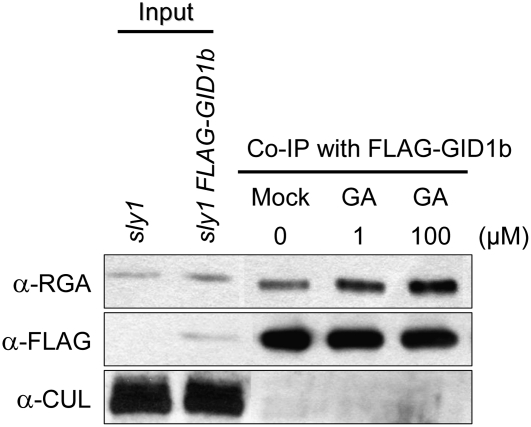
The fact that sly1 rescue by GID1 overexpression requires GA and a functional DELLA motif suggested that GID1 inactivates DELLA protein by direct protein interaction. We next confirmed the GA-dependent interaction of DELLA GAI and RGA with the GID1 receptor. Previously published yeast two-hybrid results indicated that GAI shows GA-dependent interaction with the three GID1 proteins (Nakajima et al., 2006). In an in vitro GST-GID1 pull-down assay, GST-GID1a and GST-GID1b showed interaction with GAI only in the presence of GA, whereas GST-GID1c showed some interaction in the absence of GA and increased interaction in the presence of GA (see Supplemental Figure 7 online). Previously published results showed GA-dependent GST-GID1 pull-down of DELLA RGA (Griffiths et al., 2006). Coimmunoprecipitation (co-IP) of RGA using FLAG:GID1b was used to confirm that RGA shows increasing interaction with FLAG:GID1b with increasing concentrations of GA3 in vivo (Figure 4).
Figure 4.
Increased Interaction of GID1b with DELLA RGA Protein Is Dependent on GA.
The co-IP experiment was performed using protein extracted from 12-d-old sly1-10 FLAG:GID1b seedlings. Protein extract was incubated with FLAG agarose in the presence of 0.1% ethanol (mock), 1 μM GA3, or 100 μM GA3 and loaded on an SDS-PAGE gel. Protein blot analysis was performed using anti-RGA, anti-FLAG, and anti-cullin. Forty micrograms of total sly1-10 and sly1-10 FLAG:GID1b protein were loaded (input).
GID1-OE Suppression of sly1 Dwarfism Is Dependent on the Presence of GA
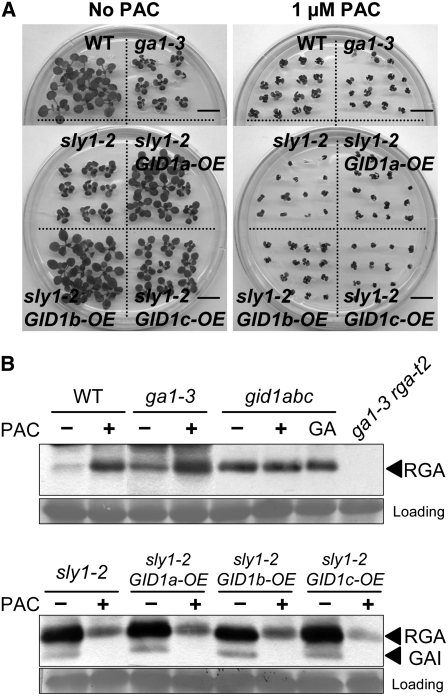
The GA receptor GID1 requires GA to efficiently bind DELLA proteins. If the intermediate plant height and fertility phenotypes of sly1 mutants result from DELLA interaction with GID1-GA, we expect the rescue of sly1 mutants by GID1 overexpression to require GA. Treatment of pregerminated sly1 GID1-OE seedlings with the GA biosynthesis inhibitor paclobutrazol (PAC) blocked the growth restoration from GID1a, GID1b, and GID1c overexpression in sly1 seedlings (Figure 5A; see Supplemental Figure 8 online). This confirms that growth stimulation by GID1 overexpression requires GA. PAC treatment also suppressed the growth of the wild type and ga1-3 (Figure 5A). PAC treatment probably slightly retards the growth of ga1-3 due to the fact that some GA biosynthesis occurs in ga1-3 (Sun et al., 1992; Zeevaart and Talon, 1992).
Figure 5.
The GA Biosynthesis Inhibitor PAC Blocks Rescue of sly1 by GID1 Overexpression and Causes Decreased RGA and GAI Accumulation.
(A) The 10-d-old Ler, ga1-3, sly1-2, and the sly1-2 GID1-OE plants were transferred to MS-agar with and without 1 μM PAC treatment and incubated for 12 d at 22°C. Bars = 1 cm.
(B) The effect of PAC treatment on the RGA and GAI protein accumulation was determined by protein blot analysis using RGA antibody. Forty micrograms of total protein from (A) was loaded and equal loading confirmed by Ponceau staining. Controls include wild-type Ler, ga1-3, ga1-3 rgat-2, and the gid1a gid1b gid1c triple mutant with and without GA treatment.
PAC treatment caused the expected increase in RGA protein accumulation in GA-treated wild type (Figure 5B). This is likely due to reduced GA-dependent proteolysis of RGA protein. PAC treatment caused no significant change in RGA protein levels in the gid1a gid1b gid1c triple mutant. By contrast, PAC treatment caused a decrease in RGA and GAI protein accumulation in sly1-10, sly1-2, and in sly1 mutants transformed with the GID1-OE constructs. Following PAC treatment of sly1 mutants, the RGA protein accumulation decreased to a level similar to that seen in PAC-treated wild type and ga1-3. These results raise two interesting notions. First, GA is necessary for GID1-OE suppression of the GA-insensitive sly1 dwarf phenotype. Second, the high level of RGA protein accumulation in sly1 mutants requires GA synthesis.
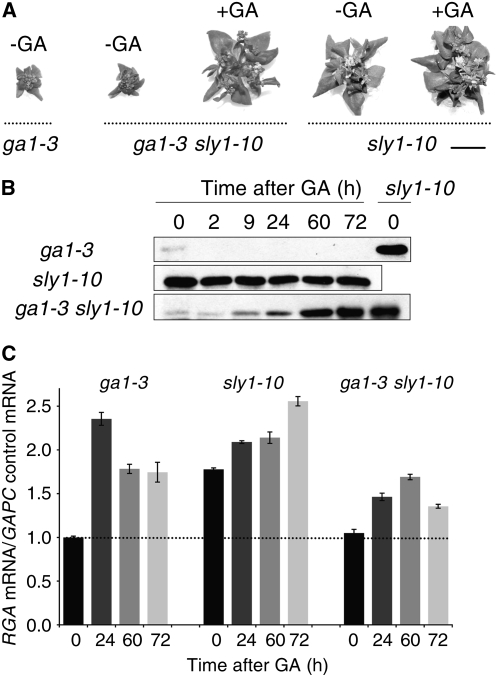
To confirm that the decrease in DELLA accumulation was not due to nonspecific effects of PAC, the accumulation of DELLA RGA was examined over a time course of GA treatment in a ga1-3 sly1-10 double mutant. The dwarf phenotype of ga1-3 sly1-10 is more severe than that of sly1-10 and is rescued by GA treatment (Figure 6A). Consistent with the PAC experiment, less DELLA RGA protein was seen in the ga1-3 sly1-10 double mutant than in the sly1-10 mutant in the absence of GA (Figure 6B). Surprisingly, GA treatment of the ga1-3 sly1-10 double mutant resulted in a gradual increase in DELLA RGA protein accumulation (Figure 6B) associated with a reproducible ∼50% increase in RGA mRNA level (Figure 6C). Thus, the severity of the dwarf phenotype does not directly correlate with the level of DELLA RGA protein accumulation in the sly1-10 background, suggesting that GID1-GA can partly inactivate DELLA RGA protein repression of stem elongation in the sly1 mutant background, even when DELLA RGA protein levels are increasing.
Figure 6.
Effect of the ga1-3 Mutation on sly1-10 Growth and RGA Protein Accumulation.
(A) The 21-d-old seedlings of ga1-3, sly1-10, and the ga1-3 sly1-10 double mutant in the absence (−GA) and presence (+GA) of 100 μM GA4. Bar = 5 mm.
(B) The effect of GA treatment (100 μM GA3 treatment) on accumulation of RGA protein in the ga1-3, sly1-10, and ga1-3 sly1-10 mutants. Plants (35 d old) were treated with GA, and time points were taken as indicated for protein blot analysis. sly1-10 is a control for equal loading. Protein was extracted from rosette leaves.
(C) RGA mRNA accumulation in rosette leaves of the ga1-3, sly1-10, and ga1-3 sly1-10 mutants was determined at time points indicated after GA treatment by quantitative RT-PCR. Mean values for at least three independent experiments are shown. Error bars show sd.
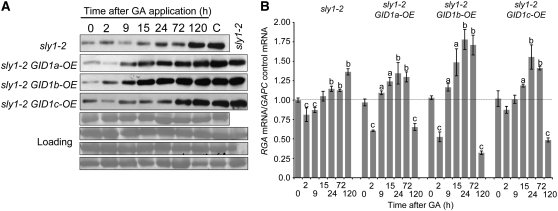
Next, we examined the effect of GID1 overexpression on GA induction of RGA protein accumulation over a time course in PAC-treated sly1-2 mutant and sly1-2 GID-OE seedlings. Pregerminated seedlings were grown in the presence of PAC and then treated with 100 μM GA3. Time points were taken for protein blot and quantitative RT-PCR analysis of RGA protein and RGA mRNA accumulation, respectively. All GA-treated lines showed a gradual increase in RGA protein levels reaching a maximum that resembled control seedlings that were not treated with PAC (Figure 7A). This increase was associated with a small but significant increase in RGA mRNA transcript level (Figure 7B), suggesting that this increase is due at least in part to a gradual induction of RGA transcription by GA. PAC treatment caused a decrease in RGA mRNA levels both in sly1-2 and in wild-type Ler (see Supplemental Figure 9 online). It appeared that GID1 overexpression significantly accelerated RGA mRNA and protein accumulation in response to GA treatment (Figure 7). Interestingly, GID1 overexpression results in a higher level of RGA mRNA accumulation associated with a more rapid increase in RGA protein accumulation.
Figure 7.
Interaction with GID1 Accelerates RGA Protein Accumulation after GA Treatment.
(A) Ten-day-old sly1-2 and sly1-2 GID1-OE seedlings were transferred to MS-agar plus 1 μM PAC for 12 d. Seedlings were sprayed with 100 μM GA3 and time points (0, 2, 9, 15, 24, 72, and 120 h) taken for protein blot analysis. Untreated sly1-2 is a loading control. C, control without PAC. Equal protein loading was confirmed by Ponceau staining.
(B) RGA mRNA accumulation was determined by quantitative RT-PCR during the time course indicated. Mean values for at least three independent experiments are shown. Error bars show sd. A t test was used to determine a statistically significance increase (a, P < 0.05; b, P < 0.01) or decrease (c, P < 0.05) compared with 0 h.
DISCUSSION
The data presented here suggest that DELLA repression activity may be regulated by a proteolysis-independent mechanism, involving protein interaction with GID1-GA. GID1 overexpression can rescue the sly1 dwarf and infertility phenotypes without leading to a decrease in DELLA protein level (Figure 1B). The rescue of the sly1 phenotype by GID1-OE constructs required the presence of GA and an intact DELLA domain (Figures 1 to 5; see Supplemental Figures 7 and 8 online). Since the interaction of GID1 with DELLA protein requires GA, this result suggested to us that GID1 rescue of sly1 mutants may result from inactivation of DELLA protein through interaction with GID1-GA via the DELLA/TVHYNP motif. In support of this, we found that GID1 overexpression failed to rescue the dwarf phenotype of DELLA deletion mutants rga-Δ17 and gai-1 (Figure 1A; see Supplemental Figure 6 online). This suggests that GID1-OE cannot rescue GA signaling without interaction with DELLA proteins through the DELLA motif. Moreover, it appears that the intermediate phenotype of sly1 mutants is due to inactivation of DELLA repressors through GA-dependent interaction of GID1 with DELLA protein (Figures 3 to 6; see Supplemental Figure 7 online).
Degree of Overlap in GID1a, GID1b, and GID1c Functions
Overexpression of GID1b caused the strongest suppression of the sly1 dwarf phenotype (Figure 1A), whereas overexpression of GID1a and GID1c gave a stronger suppression of the sly1 fertility phenotype (see Supplemental Table 2 online). By contrast, the gid1a gid1c double mutant showed more severe stem elongation and fertility phenotypes than did the gid1a gid1b and gid1b gid1c double mutants (Griffiths et al., 2006; Iuchi et al., 2007; Willige et al., 2007). The fact that the gid1a gid1c double mutant shows a GA-insensitive semidwarf phenotype suggests that GID1b cannot fully substitute for GID1a and GID1c (Griffiths et al., 2006). This may be due to the fact that the GID1b mRNA accumulates at lower levels in most tissues than do GID1a and GID1c, rather than to lower GA receptor activity (Griffiths et al., 2006). Interestingly, constitutive expression of GID1b in the ga1-3 background resulted in the largest increase in GA sensitivity in dose–response experiments (Figure 2; see Supplemental Figure 3 online). This is consistent with the data of Nakajima et al. (2006), showing that of the three Arabidopsis GID1 proteins, GID1b has the strongest GA binding affinity. In yeast two-hybrid assays, GID1b shows reduced interaction with DELLA proteins in the absence of GA (Griffiths et al., 2006; Nakajima et al., 2006). However, this GA-independent interaction may not be significant in planta since GID1b showed GA-dependent interaction with DELLAs RGA and GAI in co-IP and GST-GID1 pull-down assays (Figure 4; Griffiths et al., 2006). GA appears to be necessary for in planta downregulation of DELLA by GID1 since transformation of ga1-3 with the GID1b-OE construct resulted in increased GA sensitivity but no increase in plant height in the absence of GA (Figures 1 and 2). Thus, GID1b overexpression may result in the strongest rescue in part because GID1b interaction with DELLA protein requires less bioactive GA (Figure 2; Griffiths et al., 2006; Nakajima et al., 2006).
The Intermediate Phenotype of sly1 Mutants Is Dependent on GA and the DELLA Motif
If GA stimulates plant growth solely by stimulating proteolysis of DELLA repressors, then we would expect the level of DELLA protein accumulation to directly correlate with the severity of GA-deficient or -insensitive growth phenotypes. By contrast, this study and others have found that the gid1a gid1b gid1c triple mutant and ga1-3 GA biosynthesis mutant display far more severe plant growth phenotypes than the sly1-10 and sly1-2 mutants but accumulate far lower levels of the DELLA proteins RGA, GAI, and RGL2 (Figures 3 and 5; see Supplemental Figure 8 online; McGinnis et al., 2003; Griffiths et al., 2006; Ariizumi and Steber, 2007; Willige et al., 2007). This discrepancy suggests that the DELLA protein accumulating in sly1 mutants is less active as a repressor of GA responses. It appears that the inactivation of DELLA protein in the sly1 background requires GA and the DELLA motif since the sly1 dwarf and infertility phenotypes were rendered more severe by introduction of either a ga1-3 mutation (Figure 6) or a deletion of the DELLA/TVHYNP motif required for interaction with GID1 in RGA or GAI (rga-Δ17 or gai-1; Figure 3). These results suggest that RGA protein functions purely as a repressor of GA responses in the absence of GA and that DELLA protein inactivation in the sly1 mutant background requires the ability of GID1-GA to interact with the DELLA motif. If this model is true, increased expression of the GID1 gene might increase the ratio of inactive GID1-GA-DELLA complex to active DELLA repressor in the sly1 mutant background.
Evidence for Proteolysis-Independent Regulation of DELLA Protein in Plant Growth
Evidence presented here indicates that GID1 overexpression can rescue the sly1 dwarf and infertility phenotypes in the presence of high-level DELLA RGA accumulation. GID1-GA triggers GA responses in part by stimulating SCFSLY1-directed ubiquitination and proteolysis of DELLA proteins (Jiang and Fu, 2007; Ueguchi-Tanaka et al., 2007a). HA:GID1a, HA:GID1b, and HA:GID1c overexpression rescues the sly1 dwarf and fertility phenotype without causing RGA destruction (Figure 1). This is consistent with previously published work showing that GID1a overexpression causes increased plant height and earlier flowering in wild-type Ler (Willige et al., 2007) and showing that GID1b overexpression can partly rescue the dwarfism of the ga20ox1 ga20ox2 double mutant (Rieu et al., 2007). Rescue of the sly1 phenotype by GID1 overexpression was blocked by the GA biosynthesis inhibitor PAC, indicating that rescue was dependent on the presence of GA (Figure 5; see Supplemental Figure 8 online). Thus, GA signaling can occur in the absence of DELLA destruction.
In examining the GA dependence of GID1-OE rescue of sly1, we observed that PAC inhibition of GA biosynthesis leads to decreased RGA accumulation (Figure 5; see Supplemental Figure 8 online), whereas GA treatment of PAC-treated sly1 seedlings leads to increased DELLA RGA protein accumulation (Figure 7). This is the reverse of what occurs in the presence of a wild-type SLY1 allele. Similarly, the ga1-3 sly1-10 double mutant accumulates RGA at a lower level similar to that found in ga1-3, and GA treatment of ga1-3 sly1-10 causes increased RGA and protein accumulation (Figure 6B). This GA-induced increase in DELLA RGA protein accumulation was associated with a small but significant increase in RGA mRNA accumulation (Figures 6C and 7B). This small increase in mRNA accumulation may result in a gradual increase in RGA protein accumulation in the absence of SCFSLY1-directed proteolysis. PAC treatment also leads to a decrease in RGA mRNA accumulation in wild-type Ler, indicating that this mechanism is not unique to the sly1 mutant background (see Supplemental Figure 9 online). The RGA mRNA levels are transiently induced by GA and show a decrease at 120 h after GA treatment (Figure 7B). The fact that RGA protein remained high suggests that RGA protein is more stable in the sly1 mutant background.
The GA-dependent increase in DELLA protein may also result in part from the interaction of DELLA protein with GID1-GA. The DELLA motif deletion in gai-1 resulted in decreased accumulation of gai-1 protein in the sly1-10 mutant background (Figure 3C). Moreover, mutations in the rice GID1 gene act additively to decrease accumulation of DELLA SLR1 protein in the rice gid2 (sly1 homolog) mutant background (Ueguchi-Tanaka et al., 2008). Thus, the overaccumulation of DELLA protein in gid2/sly1 mutants depends on GID1 and the DELLA domain. Future work will need to explore whether DELLA protein accumulation is regulated by additional posttranscriptional mechanisms.
GID1 may influence DELLA accumulation or repression of GA responses through changes in DELLA protein posttranslational modification, such as phosphorylation or O-Glc-NAc modification. Phosphorylated forms of rice DELLA SLR1 and of Arabidopsis DELLA proteins gai-1 and RGL2 have been identified (Sasaki et al., 2003; Fu et al., 2004; Gomi et al., 2004; Hussain et al., 2005, 2007; Itoh et al., 2005). The phosphorylated form of RGL2 may show increased stability (Hussain et al., 2005). The O-Glc-NAc transferase SPINDLY (SPY) functions as a negative regulator of GA signaling in Arabidopsis, barley (Hordeum vulgare), and rice (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996; Swain et al., 2001; Robertson, 2004; Shimada et al., 2006). Mutations in Arabidopsis SPY lead to increased RGA protein accumulation and partly rescue the dwarfism of rga-Δ17, suggesting that SPY may provide DELLA motif-independent activation of the DELLA repressor (Silverstone et al., 2007). Silencing of rice SPY led to increased accumulation of phosphorylated DELLA SLR1, suggesting that O-Glc-NAc may compete with phosphorylation for modification of the same Ser or Thr residues (Shimada et al., 2006). It may be inferred that the O-Glc-NAc modified form is an active DELLA repressor. It is unclear whether or how GID1-GA influences these DELLA posttranslational modifications. RGA protein attains resistance to GA-induced degradation by proteolysis in the presence of other plant hormones such as auxin, ethylene, and abscisic acid (Achard et al., 2003, 2006, 2007; Fu and Harberd, 2003), but it is unknown whether these hormones alter the posttranslational modification of DELLA.
Recent work has shown that DELLA proteins RGA and GAI bind to PIF3 and PIF4 proteins, DNA binding bHLH-type transcription factors that positively regulate gene expression associated with hypocotyl elongation (de Lucas et al., 2008; Feng et al., 2008). DELLA proteins were shown to inhibit hypocotyl elongation by binding to PIF proteins in the absence of GA, thus preventing PIF proteins from binding target gene promoters. GA-stimulated DELLA protein degradation releases PIF proteins, allowing them to bind and activate target gene promoters. It is possible that GID1 proteins inactivate DELLA proteins either by competing with PIF for DELLA binding or by forming of a GID1-GA-DELLA-PIF complex that allows only intermediate levels of PIF-activated gene expression.
Model for GA Signaling
Our data suggest a new model in which DELLA repressors can be deactivated both by SCFSLY1-dependent proteolysis and by direct protein interaction with the GA receptor GID1. This proteolysis-independent mechanism is conserved in Arabidopsis and rice (Ueguchi-Tanaka et al., 2008). Previously published data supported the model in which interaction of DELLA protein with GID1-GA increases the binding of DELLA protein to the SCFSLY1 complex (Griffiths et al., 2006). SCFSLY1 catalyzes polyubiquitination of DELLA proteins leading to their destruction by the 26S proteasome. In this case, DELLA represses GA responses in the absence of GA and then is rapidly destroyed upon addition of GA thereby lifting DELLA repression (Figure 8A). The fact that GA cannot cause DELLA destruction in the sly1 mutant results in high levels of DELLA protein accumulation. However, some of the DELLA protein in sly1 mutants is inactivated by binding to GID1-GA, resulting in an intermediate phenotype (Figure 8B). GID1 overexpression in the sly1 mutant background leads to increased GA response by increasing the proportion of GID1-GA-DELLA complex to DELLA (Figure 8C). In the absence of SCFSLY1-directed ubiquitination, DELLA repression of stem elongation and flowering can still be blocked via a process that requires GID1, GA, and an intact DELLA motif. The model proposed is that GID1-GA binding to DELLA protein leads to inactivation of DELLA repressor activity. Inactivation may result directly from protein interaction or indirectly through posttranslational modification or competition with other DELLA binding proteins. This mechanism may stimulate GA responses under environmental conditions that may block DELLA degradation, such as drought or high-salinity contributing to the delicate balance of GA-regulated growth and development (Achard et al., 2006, 2007).
Figure 8.
Model for the Proteolysis-Independent Regulation of DELLA Repressor Activity.
(A) In the absence of GA, GA responses are inhibited since RGA protein levels and repressor activity is high. GA treatment relieves DELLA repression of GA responses by causing the formation of the GID1-GA-DELLA complex recognized by the SCFSLY1 E3 ubiquitin ligase. Polyubiquitination by SCFSLY1 causes DELLA proteolysis via the 26S proteasome.
(B) The sly1 mutants accumulate DELLA proteins at a higher level due to lack of DELLA ubiquitination and proteolysis. However, the DELLA protein that accumulates is a mixture of the active DELLA repressor (light gray) and inactive GID1-GA-DELLA (dark gray), resulting in an intermediate phenotype.
(C) sly1 GID1-OE plants show increased GA response due to an increase in the proportion of inactive GID1-GA-DELLA complex relative to active DELLA repressor.
METHODS
Plant Materials and Growth Conditions
Seeds of wild-type Arabidopsis thaliana Ler, sly1-10, sly1-2, gai-1, rga-Δ17, ga1-3, gai-t6, ga1-3 rga-t2, and transformants in which each GID1 gene is overexpressed were used in this study (Koornneef et al., 1985; Peng and Harberd, 1993; Peng et al., 1997; Steber et al., 1998; Dill et al., 2001; Steber and McCourt, 2001). All genotypes are in the Ler background. The rga-Δ17 line contains a wild-type copy of RGA on the chromosome and a transgene containing a 17–amino acid deletion of the DELLA motif as described by Dill et al. (2001). Germination of ga1-3 seeds was stimulated by first imbibing in 100 μM GA4 for 3 d at 4°C and then washing five times with sterile water. Seeds of Ler, sly1-10, sly1-2, gai-1, and rga-Δ17 backgrounds were sterilized and imbibed in sterile water for 3 d at 4°C. After the stratification, all seeds were transferred to the 0.5× Murashige and Skoog (MS) salts (Sigma-Aldrich)/0.8% agar (MS-agar), and they were incubated at 22°C for 10 to 14 d. Seedlings were transferred to soil and grown at 22°C under fluorescent light (16 h day; McGinnis et al., 2003) for growth rate, and fertility was compared. To determine the effect of GA treatment on the plant growth, plants grown in soil were sprayed every 3 d with 10 μM GA4.
Vector Construction and Transformation
In-frame fusions of the GID1a, GID1b, and GID1c to the hemagglutinin (HA) epitope tag were constructed under control of the constitutive CaMV 35S promoter. The DNA sequence corresponding to three repetitions of the HA epitope was amplified using HA-F and HA-3R primers (see Supplemental Table 3 online). The PCR fragment was phosphorylated with T4 polynucleotide kinase (Fermentas) and then blunt-ligated into the EcoRV site of pBluescript II KS− vector. This vector was named HA/pBluescript. PCR fragments containing the full-length coding region of the GID1a, GID1b, and GID1c genes were obtained by RT-PCR using gene-specific primer pairs (Griffiths et al., 2006; see Supplemental Table 3 online) and then directly cloned as a blunt-end fragment into HA/pBluescript at the SmaI site to obtain HA-GID1a-c/pBluescript vectors. To make the HA-only control and HA:GID1 constructs, the HindIII-SacI fragment from the HA/pBluescript and HA:GID1a-c/pBluescript plasmids were excised and cloned into the T-DNA binary vector pTA27 that had been digested with HindIII and SacI. pTA27 contains the 35S promoter and nos terminator from pBI101H (Ariizumi et al., 2002) inserted as an EcoRI-HindIII fragment into EcoRI-HindIII–digested pGPTV-HPT (Becker et al., 1992) thereby replacing the GUS gene. The in-frame N-terminal fusion of GID1b to the FLAG epitope was constructed by transferring the GID1b gene from pENTR1A (Invitrogen) to pEarleyGate202 (ABRC) using the Gateway LR Clonase enzyme (Invitrogen). The constructs were then transformed into Agrobacterium tumefaciens GV3101 by the freeze-thaw method (An et al., 1988). Constructs were transformed into Arabidopsis Ler, ga1-3, sly1-2, sly1-10, gai-1, and rga-Δ17 by the flower dip method (Clough and Bent, 1998). Transgenic plants were selected on MS-agar plates containing hygromycin (15 to 20 mg/L). Four to eight independent transformants were isolated in each background. Transformants showing similar levels of HA:GID1 protein accumulation were selected for further studies (Figure 1). The construction of the GST:GID1 vectors was previously described (Griffiths et al., 2006).
Measurement of GA Sensitivity in ga1-3 GID1-OE Lines
To examine GA sensitivity, seeds of the ga1-3 mutant and of ga1-3 GID1-OE lines were imbibed at 4°C for 3 d in the presence of 10 μM GA4, washed five times with sterile water, then transferred to the MS-agar plates containing 0, 10−10 M, 10−9 M, and 10−8 M GA4. Leaf area and root length were determined after 10 d of growth at 22°C under constant light.
PAC Experiments
Experiments were conducted to examine the effect of the GA biosynthesis inhibitor PAC on seedling growth and DELLA RGA protein accumulation. Seeds of Ler, sly1-10, and sly1-2 and corresponding transformants carrying the GID1-OE constructs were first germinated under MS-agar plates as described above. Note that ga1-3 seeds were pretreated with GA4 to stimulate germination. Ten-day-old seedlings were transferred to fresh MS-agar plates including 1 μM PAC, followed by further incubation for 12 d. For time-course experiments, the PAC-treated plants were first prepared as described above and treated with 100 μM GA3, and time points (0, 2, 9, 15, 24, 72, and 120 h) were taken for quantitative RT-PCR and protein blot analyses.
Expression Analysis
Protein blot analysis was used to examine DELLA RGA and GAI protein accumulation in 10- to 14-d-old seedlings. Transgenic homozygous T3 seeds were germinated under hygromycin selection (15 mg/L) and imbibed for 3 d at 4°C, followed by incubation at 22°C. Total plant protein was extracted as described (Silverstone et al., 2001). Forty micrograms of protein was separated on an SDS-PAGE and transferred onto a polyvinylidene difluoride membrane. The protein concentration was determined using the Bio-Rad protein assay and even loading confirmed by Ponceau staining. For RGA protein detection in the wild-type background, 150 μg of protein was loaded. Protein detection was performed using an enhanced chemiluminescence system (ECL; GE Healthcare) according to the manufacturer's protocol. RGA and GAI proteins were detected using the polyclonal RGA (1:10,000; Silverstone et al., 2001), GAI (1:1000; Willige et al., 2007), and CULLIN (1:10,000; Chen et al., 2006) as the primary antibodies. HA:GID1 fusion proteins were detected using anti-HA (1:5000; Immunology Consultants Laboratory.). The anti-rabbit IgG-horseradish peroxidase (GE Healthcare) was used as a secondary antibody (1:150,000).
RGA mRNA accumulation was analyzed by quantitative RT-PCR using total RNA extracted from GA-treated seedling tissues as indicated in Figures 6 and 7 using an RNA easy kit (Qiagen). Genomic DNA contamination was removed using the DNA-Free RNA kit (ZYMO Research). cDNA was generated from 1 μg total RNA using a first-strand cDNA synthesis kit (GE Healthcare). cDNA was then used as a template for quantitative PCR with specific primers (see Supplemental Table 3 online; Tyler et al., 2004, Griffiths et al., 2006). The quantitative PCR experiments were performed using a Roche LightCycler with LightCycler FastStart DNA Master SYBR Green I kit. PCR conditions consisted of 10 min of denaturation at 95°C, followed by 45 cycles of 10 s denaturation at 95°C, 5 s annealing at 60°C, and 10 s extension at 72°C. Transcript levels were analyzed using LightCycler Software version 3.5 to determine RGA mRNA levels relative to the GAPC control mRNA as by Griffiths et al. (2006).
Co-IP Experiment
The sly1-10 mutant was transformed with a GID1b N-terminal fusion to the FLAG epitope expressed on the constitutive 35S promoter (FLAG:GID1b). The fusion appeared to be functional since transformation of sly1-10 with 35S:FLAG:GID1b resulted in a rescue of dwarfism similar to than seen with 35:HA:GID1b (Figure 1). The 10-d-old sly1-10 FLAG:GID1b seedlings were ground in liquid N2 and suspended in buffer C (20 mM Tris, 150 mM NaCl, 0.5% Triton, and 1× complete proteinase inhibitor [Roche]). The protein extract was centrifuged at 21,000g for 15 min, and 1 mg of supernatant was incubated with FLAG M2 agarose (Sigma-Aldrich) for 12 h at 4°C in the presence of ethanol (mock), 1 μM GA3, and 100 μM GA3. The co-IP agarose was washed three times with buffer C, 6× sample buffer was added, and the sample was boiled for 5 min prior to protein blot analysis.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: SLY1 (At4g24210), RGA (At2g01570), GAI (At1g14920), GID1a (At3g05120), GID1b (At3g63010), GID1c (At5g27320).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Diagram of the Chimeric HA:GID1 Constructs.
Supplemental Figure 2. The HA Control Construct Had No Effect on Growth and Development.
Supplemental Figure 3. ga1-3 Plants Used for Measurements Shown in Figure 2 Show Enhanced GA Sensitivity when Transformed with GID1-OE Constructs.
Supplemental Figure 4. RGA Protein Accumulation after GA Treatment in ga1-3 GID1-OE Plants.
Supplemental Figure 5. HA:GID1b Levels Correlate with sly1-10 Seedling Growth.
Supplemental Figure 6. HA:GID1 Constructs Did Not Suppress Growth Defects of the gai-1 Mutant.
Supplemental Figure 7. GST-GID1 Pull-Down Assay Shows Interaction with DELLA GAI Protein.
Supplemental Figure 8. The GA Biosynthesis Inhibitor PAC Blocks Rescue of sly1-10 by GID1 Overexpression.
Supplemental Figure 9. Comparison of the Level of RGA mRNA Accumulation in Wild-Type Ler, ga1-3, sly1-2, and gid1 Mutants.
Supplemental Table 1. Final Plant Height in GID1-OE Lines.
Supplemental Table 2. Fertility in GID1-OE.
Supplemental Table 3. Primer Sequences Used for This Study.
Supplementary Material
Acknowledgments
We thank M. Matsuoka and M. Ueguchi-Tanaka for stimulating discussions, for sharing data prior to publication, and for the SLR1 antibody. Thanks are due to C. Schwechheimer for the gift of the GAI antibody and the gid1a gid1b gid1c mutant. We are also grateful to X.W. Deng for providing the cullin antibody. We thank P. Chandler for long ago suggesting the construction of a ga1-3 sly1 double mutant. Finally, we would like to thank members of the Steber and Sun laboratories for lively discussion and critical reading of the manuscript. This work was supported by Japan Society for the Promotion of Science fellowship funds to T.A. and K.M., USDA Cooperative State Research, Education, and Extension Service Grant 2005-01099 and USDA Agricultural Research Service funds to C.M.S., and the National Science Foundation (IBN-0348814) to T.-p.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Camille M. Steber (csteber@wsu.edu).
Online version contains Web-only data.
References
- Achard, P., Baghour, M., Chapple, A., Hedden, P., Van der Straeten, D., Genschik, P., Moritz, T., and Harberd, N.P. (2007). The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation ot floral meristem-identity genes. Proc. Natl. Acad. Sci. USA 104 6484–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., Van der Straeten, D., Peng, J.R., and Harberd, N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311 91–94. [DOI] [PubMed] [Google Scholar]
- Achard, P., Vriezen, W.H., Van Der Straeten, D., and Harberd, N.P. (2003). Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15 2816–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, G., Ebert, R.R., Mitra, A., and Ha, S.B. (1988). Binary vectors. In Plant Molecular Biology Manual, S.B Gelvin and R.A. Schilpeoort, eds (Dordrecht, The Netherlands: Kluwer), pp. 1–19.
- Ariizumi, T., Kishitani, S., Inatsugi, R., Nishida, I., Murata, N., and Toriyama, K. (2002). An increase in unsaturation of fatty acids in phosphatidylglycerol from leaves improves the rates of photosynthesis and growth at low temperatures in transgenic rice seedlings. Plant Cell Physiol. 43 751–758. [DOI] [PubMed] [Google Scholar]
- Ariizumi, T., and Steber, C.M. (2007). Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell 19 791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20 1195–1197. [DOI] [PubMed] [Google Scholar]
- Cao, D.N., Hussain, A., Cheng, H., and Peng, J.R. (2005). Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223 105–113. [DOI] [PubMed] [Google Scholar]
- Chen, H., Shen, Y., Tang, X., Yu, L., Wang, J., Guo, L., Zhang, Y., Zhang, H., Feng, S., Strickland, E., Zheng, N., and Deng, X.W. (2006). Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18 1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H., Qin, L.J., Lee, S.C., Fu, X.D., Richards, D.E., Cao, D.N., Luo, D., Harberd, N.P., and Peng, J.R. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131 1055–1064. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- de Lucas, M., Daviere, J.M., Rodriguez-Falcon, M., Pontin, M., Iglesias-Pedraz, J.M., Lorrain, S., Fankhauser, C., Blazquez, M.A., Titarenko, E., and Prat, S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451 480–484. [DOI] [PubMed] [Google Scholar]
- Dill, A., Jung, H.S., and Sun, T.P. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 98 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., and Sun, T.P. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., Thomas, S.G., Hu, J.H., Steber, C.M., and Sun, T.P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X.D., and Harberd, N.P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421 740–743. [DOI] [PubMed] [Google Scholar]
- Fu, X.D., Richards, D.E., Fleck, B., Xie, D.X., Burton, N., and Harberd, N.P. (2004). The Arabidopsis mutant sleepy1(gar2–1) protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16 1406–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi, K., Sasaki, A., Itoh, H., Ueguchi-Tanaka, M., Ashikari, M., Kitano, H., and Matsuoka, M. (2004). GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 37 626–634. [DOI] [PubMed] [Google Scholar]
- Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z.L., Powers, S.J., Gong, F., Phillips, A.L., Hedden, P., Sun, T.P., and Thomas, S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain, A., Cao, D.N., Cheng, H., Wen, Z.L., and Peng, J.R. (2005). Identification of the conserved serine/threonine residues important for gibberellin-sensitivity of Arabidopsis RGL2 protein. Plant J. 44 88–99. [DOI] [PubMed] [Google Scholar]
- Hussain, A., Cao, D.N., and Peng, J.R. (2007). Identification of conserved tyrosine residues important for gibberellin sensitivity of Arabidopsis RGL2 protein. Planta 226 475–483. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Matsuoka, M., and Steber, C.M. (2003). A role for the ubiquitin-26S-proteasome pathway in gibberellin signaling. Trends Plant Sci. 8 492–497. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Sasaki, A., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Hasegawa, Y., Minami, E., Ashikari, M., and Matsuoka, M. (2005). Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1. Plant Cell Physiol. 46 1392–1399. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi, S., Suzuki, H., Kim, Y.C., Iuchi, A., Kuromori, T., Ueguchi-Tanaka, M., Asami, T., Yamaguchi, I., Matsuoka, M., Kobayashi, M., and Nakajima, M. (2007). Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J. 50 958–966. [DOI] [PubMed] [Google Scholar]
- Jacobsen, S.E., Binkowski, K.A., and Olszewski, N.E. (1996). SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction Arabidopsis. Proc. Natl. Acad. Sci. USA 93 9292–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., and Olszewski, N.E. (1993). Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C., and Fu, X. (2007). GA action: Turning on de-DELLA repressing signaling. Curr. Opin. Plant Biol. 10 461–465. [DOI] [PubMed] [Google Scholar]
- King, K.E., Moritz, T., and Harberd, N.P. (2001). Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Elgersma, A., Hanhart, C.J., van Loenen-Martinet, E.P., van Rign, L., and Zeevaart, J.A.D. (1985). A gibberellin in sensitive mutant of Arabidopsis thaliana. Physiol. Plant. 65 33–39. [Google Scholar]
- Lee, S.C., Cheng, H., King, K.E., Wang, W.F., He, Y.W., Hussain, A., Lo, J., Harberd, N.P., and Peng, J.R. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis, K.M., Thomas, S.G., Soule, J.D., Strader, L.C., Zale, J.M., Sun, T.P., and Steber, C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, M., et al. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46 880–889. [DOI] [PubMed] [Google Scholar]
- Peng, J., and Harberd, N.P. (1997). Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome-deficient mutants of Arabidopsis. Plant Physiol. 113 1051–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J.R., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J.R., and Harberd, N.P. (1993). Derivative alleles of the Arabidopsis gibberellin-insensitive (gai) mutation confer a wild-type phenotype. Plant Cell 5 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J.R., et al. (1999). 'Green revolution' genes encode mutant gibberellin response modulators. Nature 400 256–261. [DOI] [PubMed] [Google Scholar]
- Richards, D.E., King, K.E., Ait-ali, T., and Harberd, N.P. (2001). How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 67–88. [DOI] [PubMed] [Google Scholar]
- Rieu, I., Ruiz-Rivero, O., Fernandez-Garcia, N., Griffiths, J., Powers, S.J., Gong, F., Linhartova, T., Eriksson, S., Nilsson, O., Thomas, S.G., Phillips, A.L., and Hedden, P. (2007). The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 53 488–504. [DOI] [PubMed] [Google Scholar]
- Robertson, M. (2004). Two transcription factors are negative regulators of gibberellin response in the HvSPY-signaling pathway in barley aleurone. Plant Physiol. 136 2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Jeong, D.H., An, G., Kitano, H., Ashikari, M., and Matsuoka, M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299 1896–1898. [DOI] [PubMed] [Google Scholar]
- Shimada, A., Ueguchi-Tanaka, M., Sakamoto, T., Fujioka, S., Takatsuto, S., Yoshida, S., Sazuka, T., Ashikari, M., and Matsuoka, M. (2006). The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 48 390–402. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L., Jung, H.S., Dill, A., Kawaide, H., Kamiya, Y., and Sun, T.P. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13 1555–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Mak, P.Y.A., Martinez, E.C., and Sun, T.P. (1997). The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Tseng, T.S., Swain, S.M., Dill, A., Jeong, S.Y., Olszewski, N.E., and Sun, T.P. (2007). Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 143 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber, C.M., Cooney, S.E., and McCourt, P. (1998). Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber, C.M., and McCourt, P. (2001). A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 125 763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T.P., and Gubler, F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55 197–223. [DOI] [PubMed] [Google Scholar]
- Sun, T.P., Goodman, H.M., and Ausubel, F.M. (1992). Cloning the Arabidopsis Ga1 locus by genomic subtraction. Plant Cell 4 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T.P., and Kamiya, Y. (1994). The Arabidopsis Ga1 locus encodes the cyclase Ent-kaurene synthetase-a of gibberellin biosynthesis. Plant Cell 6 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain, S.M., Muller, A.J., and Singh, D.P. (2004). The gar2 and rga alleles increase the growth of gibberellin-deficient pollen tubes in Arabidopsis. Plant Physiol. 134 694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain, S.M., and Singh, D.P. (2005). Tall tales from sly dwarves: Novel functions of gibberellins in plant development. Trends Plant Sci. 10 123–129. [DOI] [PubMed] [Google Scholar]
- Swain, S.M., Tseng, T.S., and Olszewski, N.E. (2001). Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiol. 126 1174–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, S.G., Rieu, I., and Steber, C.M. (2005). Gibberellin metabolism and signaling. Vitam. Horm. 72 289–338. [DOI] [PubMed] [Google Scholar]
- Tyler, L., Thomas, S.G., Hu, J.H., Dill, A., Alonso, J.M., Ecker, J.R., and Sun, T.P. (2004). DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T.Y., Hsing, Y.I.C., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). Gibberellin Insensitive Dwarf1 encodes a soluble receptor for gibberellin. Nature 437 693–698. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Hirano, K., Kitano, H., and Matsuoka, M. (2008). Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell 20 2437–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Nakajima, M., Katoh, E., Ohmiya, H., Asano, K., Saji, S., Xiang, H.Y., Ashikari, M., Kitano, H., Yamaguchi, I., and Matsuokaa, M. (2007. b). Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19 2140–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Nakajima, M., Motoyuki, A., and Matsuoka, M. (2007. a). Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 58 183–198. [DOI] [PubMed] [Google Scholar]
- Wen, C.K., and Chang, C. (2002). Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige, B.C., Ghosh, S., Nill, C., Zourelidou, M., Dohmann, E.M.N., Maier, A., and Schwechheimer, C. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H., Ito, T., Zhao, Y.X., Peng, J.R., Kumar, P.P., and Meyerowitz, E.M. (2004). Floral homeotic genes are targets of gibberellin signaling in flower development. Proc. Natl. Acad. Sci. USA 101 7827–7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart, J.A.D., and Talon, M. (1992). Gibberellin mutants in Arabidopsis thaliana. In Progress in Plant Growth Regulation, C.M. Karssen, L.C. Van Loon, and D. Vreugdenhil, eds (Dordrecht, The Netherlands: Kluwer), pp. 34–42.
- Zentella, R., Zhang, Z.L., Park, M., Thomas, S.G., Endo, A., Murase, K., Fleet, C.M., Jikumaru, Y., Nambara, E., Kamiya, Y., and Sun, T.P. (2007). Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19 3037–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.