Abstract
Plants develop various endoplasmic reticulum (ER)–derived structures, each of which has specific functions. The ER body found in Arabidopsis thaliana is a spindle-shaped structure that specifically accumulates high levels of PYK10/BGLU23, a β-glucosidase that bears an ER-retention signal. The molecular mechanisms underlying the formation of the ER body remain obscure. We isolated an ER body–deficient mutant in Arabidopsis seedlings that we termed nai2. The NAI2 gene (At3g15950) encodes a member of a unique protein family that is only found in the Brassicaceae. NAI2 localizes to the ER body, and a reduction in NAI2 gene expression elongates ER bodies and reduces their numbers. NAI2 deficiency does not affect PYK10 mRNA levels but reduces the level of PYK10 protein, which becomes uniformly diffused throughout the ER. NAI1, a transcription factor responsible for ER body formation, regulates NAI2 gene expression. These observations indicate that NAI2 is a key factor that enables ER body formation and the accumulation of PYK10 in ER bodies of Arabidopsis. Interestingly, ER body–like structures are also restricted to the Brassicales, including the Brassicaceae. NAI2 homologs may have evolved specifically in Brassicales for the purpose of producing ER body–like structures.
INTRODUCTION
Proteins that eventually enter the secretory pathway are synthesized on the rough endoplasmic reticulum (ER), where the ribosomes are attached. These newly synthesized proteins are then modified by disulfide bond formation and the attachment of oligosaccharides in the ER lumen, before being transported to their destination by vesicle trafficking. Most transport vesicles moving from the ER are coat protein II (COPII) vesicles of ∼50 nm in diameter. Plants differ from animals in that they also produce different types of ER-derived vesicles involved in the accumulation of single types of proteins (Chrispeels and Herman, 2000; Galili, 2004; Hara-Nishimura et al., 2004; Herman and Schmid, 2004). For example, the endosperm of maize (Zea mays) and rice (Oryza sativa) produces protein bodies (PBs) that store seed storage proteins (Herman and Larkins, 1999). Similarly, the maturing cotyledons of pumpkin (Cucurbita maxima) produce precursor-accumulating (PAC) vesicles that accumulate the precursors of seed proteins and mediate their transport to the protein storage vacuoles (Hara-Nishimura et al., 1998). In addition, the dying tissues of mung bean (Vigna mungo) and castor bean (Ricinus communis) seedlings produce KDEL-tailed proteinase-accumulating vesicles (KV) and ricinosomes, respectively, that accumulate a papain-type proteinase responsible for the degradation of cellular materials (Schmid et al., 1999, 2001; Toyooka et al., 2000). Interestingly, these unique vesicles are all larger (∼500 nm) than COPII vesicles, are species-specific, and appear at a specific stage in the plant's life cycle.
We have identified a distinct type of ER-derived structure in Arabidopsis thaliana that we designated the ER body (Matsushima et al., 2003a). In Arabidopsis expressing ER-targeted green fluorescent protein (GFP), the spindle-shaped 5- to 10-μm-long ER bodies can be easily detected (Hayashi et al., 2001). Electron microscopic analysis has revealed that ER bodies have a fibrous structure and are surrounded by a single ribosome-bearing membrane (Hayashi et al., 2001). ER bodies are uniformly distributed throughout the epidermis of cotyledons and hypocotyls in young seedlings and subsequently disappear with plant growth (Matsushima et al., 2002). By contrast, the majority of the root tissues constitutively accumulate ER bodies (Matsushima et al., 2003b). Interestingly, wounding or treatment with the wound hormone jasmonate induces the accumulation of ER bodies in adult leaves (Matsushima et al., 2002). This suggests that the ER body is involved in pest/pathogen resistance in Arabidopsis (Hara-Nishimura and Matsushima, 2003). Structures that are similar to the ER bodies of Arabidopsis have also been reported in the cells of various organs of other Brassicales plants, including Arabis alpina, Brassica oleracea, Raphanus sativus, Capparis spinosa, and Cleome spinosa (Iversen, 1970a, 1970b; Behnke and Eschlbeck, 1978).
The ER bodies in Arabidopsis seedlings accumulate PYK10 protein, a β-glucosidase that bears the ER retention signal KDEL (Matsushima et al., 2003b). Most ER-soluble proteins bear this KDEL sequence at the C terminus, and this sequence is required for their retention in the ER. The presence of KDEL on PYK10 suggests that the ER-retention system is responsible for the accumulation of PYK10 in the ER body. PYK10 seems to be the major component of the ER body (Matsushima et al., 2003b). Since electron microscopic analysis has revealed that there are high-density materials in the ER body, it seems likely that PYK10 accumulates as a condensed (or aggregated) material in the ER body. However, like animal cells, plant cells have ER quality control systems known as ER-associated protein degradation (ERAD) systems that digest unfolded and aggregated ER proteins (Di Cola et al., 2001; Vitale and Ceriotti, 2004). Furthermore, ER proteins become degraded during their constitutive transport to the vacuole (Tamura et al., 2004; Pimpl et al., 2006). Therefore, it appears that Arabidopsis has developed a unique system to protect PYK10 from ER-associated degradation in the ER body.
We isolated an Arabidopsis nai1 mutant that lacks ER bodies (Matsushima et al., 2003b) and found that NAI1 encodes a basic helix-loop-helix–type transcription factor (Matsushima et al., 2004). We found that NAI1 regulates the expression of PYK10, jacalin-related lectin22 (JAL22), JAL23, JAL31, JAL33, PBP1/JAL30, GDSL-lipase–like protein23 (GLL23), and GLL25 (Matsushima et al., 2004; Nagano et al., 2005, 2008). PBP1 localizes to the cytosol, while PYK10 localizes to the ER body (Matsushima et al., 2004; Nagano et al., 2005). Recently, we found that when cells are broken, PYK10 forms a large complex (from 0.65 to >70 μm in diameter) with JALs and GLLs (Nagano et al., 2008). JALs and GLLs regulate the size of the PYK10 complex and may regulate its substrate specificity. Since NAI1 deficiency caused the loss of ER bodies (Matsushima et al., 2004), NAI1 may regulate as yet unknown factors in the ER body that regulate the formation of the ER body. Here, we show, by analyzing a new Arabidopsis mutant, nai2, that NAI2 is one of these factors.
RESULTS
Isolation of an ER Body–Deficient Mutant
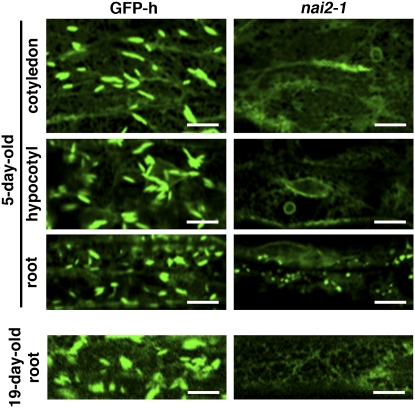
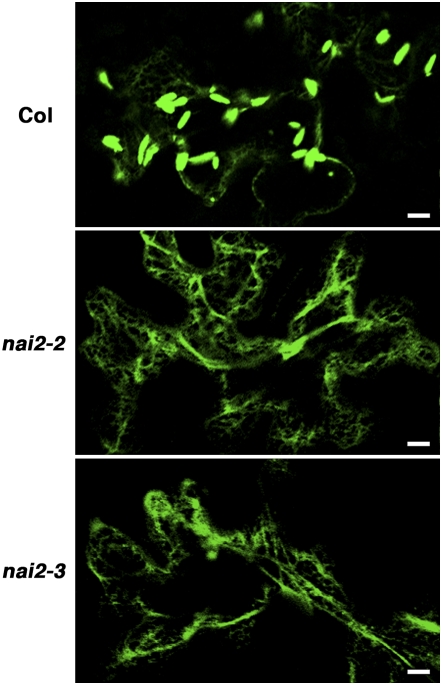
To improve our understanding of how ER bodies form, we generated activation tag–inserted Arabidopsis lines and searched for mutants that lacked ER bodies. As a background plant, we used the transgenic GFP-h Arabidopsis plant, which expresses ER-targeted GFP (see Methods). We screened ∼10,000 independent lines and isolated one ER body–deficient mutant, which we named nai2-1. The nai2-1 mutant lacks ER bodies in all parts of the seedling (Figure 1; see Supplemental Figure 1 online). While ER bodies were observed in the roots of mature GFP-h plants, they were not detected in the roots of 19-d-old nai2-1 plants (Figure 1). Thus, the nai2-1 mutant is an ER body–deficient mutant. The first filial (F1) progeny from the crossing of the nai2-1 mutant with the GFP-h plant all displayed the wild-type phenotype, while the second filial (F2) progeny segregated into wild-type and mutant phenotypes with a 3:1 ratio, indicating that nai2-1 is a single recessive mutation (Table 1). This also suggested that the nai2-1 mutant is a gene-disruption mutant rather than a gene-activation mutant. The phenotype of the nai2-1 mutant was very similar to that of the nai1-1 mutant (Matsushima et al., 2004), which also lacks ER bodies. However, unlike the nai1-1 mutant, the nai2-1 mutant has vesicle-like structures on the ER network in the roots of seedlings (Figure 1). The F1 progeny of the cross between the nai2-1 mutant and the nai1-1 mutant displayed a wild-type phenotype (Table 1), which indicates that nai1-1 and nai2-1 are not alleles of the same gene. These observations together indicate that nai2-1 is a novel ER body–deficient mutant and that the NAI2 gene is responsible for ER body formation in Arabidopsis.
Figure 1.
The nai2-1 Mutant Lacks ER Bodies.
Epidermal cells of 5-d-old seedlings and 19-d-old mature plants were inspected with a confocal laser scanning microscope. Bars = 10 μm.
Table 1.
Genetic Analysis of the nai2-1 Mutant
| F1
|
F2
|
||||
|---|---|---|---|---|---|
| Progeny | Wild Type | Mutant | Wild Type | Mutant | P (χ2)a |
| nai2-1 × GFP-h | 27 | 0 | 73 | 22 | 0.678 |
| nai2-1 × nai1-1 | 45 | 0 | – | ||
Probability was calculated by χ2 test with 3:1 segregation.
Identification of the NAI2 Gene
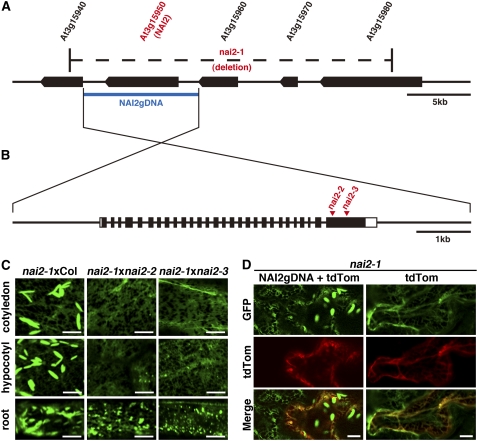
The phenotype of nai2 is similar to that of nai1. This suggested that the expression pattern of the NAI2 gene may correlate with the expression pattern of the NAI1 gene and that the expression of the NAI2 gene may be reduced in nai1. We used the Arabidopsis trans-factor and cis-element prediction database-II (ATTED-II; http://www.atted.bio.titech.ac.jp/) (Obayashi et al., 2007) to generate a list of genes that are coexpressed with the NAI1 gene (see Supplemental Table 1 online), in addition to analyzing microarray expression data of nai1-1 (Nagano et al., 2008). These analyses led us to focus on the uncharacterized gene, At3g15950. Our subsequent genome analysis revealed that we could not generate PCR products that included the At3g15950 gene when we used the nai2-1 genome as the template DNA. This indicated that the nai2-1 mutant lacks the At3g15950 gene. Further analysis revealed that the nai2-1 mutant lacks five genes in total, At3g15940, At3g15950, At3g15960, At3g15970, and At3g15980 (Figure 2A). This suggested that a large genome deletion that included these genes had occurred in the nai2-1 mutant.
Figure 2.
The NAI2 Gene Is At3g15950.
(A) The structure of the genome region around the NAI2 gene. The black boxes indicate the coding regions of each gene. The nai2-1 mutant has a putative large genome deletion indicated by the region marked by dashes. The blue bar indicates the genomic region that was used for the complementation testing of the nai2-1 mutant.
(B) The exon/intron structure of the NAI2 gene. The black boxes indicate the protein-coding regions. The white boxes indicate the untranslated regions. The triangles indicate the T-DNA insertion sites in the nai2-2 and nai2-3 mutants.
(C) Test of allelism between the nai2 mutants. Cotyledon epidermal cells of nai2-1 × nai2-2 or nai2-1 × nai2-3 F1 progeny were inspected with a confocal laser scanning microscope. Bars = 10 μm.
(D) Complementation of the nai2-1 phenotype by the introduction of a genome fragment carrying the NAI2 gene. Cotyledon epidermal cells from 6-d-old nai2-1 seedlings were bombarded with gold particles coated with plasmids containing the NAI2 genome fragment shown in (A) plus the tdTomato gene (NAI2gDNA + tdTom; left panels) or the plasmid carrying the tdTomato gene only (right panels). After germination for 2 d, the cells were inspected with a confocal laser scanning microscope. The florescence of tdTomato indicates cells that were transfected with plasmids. The filamentous structures observed by tdTomato are cytoplasmic strands. Bars = 10 μm.
To determine whether the NAI2 gene is At3g15950, we performed two experiments. First, we asked whether two T-DNA insertion mutant alleles of At3g15950 are the alleles of nai2-1. We crossed nai2-1 with either SALK_005896 (nai2-2) or SALK_043149 (nai2-3) and observed the phenotypes of the F1 progeny. Like nai2-1, the F1 progeny did not accumulate ER bodies (Figure 2C). Thus, neither of the T-DNA insertion mutants could complement the nai2-1 allele. Second, we asked whether the At3g15950 gene could complement the nai2-1 phenotype. We introduced the genome fragment of At3g15950 (Figure 2B) into epidermal cells of the nai2-1 mutant by the particle gun method. ER bodies were detected in the nai2-1 mutant when the genome fragment of At3g15950 was introduced (Figure 2D). ER bodies were not detected when the genome fragment was omitted (Figure 2D). This indicates that the presence of At3g15950 was sufficient for ER body formation in the nai2-1 mutant. These data together indicate that At3g15950 is necessary and sufficient for ER body formation in the nai2-1 mutant. Consequently, we concluded that the NAI2 gene is At3g15950.
NAI2 Homologous Genes Are Found Only in Brassicales Plants
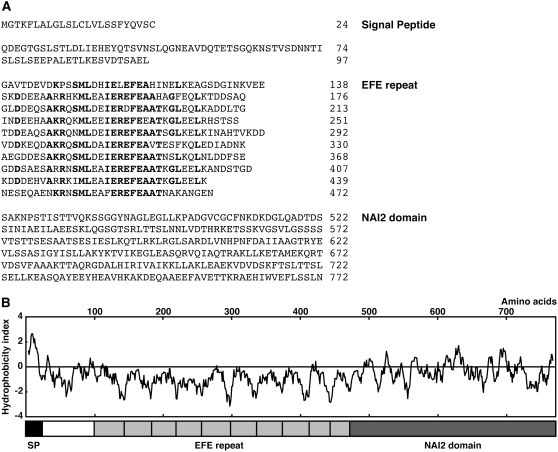
The NAI2 gene encodes a 772–amino acid protein (Figure 3A) and has a signal peptide at its N terminus, which suggests that NAI2 is a secretory protein (von Heijne, 1986). The N-terminal half of NAI2 has a Glu-Phe-Glu (EFE) motif consisting of 10 repeats of an ∼40–amino acid sequence that has a specific Glu-Phe-Glu sequence (Suzuki et al., 2005). The EFE motif region of NAI2 is hydrophilic (Figure 3B) and rich in Ala (12.4%), Asp (10.0%), and Glu (19.0%) residues. The sequence of the C-terminal region of NAI2 is unique, and we termed it the NAI2 domain (Figure 3). Two proteins in Arabidopsis are structurally related to NAI2, TSK-associating protein1 (TSA1)/At1g52410 and At3g15960 (see Supplemental Figure 2 online). TSA1, which is the closest NAI2 homolog, bears a signal peptide, 10 EFE repeats, and a NAI2 domain (see Supplemental Figures 2A and 2B online). TSA1 was found by a yeast two-hybrid assay to interact with the chromosomal regulatory protein TONSOKU(TSK)/MGOUN3/BRUSHY1 (Suzuki et al., 2005). The At3g15960 protein is smaller than NAI2 or TSA1 and has a signal peptide and a NAI2 domain but lacks EFE repeats (see Supplemental Figures 2C and 2D online). The At3g15960 gene lies next to NAI2 and is missing in the nai2-1 mutant.
Figure 3.
Structure of NAI2.
(A) The deduced amino acid sequence of NAI2. The numbers show the amino acid residue positions. NAI2 contains a signal peptide, 10 EFE repeats, and the NAI2 domain. The boldface letters show the conserved amino acids within the EFE repeats.
(B) Hydrophobicity index of NAI2. The structure of NAI2 is shown below. SP, signal peptide.
We searched GenBank/EMBL/DDBJ to identify NAI2 homologous genes in other species. None could be found in animals, fungi, or unicellular organisms. Moreover, we were unable to find any homologous genes in rice or poplar (Populus trichocarpa), for which whole genome sequences are available. We only identified NAI2 homologous genes in Brassicaceae plants. At least two NAI2 homologous genes were found in the partial genome sequences of Brassica rapa (AC189268 and AC189514). Similarly, several NAI2 homologous genes could be identified in the partial genome sequences of B. oleracea (e.g., BZ479594 and BZ006167). Since these sequences included only parts of genes, we could not assign whole protein-coding regions. In B. napus, we identified one tentative consensus (TC) sequence (TC74287) that was assembled from ESTs (http://compbio.dfci.harvard.edu/tgi/plant.html). TC74287 encodes a protein with a signal peptide, nine EFE repeats, and a NAI2 domain (see Supplemental Figure 3 online). These findings suggest that NAI2 homologous genes are unique to Brassicales plants.
NAI2 Is an ER Body Protein
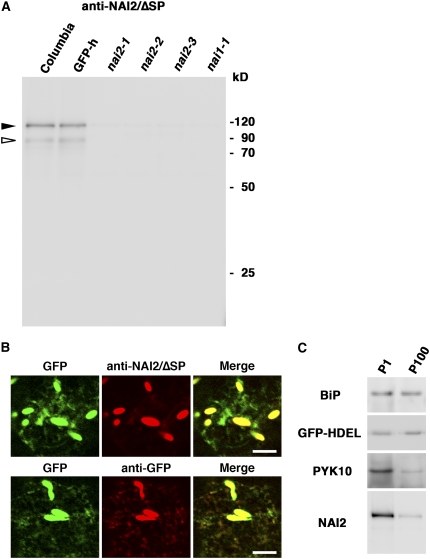
NAI2 has a signal peptide at its N terminus, suggesting that NAI2 enters the ER lumen. We raised two antibodies, one against the signal peptideless NAI2 polypeptide (anti-NAI2/ΔSP) and the other against part of the C-terminal NAI2 region (residues 636 to 772; anti-NAI2/C). These antibodies were used in immunoblot analysis of 7-d-old Arabidopsis seedlings (Figure 4A; see Supplemental Figure 4A online). Both antibodies detected an ∼120-kD polypeptide in the wild-type seedlings that could not be observed in any of the three nai2 mutants, which indicates that the band corresponds to NAI2. The calculated molecular mass of the NAI2 polypeptide without the signal peptide was 82.4 kD, which was slightly lower than the molecular mass of the band detected by immunoblot analysis. We detected an ∼120-kD polypeptide in the bacteria expressing NAI2/ΔSP polypeptides (see Supplemental Figure 5 online), suggesting that the NAI2 polypeptide migrates slowly during electrophoresis. NAI2 may be subjected to posttranslational modification, such as glycosylation, as NAI2 in Arabidopsis migrated to almost the same position as bacterially expressed NAI2/ΔSP with an ∼3-kD His tag sequence. We also observed a fainter ∼90-kD polypeptide in wild-type seedlings that was recognized by anti-NAI2/ΔSP but not by anti-NAI2/C and an ∼25-kD polypeptide in wild-type seedlings that was recognized by anti-NAI2/C but not by anti-NAI2/ΔSP. These results suggest that these polypeptides were minor degradation products of NAI2 protein.
Figure 4.
NAI2 Localizes in ER Bodies.
(A) Immunoblot analysis of 7-d-old seedlings from the indicated strains using antibody against NAI2/ΔSP (without signal peptide). Arrowheads indicate the bands corresponding to the NAI2 polypeptide: closed, major bands; open, minor bands.
(B) Immunofluorescence analysis of NAI2 in 7-d-old GFP-h seedlings. Left panels, the ER-targeted GFP signals; middle panels, the NAI2 (top) and GFP (bottom) signals, which were detected by antibodies against NAI2/ΔSP and GFP, and Cy3-labeled second antibodies, respectively; right panels, the merged images. Bars = 10 μm.
(C) Immunoblot analysis of the subcellular fractions enriched in ER bodies (P1) or ER networks (P100) using antibodies against BiP, GFP (GFP-HDEL), PYK10, and NAI2/ΔSP (NAI2).
To determine the subcellular localization of NAI2 protein, we subjected GFP-h plants to immunofluorescence analysis. The immunofluorescence signal of NAI2 was detected in the ER body but not the ER network, whereas the immunofluorescence signal of GFP was detected in both the ER network and the ER body (Figure 4B). Next, we separated ER body–rich fraction (P1) and ER network–rich fraction (P100) by the subcellular fractionation method established previously (Matsushima et al., 2003b) and subjected the fractions to immunoblot analysis (Figure 4C). BiP and GFP-HDEL were detected in both the P1 and P100 fractions, which indicates that these proteins accumulate in both the ER and the ER body. By contrast, NAI2 was mainly detected in the P1 fraction. This was also the case for PYK10, an ER body protein. These data demonstrate that NAI2 is an ER body protein that accumulates specifically in the ER body.
A Reduction of NAI2 Levels Elongates ER Bodies and Reduces Their Number
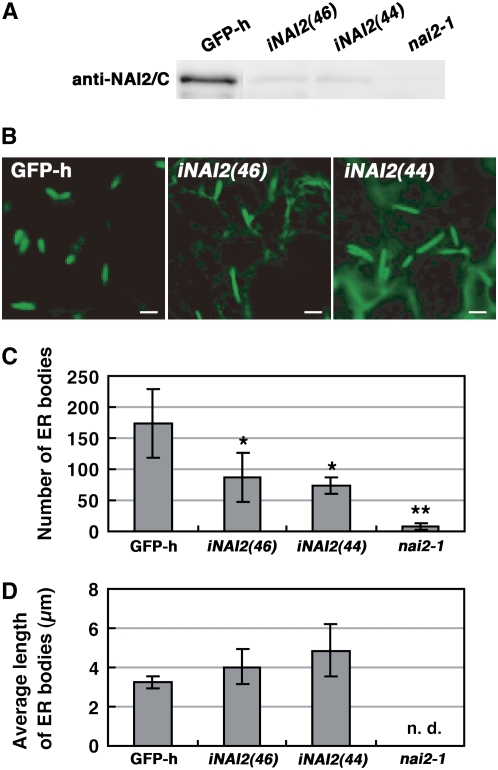
To characterize the role that NAI2 plays in ER body formation in more detail, we examined the effect of RNA interference (RNAi) on the NAI2 gene on ER body numbers and shape. For this, we used two independently derived NAI2-RNAi (iNAI2) lines that showed reduced levels of NAI2 (Figure 5A). While the seedlings of the iNAI2 lines had ER bodies (Figure 5B), they were fewer in number (Figure 5C) and were longer than the ER bodies of the GFP-h plant (Figures 5B and 5D). Thus, the reduction of NAI2 levels elongates ER bodies and reduces their number. This provides additional evidence that NAI2 is responsible for ER body formation in Arabidopsis.
Figure 5.
Reduction of NAI2 Reduces the Number of ER Bodies and Elongates Their Shape.
(A) Extracts from 8-d-old seedlings of GFP-h, two independent NAI2 RNAi lines, and nai2-1 were subjected to immunoblot analysis with anti-NAI2/C antibodies.
(B) ER bodies in 8-d-old cotyledons. Bars = 5 μm.
(C) Quantification of ER bodies. The GFP fluorescent spots in a 5.29 × 104 μm2 image area were automatically counted after shutting out weak ER fluorescence. The iNAI2 plants had significantly fewer ER bodies than GFP-h (* P < 0.05, Welch's t test). The error bars indicate sd (n = 4 to 6). ** The lower number of GFP fluorescent spots in nai2-1 shows that ER fluorescence was removed during counting analysis.
(D) Quantification of ER body length. Average ER body length was calculated from a single image. The error bars indicate sd of the averages (n = 4 to 6). The length of the ER bodies in nai2-1 was not measured, since so few ER bodies were detected (n.d., not determined).
NAI2 mRNA Levels Are Reduced in the nai1 Mutant, but NAI1 and PYK10 mRNA Levels Are Not Reduced in the nai2 Mutant
The NAI1 gene regulates ER body formation and the expression of the PYK10 gene, whose gene product is a major component of ER bodies (Matsushima et al., 2003b, 2004). Therefore, we used real-time RT-PCR to measure the PYK10 and NAI1 mRNA levels in nai2 mutant seedlings (Figure 6). The wild-type plants and the nai2 mutants did not differ in PYK10 and NAI1 mRNA levels, which indicates that NAI2 gene deficiency does not affect PYK10 and NAI1 mRNA levels in Arabidopsis seedlings. Next, we measured the NAI2 mRNA levels in the nai1-1 mutant. The nai1-1 mutant had lower NAI2 mRNA levels than the wild-type plant, which indicates that NAI1 gene deficiency reduces NAI2 mRNA levels. Thus, the NAI1 gene regulates the expression of the NAI2 gene but the NAI2 gene does not regulate the expression of the NAI1 gene. Notably, real-time RT-PCR revealed that nai1-1 and the wild-type plant had similar NAI1 mRNA levels. This can be explained by the accumulation of the aberrant form of NAI1 mRNA generated by missplicing, as reported previously (Matsushima et al., 2004).
Figure 6.
NAI2, PYK10, and NAI1 mRNA Levels in the nai2 and nai1 Mutants.
Total RNAs from 7-d-old seedlings were subjected to quantitative RT-PCR analysis. The data were normalized with respect to actin8 mRNA levels. The relative quantity of each mRNA was calibrated with the amounts in wild-type plants (Columbia [Col] accession). The relative quantity was calculated by the 2−ΔΔCt method (Levak and Schmittgen, 2001). Error bars indicate se of the threshold cycle (Ct) values, which are calculated using the formula 2−ΔΔCt±se. The data represent the results of three independent biological replications.
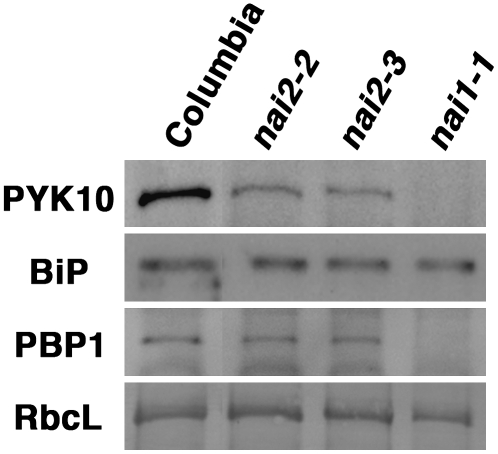
NAI2 Deficiency Reduces the Accumulation of PYK10, a Major ER Body Protein
PYK10 accumulates in ER bodies (Matsushima et al., 2003b). Therefore, we examined the PYK10 levels in nai2 mutants by immunoblot analysis (Figure 7). As a control, we also examined the accumulation of the ER chaperone protein BiP and the NAI1-regulated cytosolic lectin homolog, PYK10 binding protein1 (PBP1). While high levels of PYK10 were detected in 7-d-old wild-type plants, these levels were reduced (but not absent) in the nai2 mutants. However, there was no change in the levels of BiP. There was also no change in the levels of PBP1, which is reduced in the nai1 mutant (Matsushima et al., 2004; Nagano et al., 2005). These findings indicate that the NAI2 deficiency in Arabidopsis specifically reduces the accumulation of PYK10, a major ER body protein, but does not affect the accumulation of BiP and PBP1.
Figure 7.
PYK10, BiP, and PBP1 Levels in nai2 Mutants.
Total proteins from 7-d-old seedlings were subjected to immunoblot analysis using antibodies against PYK10, BiP, or PBP1. Coomassie blue staining shows the ribulose-1,5-bis-phosphate carboxylase/oxygenase large subunit (RbcL), which served as a loading control.
NAI2 Deficiency Alters PYK10 Localization
The nai2 mutants lacked ER bodies. However, we could still detect substantial amounts of PYK10 protein in the nai2 mutants, in contrast with the nai1 mutant, although these amounts were still lower than those in the wild-type plant (Figure 7). Therefore, we examined the localization of PYK10 protein in the nai2 mutants by transiently expressing PYK10 as a GFP fusion protein (Figure 8). For this, we constructed a vector encoding recombinant PYK10 that has GFP between its signal peptide and its enzyme region (GFP-PYK10) and introduced it into the epidermal cells of Arabidopsis cotyledons by particle bombardment. In wild-type Arabidopsis, GFP-PYK10 mainly accumulated in ER bodies (Figure 8). This is consistent with previous findings, showing that PYK10 mainly accumulates in ER bodies (Matsushima et al., 2003b). Surprisingly, in the nai2-2 and nai2-3 mutants, GFP-PYK10 was uniformly distributed throughout the ER network (Figure 8). These results indicate that NAI2 deficiency causes PYK10 to diffuse throughout the ER network. Thus, NAI2 is responsible for the accumulation of PYK10 in ER bodies.
Figure 8.
Localization of GFP-PYK10 in nai2 Mutants.
Six-day-old wild-type (Columbia [Col] accession), nai2-2, and nai2-3 seedlings were bombarded with gold particles coated with plasmids carrying the GFP-PYK10 gene and germinated for 1 d. Cotyledon epidermal cells were inspected with a confocal laser scanning microscope to observe the distribution of GFP-PYK10. Bars = 10 μm.
DISCUSSION
We isolated an ER body–deficient mutant, nai2 (Figure 1), and found that the NAI2 gene encodes a unique protein (Figures 2 and 3) whose homologs occur only in Brassicaceae plants. We found that NAI2 is localized to ER bodies (Figure 4) and that a reduction of NAI2 levels reduces the number of ER bodies and alters their shape (Figure 5). NAI2 deficiency reduced PYK10 levels and dispersed PYK10 throughout the ER (Figures 7 and 8), although it did not affect PYK10 mRNA levels (Figure 6). These findings indicate that NAI2 is an ER body component that enables ER body formation and the accumulation of PYK10 in Arabidopsis.
Unlike Other ER-Derived Vesicles, ER Body Formation Requires a Specific Component
Plants produce various types of ER-derived structures that are involved in bulk protein transport/storage, such as PBs, PAC vesicles, ricinosomes, and KVs (Hara-Nishimura et al., 1998; Herman and Larkins, 1999; Chrispeels and Herman, 2000; Toyooka et al., 2000; Schmid et al., 2001). It has been shown that overproduction of the major contents of the PB, PAC vesicle, or KV results in the formation of PBs, PAC vesicles, and KVs in tobacco (Nicotiana tabacum) and Arabidopsis leaves (Bagga et al., 1995; Hayashi et al., 1999; Okamoto et al., 2003). This indicates that the accumulation of the major vesicle components in the ER is sufficient for the induction of these vesicles. While the ER body is also an ER-derived structure, it is larger and longer than these vesicles (Hara-Nishimura and Matsushima, 2003; Matsushima et al., 2003a; Hara-Nishimura et al., 2004) and accumulates the β-glucosidase PYK10 (Matsushima et al., 2003a). However, the overproduction of GFP-PYK10 fusion proteins did not induce the formation of ER bodies in the nai2 mutants (Figure 8), which indicates that the accumulation of PYK10 in the ER is not sufficient for the induction of ER body formation. Instead, NAI2, an ER body component, is required for ER body formation, since reduction of NAI2 levels by RNAi elongates the ER bodies and reduces their numbers (Figure 5). In addition, complete loss of NAI2 abolishes ER bodies in Arabidopsis (Figures 1 and 2). Thus, the formation of the ER body differs from the formation of other ER-derived structures in that it depends on a specific component but not on its main component. It was revealed recently that maize Floury1 (FL1) facilitates the formation of intact PB in maturating maize seed (Holding et al., 2007). FL1 is an ER protein with four transmembrane regions. Arabidopsis lacks PBs but has FL1 homologs (Holding et al., 2007), while rice lacks both ER bodies and a NAI2 homolog. These observations suggest that FL1 and NAI2 are functionally different.
NAI2 Is an ER Body Component That Is Responsible for ER Body Formation
Our immunofluorescence analysis revealed that NAI2 is an ER body protein (Figure 4). Furthermore, NAI2 seems to accumulate specifically in ER bodies, since immunofluorescence analysis failed to detect a fluorescence signal of NAI2 in the ER (Figure 4B) and NAI2 was enriched in the ER body fraction (Figure 4C). The hydrophobicity index of NAI2 suggests that it lacks a transmembrane region (Figure 3B), which in turn suggests that NAI2 is a soluble ER body component that is responsible for ER body formation. We speculated that NAI2 interacts with PYK10 and assists PYK10 condensation, thereby generating a subdomain in the ER that eventually leads to the formation of an ER body. However, coimmunoprecipitation experiments with anti-NAI2 antibody or anti-PYK10 antibody failed to detect an interaction between NAI2 and PYK10 (see Supplemental Figure 6 online), and NAI2 proteins were not detected in the PYK10 complex (Nagano et al., 2008). This suggests that NAI2 may not interact with PYK10 directly, although we could not exclude the possibility that the interaction of PYK10 and NAI2 was weak or transient. Alternatively, NAI2 may interact with specific ER body membrane protein(s) to form an ER body framework, thereby facilitating the subsequent accumulation of PYK10. ATTED-II analysis revealed that the NAI2 gene is coexpressed with At4g27860 and At5g24290, which encode integral membrane proteins (see Supplemental Table 2 online). Interestingly, according to the Localization of Organelle Proteins by Isotope Tagging data of Arabidopsis (Dunkley et al., 2006), At5g24290 protein cofractionates with NAI2 (see Supplemental Table 3 online), suggesting that At5g24290 is an ER body membrane protein.
Most ER-soluble proteins bear the KDEL ER retention signal at their C terminus, a signal required for their retention in the ER. However, some ER-soluble proteins lack the ER retention signal. For example, ER protein disulfide isomerases, including the soybean (Glycine max) protein disulfide isomerase, lack canonical ER retention signals (Wadahama et al., 2007). NAI2 also lacks a typical ER retention signal at its C terminus (Figure 3A). This indicates that NAI2 is retained in the ER and accumulates in ER bodies independent of known ER retention signal–mediated mechanisms. NAI2 may interact with other ER resident proteins in order to be retained in the ER.
The NAI1 Gene Regulates the NAI2 Gene Expression Needed for ER Body Formation
The NAI1 gene encodes a basic helix-loop-helix–type transcription factor that is responsible for ER body formation and regulates PYK10 and PBP1 gene expression (Matsushima et al., 2004; Nagano et al., 2005). However, until now, the mechanisms operating downstream of the NAI1 gene activity that leads to ER body formation in Arabidopsis were unclear. We found that NAI2 gene expression was reduced in the nai1-1 mutant (Figure 6), which indicates that NAI2 is a downstream factor of the NAI1 gene and serves to induce ER body formation in Arabidopsis.
Do ER body numbers and PYK10 levels regulate NAI1 activity in a feedback loop? The nai2 mutant lacked ER bodies and showed reduced PYK10 levels. However, the expression of NAI1 and PYK10 genes was unchanged in the nai2 mutants (Figure 6). Thus, the ER body deficiency and reduction in PYK10 levels in the nai2 mutants did not induce NAI1 and PYK10 gene expression. This suggests that ER body formation and/or PYK10 levels do not regulate NAI1 activity in Arabidopsis seedlings.
High Accumulation of PYK10 Requires NAI2-Mediated ER Body Formation
We previously identified PYK10, a β-glucosidase, as a major component of ER bodies (Matsushima et al., 2003b). In the nai2 mutant, we found normal PYK10 mRNA levels (Figure 6) and reduced but not completely abolished PYK10 protein levels (Figure 7). Confocal microscopy revealed that GFP-PYK10 accumulates uniformly throughout the ER network of the nai2 mutants, while in wild-type plants it condenses into ER bodies (Figure 8). Thus, PYK10 accumulates in the ER in the absence of ER bodies but condenses highly in ER bodies in the presence of ER bodies. PYK10 has the KDEL ER retention signal (Matsushima et al., 2003b) and therefore naturally accumulates in the ER. Electron microscopic analysis showed that there are large condensations of protein in ER bodies (Hayashi et al., 2001). These observations together suggest that NAI2 enables the formation of ER bodies in Arabidopsis and that this facilitates PYK10 accumulation in ER bodies. In other words, the NAI2-mediated ER body formation is responsible for the stable and high-level accumulation of PYK10 in Arabidopsis seedlings.
The reduction of PYK10 levels in the nai2 mutants may reflect a reduction of PYK10 translation efficiency. However, given that NAI2 is predicted to be a luminal protein (Figure 3), it is hard to imagine how it could stimulate PYK10 translation. Alternatively, NAI2 may stimulate the stabilization of PYK10 by promoting its sequestration in ER bodies. In the nai2 mutants, PYK10 is dispersed throughout the ER network and may be degraded by mechanisms used to degrade ER proteins, such as ERAD (Di Cola et al., 2001; Vitale and Ceriotti, 2004) or transportation to the lytic vacuole (Tamura et al., 2004; Pimpl et al., 2006).
NAI2 Homologous Genes Have Different Functions in Arabidopsis
Of the two NAI2 homologs in Arabidopsis, TSA1 has been identified as a factor that interacts with TSK/MGO3/BRU, a key factor acting in cell division control and plant morphogenesis (Suzuki et al., 2005). However, the in vivo relationship between TSA1 and TSK/MGO3/BRU remains unclear. NAI2 is the closest homolog of TSA1, which suggests that both proteins may have similar function(s). Indeed, like NAI2, TSA1-GFP fusion protein localizes in vesicle-like structures that are presumably ER bodies when overproduced in Arabidopsis epidermal cells (Suzuki et al., 2005). This suggests that TSA1 functions in ER bodies. However, we did not detect any morphological changes in nai2 mutants, which suggests that the NAI2 gene is not involved in cell division control. On the other hand, ER bodies in seedlings were eliminated by the nai2 mutation. Thus, NAI2 has a specific function that is not complemented by the TSA1 gene. The other NAI2 homolog, At3g15960, is a neighboring gene of NAI2, which suggests that At3g15960 and NAI2 were generated by gene duplication. However, At3g15960 protein lacks EFE motifs, which suggests that the function of the At3g15960 gene also differs from that of the NAI2 gene. Thus, NAI2 homologous genes have different functions in Arabidopsis.
Are NAI2 Homologs Key Factors Enabling ER Body Formation in Brassicales Plants?
We identified ER bodies in Arabidopsis plants that expressed ER-targeting GFP (Hayashi et al., 2001). Previously, Iversen (1970a) also observed ER-derived structures in Brassicales plants that he termed dilated cisternae. Moreover, he observed that these dilated cisternae accumulated myrosinase (β-thioglucosidase), had a spindle-shaped structure (Iversen, 1970a), and occurred in Brassicales plants but not in other orders (Iversen, 1970b; Behnke and Eschlbeck, 1978). Interestingly, we also found that NAI2 homologous genes were present in the Arabidopsis genome but not in the rice or poplar genome. Our search for NAI2-related sequences in databases of ESTs from various plant species revealed that NAI2 homologous genes exist only in Brassicaceae plants, such as Arabidopsis (Figure 3; see Supplemental Figure 2 online), B. rapa, B. oleracea, and B. napus (see Supplemental Figure 3 online). We also did not find any NAI2 homologous genes in animals, fungi, and unicellular organism genomes. Thus, it appears that the NAI2 gene family has rapidly evolved in Brassicales plants for the purpose of ER body formation.
The production of ER bodies is induced by wounding and the wounding hormone jasmonate, which suggests that ER bodies may participate in pest/pathogen resistance (Matsushima et al., 2002; Hara-Nishimura and Matsushima, 2003). Recently, Sherameti et al. (2008) reported that nai1 and pyk10 mutants are hyperinfected by the entophytic fungus Piriformospora indica and show reduced growth. This suggests that the ER body may function in plant resistance against fungus infection. Since the NAI2 homologous genes seem to enable ER body formation, their ultimate function may be to promote pest/pathogen resistance in Brassicales plants. Further attempts to identify NAI2 homologous genes in other Brassicales plants and neighboring orders may reveal how and why Brassicales plants developed NAI2 homologous genes along with ER bodies. This in turn may provide new insights into the evolutionary pathways that result in the appearance of new cellular structures by specific genes.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana (Columbia accession) served as the wild-type plant. We generated a kanamycin-resistant and hygromycin-sensitive transgenic plant that accumulates GFP in its ER and ER bodies (GFP-h). For this, we inserted the SP-GFP-HDEL gene into the pBI121 binary vector (Takara). The SP-GFP-HDEL gene encodes a fusion protein composed of the pumpkin (Cucurbita maxima) 2S albumin signal peptide, GFP, and the ER-retention signal HDEL (Mitsuhashi et al., 2000; Hayashi et al., 2001). Wild-type Arabidopsis was then transformed with Agrobacterium tumefaciens strain C58C1Rif containing this recombinant vector. We also used the nai1-1 mutant (Matsushima et al., 2004). From the ABRC, we obtained two T-DNA insertion mutants of At3g15950 that had been identified by the Salk Institute Genomic Analysis Laboratory and were denoted SALK_005896 (nai2-2) and SALK_043149 (nai2-3) (Alonso et al., 2003). From the European Arabidopsis Stock Center, we obtained two RNAi lines denoted N207544 [iNAI2(44)] and N207546 [iNAI2(46)] that had been generated by the Arabidopsis genomic RNAi knockdown line analysis project (Hilson et al., 2004). We crossed these RNAi lines with the GFP-h plant. All plants were germinated aseptically at 22°C under continuous light (∼100 μE·m−2·s−1) on 0.5% Gellun Gum (Wako) plates containing 0.5% (w/v) MES-KOH buffer, pH 5.7, and 1× Murashige and Skoog (MS) salt mixture (Wako).
Mutant Screening
The GFP-h plant was transformed with Agrobacterium strain GV3101 (pMP90RK) (Deutsche Sammlung von Mikroorganismen und Zellkulturen) containing the activation-tagging vector pPCVICEn4HPT (Kakimoto, 1996). T2 seeds from 100 to 150 T1 transformants were assembled and seeded aseptically on 0.5% Gellun Gum (Wako) plates containing 1% (w/v) sucrose, 0.5% (w/v) MES-KOH buffer, pH 5.7, 1× MS salt mixture, and 50 mg/L hygromycin sulfate (Wako). The plants were germinated at 22°C for 6 d in dark and then for 3 to 6 d under continuous light. The hypocotyls of the T2 seedlings were inspected using a fluorescence stereomicroscope (SteREO Lumer; Carl Zeiss) to identify ER body–deficient mutants.
Confocal Laser Scanning Microscopy
We used a confocal laser scanning microscope (LSM510; Carl Zeiss) to observe fluorescent proteins and dyes. To observe GFP, we used an argon laser (488 nm) and a 505/530-nm band-pass filter. To observe tdTomato and Cy3 dye, we used a helium–neon laser (543 nm) and a 560/615-nm band-pass filter.
DNA Constructs
A genome fragment of the NAI2 gene was amplified by PCR and introduced into the Gateway entry vector pCR8/GW/Topo (Invitrogen) to produce pCR8/gNAI2. The tdTomato-expressing vector ptdGW (a kind gift from S. Mano) served as a control (Shaner et al., 2004). A cDNA fragment of PYK10 was amplified by RT-PCR using a gene-specific primer set (see Supplemental Table 4 online) and introduced into pCR8/GW/Topo to produce pCR8/PYK10. To produce GFP-PYK10, we first generated a SalI restriction site in PYK10 cDNA by modifying the nucleotide sequence that encodes the amino acids neighboring the signal peptide cleavage site. Briefly, we amplified a DNA fragment from pCR8/PYK10 using specific primers that bear half of the SalI site (see Supplemental Table 4 online). The amplified DNA fragment was subsequently self-ligated to produce pCR8/SaI-PYK10. The GFP gene was amplified with specific primers that bear the SalI site (see Supplemental Table 4 online) and then inserted into a SalI site of pCR8/SaI-PYK10, thereby generating the Gateway entry clone pCR8/GFP-PYK10. The protein-coding region of pCR8/GFP-PYK10 was transferred to pUGW2 (Nakagawa et al., 2007) to generate pUGW2/GFP-PYK10. pUGW2/GFP-PYK10 carries the Pro35S:GFP-PYK10 gene and encodes a fusion protein in which GFP is located between the signal peptide of PYK10 and its enzyme region.
Particle Bombardment
The nai2-1 plants were germinated aseptically on a 0.5% Gellun Gum plate containing 0.5% (w/v) MES-KOH buffer, pH 5.7, and 1× MS salt mixture. The plasmid DNAs were bombarded into 5-d-old seedlings using the Biolistic Particle Delivery system (Bio-Rad Laboratories) according to the manufacturer's instructions (Yamada et al., 2007).
Quantitative RT-PCR
Arabidopsis seedlings were germinated aseptically at 22°C under continuous light (∼100 μE·m−2·s−1) on 0.5% Gellun Gum (Wako) plates containing 0.5% (w/v) MES-KOH buffer, pH 5.7, and 1× MS salt mixture (Wako). We isolated total RNAs from 20 plants of each 7-d-old wild-type or mutant line using 700 μL of Isogen (Nippon Gene) and dissolved them in 175 μL of distilled water. First-strand cDNAs were synthesized from 5-μL RNA solutions using Ready-to-Go RT-PCR beads (GE Healthcare Life Science) and random oligomers. Real-time PCR was performed using the 7500 Fast Real-Time PCR system (Applied Biosystems) and the TaqMan gene expression assay kit (Applied Biosystems) according to the manufacturer's instructions. The assay identifiers are At02290764_gH for PYK10, At02252556_g1 for NAI2, At02251356_g1 for NAI1, and At02270958_gH for actin8. We used the threshold cycle method for relative quantification (Levak and Schmittgen, 2001).
Antibody Preparation
Two partial cDNA fragments of NAI2, which encode amino acids 25 to 772 (NAI2/ΔSP) and 636 to 772 (NAI2/C), were amplified by PCR using gene-specific primer sets (see Supplemental Table 4 online) and a NAI2 EST clone (U16255). The EST clone is a Salk/Stanford/Plant Gene Expression Center Consortium full-length cDNA/open reading frame clone and was obtained from the ABRC. The amplified fragments were introduced into pCR8/GW/Topo. The protein-coding region was then transferred to pDEST17 (Invitrogen) to produce His-tagged fusion proteins in Escherichia coli (BL21-AI; Invitrogen). Recombinant NAI2/ΔSP protein was dissolved in 50 mM sodium phosphate buffer, pH 7.2, 0.3 M NaCl, and 50 mM imidazole and purified by nickel nitrilotriacetic acid agarose (Qiagen) column chromatography according to the manufacturer's instructions. The recombinant NAI2/C protein in inclusion bodies was dissolved in 100 mM sodium phosphate buffer, pH 7.2, and 8 M urea and also purified by nickel column chromatography. The purified proteins were injected into rabbits to raise antibodies (by Shibayagi). We also used anti-PYK10/IM (Matsushima et al., 2004), anti-BiP (Hara-Nishimura et al., 1998), anti-PBP1/C (Nagano et al., 2005), and anti-GFP (Mitsuhashi et al., 2000) antibodies, which were prepared previously.
Immunoblot Analysis
We isolated total proteins from 20 plants of 7-d-old seedlings using 200 μL of 2× sample buffer (20 mM Tris-HCl buffer, pH 6.8, 40% [v/v] glycerol, 2% [w/v] SDS, and 2% [v/v] 2-mercaptoethanol). The extracts (10 μL) were subjected to SDS-PAGE (12.5% [w/v] acrylamide gel). The separated proteins were transferred to a nylon membrane and subjected to immunoblot analysis using anti-NAI2/ΔSP (1:2000 dilution), anti-NAI2/C (1:2000 dilution), anti-PYK10/IM (1:10,000 dilution), anti-BiP (1:5000 dilution), anti-GFP (1:5000 dilution), and anti-PBP1/C (1:15,000 dilution) antibodies. Alternatively, the proteins were stained with Coomassie Brilliant Blue R 250.
Immunofluorescence Staining
We infiltrated 7-d-old Arabidopsis seedlings with fixing solution (25 mM sodium phosphate buffer, pH 7.2, 100 mM sucrose, 4% [w/v] paraformaldehyde, and 0.5% [v/v] glutaraldehyde). The samples were fixed for 2 h at room temperature and then washed four times with phosphate–sucrose buffer (100 mM sodium phosphate buffer, pH 7.2, and 100 mM sucrose). We performed several freeze–thaw cycles to break the cell walls, and then treated the samples with primary antibody for ∼16 h (anti-NAI2/ΔSP and anti-GFP; 1:2000 dilution in phosphate–sucrose buffer containing 0.1% [v/v] Triton X-100). We then washed the samples with phosphate–sucrose buffer and treated them for 2.5 h with secondary antibody (anti-rabbit IgG, Cy3-conjugated; 1:1000 dilution in phosphate–sucrose buffer containing 0.1% [v/v] Triton X-100). Finally, we washed the samples with phosphate–sucrose buffer containing 0.1% (v/v) Triton X-100 and then inspected them with a confocal laser scanning microscope.
Subcellular Fractionation
The subcellular fractionation method was as described previously (Matsushima et al., 2003b). Briefly, 0.83 g of 8-d-old seedlings was chopped on ice in 2.5 mL of chopping buffer that contained 50 mM HEPES-NaOH, pH 7.5, 5 mM EDTA, 0.4 M sucrose, and protease inhibitor cocktail (one tablet per 50 mL; Boehringer Mannheim). The homogenate was filtered through cheesecloth and then centrifuged at 1000g at 4°C for 20 min. The pellet was designated the P1 fraction. The supernatant was centrifuged once at 8000g at 4°C for 20 min and then again at 100,000g at 4°C for 1 h. The pellet after ultracentrifugation was designated the P100 fraction. The P1 and P100 fractions were resuspended in the same volume of chopping buffer and subjected to immunoblot analysis.
Analysis of Protein Structure
The signal peptide cleavage site was predicted using SignalP (http://www.cbs.dtu.dk/services/SignalP/). The hydrophobicity index was calculated using the Kyte–Doolittle method on Genetyx. The plot was generated with a window setting at 10.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: NAI1 (At2g22770, NM_179700), NAI2 (At3g15950, NM_112465), the NAI2 EST clone (U16255, BT001207), PYK10 (At3g09260, NM_111760), TSA1 (At1g52410, NM_179466), At3g15960 (NM_112466), tdTomato (EU855182), two genome sequences of B. rapa (AC189268, gi:110744053 and AC189514, gi:110797194), and two genome sequences of B. oleracea (BZ479594, gi:26781992 and BZ006167, gi:23554425). The identifier of the tentative consensus sequence in B. napus is TC74287. The accession numbers of the SALK T-DNA insertion mutants are SALK_005896 for nai2-2 and SALK_043149 for nai2-3. The accession numbers of the European Arabidopsis Stock Center RNAi lines are N207544 for iNAI2(44) and N207546 for iNAI2(46).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. GFP Fluorescence and Bright-Field Images of the GFP-h Plant and nai2-1 Mutant.
Supplemental Figure 2. Structure of the Two NAI2 Homologs in Arabidopsis.
Supplemental Figure 3. Structure of the NAI2 Homolog in B. napus.
Supplemental Figure 4. Immunoblot Analysis Using Antibody against NAI2/C.
Supplemental Figure 5. SDS-PAGE of E. coli Expressing NAI2.
Supplemental Figure 6. Immunoprecipitation of NAI2 and PYK10.
Supplemental Table 1. Genes That Are Coexpressed with the NAI1 Gene According to ATTED-II.
Supplemental Table 2. Genes That Are Coexpressed with the NAI2 Gene According to ATTED-II.
Supplemental Table 3. Proteins That Colocalize with NAI2 According to Localization of Organelle Proteins by Isotope Tagging Data.
Supplemental Table 4. Nucleotide Sequences of Oligonucleotide Primers Used in This Study.
Supplemental Methods. Immunoprecipitation.
Supplementary Material
Acknowledgments
We thank Tatsuo Kakimoto (Osaka University), Roger Y. Tsien (University of California), Detlef Weigel (Max Plank Institute), and Shoji Mano (National Institute for Basic Biology) for the donation of vectors and bacterial strains. We thank Tadashi Kunieda (Kyoto University) and Iku Suzuki (National Institute for Basic Biology) for helpful experimental techniques. We also thank the National Institute for Basic Biology Center for Analytical Instruments for providing the instruments. This work was supported by Grants-in-Aid for Scientific Research to K.Y. (Grant 19770040), I.H.-N. (Grants 16085203 and 17107002), and M. Nishimura (Grant 1685209) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT), by the Global Center of Excellence Program Formation of a Strategic Base for Biodiversity and Evolutionary Research: from Genome to Ecosystem of MEXT, and by a Grant-in-Aid for JSPS Fellows to A.J.N. (Grant 18003246) from the Japan Society for the Promotion of Science.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Mikio Nishimura (mikosome@nibb.ac.jp).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Bagga, S., Adams, H., Kemp, J.D., and Sengupta-Gopalan, C. (1995). Accumulation of 15-kilodalton zein in novel protein bodies in transgenic tobacco. Plant Physiol. 107 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke, H.-D., and Eschlbeck, G. (1978). Dilated cisternae in Capparales: An attempt towards the characterization of a specific endoplasmic reticulum. Protoplasma 97 351–363. [Google Scholar]
- Chrispeels, M.J., and Herman, E.M. (2000). Endoplasmic reticulum-derived compartments function in storage and as mediators of vacuolar remodeling via a new type of organelle, precursor protease vesicles. Plant Physiol. 123 1227–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cola, A., Frigerio, L., Lord, J.M., Ceriotti, A., and Roberts, L.M. (2001). Ricin A chain without its partner B chain is degraded after retrotranslocation from the endoplasmic reticulum to the cytosol in plant cells. Proc. Natl. Acad. Sci. USA 98 14726–14731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley, T.P., et al. (2006). Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. USA 103 6518–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili, G. (2004). ER-derived compartments are formed by highly regulated processes and have special functions in plants. Plant Physiol. 136 3411–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura, I., and Matsushima, R. (2003). A wound-inducible organelle derived from endoplasmic reticulum: A plant strategy against environmental stress? Curr. Opin. Plant Biol. 6 583–588. [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura, I., Matsushima, R., Shimada, T., and Nishimura, M. (2004). Diversity and formation of endoplasmic reticulum-derived compartments in plants. Are these compartments specific to plant cells? Plant Physiol. 136 3435–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura, I., Shimada, T., Hatano, K., Takeuchi, Y., and Nishimura, M. (1998). Transport of storage proteins to protein-storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell 10 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, M., Toriyama, K., Kondo, M., Hara-Nishimura, I., and Nishimura, M. (1999). Accumulation of a fusion protein containing 2S albumin induces novel vesicles in vegetative cells of Arabidopsis. Plant Cell Physiol. 40 263–272. [DOI] [PubMed] [Google Scholar]
- Hayashi, Y., Yamada, K., Shimada, T., Matsushima, R., Nishizawa, N., Nishimura, M., and Hara-Nishimura, I. (2001). A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol. 42 894–899. [DOI] [PubMed] [Google Scholar]
- Herman, E.M., and Larkins, B.A. (1999). Protein storage bodies and vacuoles. Plant Cell 11 601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, E.M., and Schmid, M. (2004). Endoplasmic reticulum to vacuole trafficking of endoplasmic reticulum bodies an alternate pathway for protein transfer to the vacuole. Plant Physiol. 136 3440–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilson, P., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res. 14 2176–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding, D.R., Otegui, M.S., Li, B., Meeley, R.B., Dam, T., Hunter, B.G., Jung, R., and Larkins, B.A. (2007). The maize Floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. Plant Cell 19 2569–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen, T.-H. (1970. a). Cytochemical localization of myrosinase (β-thioglucosidase) in root tips of Sinapis alba. Protoplasma 71 451–466. [Google Scholar]
- Iversen, T.-H. (1970. b). The morphology, occurrence, and distribution of dilated cisternae of the endoplasmic reticulum in tissues of plants of the Cruciferae. Protoplasma 71 467–477. [Google Scholar]
- Kakimoto, T. (1996). CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274 982–985. [DOI] [PubMed] [Google Scholar]
- Levak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Matsushima, R., Fukao, Y., Nishimura, M., and Hara-Nishimura, I. (2004). NAI1 gene encodes a basic-helix-loop-helix-type putative transcription factor that regulates the formation of an endoplasmic reticulum-derived structure, the ER body. Plant Cell 16 1536–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima, R., Hayashi, Y., Shimada, T., Nishimura, M., and Hara-Nishimura, I. (2002). An endoplasmic reticulum-derived structure that is induced under stress conditions in Arabidopsis. Plant Physiol. 130 1807–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima, R., Hayashi, Y., Yamada, K., Shimada, T., Nishimura, M., and Hara-Nishimura, I. (2003. a). The ER body, a novel endoplasmic reticulum-derived structure in Arabidopsis. Plant Cell Physiol. 44 661–666. [DOI] [PubMed] [Google Scholar]
- Matsushima, R., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (2003. b). A novel ER-derived compartment, the ER body, selectively accumulates β-glucosidase with an ER-retention signal in Arabidopsis. Plant J. 33 493–502. [DOI] [PubMed] [Google Scholar]
- Mitsuhashi, N., Shimada, T., Mano, S., Nishimura, M., and Hara-Nishimura, I. (2000). Characterization of organelles in the vacuolar-sorting pathway by visualization with GFP in tobacco BY-2 cells. Plant Cell Physiol. 41 993–1001. [DOI] [PubMed] [Google Scholar]
- Nagano, A.J., Fukao, Y., Fujiwara, M., Nishimura, M., and Hara-Nishimura, I. (2008). Antagonistic jacalin-related lectins regulate the size of ER body-type β-glucosidase complexes in Arabidopsis thaliana. Plant Cell Physiol. 49 969–980. [DOI] [PubMed] [Google Scholar]
- Nagano, A.J., Matsushima, R., and Hara-Nishimura, I. (2005). Activation of an ER-body-localized β-glucosidase via a cytosolic binding partner in damaged tissues of Arabidopsis thaliana. Plant Cell Physiol. 46 1140–1148. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., Toyooka, K., Matsuoka, K., Jinbo, T., and Kimura, T. (2007). Development of series of Gateway binary vectors, pGWBs for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007 34–41. [DOI] [PubMed] [Google Scholar]
- Obayashi, T., Kinoshita, K., Nakai, K., Shibaoka, M., Hayashi, S., Saeki, M., Shibata, D., Saito, K., and Ohta, H. (2007). ATTED-II: A database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 35 D863–D869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, T., Shimada, T., Hara-Nishimura, I., Nishimura, M., and Minamikawa, T. (2003). C-terminal KDEL sequence of a KDEL-tailed cysteine proteinase (sulfhydryl-endopeptidase) is involved in formation of KDEL vesicle and in efficient vacuolar transport of sulfhydryl-endopeptidase. Plant Physiol. 132 1892–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpl, P., Taylor, J.P., Snowden, C., Hillmer, S., Robinson, D.G., and Denecke, J. (2006). Golgi-mediated vacuolar sorting of the endoplasmic reticulum chaperone BiP may play an active role in quality control within the secretory pathway. Plant Cell 18 198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., Simpson, D., and Gietl, C. (1999). Programmed cell death in castor bean endosperm is associated with the accumulation and release of a cysteine endopeptidase from ricinosomes. Proc. Natl. Acad. Sci. USA 96 14159–14164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., Simpson, D.J., Sarioglu, H., Lottspeich, F., and Gietl, C. (2001). The ricinosomes of senescing plant tissue bud from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 98 5353–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner, N.C., Campbell, R.E., Steinbach, P.A., Giepmans, B.N., Palmer, A.E., and Tsien, R.Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22 1567–1572. [DOI] [PubMed] [Google Scholar]
- Sherameti, I., Venus, Y., Drzewiecki, C., Tripathi, S., Dan, V.M., Nitz, I., Varma, A., Grundler, F.M., and Oelmüller, R. (2008). PYK10, a β-glucosidase located in the endoplasmic reticulum, is crucial for the beneficial interaction between Arabidopsis thaliana and endophytic fungus Piriformospora indica. Plant J. 54 428–439. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Nakajima, S., Morikami, A., and Nakamura, K. (2005). An Arabidopsis protein with a novel calcium-binding repeat sequence interacts with TONSOKU/MGOUN3/BRUSHY1 involved in meristem maintenance. Plant Cell Physiol. 46 1452–1461. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Yamada, K., Shimada, T., and Hara-Nishimura, I. (2004). Endoplasmic reticulum-resident proteins are constitutively transported to vacuoles for degradation. Plant J. 39 393–402. [DOI] [PubMed] [Google Scholar]
- Toyooka, K., Okamoto, T., and Minamikawa, T. (2000). Mass transport of proform of a KDEL-tailed cysteine proteinase (SH-EP) to protein storage vacuoles by endoplasmic reticulum-derived vesicle is involved in protein mobilization in germinating seeds. J. Cell Biol. 148 453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale, A., and Ceriotti, A. (2004). Protein quality control mechanisms and protein storage in the endoplasmic reticulum. A conflict of interests? Plant Physiol. 136 3420–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne, G. (1986). A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14 4683–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadahama, H., Kamauchi, S., Ishimoto, M., Kawada, T., and Urade, R. (2007). Protein disulfide isomerase family proteins involved in soybean protein biogenesis. FEBS J. 274 687–703. [DOI] [PubMed] [Google Scholar]
- Yamada, K., Fukao, Y., Hayashi, M., Fukazawa, M., Suzuki, I., and Nishimura, M. (2007). Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J. Biol. Chem. 282 37794–37804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.