Abstract
COP1 (for CONSTITUTIVELY PHOTOMORPHOGENIC1) and the four partially redundant SPA (for SUPPRESSOR OF PHYA) proteins work in concert to repress photomorphogenesis in Arabidopsis thaliana by targeting key transcription factors and phytochrome A for degradation via the 26S proteasome. Here, we report a detailed biochemical characterization of the SPA-COP1 complexes. The four endogenous SPA proteins can form stable complexes with COP1 in vivo regardless of light conditions but exhibit distinct expression profiles in different tissues and light conditions. The SPA proteins can self-associate or interact with each other, forming a heterogeneous group of SPA-COP1 complexes in which the exact SPA protein compositions vary depending on the abundance of individual SPA proteins. The four SPA proteins could be divided into two functional groups depending on their interaction affinities, their regulation of ELONGATED HYPOCOTYL5 degradation, and their opposite effects on COP1 protein accumulation. Loss-of-function mutations in a predominant SPA protein may cause a significant reduction in the overall SPA-COP1 E3 ligase activity, resulting in a partial constitutive photomorphogenic phenotype. This study thus provides an in-depth biochemical view of the SPA-COP1 E3 ligase complexes and offers new insights into the molecular basis for their distinct roles in the light control of plant development.
INTRODUCTION
Being sessile organisms, plants have evolved the capacity to optimize their growth in response to their light environment. Arabidopsis thaliana possesses several photoreceptors to mediate the perception of light signals (Neff et al., 2000), including the far-red and red light (600 to 750 nm)–sensing phytochromes (PHYA to PHYE), the UV-A/blue light (320 to 500 nm)–perceiving cryptochromes and phototropins, and the not-yet identified UV-B light receptors (Ulm and Nagy, 2005; Rockwell et al., 2006; Li and Yang, 2007). Activation of these photoreceptors, through distinct signal transduction pathways, leads to altered gene expression and photomorphogenic development.
A group of 11 CONSTITUTIVELY PHOTOMORPHOGENIC/DE-ETIOLATED/FUSCA (COP/DET/FUS) genes has been identified by screening for mutants that display a pleiotropic constitutive photomorphogenic phenotype in darkness (Schwechheimer and Deng, 2000; Yi and Deng, 2005). These genes act as negative regulators of the light control of plant development (Hoecker, 2005). Biochemically, these COP/DET/FUS proteins belong to three groups of protein complexes: the COP1 complex(es), the COP9 signalosome (CSN), and the CDD complex (COP10, DDB1, and DET1). It has been hypothesized that all three complexes function collectively in the ubiquitination/proteasome-mediated degradation of key photomorphogenesis-promoting transcription factors, thus achieving their repression of photomorphogenesis (Yanagawa et al., 2004; Chen et al., 2006).
Among these COP/DET/FUS proteins, COP1 is a point of convergence downstream of multiple light signals (von Arnim and Deng, 1994; Osterlund and Deng, 1998), acting as a central repressor of photomorphogenesis that can be inactivated by visible light and UV-A light. However, COP1 also acts as a positive regulator for several specific responses in tolerance toward low levels of UV-B light and possibly in a response mediated by PHYB (Boccalandro et al., 2004; Oravecz et al., 2006). Molecularly, COP1 acts as an E3 ligase to target several photomorphogenesis-promoting transcription factors, such as ELONGATED HYPOCOTYL5 (HY5) (Osterlund et al., 2000b), LONG AFTER FAR-RED LIGHT1 (Seo et al., 2003), and LONG HYPOCOTYL IN FAR-RED1 (HFR1) (Duek et al., 2004; Jang et al., 2005; Yang et al., 2005; Lin and Wang, 2007), for degradation via the 26S proteasome. In addition, the far-red light photoreceptor phyA (Seo et al., 2004) and the blue light photoreceptor cry2 (Wang et al., 2001; Shalitin et al., 2002) are also likely targets of COP1. COP1 contains three structural domains: an N-terminal RING finger domain, a central coiled-coil domain, and a C-terminal WD-repeat domain.
SUPPRESSOR OF PHYA (SPA1) was first identified as a repressor of a weak phyA mutation (Hoecker et al., 1998, 1999), and it encodes a 115-kD protein that contains an N-terminal kinase-like domain, a central coiled-coil domain, and a C-terminal WD-repeat domain (Hoecker et al., 1999). In Arabidopsis, there are three SPA1-like proteins: SPA2, SPA3, and SPA4. Mutation in each individual SPA protein causes a weak or no phenotype defect, while the quadruple spa mutant displays a constitutive photomorphogenic phenotype similar to that of a strong cop1 mutant allele (cop1-5). Systematic phenotypic analysis of various combinations of spa mutants indicated that the four SPA genes confer partially redundant but not completely overlapping functions in mediating light responses at both seedling and adult stages (Laubinger and Hoecker, 2003; Hoecker et al., 2004; Laubinger et al., 2004, 2006; Ishikawa et al., 2006).
Genetic interaction between cop1 and spa mutations and the detected physical interaction between the proteins led to the proposition that COP1 works in concert with the SPA protein family to repress photomorphogenesis (Hoecker and Quail, 2001; Baumgardt et al., 2002; Laubinger and Hoecker, 2003; Lin and Wang, 2007). However, the exact nature of their interaction and relationship in response to distinct light signals remains largely unknown. Among these SPA proteins, SPA1 is the most extensively characterized, and its biochemical interaction with COP1 has been demonstrated both in vitro and in vivo. It has been shown that SPA1 cofractionates with COP1 in high-molecular-weight complex(es) and that SPA1 can modulate the COP1 E3 ligase activity in vitro (Saijo et al., 2003; Seo et al., 2003). However, it is not clear exactly how SPA1 and other SPA proteins contribute to the architecture of COP1 complex(es). In addition, the molecular basis for the partially overlapping as well as distinct functions of SPA1 to SPA4 remains to be elucidated. Due to the central role of the COP1 E3 ligase activity in regulating light signaling, a detailed biochemical characterization of SPA proteins and their functional relationship with COP1 is highly desirable.
Here, by generating SPA member–specific antibodies and transgenic plants expressing epitope-tagged SPA fusion proteins, we have characterized each member of the SPA family at the protein level and have elucidated their interactions in a systematic manner. We have documented the distinct expression profiles of each endogenous SPA protein under various light conditions and at different developmental stages. Our biochemical analysis has demonstrated that COP1 and all four SPA proteins are interdependent in forming a heterogeneous group of functional SPA-COP1 complexes in Arabidopsis. Our data suggest that each complex may contribute unequally to the regulated proteolysis of substrates such as HY5, thus contributing to the fine-tuning of the light control of plant development.
RESULTS
Arabidopsis SPA Proteins Are Differentially Expressed in Most Tissues
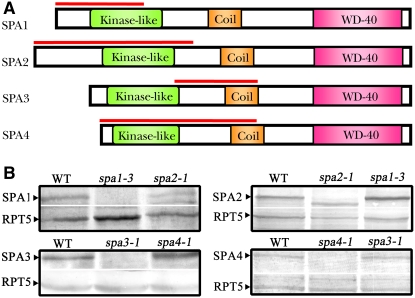
To monitor the expression pattern of endogenous SPA1 to SPA4 proteins, we raised member-specific polyclonal antibodies from rabbits for each SPA protein. To ensure the specificity of these antibodies, fragments containing ∼300 to 500 amino acids in the relatively less conserved N-terminal regions of SPA1, SPA2, and SPA4 were chosen to produce recombinant proteins as antigens (Figure 1A). In the case of SPA3, which shares 76% amino acid identity with SPA4, a 120–amino acid fragment from the most divergent region of SPA3 was chosen to raise its antibodies. As expected, the protein gel blot of the total protein extracts from wild-type Arabidopsis seedlings and individual spa mutants revealed that each of the four SPA proteins migrated at the predicated size and showed no observable cross-detection among the four individual SPA protein antibodies (Figure 1B). It is worthwhile to note that the spa2 mutant allele produces a truncated SPA2 protein at a level comparable to that of the endogenous wild-type protein; thus, a truncated protein band was observed in spa2 mutants together with a faint slightly higher cross-reacting background band.
Figure 1.
Generation and Characterization of Antibodies Specific to Each of the Four SPA Proteins.
(A) Structural diagrams of SPA1, SPA2, SPA3, and SPA4 proteins showing conserved domains. The red lines indicate the regions used as antigens for raising antibodies.
(B) Protein gel blots showing the specificity of the anti-SPA antibodies. Extracts from 6-d-old white light–grown plants were applied. Each SPA protein's specific band is indicated by a black arrowhead. RPT5 was used as a loading control.
Next, with these specific antibodies, we investigated the expression levels of endogenous SPA1 to SPA4 proteins in different tissue types. In 4-week-old Arabidopsis plants, SPA1 protein was highly abundant in leaves and flowers, but lesser amounts were detected in other organs (Figure 2A; see Supplemental Figure 1A online). Notably, the highest level of expression was detected in root tissues for SPA2, which may implicate a role of SPA2 in regulating root development in darkness under the soil. SPA3 is expressed in stems, leaves, and, more copiously, in flowers, but it is almost undetectable in roots. Abundant expression of SPA4 was detected in most tissues examined (root, leaf, stem, and flower), but only a trace amount was detected in siliques. Moreover, the comparable expression levels of SPA proteins in both white light–grown seedlings and 4-week-old adult plants suggest that these SPA proteins are required for Arabidopsis development throughout different stages (Figure 2B). Together, our data suggest that the four SPA proteins are expressed in all developmental stages, but with distinct tissue-specific expression patterns.
Figure 2.
Tissue-Specific Expression of Endogenous SPA Proteins and Their Regulation by Light.
(A) Protein gel blots showing organ-specific accumulation of the four SPA proteins in 4-week-old wild-type Arabidopsis plants. RPT5 is shown as a control for the amount of protein loaded.
(B) Protein gel blots showing accumulation of the four SPA proteins in 6-d-old Arabidopsis seedlings under different light conditions and in adult plants. Wc, continuous white light; FRc, continuous far-red light; Rc, continuous red light; Bc, continuous blue light; Dk, continuous darkness.
(C) Protein gel blots showing accumulation of the four SPA proteins during a darkness-to-light transition. Four-day-old dark-grown seedlings were transferred to white light for the indicated periods of time. The COP1 abundance in the same samples is shown below for comparison.
The Four SPA Proteins Exhibit Distinct Accumulation Patterns in Response to Light
Previous studies demonstrated that the transcript levels of SPA1, SPA3, and SPA4 are upregulated by various light signals, but the mRNA level of SPA2 is not altered in response to light (Hoecker et al., 1999; Laubinger et al., 2004; Fittinghoff et al., 2006). Here, we examined the protein expression patterns of these SPA genes under different light conditions. Consistent with the reported changes in transcript levels, the expression of SPA1 and SPA3 endogenous proteins is more abundant in 6-d-old seedlings grown under various continuous light conditions (white, far-red, red, and blue light) than seedlings grown in complete darkness (Figure 2B; see Supplemental Figure 1B online), while the expression of SPA4 is more abundant in both far-red light and blue light conditions than in darkness. By contrast, the level of SPA2 protein is approximately sixfold higher in dark-grown seedlings than in white light-grown seedlings. This result is in agreement with the previous finding that SPA2 is a predominant player in darkness (Laubinger et al., 2004).
We next investigated light regulation on SPA protein accumulation during a dark-to-light transition. For this experiment, 4-d-old dark-grown wild-type seedlings were transferred to white light for different periods of time and the abundance of individual SPA proteins was examined. As shown in Figure 2C, the abundance of SPA1, SPA3, and SPA4 proteins was upregulated by light. Consistent with the change in transcript level, the increase in SPA1 protein level was fast and could be detected after 1 h of light exposure, while the upregulation of SPA3 and SPA4 could only be clearly seen after 6 h of white light irradiation. By contrast, the endogenous SPA2 level was downregulated by light, which is evident after 1 h of white light irradiation for seedlings transferred from darkness. As reported previously (von Arnim and Deng, 1994), the cellular pool of COP1 remained constant during the dark-to-light transition (Figure 2C; see Supplemental Figure 1C online).
SPA Proteins Are Capable of Self-Association and Pairwise Interaction in Vitro
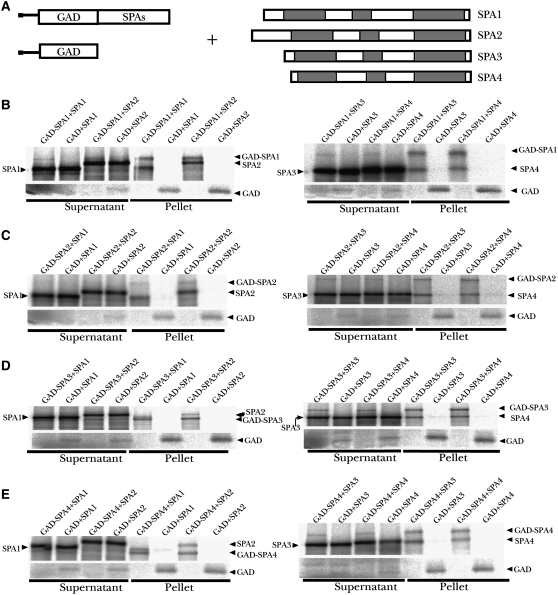
Since the most pronounced constitutive hyperphotomorphogenic phenotype can only be seen in the absence of all four SPA genes, these SPA genes must function redundantly in repressing photomorphogenesis (Laubinger et al., 2004). To help understand the molecular basis of this functional redundancy, we first tested SPA protein self-association and interaction by an in vitro pull-down assay. In this assay (Figure 3A), recombinant GAD-SPA fusion proteins (fused with the GAL4 activation domain at the N terminus) were used to pull down [35S]Met-labeled SPA proteins produced by coupled in vitro transcription/translation reactions (Hoecker and Quail, 2001). As shown in Figure 3B, GAD-SPA1 can pull down SPA1 itself as well as the other three SPA proteins. Reciprocal experiments using GAD-SPA2, GAD-SPA3, and GAD-SPA4 fusion proteins as baits confirmed the self-association and pairwise interaction of these SPA proteins in vitro (Figures 3C to 3E). Interestingly, a quantitative analysis of the percentage of preys bound to each of the four GAD-SPA proteins revealed that SPA1 and SPA2 have the highest affinity toward each other among all possible hetero–pairwise interactions (see Supplemental Figure 2 online). Furthermore, self-association of SPA1 and SPA2 was also significantly stronger than that of SPA3 and SPA4. The unequal strength of self-association or interaction among these SPA proteins may affect the formation and relative abundance of various SPA-COP1 protein complexes in vivo (see below).
Figure 3.
In Vitro Interactions among the Four SPA Proteins.
(A) Diagram of constructs used in the in vitro binding assays.
(B) In vitro coimmunoprecipitation of SPA proteins by GAD-SPA1. 35S-labeled SPA1 to SPA4 were incubated with GAD-SPA1 or GAD and subsequently coimmunoprecipitated with anti-GAD antibodies. Supernatant fractions and pellet fractions were resolved by SDS-PAGE and visualized by autoradiography using a phosphor imager.
(C) to (E) A series of reciprocal in vitro coimmunoprecipitation assays of SPA1 to SPA4 by GAD-SPA2, GAD-SPA3, and GAD-SPA4, respectively. 35S-labeled SPA1 to SPA4 were incubated with GAD-SPA2, GAD-SPA3, GAD-SPA4, or GAD, respectively, and subsequently coimmunoprecipitated with anti-GAD antibodies. Supernatant fractions and pellet fractions were collected using the same method as in (B).
The N Terminus of SPA1 Is Responsible for Mediating SPA1 Self-Association and the SPA1–SPA2 Interaction
Previous studies have shown that the coiled-coil domain of SPA1 mediates its interaction with COP1 (Hoecker and Quail, 2001; Yang et al., 2005) and that the WD-repeat domain is involved in binding the substrate proteins, such as HY5 and HFR1 (Hoecker et al., 1999; Saijo et al., 2003; Yang and Wang, 2006). To determine which region within SPA1 is responsible for mediating its self-association and interaction with other SPA proteins, we tested a series of deletion derivatives of SPA1 using an in vitro binding assay (Figure 4A). 35S-labeled full-length SPA1 and deletion derivatives SPA1-NT696, SPA1-NT545, and SPA1-CC were used as the preys. GAD-SPA1, GAD-SPA2, and GAD were used as the baits. SPA1-NT696, which lacks the C-terminal WD-repeat domain, bound to both SPA1 and SPA2 at least as efficiently as the SPA1 full-length protein, indicating that the WD-repeat domain is not essential for SPA1 self-association or SPA1–SPA2 interaction (Figure 4B; see Supplemental Figure 3 online). Deletion of the coiled-coil domain strongly reduced but did not fully abolish these interactions. Furthermore, the coiled-coil domain by itself also supported weak interactions (Figure 4B; see Supplemental Figure 3 online). Hence, we conclude that the N-terminal region of SPA1 containing the kinase-like domain and the coiled-coil domain is sufficient for strong self-interaction and SPA2 binding, while the C-terminal WD-repeat domain of SPA1 is dispensable for these interactions.
Figure 4.
The N Terminus of SPA1 Is Essential for SPA1 Self-Association and SPA2 Binding.
(A) Schematic diagram of SPA1 and SPA1 deletion derivatives.
(B) In vitro coimmunoprecipitation assay showing that the N-terminal domain of SPA1 is essential for SPA1 self-association and SPA2 binding. 35S-labeled full-length SPA1, SPA1-NT696, SPA1-NT545, and SPA1-CC were used as the prey molecules and incubated with unlabeled GAD-SPA1, GAD-SPA2, or GAD. Anti-GAD antibodies were used for immunoprecipitation. Supernatant and pellet fractions were resolved by SDS-PAGE and visualized by autoradiography using a phosphor imager. The samples were separated on two gels as shown. The lower portion of the right gel is also shown at bottom to indicate the SPA1-CC mutant protein.
Colocalization of the Four SPA Proteins
To investigate whether these SPA proteins can interact directly with each other in plant cells, we first used bimolecular fluorescence complementation (BIFC) (Walter et al., 2004) to investigate their interactions. Successfully transfected cells were monitored in the cyan fluorescent protein (CFP) channel with a cotransformed CFP-talin construct. Coexpression of a yellow fluorescent protein (YFP) N-terminal fragment fused to SPA1 with a YFP C-terminal fragment also fused to SPA1 in onion (Allium cepa) epidermal cells resulted in strong YFP fluorescence in the nuclei, indicating that SPA1 can self-interact (Figure 5A). The BIFC system could only be reliably used for the SPA1–SPA1 interaction. For unknown reasons, the SPA2, SPA3, and SPA4 fusion proteins produced weak YFP signals when coexpressed with negative controls (data not shown).
Figure 5.
Interactions among the Four SPA Proteins in Plant Cells.
(A) BIFC analysis of the SPA1 homodimeric interaction. YN-SPA1 and YC-SPA1 fusion proteins were transiently expressed in onion epidermal cells. Cytoplasmically localized CFP-talin was used as a marker for successfully transfected cells. The YN and YC fusions of the nuclear protein CPRF1 were used as negative controls. The magnification is the same in all images. Bar = 100 μm.
(B) Nuclear images showing colocalization of SPA proteins in plant cells. CFP and YFP fusions of SPA proteins were transiently coexpressed in Arabidopsis leaf epidermal cells and visualized using fluorescence microscopy. The magnification is the same in all images. Bars = 10 μm.
We next used an alternative approach to examine the colocalization of SPA proteins by coexpressing CFP-SPA and YFP-SPA fusion proteins in transiently transfected Arabidopsis leaf epidermal cells. Figure 5B shows that CFP-SPA1 colocalized with YFP-SPA1, YFP-SPA2, and YFP-SPA4 in nuclear speckles. Interestingly, YFP-SPA4 only produced speckles when coexpressed with CFP-SPA1, indicating that SPA1 can recruit SPA4 into nuclear speckles. This colocalization suggests that SPA1 can interact with itself as well as with SPA2 and SPA4 in plant cell nuclei. We also observed colocalization of CFP-SPA2 and YFP-SPA2 (Figure 5B), thus supporting the conclusion that SPA2 can self-associate in plant cell nuclei as well. However, we did not observe any recruitment of SPA3 into the nuclear speckles of SPA1 (data not shown).
SPA Proteins Self-Associate in Vivo
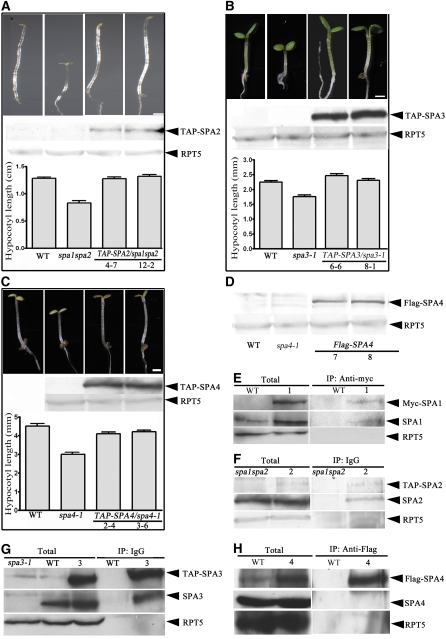
To test for in vivo homodimerization of these SPA proteins, we generated transgenic Arabidopsis plants expressing 35S promoter–driven N-terminal TAP (for tandem affinity purification)–tagged full-length SPA2, SPA3, and SPA4 transgenes in the spa1 spa2, spa3-1, and spa4-1 mutant backgrounds, respectively. The progeny homozygous for the respective transgene were selected, and the transgenes were subsequently introduced into the wild-type background by genetic cross for further biochemical assays. As shown in Figures 6A to 6C, the TAP-SPA2, TAP-SPA3, and TAP-SPA4 transgenes complemented their corresponding mutant phenotypes, suggesting that these TAP-SPA fusion proteins are functional. The expression of these SPA fusion proteins was confirmed by protein gel blot using anti-myc monoclonal antibody. We also generated transgenic plants expressing 35S promoter–driven full-length SPA4 fused with three copies of the flag epitope tag at the N terminus (3× flag-SPA4).
Figure 6.
Self-Association of SPA Proteins in Vivo.
(A) to (C) Seedling images of the wild type, spa1 spa2, and two lines of spa1 spa2/35S:TAP-SPA2 grown in darkness ([A], top), the wild type, spa1 spa2, and two lines of spa3-1/35S:TAP-SPA3 grown in continuous red light ([B], top), or the wild type, spa1 spa2, and two lines of spa4-1/35S:TAP-SPA4 grown in continuous far-red light ([C], top). All photographs were taken under the same magnification. Bars = 1 mm. The middle panels of (A) to (C) are protein gel blots showing the accumulation of TAP-SPA fusion proteins, with RPT5 shown as a control for protein levels. The bottom panels of (A) to (C) show the hypocotyl lengths (average of 20 seedlings) with the mean and sd (error bars).
(D) Protein gel blot of the flag-SPA4 fusion protein in the wild type, spa4-1, and two lines of spa4-1 /35S:TAP-SPA4.
(E) to (H) Coimmunoprecipitation and protein gel blot analysis showing self-association of SPA proteins in vivo. The affinity beads used for immunoprecipitation (IP) are indicated at top, and the antibodies used for detection are shown at right. The various SPA proteins were detected using their respective antibodies by protein gel blot analyses. Lane 1, Col/35S:myc-SPA1; lane 2, spa1 spa2/35S:TAP-SPA2; lane 3, Col/35S:TAP-SPA3; lane 4, Col/35S:flag-SPA4.
Using these and the previously generated 35S:myc-SPA1 transgenic lines, we examined the interaction of these tagged SPA proteins with their endogenous counterparts by coimmunoprecipitation. Wild-type Arabidopsis, spa control mutants, and transgenic lines were grown under continuous white light for 6 d. Protein extracts prepared from the seedlings were subjected to pull-down assays using an anti-myc affinity matrix (Figure 6E), IgG-agarose beads (Figures 6F and 6G), or anti-flag affinity matrix (Figure 6H). In the case of SPA1 (Figure 6E), the agarose beads conjugated with anti-myc antibody efficiently pulled down myc-SPA1 and a similar amount of endogenous SPA1. Similarly, both TAP-SPA3 and endogenous SPA3, but not the negative control RPT5, were pulled down in the immunoprecipitates from the 35S:TAP-SPA3 transgenic plants (Figure 6G). For SPA2, we used spa1 spa2/35S:TAP-SPA2 transgenic plants. Protein gel blot analysis from IgG precipitates using SPA2-specific antibody resulted in the identification of the TAP-SPA2 protein as well as the slightly fast-migrating truncated SPA2 protein that lacks the last 64 amino acids at the C terminus (Figure 6F). Of note, the truncated SPA2 protein is still able to coimmunoprecipitate and cofractionate with COP1 in vivo (see Supplemental Figure 4 online). In the case of SPA4, the endogenous SPA4 was not successfully detected in the immunoprecipitates using the anti-flag affinity matrix (Figure 6H). Instead, a slightly lower mobility band was recognized by both anti-flag and anti-SPA4 antibodies (see Supplemental Figure 5 online). The identity of this band is not clear, but it is possible that it may represent a mixture of the endogenous SPA4 and 1× flag-tagged SPA4 (due to a separate ATG start codon in the third flag copy in the 3× flag-SPA4 construct). Regardless, our data confirmed the in vivo homodimeric interaction for at least SPA1, SPA2, and SPA3.
Multiple Heteromeric Interactions among SPA Proteins in Vivo
We next examined the possible heteromeric association among these SPA proteins and the effect of light on these associations in vivo. As shown in Figure 7A, immunoprecipitation of TAP-SPA1 fusion protein from 6-d-old far-red light–grown and dark-grown spa1-3/35S:TAP-SPA1 seedlings also pulled down three other SPA proteins and COP1, but not the RPT5 negative control (Figure 7A). In similar experiments with TAP-SPA3 (Figure 7B) and TAP-SPA4 (Figure 7C), anti-myc antibody–conjugated beads immunoprecipitated not only the TAP fusion proteins but also the other SPA proteins and COP1. These results support the notion that these SPA proteins are able to form all possible heteromeric interactions and that all members of the SPA family of proteins associate with COP1 in vivo.
Figure 7.
Heteromeric Interaction of SPA Proteins in Vivo.
(A) Coimmunoprecipitation and protein gel blot analysis showing that TAP-SPA1 associates with SPA2, SPA3, SPA4, and COP1 in vivo. Continuous far-red light (FR)–grown and continuous dark (Dk)–grown spa1-3/35S:TAP-SPA1 were used; continuous far-red light–grown control spa1-3 seedlings were also used.
(B) and (C) Coimmunoprecipitation and protein gel blot analysis showing that TAP-SPA3 and TAP-SPA4 interact with other SPA proteins and COP1 in vivo. Continuous far-red light–grown spa3-1 and spa4-1 mutants were used as controls.
IgG-conjugated agarose beads were used for the immunoprecipitation experiments with samples of spa1-3 and spa1-3/35S:TAP-SPA1 (A), spa3-1 and spa3-1/35S:TAP-SPA3 (B), and spa4-1 and spa4-1/35S:TAP-SPA4 (C).
It is worth noting that in each pull-down assay, although essentially the same amount of COP1 was precipitated by TAP-SPA1, TAP-SPA3, and TAP-SPA4, the yields of TAP-SPA proteins in the immunoprecipitates were slightly higher under light conditions compared with darkness (Figure 7). This is most likely due to the elevated levels of total TAP-SPA proteins under light conditions. On the contrary, the yield of coimmunoprecipitated SPA2 protein seems to be downregulated by light (Figure 7), again consistent with the accumulation of SPA2 being highest in darkness.
COP1 Interacts with All Four SPA Proteins in Vivo
We next performed reciprocal immunoprecipitation to test the ability of COP1 to pull down each of the four SPA proteins in vivo. As shown in Figure 8A, COP1 was able to pull down each of the four SPA proteins effectively in various light conditions tested. Interestingly, the yields of endogenous COP1 precipitated by anti-COP1 antibody are similar regardless of the light signals, while the yields of individual SPA proteins pulled down by anti-COP1 antibody differ. A quantification of the ratios of each precipitated SPA protein versus COP1 suggests that the amounts of SPA proteins coimmunoprecipitated with COP1 were correlated with their expression levels in different light conditions (Figures 8B to 8E), with more SPA1, SPA3, and SPA4 and less SPA2 proteins being coimmunoprecipitated with COP1 under all light conditions. Furthermore, it is also possible that light may affect the affinity of the SPA–COP1 interaction.
Figure 8.
COP1 Associates with the Four SPA Proteins.
(A) Coimmunoprecipitation and protein gel blot analysis showing that endogenous COP1 coimmunoprecipitates with each of the four SPA proteins using the wild-type seedlings under various light conditions. Six-day-old white light–grown wild-type and cop1-6 seedlings were used for total samples. The cop1-6 mutant was used as a negative control. Anti-COP1 antibodies were used for immunoprecipitation.
(B) to (E) Quantification of the yields of SPA proteins that copurified with COP1 in the indicated light conditions. Relative SPA1 to SPA4 (SPA/COP1) protein levels in (A) were quantified, with the level of COP1 in immunoprecipitates set to 100%. Error bars represent se of the values collected from three repeated experiments.
(F) to (H) Protein gel blot analyses showing the gel filtration patterns of COP1 and SPA proteins in wild-type (F), cop1-5 null allele (G), and spa1 spa2 spa3 and the quadruple spa seedlings (H). Fraction numbers and molecular weights are indicated at top. T, total unfractionated extracts.
The Formation of Normal SPA-COP1 Protein Complex(es) Is Dependent on the Presence of SPA Proteins and COP1
To test whether SPA2, SPA3, and SPA4 are indeed components of COP1 complexes, as previously shown for SPA1 (Saijo et al., 2003), we performed gel filtration analysis using 6-d-old seedlings grown in both light and dark conditions. All four endogenous SPA proteins roughly cofractionated with each other and with COP1, regardless of light conditions (Figure 8F). The gel filtration patterns of all four SPA proteins were significantly altered in the cop1-5 mutant (Figure 8G), indicating that COP1 is essential for the integrity of the SPA-COP1 complexes. The profiles of the four SPA proteins can be classified into two groups: in the first group, SPA1 and SPA2 cofractionated at around 440 kD in the absence of COP1; the other group, including SPA3 and SPA4, cofractionated and migrated at much smaller fractions (around 200 kD) in the absence of COP1. Thus, we concluded that COP1 is required for proper complex formation for all SPA proteins.
We also examined the gel filtration pattern of COP1 in a series of spa mutants. We did not see any notable change in the COP1 fractionation pattern between the wild type and a selected group of spa double mutants. However, both the quantity and integrity of the COP1 complexes were affected in the triple mutant spa1 spa2 spa3, although this was not observed in other triple mutants (Figure 8H). Strikingly, in the spa quadruple mutant, the COP1 complex was split into two parts, one eluting at around 700 kD and the other eluting at around 440 kD. Thus, the integrity of the SPA-COP1 association depends on the SPA proteins, although COP1 can aggregate into some complexed forms (the 700-kD fractions) in the absence of all four SPA proteins.
COP1 and the Four SPA Proteins Most Likely Form a Family of Heterogeneous SPA-COP1 Complexes in Arabidopsis
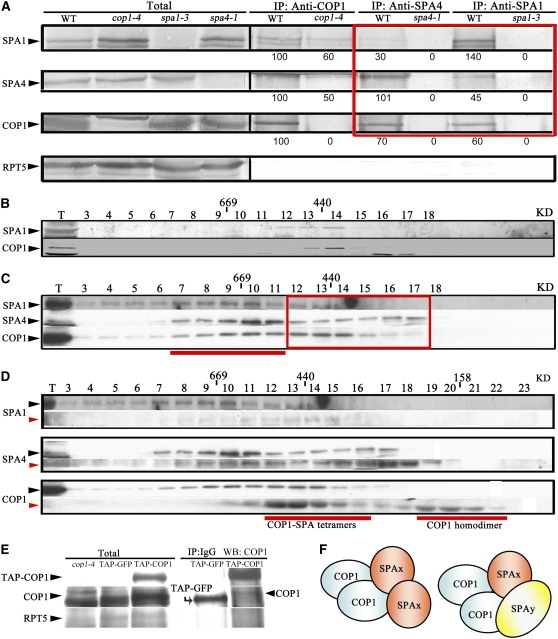
To characterize the protein–protein interactions further among the four endogenous SPA proteins and COP1, we performed coimmunoprecipitation experiments using anti-COP1, anti-SPA1, and anti-SPA4 antibodies. As shown in Figure 9A, these antibodies consistently pulled down their respective endogenous protein as well as the other two proteins. It is interesting that the COP1 antibody can still bring down a reduced amount of SPA1 and SPA4 in the cop1-4 mutant seedlings. This is because cop1-4 produces a truncated COP1 protein containing the N-terminal 282 amino acids, including the SPA-interacting coiled-coil domain. This result supports the conclusion that the coiled-coil domain of COP1 is involved in mediating the interactions between COP1 and SPA proteins.
Figure 9.
Evidence for the Presence of Multiple SPA-COP1 Complexes in Arabidopsis.
(A) Coimmunoprecipitation and protein gel blot analysis showing that endogenous COP1, SPA1, and SPA4 associate with each other in vivo. Anti-COP1, anti-SPA1, and anti-SPA4 antibodies were used for immunoprecipitation. Protein A–agarose beads were subsequently added to each of the protein samples, and the immunoprecipitates were collected for further protein blot analysis. The genotypes are indicated above each lane. Numbers below the lanes indicate relative band intensities that are quantified and normalized for each panel. The red box highlights the differences among the amounts of immunoprecipitated SPA proteins using anti-SPA4 and anti-SPA1 antibodies.
(B) to (D) Protein gel blot analyses of gel filtration fractions collected from the inflorescence of cauliflower (B), 6-d-old light-grown wild-type Arabidopsis seedlings (C), and 6-d-old light-grown wild-type Arabidopsis seedlings using low-salt (150 mM NaCl; black arrowheads) and high-salt (450 mM NaCl; red arrowheads) washing, respectively (D). Fraction numbers are indicated at top. T, total extracts before fractionation. The red box in (C) highlights the fractions of the core SPA-COP1 complexes in wild-type plants. The red line in (C) indicates the larger fractions of SPA-COP1 complexes in wild-type plants. The two red lines in (D) indicate the core SPA-COP1 complexes and COP1 monomers, respectively.
(E) Protein gel blot analysis showing coimmunoprecipitation of TAP-COP1 and endogenous COP1 from white light–grown 35S:TAP-COP1 transgenic plants.
(F) Possible compositional configurations of the core SPA-COP1 complexes. The core SPA-COP1 complexes are likely tetramers, including both homogeneous complexes (two COP1 plus two homogenous SPAs; indicated by SPAx) and heterogeneous complexes (two COP1 plus two heterogeneous SPAs, indicated by SPAx and SPAy).
It is important to note that in both the SPA1 and SPA4 immunoprecipitates, the yields of COP1 were similar but the amounts of other SPA proteins pulled down were much reduced (Figure 9A, red box; see Supplemental Figure 6 online). This unequal efficiency of immunoprecipitation of the different SPA proteins strongly argues against the presence of all SPA proteins in the same SPA-COP1 complex. Instead, the data suggest the coexistence of a group of heterogeneous SPA-COP1 complexes, all of which contain COP1 but that differ in the composition of SPA proteins.
A Possible Configuration of the SPA-COP1 Heterogeneous Complexes in Arabidopsis
To gain further insight into the possible compositional configuration of the SPA-COP1 complexes, we took advantage of a unique distinction in the COP1 and SPA fractionation patterns of Arabidopsis seedlings and floral tissue of cauliflower (Brassica oleracea), a closely related species. Both anti-COP1 and anti-SPA1 antibodies recognize their respective counterparts from cauliflower floral extracts (Figure 9B). Strikingly, the size of the SPA1-COP1 complex in cauliflower floral tissue is around 440 kD in a gel filtration assay (Figure 9B), whereas Arabidopsis COP1 and all four SPA proteins show a very broad distribution toward larger fractions (Figure 9C). Thus, the cauliflower SPA-COP1 complexes could represent a stripped-down version, or core complex, while the Arabidopsis complexes may contain other peripheral subunits or represent an oligomerized form of the core complexes.
To test this possibility, we performed size-fractionation analysis of Arabidopsis SPA-COP1 complexes with a more stringent high-salt wash (450 mM NaCl), which should strip away the peripheral subunits or disassociate the oligomerized complexes, thus reducing the apparent complex size. Indeed, under such a stringent condition, COP1, SPA1, and SPA4 still cofractionated with each other (Figure 9D), with the peak fractions changed from high molecular weight around 700 kD to smaller fractions around 400 kD. In addition, some COP1 fractionated to the size range representing COP1 dimers (Torii et al., 1998; Subramanian et al., 2006; Xu et al., 2007).
To examine whether each SPA-COP1 complex contains at least two COP1 molecules (dimer), we tested the coimmunoprecipitation of endogenous COP1 with a TAP-tagged COP1 (TAP-COP1). As shown in Figure 9E, IgG agarose beads effectively pulled down TAP-COP1 and the endogenous COP1 from both dark- and light-grown seedlings. Thus, our data indicated that there are at least two copies of COP1 in a SPA-COP1 complex. It is worth pointing out that under normal salt conditions, no endogenous COP1 was detected in the dimer fractions. Thus, the COP1 dimer most likely exists within the heterogeneous SPA-COP1 complexes under normal physiological conditions. Overall, our data point to a possible tetrameric configuration of the heterogeneous SPA-COP1 core complexes that contain two COP1 molecules and two SPA molecules (Figure 9F). Theoretically, there are 10 possible compositional configurations for two SPA proteins within a complex, considering all possible homomeric and heteromeric associations among the four SPA proteins.
COP1 and SPA Proteins Work Together to Mediate HY5 Degradation in Vivo
As reported previously (Laubinger et al., 2004; Fittinghoff et al., 2006), various spa mutant combinations exhibited a varying range of hyperphotomorphogenic phenotypes (Figure 10A; see Supplemental Figure 7 online). The spa quadruple mutant displayed a strong cop-like phenotype in darkness. All other spa mutants could be divided into two subgroups according to their dark-grown phenotypes: (1) mutants lacking both SPA1 and SPA2 proteins exhibited partial photomorphogenic development in darkness; (2) mutants lacking both SPA3 and SPA4 displayed a normal etiolation phenotype in darkness (Figure 10A). To determine whether their phenotypic differences can be correlated with the overall activity of the SPA-COP1 complexes, we examined HY5 accumulation in a series of spa mutants, as the abundance of HY5 can be used as a hallmark for COP1 activity. Interestingly, we noticed that HY5 abundance was higher in all spa mutants tested than in the wild-type background (Figure 10A; see Supplemental Figure 8 online), indicating that all SPA proteins contributed toward the targeted degradation of HY5. In particular, we detected high levels of HY5 protein in the spa1 spa2, spa1 spa2 spa3, and spa1 spa2 spa4 mutants that show a deetiolated phenotype in darkness (Figure 10B). By contrast, loss of spa3 and spa4 resulted in relatively low levels of HY5 accumulation. Additionally, loss of SPA1 or SPA2 in the spa3 spa4 background increased HY5 accumulation. Together, these data suggest a strict correlation between HY5 level and the severity of the spa mutant phenotype in darkness (see Supplemental Figure 7 online). Furthermore, our data suggest that SPA1 and SPA2 contribute more to targeted HY5 degradation in darkness than do SPA3 and SPA4 in vivo.
Figure 10.
COP1 and SPA Proteins Function Together to Mediate HY5 Degradation in Darkness.
(A) Morphology of 7-d-old dark-grown wild-type and spa mutant seedlings.
(B) and (C) Protein gel blot analysis of HY5 and COP1 in a series of spa mutants. Six-day-old dark-grown wild-type and spa mutant seedlings were used.
(D) to (G) Expression of SPA proteins in a series of spa mutants. Six-day-old wild-type and spa mutant seedlings were used.
To gain further insights into the relationship between COP1 and SPA proteins, we examined COP1 protein accumulation in various spa mutants. Interestingly, elevated COP1 accumulation was observed in spa1 spa2 double mutants. On the contrary, COP1 abundance was slightly reduced in the normal etiolated spa3 spa4 double mutants (Figure 10C; see Supplemental Figure 9A online). Although additional lesions in SPA1 or SPA2 resulted in an increase of COP1 accumulation, the levels of COP1 in spa1 spa3 spa4 and spa2 spa3 spa4 mutants were still much lower than the levels in spa1 spa2 spa3 and spa1 spa2 spa4 mutants. It is also interesting that COP1 level was decreased by almost twofold in the spa quadruple mutant. Therefore, it appears that these SPA proteins might have opposite effects on COP1 accumulation. It is worth noting that the endogenous COP1 transcript levels are very similar in the wild type and the representative spa1 spa2 and spa3 spa4 mutants (see Supplemental Figure 9A online). Our data suggested that translational or posttranslational regulation may be involved in regulating the abundance of COP1 by SPA proteins. This conclusion is supported by the observation that treatment of wild-type and spa mutant seedlings with a proteasome inhibitor (50 μM MG132) resulted in increased COP1 protein amounts but not transcript levels (see Supplemental Figure 9A online). Furthermore, SPA1 and SPA2 play a negative role in regulating COP1 accumulation, whereas SPA3 and SPA4 are required to promote COP1 accumulation. Indeed, continuous treatment of the 5-d-old dark-grown seedlings with the proteasome inhibitor MG132 for 9 h enabled detection of polyubiquitinated forms of COP1 in the coimmunoprecipitates using spa3 spa4 double mutants (see Supplemental Figure 9B online).
Our data also suggest that the high levels of HY5 accumulation in spa mutants cannot be solely due to the changes in COP1 levels, since even in the spa quadruple mutant, which shows a constitutive cop phenotype, there is ∼50% as much COP1 as in the wild type (see Supplemental Figure 10 online), a situation similar to that in heterozygous cop1 mutant plants. As cop1 mutations are strictly recessive, our data provided another layer of evidence to support the notion that COP1 and SPA proteins must work cooperatively to mediate HY5 degradation in darkness and that loss of a predominant SPA-COP1 complex may result in changed functional output, such as the degradation of HY5 and photomorphogenic development.
Additionally, we checked whether the difference in COP1 and HY5 protein levels seen in spa triple mutants could be caused by altered levels of the remaining SPA protein. The abundance of SPA1 and SPA4 was not obviously altered even in the absence of the other three SPA proteins (Figures 10D and 10G). However, accumulation of both SPA2 and SPA3 increased in the relevant triple mutants (Figures 10E and 10F). Thus, increased accumulation of one SPA protein in the absence of the other three SPA proteins is not sufficient for normal degradation of HY5 in vivo, further supporting our idea that COP1 and the four SPA proteins function cooperatively.
Loss of the Pleiotropic COP/DET/FUS Proteins Results in a Significant Increase in SPA Protein Accumulation
The COP/DET/FUS proteins are known to be important negative regulators of photomorphogenesis (Kim et al., 2002; Serino and Deng, 2003). To test the possible involvement of other COP/DET/FUS proteins in regulating SPA proteins, we examined the expression of SPA proteins in dark-grown cop1-6, cop9-1, cop10-1, and det1-1 mutants. We observed that both COP1 and SPA proteins accumulated at high levels in both cop9-1 and det1-1 backgrounds (Figure 11). In the absence of COP10, accumulation of SPA2 and COP1 was slightly decreased compared with the wild-type. However, the abundance of SPA1, SPA3, and SPA4 was upregulated in the cop10-1 lethal allele. Our data suggest that these COP/DET/FUS proteins also play a role in regulating the steady state levels of SPA proteins.
Figure 11.
The Normal Accumulation of SPA Proteins Requires Functional COP/DET/FUS Proteins.
Protein gel blot analysis showing the effects of cop1-6, cop9-1, cop10-1, and det1-1 mutations on the accumulation of COP1 and SPA proteins. Six-day-old dark-grown seedlings were used.
DISCUSSION
Prior to this investigation, our knowledge about the SPA gene family was based on genetic and molecular characterization of these genes (Hoecker, 2005; Yi and Deng, 2005). Although there were data from several groups demonstrating the functional interdependence of COP1 and SPA1 (Saijo et al., 2003; Laubinger et al., 2004; Yang and Wang, 2006), the biochemical relationship between COP1 and the whole SPA protein family remained unclear. Therefore, we generated member-specific antibodies against SPA proteins as well as epitope-tagged SPA transgenic plants for biochemical characterization of the interdependence of those five proteins. Our analyses using these resources provide evidence to support the conclusion that COP1 and SPA proteins form a heterogeneous group of complexes in vivo and also provide critical insight into the compositional configuration of these complexes. Thus, our work adds an important dimension to prior genetic analyses of the SPA protein family and builds a platform for further understanding the molecular action mechanisms of SPA-COP1 protein complexes.
COP1 and SPA Proteins Form a Group of Heterogeneous Complexes in Planta
In this study, we first documented the existence of SPA-COP1 complex(es) by in vivo coimmunoprecipitation and gel filtration analyses (Figures 7 and 8). There are two possibilities regarding the configuration of the SPA-COP1 complex or complexes. First, all SPA proteins could form a single supercomplex with COP1. In this scenario, immunoprecipitation of any SPA protein should pull down other SPA proteins more or less equally. Alternatively, there might be multiple SPA-COP1 complexes, with variable SPA protein composition in each complex. This later scenario would predict that immunoprecipitation of one SPA would bring down much less of the other SPA proteins than COP1 and itself. Three lines of evidence support the latter scenario. First, there are striking differences among the amounts of immunoprecipitated SPA proteins using anti-SPA4 and anti-SPA1 antibodies (Figure 9A). This finding supports the presence of multiple SPA-COP1 complexes and argues against all SPA proteins being in the same complex. Second, no observable changes were detected in COP1 gel filtration patterns in wild-type and spa single and double mutant plants (Figure 8H). Third, the similar gel filtration pattern of all four SPA proteins with COP1 under different light conditions again argues for the existence of multiple SPA-COP1 core complexes in vivo (Figure 8F).
Our studies also provide several clues to the compositional nature of the SPA-COP1 complexes. First, COP1 physically associates with itself and with each of the four SPA proteins in vitro and in vivo (Figures 8 and 9E). Second, all SPA proteins appear to be able to self-associate (Figure 6) and to interact with each other in a pairwise fashion (Figure 7). Third, higher salt wash could reduce the size fractionation patterns of both COP1 and SPA proteins (but they still cofractionate) to around 440 kD, a size range consistent with a tetrameric configuration with two COP1 and two SPA proteins. These observations together support the notion that each of these heterogeneous SPA-COP1 complexes contains a core of two molecules of COP1 and two molecules of SPA proteins, in which the SPA protein could be a homodimer or a heterodimer of any combination of the four SPA proteins.
SPA Proteins Are Critical for the Activity of COP1 in Vivo
It has been documented that COP1 plays a critical role in mediating the light control of plant development. COP1 was shown to be modulated by light-activated photoreceptors, the COP9 signalosome, and the CDD complex (Saijo et al., 2003; Yanagawa et al., 2004; Chen et al., 2006; Yang and Wang, 2006). In this study, we showed that not only SPA1 but all four SPA proteins behave as components of COP1-containing complexes and that they collectively mediate HY5 degradation in darkness (Figure 10). Presumably, all of these proposed SPA-COP1 complexes may possess a similar E3 ligase activity, but different complexes may also have somewhat functional distinctions. Our biochemical data, together with prior genetic analyses, indicate that the SPA-COP1 complexes containing SPA1 and/or SPA2 play a more predominant role in controlling HY5 (and possibly other substrates) degradation in darkness and possibly maintaining COP1 homeostasis in vivo (see Supplemental Figure 9 online). By contrast, the SPA-COP1 complexes containing SPA3 and/or SPA4 may play a more minor role in regulating HY5 degradation but an essential role in maintaining COP1 abundance, since a reduced amount of COP1 was seen in spa3 spa4 mutants, which may be caused by enhanced ubiquitination of COP1 itself (see Supplemental Figure 9B online).
Possible Significance of the Presence of Heterogeneous SPA Protein Complexes
As the prior genetic analyses established the largely redundant function of the four SPA proteins in repressing photomorphogenesis (Laubinger et al., 2004; Fittinghoff et al., 2006), it must be asked what is the possible advantage for having four SPA proteins. It is conceivable that each of the four SPA proteins may contribute differentially to the overall SPA-COP1 complex functions at different light conditions or developmental stages. Indeed, we showed here that the four SPA proteins have distinct light-regulated and organ-specific expression patterns (Figure 2), largely reflecting their mRNA level patterns (Fittinghoff et al., 2006), although the discrepancy in their protein affinity may not strictly correspond to their expression pattern. Furthermore, all four SPA proteins form complexes with COP1. The large number of possible SPA-SPA configurations (homodimeric or heterodimeric) in a single SPA-COP1 complex may confer an exquisite regulatory mechanism for fine-tuning SPA-COP1 E3 ligase activity at different light conditions or developmental stages. The absence of a prominent SPA at a particular stage under a specific light condition could lead to a major defect in the overall SPA-COP1 E3 activity, leading to an observable phenotype under specific light conditions or at a particular developmental stage. It is also possible that the SPA-COP1 complexes with distinct SPA protein compositions may confer unique biochemical properties, such as unique affinities for various substrates under distinct light conditions or at different stages. Together, dynamic SPA-COP1 protein complex formation and their function may represent an important mechanism underlying signal transduction during photomorphogenesis. Further studies will be needed to unravel the relative relationship of these distinct SPA-COP1 complexes in the light control of plant development.
METHODS
Plant Materials and Growth Conditions
The wild-type Arabidopsis thaliana plants used in the study are the Columbia-0 and RLD ecotypes unless otherwise stated. Some of the mutants and transgenic lines used in this study were described previously: spa1-3 (Hoecker et al., 1998), spa2-1, spa1 spa2, spa1 spa2 spa3, spa1 spa2 spa4, and spa1 spa3 spa4 (Laubinger et al., 2004), spa3-1, spa4-1, and spa3 spa4 (Laubinger and Hoecker, 2003), spa2 spa3 spa4 (Fittinghoff et al., 2006), cop1-4, cop1-5, and cop1-6 (McNellis et al., 1994), cop9-1 (Wei and Deng, 1992), det1-1 (Chory et al., 1989), cop10-1 (Suzuki et al., 2002), 35S:TAP-SPA1 (Saijo et al., 2003), 35S:myc-SPA1 (Yang and Wang, 2006), and 35S:TAP-COP1 (Rubio et al., 2005).
The Arabidopsis seeds were surface-sterilized and cold-treated at 4°C for 2 to 3 d, and then the plants were grown on solid 1× Murashige and Skoog medium supplemented with 1% sucrose for biochemical assays or with 0.3% sucrose for phenotype analysis. Cold-treated seeds were exposed to white light for 12 to 14 h and then transferred to continuous light conditions (0.5 μmol·m−2·s−1 for far-red light, 10 μmol·m−2·s−1 for red light, 4 μmol·m−2·s−1 for blue light, and 30 μmol·m−2·s−1 for white light) or continuous darkness, unless otherwise stated.
Plasmid Construction and Generation of Transgenic Arabidopsis Plants
The full-length open reading frames (ORFs) of SPA2, SPA3, and SPA4 were amplified from RNA isolated from wild-type Arabidopsis seedlings by RT-PCR with the following gene-specific primers: SPA2 forward primer (5′-ACTAACCGCGGGGACAAAATGGATGAGGGATCAGTAGGGGATGTG-3′) and SPA2 reverse primer (5′-ACTAAGGCGCGCCTCAGACCAACTGTAGAACTTTGATTGA-3′); SPA3 forward primer (5′-ACTAACCGCGGGGACAAAATGGAAGGTTCTTCAAATTCTAACTCT-3′) and SPA3 reverse primer (5′-ACTAAGGCGCGCCTCAAGTCATCATCTCCAGAATTTTTAT-3′); SPA4 forward primer (5′-ACTAACCGCGGGGACAAAATGAAGGGTTCTTCAGAATCTAGTTCC-3′) and SPA4 reverse primer (5′-ACTAAGGCGCGCCTCATACCATCTCCAAAATCTTGATATT-3′). All primers contain an in-frame AscI site at the 5′ end of each ORF and a SacII site at the 3′ end of each ORF. The resulting PCR products were cloned into the pENTR/D-TOPO vector (Invitrogen). These constructs were used as LR reaction templates for subsequent cloning of SPA2 to SPA4 ORFs into the plant binary vector pN-TAPa (Rubio et al., 2005) using Gateway LR Clonase enzyme mix (Invitrogen). The transgenes are under the control of the 35S promoter.
To generate 3× flag-tagged SPA4 transgene, we amplified a PCR fragment using SPA4 forward primer (5′-TTTGGGGTACCATGAAGGGTTCTTCAGAAT-3′) and SPA4 reverse primer (5′-AGCCAGAGCTCACTAGTTCATACCATCTCCAAAAT-3′). The PCR product (an in-frame KpnI/SacI fragment) was cloned into the binary vector pF3PZPY122 (Feng et al., 2003). For stable transformation, the binary DNA constructs were electroporated into Agrobacterium tumefaciens strain GV3101 (pMP90). 35S:TAP-SPA2, 35S:TAP-SPA3, and 35S:TAP-SPA4 were transformed into spa1 spa2, spa3-1, and spa4-1, respectively. The 35S:flag-SPA4 construct was introduced into the previously generated 35S:TAP-SPA1 transgenic background (Saijo et al., 2003). All transformation was done via A. tumefaciens–mediated transformation using the floral dip method (Clough and Bent, 1998). Transgenic plants were selected using gentamycin (200 mg/L; Sigma-Aldrich).
Production of Specific SPA Antibodies and Other Antibodies Used
To generate antibodies specific to each SPA protein, NotI/BamHI fragments containing amino acid residues 1 to 273 of SPA1, amino acid residues 1 to 500 of SPA2, amino acid residues 359 to 495 of SPA3, and amino acid residues 1 to 500 of SPA4 were cloned into pET28a (Novagen). Recombinant proteins were expressed in the bacterial strain BL21 (DE3) (Novagen). The soluble recombinant His6-SPA1 protein was purified using nickel-nitrilotriacetic acid beads (Qiagen). The insoluble His6-SPA2, His6-SPA3, and His6-SPA4 recombinant proteins were separated by SDS-PAGE and electroeluted. Polyclonal SPA antibodies were raised by immunizing rabbits using purified recombinant proteins as antigens. The antibodies were affinity-purified using the purified SPA antigen proteins immobilized on polyvinylidene difluoride membranes. The polyclonal antibodies against COP1, CSN3, CSN6, RPT5, and HY5 antibodies and the monoclonal antibodies against the flag and myc epitopes were described previously (Kwok et al., 1999; Osterlund et al., 2000a; Saijo et al., 2003; Yang et al., 2005).
In Vitro Interaction Assay
Prey proteins of full-length SPA1 to SPA4, SPA1-NT696 (amino acid residues 1 to 696), SPA1-CC (amino acid residues 521 to 696), and SPA1-NT545 (amino acid residues 1 to 545) were synthesized in the presence of [35S]Met. Bait proteins GAD-SPA1, GAD-SPA2, GAD-SPA3, and GAD-SPA4 were synthesized in the presence of a mixture of [35S]Met and unlabeled Met. Both the prey and bait proteins were produced by coupled in vitro transcription/translation reactions (Promega), and the subsequent coimmunoprecipitation assays were performed as described previously (Hoecker and Quail, 2001). Immunoprecipitated proteins were detected using film in the linear range of detection, and the signal was quantified using image-analysis software (http://rsb.info.nih.gov/ij/). Three biological replicates were used for the quantification of the percentages of prey SPA proteins bound to baits.
Colocalization, BIFC Experiments, and Fluorescence Microscopy
For BIFC analysis, the ORF of SPA1 was amplified by PCR and cloned by Gateway technology (Walhout et al., 2000) into the Gateway vectors pCL112 and pCL113, which carry N-terminal (YN) and C-terminal (YC) fragments of YFP, respectively.
For colocalization experiments, the ORFs of various SPA genes were cloned into the pENSG-YFP and pENSG-CFP vectors using Gateway technology (Laubinger et al., 2006). All cDNAs were expressed from the constitutive 35S promoter. For transient expression, these constructs were introduced into onion (Allium cepa) epidermal cells or Arabidopsis leaf epidermal cells by particle bombardment using a helium Helios gun (Bio-Rad) according to the manufacturer's instructions. For the BIFC experiments, CFP-mTalin, under the control of the 35S promotor, was used as a marker for successful transfection (Saedler et al., 2004). Talin is an actin binding protein. CPRF1 fusion proteins were used as negative controls (Stolpe et al., 2005).
Fluorescence microscopy was performed using a Leica DMRE microscope equipped with a high-resolution KY-F70-3CCD JVC camera, differential interference contrast (Nomarski) optics, and epifluorescence optics.
Protein Extraction, Immunoblot Analysis, and Gel Filtration Chromatography
Protein extraction, protein gel blot, and gel filtration analyses were performed as described previously (Hardtke et al., 2002) with a modified lysis buffer: 50 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10 mM NaF, 25 mM β-glycerophosphate, 2 mM Na3VO4, 10 mM NaF, 10% glycerol, 0.1% Tween 20, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and 1× complete protease inhibitor cocktail (Roche).
For gel filtration analysis, 6-d-old Arabidopsis seedlings were homogenized in the lysis buffer, and the extract was centrifuged at 14,000 rpm for 15 min at 4°C, subsequently filtered though a 0.22-mm syringe filter, and fractionated using a Superose 6 column (Amersham Biosciences). After the void volume (7 mL) was achieved, consecutive fractions of 0.5 mL were collected. Protein samples were concentrated using Strataclean resin (Stratagene) and subsequently analyzed by protein gel blot.
In Vivo Coimmunoprecipitation Assay
For pull-down assays, 1 mg of total proteins from Arabidopsis tissues was incubated with 40 μL of IgG-agarose beads (Amersham), 40 μL of anti-flag antibody–conjugated agarose beads (Sigma-Aldrich), or 40 μL of anti-myc–conjugated Sepharose beads (9E10 affinity matrix; Covance) in the total protein extraction lysis buffer without DTT for 3 h at 4°C. Next, the beads were washed three times in the lysis buffer containing 200 or 250 mM NaCl. The immunoprecipitates were eluted into a buffer containing 100 mM Gly at pH 2.5 and 100 mM NaCl. The samples were immediately neutralized by adding buffer containing Tris (2 M, pH 9.0) and 100 mM NaCl and then concentrated using Strataresin (Stratagene). Protein blot analyses were performed as described previously (Gusmaroli et al., 2004). Three independent biological replicates were subjected to protein gel blot analysis. Protein gel blot signals were detected by film, with exposures in the linear range of detection. RPT5 was used as a loading control.
For immunoprecipitation, 500 μg of total proteins was incubated with 2 μL of polyclonal anti-COP1 or anti-SPA antibodies for 2.5 h at 4°C. Then, 10 μL of protein A–agarose beads (Sigma-Aldrich) was subsequently added to each of the protein samples and incubated for 2 h at 4°C. The antigen-antibody-protein A-agarose conjugates were precipitated by centrifugation and washed twice with the lysis buffer containing 250 mM NaCl, followed by wash using the lysis buffer containing 150 mM NaCl two times. Then, the immunoprecipitated samples were released into the acid elution buffer (100 mM Gly, pH 2.5, and 100 mM NaCl), neutralized, and subjected to protein blot analysis as described above.
Effects of spa Mutants on COP1 Level in Vivo
For examination of COP1 levels in wild-type, spa1 spa2, and spa3 spa4 seedlings, dark-grown 5-d-old seedlings were vacuum-infiltrated with 50 μM MG132 (Calbiochem) dissolved in DMSO or DMSO alone for 10 min and kept immersed in the same solution for 9 h.
Samples were extracted and analyzed by protein gel blotting using anti-COP1 antibody. Three independent plant samples were also processed for RNA gel blotting as described previously (Deng et al., 2006). 28s rRNA was stained with SYBR Green for linearity detection.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At2g46340 (NM_130197), At4g11110 (NM_117181), At3g15354 (NM_148725), At1g53090 (NM_104188), and At2g32950 (NM_128855).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Quantification of Tissue-Specific Expression of Endogenous SPA Proteins and Their Regulation by Light.
Supplemental Figure 2. Quantification of the Percentages of Prey SPA Proteins Bound to Baits.
Supplemental Figure 3. Quantification of the Percentages of Full-Length SPA1 and Its Domain Deletions Bound to Baits.
Supplemental Figure 4. The Truncated SPA2 Protein in spa2-1 Interacts with COP1 in Vivo.
Supplemental Figure 5. Detection of a Slightly Fast-Mobility Form of flag-SPA4.
Supplemental Figure 6. Quantification of the Unequal Efficiency of Immunoprecipitation of SPA1 and SPA4 Proteins.
Supplemental Figure 7. Correlation between HY5 Level and the Severity of the spa Mutant Phenotype in Darkness.
Supplemental Figure 8. HY5 Accumulates at a High Level in the spa Quadruple Mutant in Continuous White Light.
Supplemental Figure 9. Regulation of COP1 Levels by SPA Proteins in Vivo.
Supplemental Figure 10. Correlation between HY5 Level and COP1 Level in the spa Mutant Series.
Supplementary Material
Acknowledgments
We thank several former and current Deng laboratory members, Jim Sullivan, Vicente Rubio, Giuliana Gusmaroli, and Kun He, for providing reagents, helpful advice, and critical comments. We are grateful to Stefan Kircher (Institut für Biologie II, Universität Freiburg) for the gift of the CPRF1 plasmids. The Gateway vectors pCL112 and pCL113 were kind gifts from Christian Lauterbach (Scottish Crop Research Institute, Invergowrie, Dundee). This research was supported by funds from the National Institutes of Health (Grant GM47850) to X.W.D. and in part by the National Institute of Biological Sciences at Beijing and the German Research Foundation (Grant SFB 635 to U.H.). D.Z. was supported by a Monsanto Fellowship through the Peking–Yale Joint Research Center during her visit to Yale University (2004 to 2006).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Xing Wang Deng (xingwang.deng@yale.edu).
Online version contains Web-only data.
References
- Baumgardt, R.L., Oliverio, K.A., Casal, J.J., and Hoecker, U. (2002). SPA1, a component of phytochrome A signal transduction, regulates the light signaling. Planta 215 745–753. [DOI] [PubMed] [Google Scholar]
- Boccalandro, H.E., Rossi, M.C., Saijo, Y., Deng, X.W., and Casal, J.J. (2004). Promotion of photomorphogenesis by COP1. Plant Mol. Biol. 56 905–915. [DOI] [PubMed] [Google Scholar]
- Chen, H., Shen, Y., Tang, X., Yu, L., Wang, J., Guo, L., Zhang, Y., Zhang, H., Feng, S., Strickland, E., Zheng, N., and Deng, X.W. (2006). Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18 1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory, J., Peto, C., Feinbaum, R., Pratt, L., and Ausubel, F. (1989). Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58 991–999. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Deng, W., et al. (2006). Organization of the Caenorhabditis elegans small non-coding transcriptome: Genomic features, biogenesis, and expression. Genome Res. 16 20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek, P.D., Elmer, M.V., van Oosten, V.R., and Fankhauser, C. (2004). The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr. Biol. 14 2296–2301. [DOI] [PubMed] [Google Scholar]
- Feng, S., Ma, L., Wang, X., Xie, D., Dinesh-Kumar, S.P., Wei, N., and Deng, X.W. (2003). The COP9 signalosome interacts physically with SCF COI1 and modulates jasmonate responses. Plant Cell 15 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fittinghoff, K., Laubinger, S., Nixdorf, M., Fackendahl, P., Baumgardt, R.L., Batschauer, A., and Hoecker, U. (2006). Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J. 47 577–590. [DOI] [PubMed] [Google Scholar]
- Gusmaroli, G., Feng, S., and Deng, X.W. (2004). The Arabidopsis CSN5A and CSN5B subunits are present in distinct COP9 signalosome complexes, and mutations in their JAMM domains exhibit differential dominant negative effects on development. Plant Cell 16 2984–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., Okamoto, H., Stoop-Myer, C., and Deng, X.W. (2002). Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8). Plant J. 30 385–394. [DOI] [PubMed] [Google Scholar]
- Hoecker, U. (2005). Regulated proteolysis in light signaling. Curr. Opin. Plant Biol. 8 469–476. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., and Quail, P.H. (2001). The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J. Biol. Chem. 276 38173–38178. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., Tepperman, J.M., and Quail, P.H. (1999). SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284 496–499. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., Toledo-Ortiz, G., Bender, J., and Quail, P.H. (2004). The photomorphogenesis-related mutant red1 is defective in CYP83B1, a red light-induced gene encoding a cytochrome P450 required for normal auxin homeostasis. Planta 219 195–200. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., Xu, Y., and Quail, P.H. (1998). SPA1: A new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell 10 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, M., Kiba, T., and Chua, N.H. (2006). The Arabidopsis SPA1 gene is required for circadian clock function and photoperiodic flowering. Plant J. 46 736–746. [DOI] [PubMed] [Google Scholar]
- Jang, I.C., Yang, J.Y., Seo, H.S., and Chua, N.H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.H., Kim, B.H., and von Arnim, A.G. (2002). Repressors of photomorphogenesis. Int. Rev. Cytol. 220 185–223. [DOI] [PubMed] [Google Scholar]
- Kwok, S.F., Staub, J.M., and Deng, X.W. (1999). Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J. Mol. Biol. 285 85–95. [DOI] [PubMed] [Google Scholar]
- Laubinger, S., Fittinghoff, K., and Hoecker, U. (2004). The SPA quartet: A family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger, S., and Hoecker, U. (2003). The SPA1-like proteins SPA3 and SPA4 repress photomorphogenesis in the light. Plant J. 35 373–385. [DOI] [PubMed] [Google Scholar]
- Laubinger, S., Marchal, V., Gentilhomme, J., Wenkel, S., Adrian, J., Jang, S., Kulajta, C., Braun, H., Coupland, G., and Hoecker, U. (2006). Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133 3213–3222. [DOI] [PubMed] [Google Scholar]
- Li, Q.H., and Yang, H.Q. (2007). Cryptochrome signaling in plants. Photochem. Photobiol. 83 94–101. [DOI] [PubMed] [Google Scholar]
- Lin, R., and Wang, H. (2007). Targeting proteins for degradation by Arabidopsis COP1: Teamwork is what matters. J Integr Plant Biol 49 35–42. [Google Scholar]
- McNellis, T.W., von Arnim, A.G., Araki, T., Komeda, Y., Misera, S., and Deng, X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., Fankhauser, C., and Chory, J. (2000). Light: An indicator of time and place. Genes Dev. 14 257–271. [PubMed] [Google Scholar]
- Oravecz, A., Baumann, A., Mate, Z., Brzezinska, A., Molinier, J., Oakeley, E.J., Adam, E., Schafer, E., Nagy, F., and Ulm, R. (2006). CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18 1975–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund, M.T., and Deng, X.W. (1998). Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 16 201–208. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000. a). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 462–466. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Wei, N., and Deng, X.W. (2000. b). The roles of photoreceptor systems and the COP1-targeted destabilization of HY5 in light control of Arabidopsis seedling development. Plant Physiol. 124 1520–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell, N.C., Su, Y.S., and Lagarias, J.C. (2006). Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57 837–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio, V., Shen, Y., Saijo, Y., Liu, Y., Gusmaroli, G., Dinesh-Kumar, S.P., and Deng, X.W. (2005). An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41 767–778. [DOI] [PubMed] [Google Scholar]
- Saedler, R., Mathur, N., Srinivas, B.P., Kernebeck, B., Hulskamp, M., and Mathur, J. (2004). Actin control over microtubules suggested by DISTORTED2 encoding the Arabidopsis ARPC2 subunit homolog. Plant Cell Physiol. 45 813–822. [DOI] [PubMed] [Google Scholar]
- Saijo, Y., Sullivan, J.A., Wang, H., Yang, J., Shen, Y., Rubio, V., Ma, L., Hoecker, U., and Deng, X.W. (2003). The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C., and Deng, X.W. (2000). The COP/DET/FUS proteins—Regulators of eukaryotic growth and development. Semin. Cell Dev. Biol. 11 495–503. [DOI] [PubMed] [Google Scholar]
- Seo, H.S., Watanabe, E., Tokutomi, S., Nagatani, A., and Chua, N.H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H.S., Yang, J.Y., Ishikawa, M., Bolle, C., Ballesteros, M.L., and Chua, N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423 995–999. [DOI] [PubMed] [Google Scholar]
- Serino, G., and Deng, X.W. (2003). The COP9 signalosome: Regulating plant development through the control of proteolysis. Annu Rev Plant Biol. 54 165–182. [DOI] [PubMed] [Google Scholar]
- Shalitin, D., Yang, H., Mockler, T.C., Maymon, M., Guo, H., Whitelam, G.C., and Lin, C. (2002). Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417 763–767. [DOI] [PubMed] [Google Scholar]
- Stolpe, T., Susslin, C., Marrocco, K., Nick, P., Kretsch, T., and Kircher, S. (2005). In planta analysis of protein-protein interactions related to light signaling by bimolecular fluorescence complementation. Protoplasma 226 137–146. [DOI] [PubMed] [Google Scholar]
- Subramanian, C., Woo, J., Cai, X., Xu, X., Servick, S., Johnson, C.H., Nebenfuhr, A., and von Arnim, A.G. (2006). A suite of tools and application notes for in vivo protein interaction assays using bioluminescence resonance energy transfer (BRET). Plant J. 48 138–152. [DOI] [PubMed] [Google Scholar]
- Suzuki, G., Yanagawa, Y., Kwok, S.F., Matsui, M., and Deng, X.W. (2002). Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 16 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii, K.U., McNellis, T.W., and Deng, X.W. (1998). Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J. 17 5577–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm, R., and Nagy, F. (2005). Signalling and gene regulation in response to ultraviolet light. Curr. Opin. Plant Biol. 8 477–482. [DOI] [PubMed] [Google Scholar]
- von Arnim, A.G., and Deng, X.W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79 1035–1045. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J., Temple, G.F., Brasch, M.A., Hartley, J.L., Lorson, M.A., van den Heuvel, S., and Vidal, M. (2000). GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328 575–592. [DOI] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Wang, H., Ma, L.G., Li, J.M., Zhao, H.Y., and Deng, X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294 154–158. [DOI] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.W. (1992). COP9: A new genetic locus involved in light-regulated development and gene expression in Arabidopsis. Plant Cell 4 1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Soutto, M., Xie, Q., Servick, S., Subramanian, C., von Arnim, A.G., and Johnson, C.H. (2007). Imaging protein interactions with bioluminescence resonance energy transfer (BRET) in plant and mammalian cells and tissues. Proc. Natl. Acad. Sci. USA 104 10264–10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa, Y., Sullivan, J.A., Komatsu, S., Gusmaroli, G., Suzuki, G., Yin, J., Ishibashi, T., Saijo, Y., Rubio, V., Kimura, S., Wang, J., and Deng, X.W. (2004). Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 18 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Lin, R., Sullivan, J., Hoecker, U., Liu, B., Xu, L., Deng, X.W., and Wang, H. (2005). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17 804–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., and Wang, H. (2006). The central coiled-coil domain and carboxyl-terminal WD-repeat domain of Arabidopsis SPA1 are responsible for mediating repression of light signaling. Plant J. 47 564–576. [DOI] [PubMed] [Google Scholar]
- Yi, C., and Deng, X.W. (2005). COP1—From plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15 618–625. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.