Abstract
Purpose
One of the major obstacles in understanding the molecular mechanisms underlying the transition of prostate cancer growth from androgen dependency to hormone-refractory state is the lack of androgen-regulated and tumorigenic human prostate cancer cell lines.
Experimental Design
We have established and characterized a new human prostate cancer cell line, CWR22Pc, derived from the primary CWR22 human prostate xenograft tumors.
Results
The growth of CWR22Pc cells is induced markedly by dihydrotestosterone (DHT), and CWR22Pc cells express high levels of androgen receptor (AR) and prostate specific antigen (PSA). Importantly, PSA expression in CWR22Pc cells is regulated by androgens. Stat5a/b, Stat3, Akt and MAPK were constitutively active or cytokine-inducible in CWR22Pc cells. The AR in CWR22Pc cells contains the H874Y mutation, but not the exon 3 duplication or other mutations. When inoculated subcutaneously into DHT-supplemented castrated nude mice, large tumors formed rapidly in 20/20 mice whereas no tumors developed in mice without circulating DHT. Moreover, the serum PSA levels correlated with the tumor volumes. When androgens were withdrawn from the CWR22Pc tumors grown in nude mice, the tumors initially shrank but re-grew back as androgen-independent tumors.
Conclusions
This androgen-regulated and tumorigenic human prostate cancer cell line provides a valuable tool for studies on androgen-regulation of prostate cancer cells and on the molecular mechanisms taking place in growth promotion of prostate cancer when androgens are withdrawn from the growth environment. CWR22Pc cells provide a model system for studies on the regulation of transcriptional activity of mutated H874YAR in a prostate cancer cell context.
Keywords: prostate cancer, androgen-regulation, PSA, tumor growth, experimental model system
Introduction
The median duration of response to androgen deprivation therapy of primary prostate cancer is less than three years (1, 2). The molecular mechanisms underlying development of androgen-independent growth of prostate cancer are largely unknown, and no effective therapies for hormone-refractory prostate cancer exist at present. One of the key problems in conducting studies to identify growth factors and signaling pathways that can replace androgens in the growth control of prostate cancer cells is the lack of androgen-receptor (AR) positive human prostate cancer cell lines that are regulated by androgens and would be tumorigenic in nude mice. LNCaP cells respond by accelerated growth rate to androgens (3), but are able to grow relatively well in the absence of androgens (4). Moreover, tumor incidence of LNCaP cells after subcutaneous inoculation into nude mice is low and the tumors grow slowly (3).
We have characterized a new human prostate cancer cell line established from primary transplantable human CWR22 prostate tumors. CWR22 tumors were originally derived from a Gleason score 9 primary prostate cancer with bone metastases (5, 6). The primary CWR22 prostate tumors are highly responsive to androgen deprivation with marked tumor regression after castration, mimicking the course of the human disease. After androgen deprivation-induced regression of the original tumor, CWR22 prostate cancer recurs within 7-9 months and the recurrent CWR22 tumors (CWR22R) are not dependent on androgens for growth (7). CWR22Rv1 is an androgen-independent human prostate cancer cell line that has been established from one of the hormone-refractory recurrent CWR22R tumors (8). However, no cell lines displaying the growth characteristics of the original androgen-regulated primary CWR22 tumors have existed until now.
In this work, we have established an androgen-regulated human prostate cancer cell line named CWR22Pc from the primary CWR22 tumors. We demonstrate by comparative genomic hybridization and by DNA fingerprinting that the CWR22Pc cell line genetically originates from the primary CWR22 tumors. We further show that the growth of CWR22Pc cells is regulated by androgens in cell culture. Importantly, CWR22Pc cells form androgen-regulated, rapidly growing tumors at high incidence in nude mice. Moreover, CWR22Pc cells produce prostate specific antigen (PSA) in an androgen-regulated manner in culture, and the serum PSA levels correlate with the CWR22Pc xenograft tumor volumes in mice. Similar to the parental primary CWR22 tumors, CWR22Pc cells express androgen receptor (AR) containing the H874Y mutation without the tandem duplication of exon 3. We show that several key growth factor-related signaling pathways are cytokine-inducible or constitutively active in CWR22Pc cells. Finally, androgen-deprivation of the CWR22Pc tumors in nude mice induced tumor regression which was followed by androgen-independent recurrence of the CWR22Pc tumors. The androgen-regulated CWR22Pc cell line provides a new experimental model system for studies on androgen-regulated growth of prostate cancer cells and for research on genetic changes and kinase signaling pathways involved in growth promotion and dedifferentiation of androgen-deprived prostate cancer cells.
Materials and Methods
Cell culture
Suspension of CWR22Pc cells was prepared from androgen-dependent human primary prostate tumor xenograft, CWR22P (a gift from Dr. Thomas Pretlow, Case Western Reserve University) using a method described previously for the dissociation of human prostate cancer tissue (9). Briefly, under sterile conditions, six tumors were dissected from the mice and minced. The minced tumor was washed three times in RPMI 1640 (Life Technologies, Inc., Gaithersburg, MD) supplemented with 20% fetal bovine serum (FBS) (Life Technologies) and the tumor tissue was digested serially with 0.1% Pronase E (EMD Pharmaceuticals, Gibbstown NJ) in Joklik-modified MEM (Sigma, St. Louis, MO). The Pronase E digest fractions were filtered through a single layer of a NITEX 250-micron porosity membrane (Safer America, Depew, NY), and centrifuged at 97 x g for 7.5 minutes at 4°C. The pellets were resuspended in RPMI 1640 supplemented with 20% fetal bovine serum. The cells were counted and the cell viability was confirmed by trypan blue exclusion. The cells were cultured in RPMI 1640 medium containing 10% FBS, 2.5 mM L-glutamine and penicillin-streptomycin (100 IU/ml and 100 μg/ml, respectively) in the presence of 0.8 nM dihydrotestosterone (DHT) (5α-androstan-17β-ol-3-one, Sigma) at 37 °C with 5% CO2. CWR22Rv1, LNCaP and DU145 cells (ATCC, Manassas, VA) were cultured in RPMI 1640 (Biofluids) containing 10% FBS, 2.5 mM L-glutamine, and penicillin-streptomycin (100 IU/ml and 100 μg/ml, respectively) at 37 °C with 5% CO2. LNCaP cells were cultured in the presence of 0.8 nM DHT. For testing of inducibility of Stat5a/b and Stat3 by cytokines, the cell lines were serum-starved 16 h after which the cells were stimulated with 10 nM human prolactin (hPrl) or 4 nM interleukin-6 (IL6) (Upstate, Lake Placid, NY) before harvesting the cells for immunoprecipitations.
Growth of CWR22Pc cells as xenograft tumors in athymic nude mice
Castrated male athymic mice were purchased from Taconic (Germantown, NY) and cared for according to the institutional guidelines. Briefly, 20 × 106 CWR22Pc cells were mixed with one half of the total injection volume of 0.2 ml with Matrigel (BD Bioscience, Palo Alto, CA). One week before the tumor cell inoculation (2 sites/mouse), sustained-release DHT pellets (12.5 mg/pellet, 1 pellet/mouse; Innovative Research of America, Sarasota, FL) were implanted s.c. in half of the mice. The tumor sizes were measured twice a week, and the tumor volumes were calculated using the formula (3.14 × length × width × height/6) (10). When the tumors reached 15–20 mm in diameter, mice were sacrificed, the tumor tissues were harvested and the blood samples were collected. In the second set of experiments, CWR22Pc cells were inoculated subcutaneously to the flanks of castrated athymic nude mice (n=34) supplied with DHT pellets as described above (1 site/mouse). When the tumors reached 10 mm in diameter, the DHT-pellets were removed and the tumor sizes were measured twice a week.
Serum PSA determinations
Serum PSA levels were determined using the commercial kit DSL-10-9700 ACTIVE® PSA (Diagnostic Systems Laboratories, Inc, Webster, TX) according to the manufacturer’s instructions. Briefly, the standards, the controls and the serum samples were incubated in microtitration wells coated with an anti-PSA monoclonal antibody. The antigen binding was quantified by a polyclonal anti-PSA antibody labelled with horseradish peroxidase enzyme and subsequent addition of the horseradish peroxidase substrate tetramethylbenzidine. The degree of enzymatic turnover of the substrate was determined by dual wavelength absorbance measurement at 450 and 620 nm.
Protein solubilization and immunoblotting
Pellets of CWR22Pc cells were solubilized in lysis buffer (10 mM Tris-HCl, pH 7.6, 5 mM EDTA, 50 mM sodium chloride, 30 mM sodium pyrophosphate, 50 mM sodium fluoride, 1 mM sodium orthovanadate, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 5 μg/ml aprotinin, 1 μg/ml pepstatin A, and 2 μg/ml leupeptin), rotated end-over-end at 4 °C for 60 min and insoluble material was pelleted at 12,000 x g for 30 min at 4 °C. The protein concentrations of clarified cell lysates were determined by simplified Bradford method (Bio-Rad Laboratories, Hercules, CA). For immunoblotting of prostate-specific antigen (PSA) and androgen receptor (AR), we used anti-PSA pAb (1:1000, DakoCytomation, Carpinteria, CA) and anti-AR mAb (1:1000, BioGenex, San Ramon, CA). For Akt and MAPK immunoblottings, primary antibodies were used at the following concentrations: anti-phospho-Akt (Thr 308) pAb (1:500), anti-phospho-Akt (Ser 473) pAb (1:500), anti-Akt pAb (1:500), anti-phospho-42/44 MAPK (T202/Y204) mAb (1:500) (Cell Signaling, Boston, MA) and anti-pan ERK mAb (1:1000) (Transduction Laboratories, Inc.) and anti-actin pAb (1:4000) (Sigma, Saint Louis, MO). In addition, whole cell lysates were immunoprecipitated for 1 h at 4 °C with polyclonal rabbit antisera against either Stat5a or Stat5b (4 μl/ml; Advantex Bioreagents, Conroe, TX) or Stat3 (1 μg/ml, K-15; Santa Cruz Biotechnologies). Antibodies were captured by incubation for 60 min with protein A-Sepharose beads (Pharmacia Biotech, Piscataway, NJ). Samples were run on a 4-12% SDS-PAGE under reducing conditions. For western blotting of the immunoprecipitations the primary antibodies were used at the following concentrations: anti-phosphotyrosine-Stat5a/b (Y694/Y699) mAb (1 μg/ml, Advantex BioReagents), anti-Stat5ab mAb (1:250) (Transduction Laboratories, Inc.), anti-phosphotyrosine-Stat3 (Y705) pAb (1:1000) (Cell Signaling) and anti-Stat3 pAb (1:1000) (K-15; Santa Cruz Biotechnologies), and detected by horseradish peroxidase-conjugated secondary antibodies in conjunction with enhanced chemiluminescence.
Cell viability assay
The cell viability was determined by counting attached cells by hemacytometer and trypan blue exclusion. For comparison of other prostate cancer cell lines, CWR22Pc, LNCaP and CWR22Rv1 cells were grown in the presence or absence of 0.8 DHT in 3% CS-FBS containing medium for 9 days. Medium was changed every other day and cells were counted manually every third day. Representative photographs were taken nine days after cell plating.
DNA fragmentation ELISA assay
Fragmentation of DNA was determined by photometric enzyme immunoassay according to the manufacturers’ instructions (cell death detection ELISAPLUS; Roche Molecular Biochemicals, Indianapolis, IN). Briefly, cells were centrifuged at 200 x g, and cytoplasmic fractions containing fragmented DNA were transferred to streptavidin-coated microtiter plates that had been incubated with biotinylated monoclonal anti-histone antibody. The amount of fragmented DNA bound to anti-histone antibody was evaluated by peroxidase-conjugated monoclonal anti-DNA antibody using ABTS as a substrate at 405 nm.
Conventional cytogenetic analysis
Chromosome preparation and G-banding were done using standard protocols (11). Chromosomes were identified and classified according to standard cytogenetic nomenclature proposed by the International System for Human Cytogenetic Nomenclature (ISCN) (12).
Comparative genomic hybridization
Total genomic DNA was extracted from primary CWR22 tumor tissue, CWR22Pc cells from two different passages (test DNA), and from the lymphocytes of a karyotypically normal female control (control DNA), using Wizard® Genomic DNA Purification Kit (Promega, Madison, WI). Comparative genomic hybridization (CGH) analysis was performed using standard protocols that we have previously published (13). Quantitative evaluation of the hybridization was completed using a commercially available software package (Applied Imaging, Pittsburgh, PA, USA). Average ratio profiles were computed as the mean value of 8 ratio images and were used to identify changes in chromosome copy number.
Cell cycle and ploidy analysis
Cells were stained with propidium iodide (50 μg/ml) with RNAase A (50 μg/ml). Duplicate samples were run with and without human peripheral blood lymphocytes added as a standard. Cells were run on a Becton Dickinson FACSort. Twenty thousand cells were collected. Data were modeled with ModFit Software (Verity Softwarehouse, Topsham, ME)
DNA fingerprinting
Total genomic DNA was extracted from primary CWR22 tumor tissue and CWR22Pc cells from two different passages using Wizard® Genomic DNA Purification Kit (Promega, Madison, WI). DNA fingerprinting was performed using the commercially available kit, PowerPlex® 1.2 System (Promega). This system allows the co-amplification and two-color detection of nine loci (eight short tandem repeat loci (STR) and the Y-specific Amelogenin). This approach provides a powerful level of discrimination in excess of 1 in 109. The following markers were tested: D5S818, D13S317, D7S820, D16S539, vWA, TH01, Amelogenin, TPOX, and CSF1PO. The PCR amplification was performed according to the manufacturer’s recommended protocol. Allele size was determined by electrophoresis of the PCR products in 6% denaturing polyacrylamide gels and compared to ROX 500 size standards (Applied Biosystems, Foster City, CA), using the automated sequencer, ABI 377 (Applied Biosystems). The fluorescent signals from the different size alleles were recorded and analyzed using GENESCAN version 3.1 and GENOTYPER version 2.1 software, respectively (Applied Biosystems). Allele size evaluation and data analysis were done by two independent observers. The experiments were repeated twice.
The AR sequence analysis
Total RNA from Primary CWR22 xenograft tumors, CWR22Rv1, CWR22Pc and LNCaP cells was isolated using TRIZOL Reagent (Invitrogen, Carlsbad, CA) and reverse transcribed with Super-ScriptII reverse transcriptase (Invitrogen) using oligodeoxythymidylic acid primers. The conditions for PCR for all reactions were 94°C for 2 min, followed by 30 s denaturation at 94°C, 30 s annealing at 60°C, 30 s extension at 72°C, and final extension period of 10 min. The PCR products were size-separated on a 2% Tris-Borate EDTA-agarose gel. To analyze the AR cDNA, primer pairs were designed to amplify four overlapping segments (I, II, III and IV) encompassing the entire AR coding region plus 5′ and 3′ untranslated sequences. Primers for PCR amplification and sequencing for segments were designated based on the human AR mRNA reference sequence (NM_000044) deposited in the GeneBank database: segment I (forward 5′-GCCAAGCTCAAGGATGGA-3′ and reverse 5′-ATCTTCAGTGCTCTTGCCTG-3′); segment II (forward 5′-CATTGGCCGAATGCAAAGGT-3′ and reverse 5′-CGGCTCTTTTGAAGAAGACC-3′); segment III (forward 5′-GAAGACCTGCCTGATCTGTG-3′ and reverse 5′-ACATCCGGGACTTGTGCATG-3′); segment IV (forward 5′-CCTTCACCAATGTCAACTCC-3′ and reverse 5′-AGTGCAGAGTTATAACAGGC-3′). Sequencing was performed on automatic sequencer ABI 377 (Applied Biosystems) and sequences were analyzed using ClustalW multiple sequence alignment tool (http://www.ebi.ac.uk/Tools/clustalw/).
Results and Discussion
CWR22Pc cell line is derived from the androgen-dependent re-transplantable primary CWR22 tumors
CWR22 prostate tumor system was originally established from a Gleason score 9 human prostate cancer obtained from prostatectomy. The prostate cancer tissue pieces were grown in male nude mice as subcutaneous tumors and the tumor system is maintained by serial re-grafting. The tumors were named primary CWR22 tumors, and they are dependent on androgens for growth. In response to long-term androgen deprivation, the tumors regress but recur back within 7-9 months as androgen-independent tumors. The androgen-independent secondary tumors were named recurrent CWR22 tumors and a cell line established from the recurrent CWR22 tumors was named CWR22Rv1.
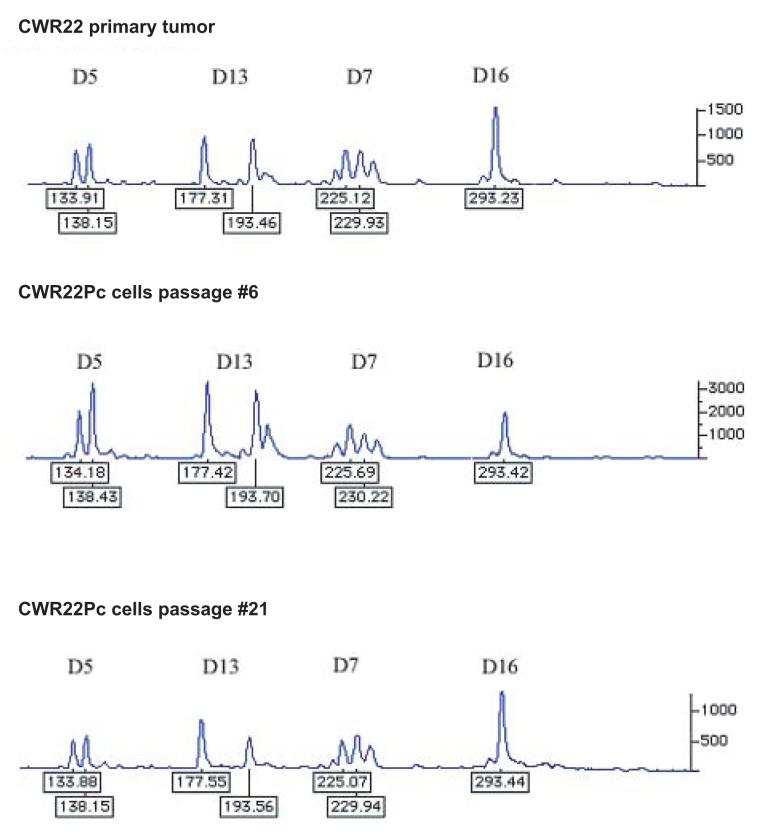
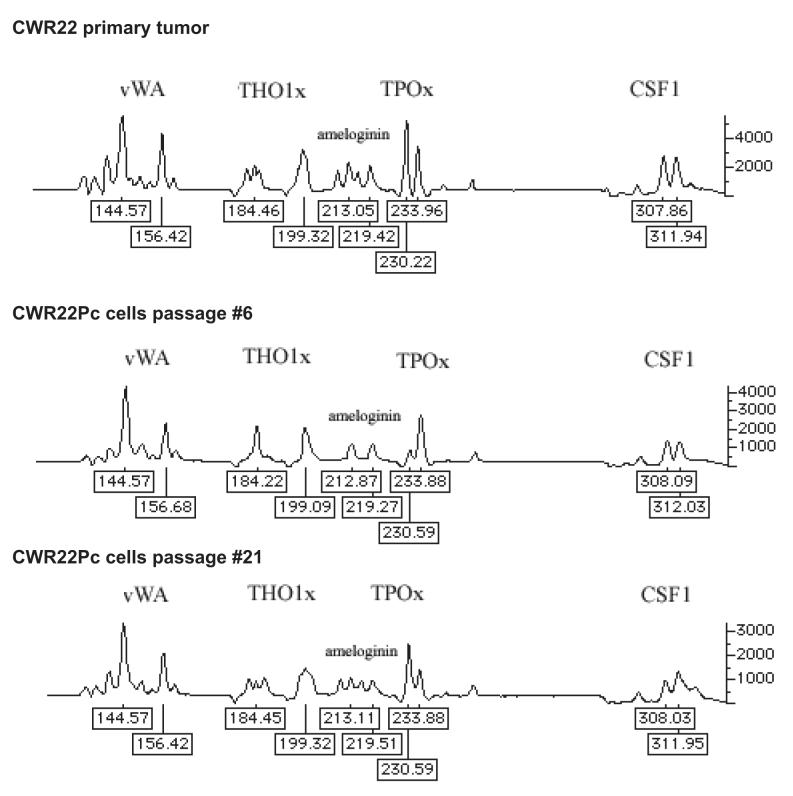
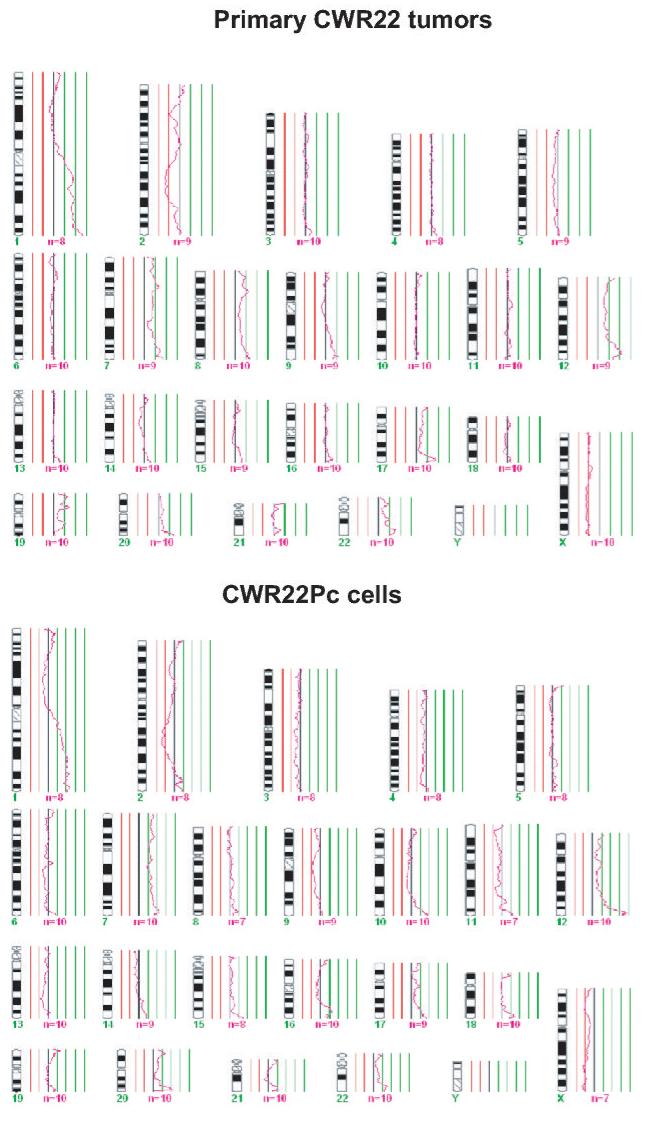
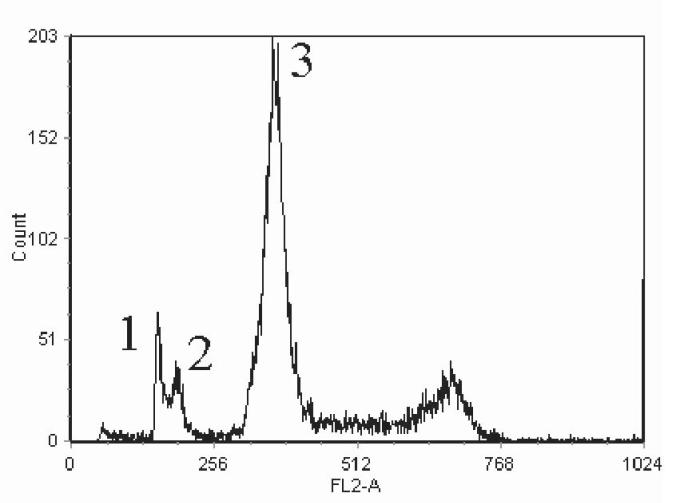
In this work, we established a new prostate cancer cell line from the primary CWR22 tumors. In order to confirm the genetic lineage identity of CWR22Pc cell line as a derivative of the primary CWR22 tumors, we compared the DNA fingerprinting pattern of two passages of CWR22Pc line with that of the CWR22 primary tumors at nine different genetic markers (8 STR markers and the Amelogenin locus). CWR22Pc cell line showed an identical DNA fingerprinting pattern to the primary CWR22 tumors (number of alleles and allele size) at all 8 STR markers. The Y-specific Amelogenin locus showed two alleles (size 213 and 219 bp) in both CWR22Pc cells and in the primary CWR22 tumors indicating the presence of an X and a Y chromosome. Figures 1A and B show a comparison between the results of DNA fingerprinting analysis of the primary CWR22 tumor and the two passages of CWR22Pc cells (#6 and #21) at the 9 loci studied. Moreover, we compared the chromosomal alterations in the CWR22Pc line with the ones in the primary CWR22 tumor. Specifically, we evaluated DNA obtained from two passages of CWR22Pc cells and DNA from the primary CWR22 tumor using comparative genomic hybridization (CGH). CGH analysis is an ideal method to detect overall chromosomal gains and losses in the genome. The CGH analysis showed that the pattern of chromosomal alteration of the CWR22Pc cell line was very similar to that of the primary CWR22 tumor. Chromosomal gains included gains at 1q, 7, 8p and 12, and losses of chromosomes 2 and X (Fig. 1C). The findings of the DNA fingerprinting analysis and CGH confirmed that the CWR22Pc cell line was indeed derived from the primary CWR22 tumors. In order to evaluate the ploidy status of the CWR22Pc cells, we performed a G-banding analysis of over 300 metaphase spreads and a ploidy analysis by flow cytometry. Metaphase analysis showed that approximately 80% of the cells were near tetraploid (chromosomal count approximately 100), and 20% of the cells showed a chromosomal count of approximately 50 chromosomes. These findings were consistent with the cell cycle and ploidy analysis which showed a mixed population of cells with DNA content of 1.11 and 2.23 (Fig. 1D).
Fig. 1. The genetic lineage identity of CWR22Pc cells is similar to that of CWR22 primary prostate tumors.
DNA fingerprinting analysis of the CWR22 primary tumor, and the cell line CWR22Pc (passages 6 and 21) show a similar pattern for all nine markers analyzed. A, Patterns of alleles at markers D5S818 (D5), D13S317 (D13), D7S820 (D7), and D16S539 (D16). B, The results at markers vWA (vWA), TH01 (THO1x), Amelogenin, TPOX (TPOx), and CSF1PO (CSF1). The allele sizes are indicated under each allele. C. The comparative genomic hybridization analysis shows gains at 1q, 7, 8p and 12, and losses of chromosomes 2 and X in both primary CWR22 tumors and in CWR22Pc cells. D. Flow cytometric analysis of CWR22Pc cells shows that the cell line consists of a mixed population of cells with DNA indices of 1.11 and 2.23. #1 represents the G1 peak of the human peripheral blood lymphocytes (added as a control). #2 represents the G1 peak of the cells with a lower ploidy (DNA index = 1.1). #3 represents the G1 peak of the cells with a higher ploidy (DNA index = 2.23).
In summary, our results of the G-banding and the ploidy analysis of the CWR22Pc cells are in line with the mosaic karyotype reported previously for the primary CWR22P tumors (6, 14), the recurrent CWR22R xenografts (14) and the CWR22Rv1 cell line established from the recurrent CWR22R tumors (8). In detail, primary CWR22 xenograft tumors were reported to have a mixture of two karyotypes where part of the cells displayed an unidentified marker chromosome (6). In addition, the relapsed strains of CWR22R tumors after androgen deprivation showed each a different karyotype (14), and the CWR22Rv1 cell line consists of a mixed population of hyperdiploid (90%) and near-tetraploid (10%) cells (15). The results of our study reported here indicated a mosaic karyotype of CWR22Pc cells. It will be both important and interesting for the future studies to determine whether the ploidy status of CWR22Pc cells will change during a long term androgen deprivation in vitro and when CWR22Pc cells are grown as xenograft tumors in nude mice in vivo.
Growth and PSA protein expression of CWR22Pc cells is regulated by androgens
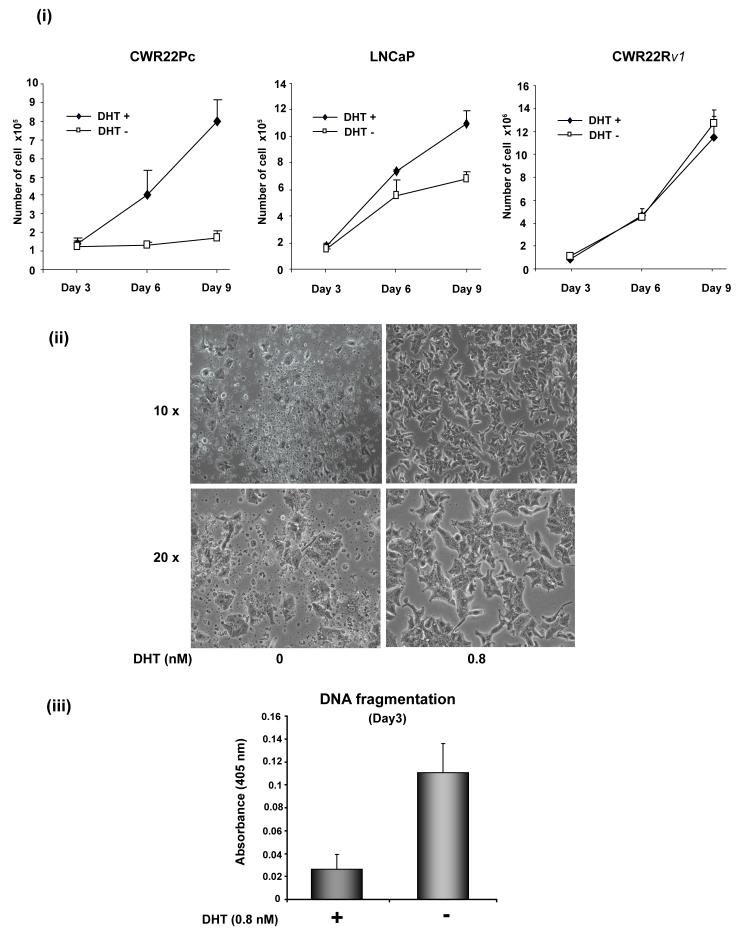
Given that the CWR22Pc cell line was established from primary CWR22 tumors, which are regulated by androgens, we first aimed to determine the effects of androgens on the growth of CWR22Pc cells in culture. CWR22Pc cells, LNCaP cells and androgen-independent CWR22Rv1 cells were cultured in the presence or absence of 0.8 nM dihydrotestosterone (DHT) for nine days, and the number of attached viable cells was determined every third day. On day 6 of the experiment, the number of CWR22Pc cells was increased by 3-fold, whereas the number of LNCaP cells was increased only by 40% (Fig. 2A, panel i). Moreover, on day 9 of the experiment, the number of CWR22Pc cells was increased by 6-fold by androgens while the number of LNCaP cells was increased only by 60% (Fig. 2A, panel i). At the same time, the growth rate of CWR22Rv1 cells was not significantly affected by DHT during a 9-day period. Deprivation of CWR22Pc cells from androgens induced apoptotic death of the cells as demonstrated by cell morphology on the day 9 of the experiment (Fig. 2A, panel ii). Specifically, androgen deprivation induced extensive detachment of the cells, cell fragmentation, shrinkage and blebbing, all of which are morphological changes consistent with apoptotic cell death (Fig. 2A, panel ii). Moreover, DNA fragmentation was increased by 5-fold on the day 3 of the experiment in the cells cultured in the absence of DHT (Fig. 2A, panel iii).
Fig. 2. The growth of CWR22Pc cells is increased by androgens.
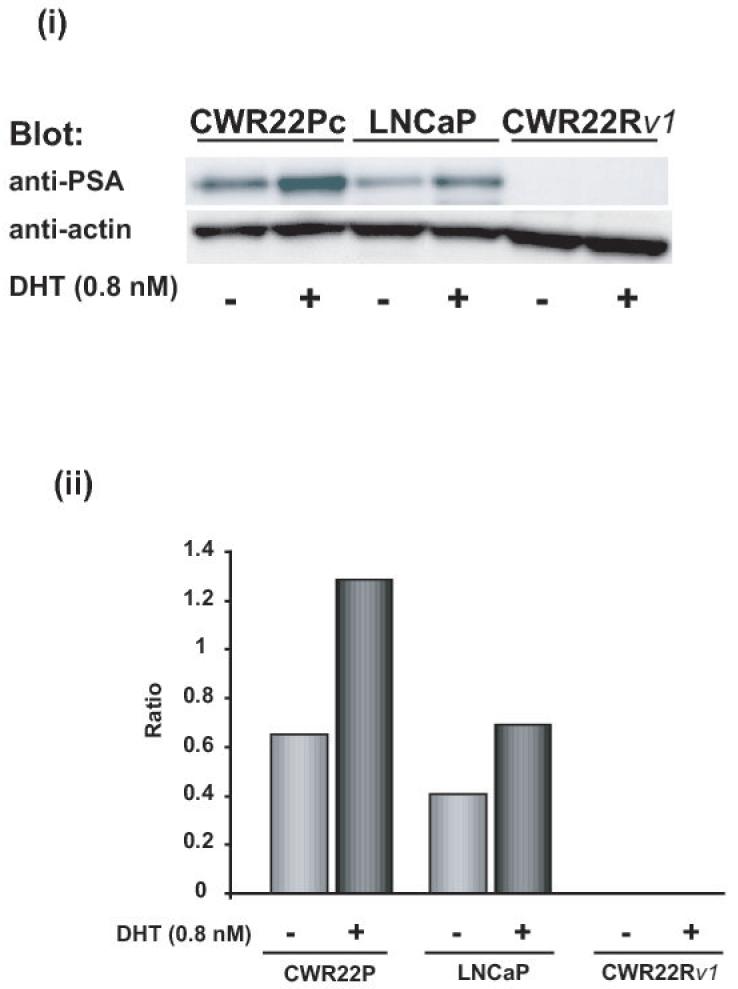
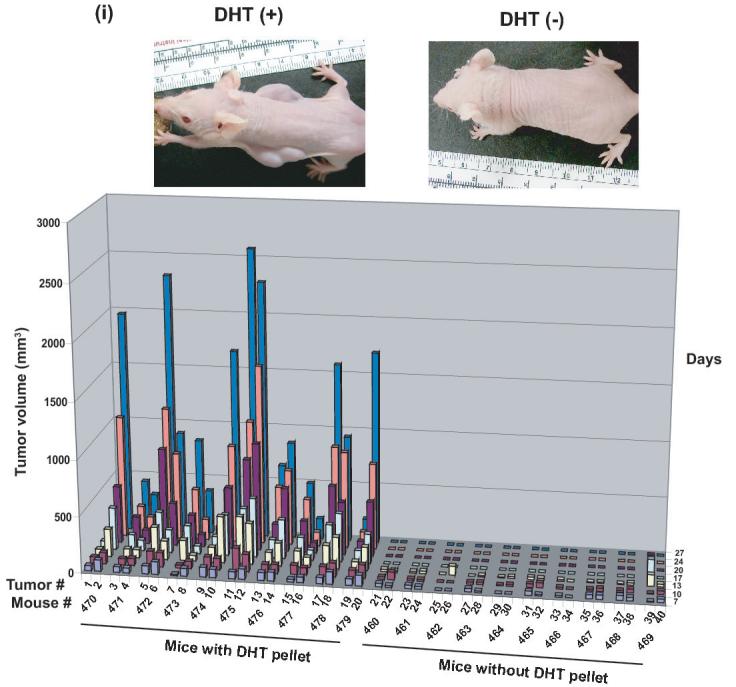
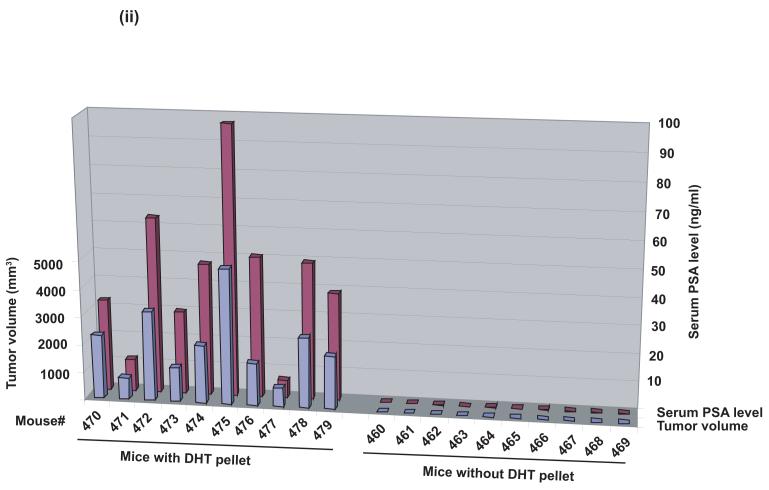
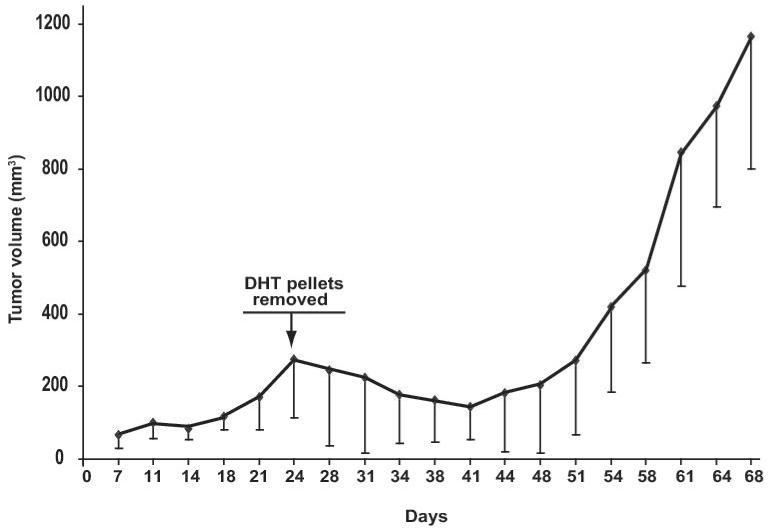
A, CWR22Pc, LNCaP and CWR22Rv1 cells were grown in the medium containing 3% CS-FBS in the presence or absence of 0.8 nM dihydrotestosterne (DHT) for 9 days. Cells were counted every third day. The means of three independent experiments are presented with SDs (panel i). The absence of DHT had marked effects on the cell morphology of CWR22Pc cells. In the absence of DHT, stereomicroscope photographs show that the majority of the cells were dead and floating (panel ii) with increased DNA fragmentation (panel iii). B, Prostate specific antigen (PSA) expression is regulated by androgens in CWR22Pc and LNCaP cells. CWR22Pc, LNCaP and CWR22Rv1 cell lines were cultured in RPMI with or without 0.8 nM DHT for 12 days. Cells were harvested, lysed and immunoblotted with anti-PSA pAb. Stripped filters were re-blotted with anti-actin pAb to demonstrate equal loading (panel i). Densitometric normalization and comparison of the PSA levels (panel ii). C, The growth of CWR22Pc cells as subcutaneous xenograft tumors in athymic nude mice is regulated by androgens. CWR22Pc cells were inoculated subcutaneously into flanks of castrated nude mice supplied with sustained-release 5α-dihydrotestosterone (DHT)-pellets (n=10/group, 2 tumors/mouse, 20 × 106 CWR22Pc cells per site). The tumor incidence and growth were measured twice a week for 36 days. Tumor volumes were calculated using the formula (3.14 x length x width x depth/6) (panel i). Serum PSA levels in mice carrying CWR22Pc tumors correlated with the volumes of the tumors (panel ii). PSA levels in the sera of mice carrying CWR22Pc xenograft tumors were determined using a PSA ELISA assay. The total tumor burden (the sum of both right and left tumor volumes) were compared to the serum PSA levels. D, CWR22Pc tumors recur after androgen-deprivation induced regression of the tumors. CWR22Pc cells were inoculated subcutaneously into flanks of castrated athymic nude mice supplied with sustained-release 5α-DHT-pellets (n=4 mice, 1 tumor/mouse, 20 ×106 CWR22Pc cells per site). Once the tumors reached 10 mm in diameter in size, the DHT-pellets were removed and the tumor growth was measured twice a week. Tumor volumes were calculated as described above.
To further examine androgen regulation of CWR22Pc cells in culture, we compared the effect of DHT on PSA protein expression in CWR22Pc vs. LNCaP or CWR22Rv1 cells. The promoter region of PSA is known to contain an androgen response element. The cells were cultured in the presence or absence of 0.8 nM DHT, and the levels of PSA protein in whole cell lysates were determined by western blotting. The PSA protein levels were increased by 3-fold in CWR22Pc cells when the cells were cultured in the presence of DHT (Fig. 2B). DHT increased PSA protein expression in LNCaP cells by approximately 2-fold. The levels of PSA protein in CWR22Rv1 cells were undetectable when compared to both CWR22Pc and LNCaP cells (Fig. 2B).
CWR22Pc cells are highly tumorigenic in nude mice and the tumor growth is regulated by androgens
Once we had established that CWR22Pc cells were genetically similar to the primary CWR22 tumors and the growth of CWR22Pc cells in culture is regulated by androgens, we next focused on evaluating whether CWR22Pc cells are tumorigenic in athymic nude mice and whether in vivo growth of CWR22Pc cells is regulated by androgens. To investigate whether CWR22Pc cells would grow as xenograft tumors in nude mice, we injected CWR22Pc cells (20 x 106 cells per site) subcutaneously to the flanks of athymic nude mice (n=20) (two tumors per mouse). The nude mice were castrated and half of the mice (n=10) were implanted with sustained-release 5α-DHT-pellets to normalize the circulating androgen levels. In the mice supplied with DHT-pellets, tumors started to form on day 7 with a 100% incidence (Fig. 2C, panel i). Importantly, the tumors in mice supplied with the DHT pellets grew rapidly, while both the tumor incidence and the growth rate were low in mice without DHT pellets (Fig. 2C, panel i). These results suggested that CWR22Pc human prostate cancer cells are highly tumorigenic in athymic nude mice and the tumor growth is regulated by androgens.
Since CWR22Pc cells in culture produced high levels of PSA, and the PSA protein expression was regulated by androgens, our next aim was to investigate whether serum PSA levels would correspond with the volumes of the CWR22Pc xenograft tumors in mice. Serum PSA levels were determined from the blood samples collected on day 27 of the experiment (Fig. 2C, panel ii) from the athymic nude mice carrying CWR22Pc tumors presented in Figure 2C, panel i. The results of the serum PSA assay showed that PSA protein levels in serum of the mice closely correlated with the volumes of the CWR22Pc subcutaneous xenograft tumors (Fig. 2C, panel ii). Specifically, serum PSA levels were almost undetectable in mice without DHT pellets carrying small tumors at low incidence. In contrast, serum PSA levels closely followed tumor volumes in DHT-supplied mice which developed large CWR22Pc xenograft tumors with a high incidence (Fig. 2C, panel ii; the tumor volume is the mean of the two tumors in each mouse). Collectively, the results presented here indicate that the cells in subcutaneous CWR22Pc xenograft tumors secrete high levels of PSA to the circulation of the mice. Moreover, the serum PSA levels in these mice correlate with the volumes of the xenograft tumors.
CWR22Pc tumors recur after androgen withdrawal
In the next set of experiments, we first grew CWR22Pc cells as subcutaneous athymic nude mice supplied with DHT-pellets two consecutive experiments (n=34 mice, 1 tumor per mouse and n=30, 2 tumors per mouse). When the tumors reached 10 mm in diameter in size, the DHT pellets were removed and the tumor growth was followed by consecutive tumor volume measurements. The removal of the DHT pellets resulted in both experiments in a regression which reached the maximum in 15 days (Fig 2D). Importantly, the tumors re-grew back in the androgen-deprived nude mice with the following 20-30 days (Fig. 2D).
The human prostate cancer cell line CWR22Pc established from the primary CWR22 tumors provides several valuable advantages as a model system for androgen-regulated growth and development of androgen-independence of prostate cancer cells. First, androgen-promoted tumor growth of CWR22Pc cells inoculated as subcutaneous tumors in nude mice occurs fast (in 3 weeks) with 100% tumor incidence. Second, development of hormone-refractory tumors is highly reproducible and the tumor re-growth takes place rapidly. Third, the CWR22Pc tumor system allows easy genetic in vitro genetic manipulation of the cells that form the tumors.
CWR22PC cells express androgen receptors
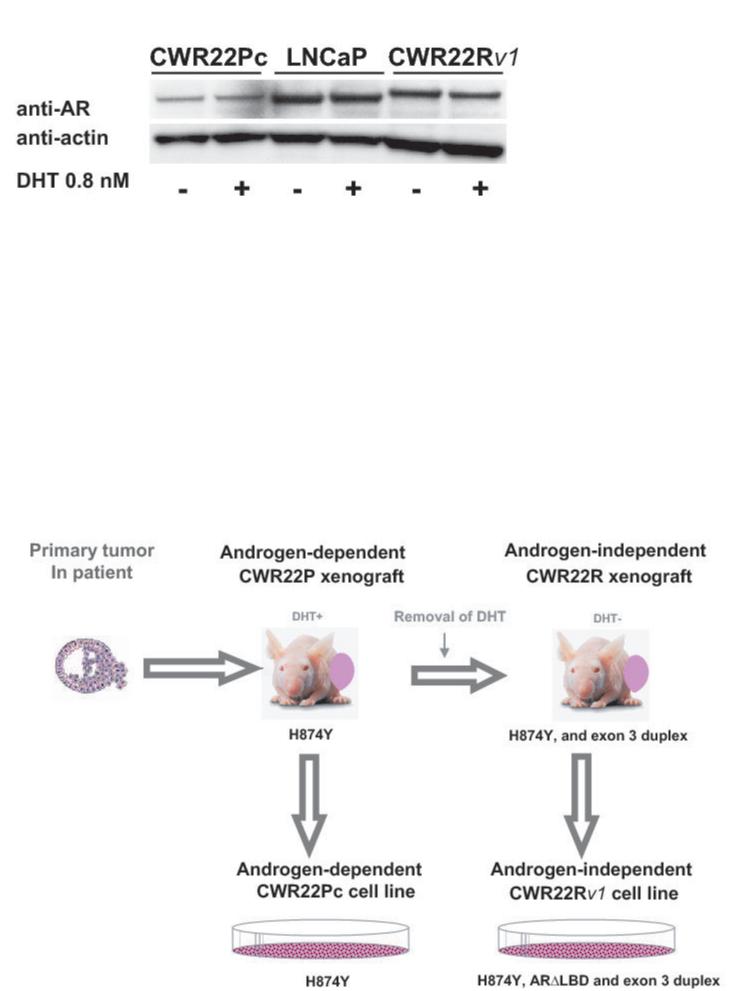
To analyze the AR protein expression level in CWR22Pc cells, we immunoblotted CWR22Pc, LNCaP and CWR22Rv1 cell lysates for androgen receptor using an anti-human AR mAb (Fig. 3A). The filters were stripped and re-blotted with anti-actin pAb to demonstrate the loading of total proteins. The results show that AR protein is expressed at a high level in all three cell lines. Moreover, the results demonstrate that the size of AR in CWR22Rv1 cells is slightly larger compared to the AR in LNCaP and CWR22Pc cells. This was the expected result since the relapsed CWR22 tumors (CWR22R) and the CWR22Rv1 cell lines established from the relapsed tumors are known to express an AR gene that contains an in-frame tandem duplication of the exon 3. This region encodes the second zinc finger of the AR DNA-binding domain and the AR protein product has an approximately 5-kDa increase in protein size relative to the LNCaP AR (16). Moreover, the AR gene in the relapsed CWR22 tumors and in CWR22Rv1 cells has the H874Y mutation in the ligand binding domain of AR, which enables the receptor to bind adrenal androgen dehydroepiandrosterone in addition to estradiol, progesterone and hydroxyflutamide (17) (Fig. 3B). The AR gene also in the primary CWR22 tumors contains the H874Y (histidine to tyrosine) mutation. However, the AR gene in the primary CWR22 tumors does not have the tandem duplication in the exon 3 present in CWR22Rv1 (Fig. 3B).
Fig. 3. Androgen receptor expression in CWR22Pc cells.
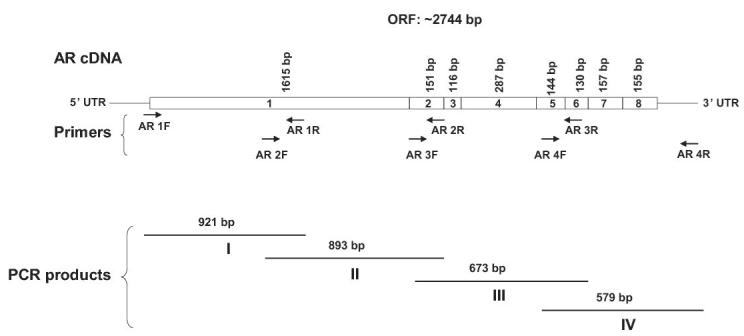
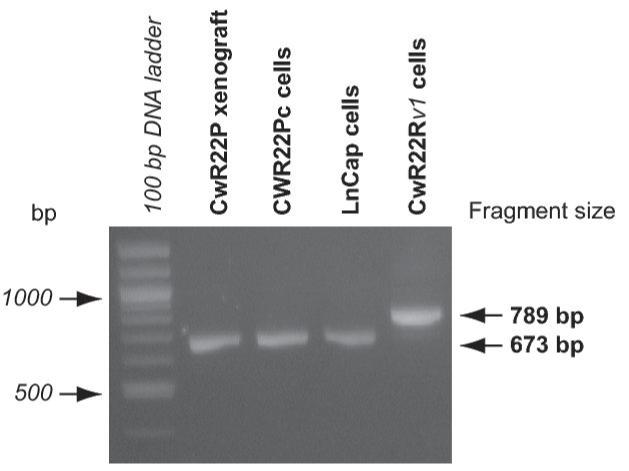
A, Androgen receptor (AR) expression in CWR22Pc, LNCaP and CWR22Rv1 cells. Cells were cultured in the presence or absence of 0.8 nM dihydrotestosterone (DHT), lysed and immunoblotted with an anti-AR mAb. Filters were re-blotted with anti-actin pAb to demonstrate the protein loading. AR protein expressed in CWR22Rv1 is of higher molecular weight than in CWR22Pc and LNCaP cells. B, Schematic presentation of AR mutations in primary CWR22 prostate tumors (CWR22P), CWR22Pc cells, recurrent CWR22 tumors (CWR22R) and CWR22Rv1 cells. C, AR gene sequencing primers. PCR primers were designed to amplify and sequence four overlapping segments (I, II, III, IV) encompassing the entire AR coding region plus 5′ and 3′ untranslated sequences. The Segment I covers a part of the 5′- UTR and the 5-prime-end part of the exon 1, the segment II covers the 3-prime-end of the exons 1 and 2, the segment III covers the exons 2, 3, 5 and a part of the exon 6. The segment IV covers a part of the exons 5, 6, 7, 8 and a part of 3′- UTR. D, RT-PCR products of the AR segment III in different human prostate cancer xenograft tumors and prostate cancer cell lines. The RT-PCR product for the segment III yielded the expected amplicon size of 673 bp in CWR22PC cells, primary CWR22 xenograft tumors and in LNCaP cells, while the CWR22Rv1 RT-PCR reaction yielded an approximately 100-bp larger RT-PCR product.
As the next step, we wanted to characterize the AR transcript in CWR22Pc cells by RT-PCR mapping and by sequencing to determine whether the AR gene in CWR22Pc cells contains the H874Y mutation found in the AR in the primary CWR22 tumors. Moreover, we wanted to determine whether the AR gene in CWR22Pc cells contains any additional mutations. To that end, we performed RT-PCR mapping using primer pairs designated to amplify overlapping AR mRNA segments of approximately 600-900 bp and spanning the entire length of the AR coding sequence. The exons and the 5′- and 3′-untranslated sequences amplified by each of the primer sets are described in Fig. 3C. RT-PCR reactions performed using RNA isolated from primary CWR22Pc cells, primary CWR22 xenograft tumors (CWR22P), LNCaP cells and CWR22Rv1 cell line yielded expected sizes for the amplification products of the segments I, II, and IV of the AR gene. The RT-PCR product for the segment III, which spans the exon 3 of the AR gene, yielded the expected amplicon size of 673 bp in CWR22Pc cells, primary CWR22 xenograft tumors and in LNCaP cells, while the CWR22Rv1 RT-PCR reaction yielded an approximately 100-bp larger RT-PCR product due to the exon 3 tandem duplication as reported previously (16) (Fig. 3D). We then purified the segments I-IV amplification products obtained from CWR22Pc cells and analyzed them automated DNA sequencing followed by ClustalW-driven pairwise alignment with the reference AR cDNA sequence in GenBank (National Center for Biotechnology Information). The sequence comparison demonstrated the H874Y mutation in the AR gene in CWR22Pc cells, while no additional mutations in the AR gene were found in CWR22Pc cells. In summary, these results demonstrated that the sequence of the AR gene in the CWR22Pc cells has the H874Y mutation but not the exon 3 duplication. Moreover, the data indicate that the AR gene is identical to that in the primary CWR22 xenograft tumors, which indirectly supports the notion that CWR22Pc cell line is derived from the primary CWR22 tumors, and is genetically different from the androgen-independent CWR22Rv1 cell line.
Currently, only few androgen-dependent human prostate cancer cell lines exist such as LNCaP (3), LAPC-4 (18) and MDA PCa 2b (13, 19). Androgen-independent cell lines PC-3 (20) and DU145 (21) were derived from metastatic lesions to the bone and brain, respectively. Moreover, CWR22Rv1 human prostate cancer cell line (8) is independent of androgens for growth and was established from a xenograft tumor derived from an untreated primary prostate cancer (5). It is important to note that AR expression persists in clinical prostate cancer despite progression to hormone-refractory state (22). AR in prostate cancer typically undergoes genetic alterations including AR gene amplification during hormone therapy. Moreover, 10-30% of prostate cancers acquire a point mutation in the AR gene (23). Both types of known AR gene alterations lead to increased sensitivity of the receptor to low levels of circulating androgens and also to the receptor’s ability to recognize a broadened spectrum of ligands as potent agonists of AR action. DU145 and PC-3 cells are both AR-negative prostate cancer cell lines, whereas LNCaP, MDA pCa 2b, LAPC-4 and CWR22Rv1 cells express AR protein. In LNCaP cells, the androgen receptor has a point mutation T877A which makes the AR more sensitive to flutamide, estradiol and progesterone (24). LAPC-4 cells express wild-type AR, while AR in MDA PCa 2b cells contains two mutations, T877A and L701H, which reduces the AR affinity to androgens but enhances binding of adrenal corticosteroids to the AR (25, 26). CWR22Rv1 cells are AR-positive, but the AR contains a mutation H874Y in addition to a tandem duplication in the exon 3 (16). The AR(H874Y) in CWR22Rv1 cells has been reported to be more sensitive to adrenal androgen dehydroepiandrosterone and anti-androgen hydroxyflutamide. Our work presented here characterizes a new androgen-dependent human prostate cancer cell line that expresses AR having the H874Y mutation without the tandem duplication in the exon 3. This is the first time such a human prostate cancer cell line is reported. CWR22Pc cells will provide a valuable experimental tool to investigate the importance of H874Y mutation in the AR for androgen regulation of prostate cancer cells as well as for development of androgen-independence.
Cytokine signaling pathways are active in CWR22Pc cells
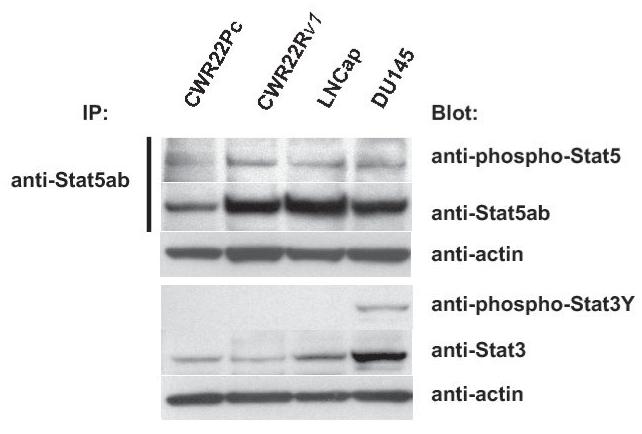
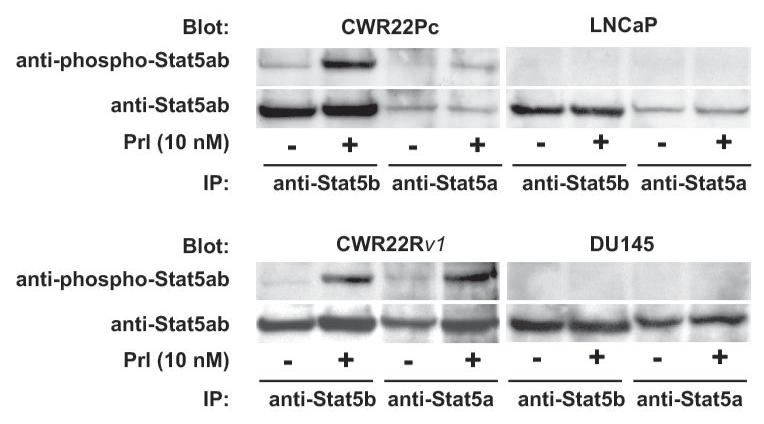
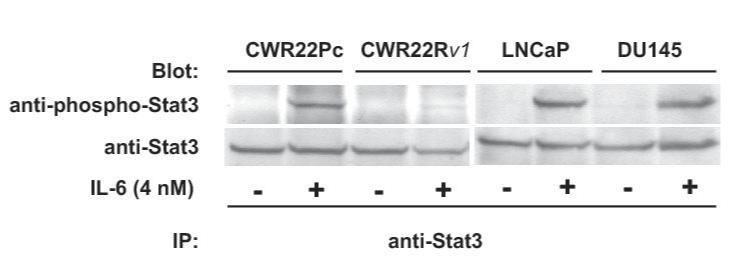
One of the key molecular mechanisms that promote prostate cancer cell growth involves protein kinase growth factor signaling pathways. These cell signaling pathways regulate prostate cancer cell survival, proliferation and/or differentiation independently of AR or by affecting the transcriptional activity of AR. To investigate the activation status of the key known signaling pathways influencing growth and AR activity in prostate cancer cells, phosphorylation and total protein levels of Stat5a/b, Akt, MAPK and Stat3 (27-36) were examined in exponentially growing CWR22Pc, CWR22Rv1, LNCaP and DU145 cells. Immunoprecipitation and immunoblotting of Stat5a/b shows that Stat5a/b is active in CWR22Pc cell line whereas Stat3 is constitutively active only in DU145 cells but not in CWR22Pc cells (Fig. 4A, upper panel). The filters were stripped and re-blotted for total Stat5a/b and Stat3 protein levels. In addition, parallel samples of the cell lysates were immunoblotted with anti-actin to demonstrate the total protein level in each lane. Immunoblotting of whole cell lysates with anti-phospho-MAPK-p44/42 antibody showed that MAPK is activated at high level in exponentially growing CWR22Pc cells, while phosphorylated at a lower level in LNCaP, CWR22Rv1 and DU145 cells (Fig. 4B). Furthermore, also Akt is phosphorylated in CWR22PC cells (Fig. 4B). Parallel samples were immunoblotted to demonstrate total ERK and AKT levels as well as actin expression to demonstrate equal protein loading.
Fig. 4. Protein kinase signaling pathways are activated in CWR22Pc cells.
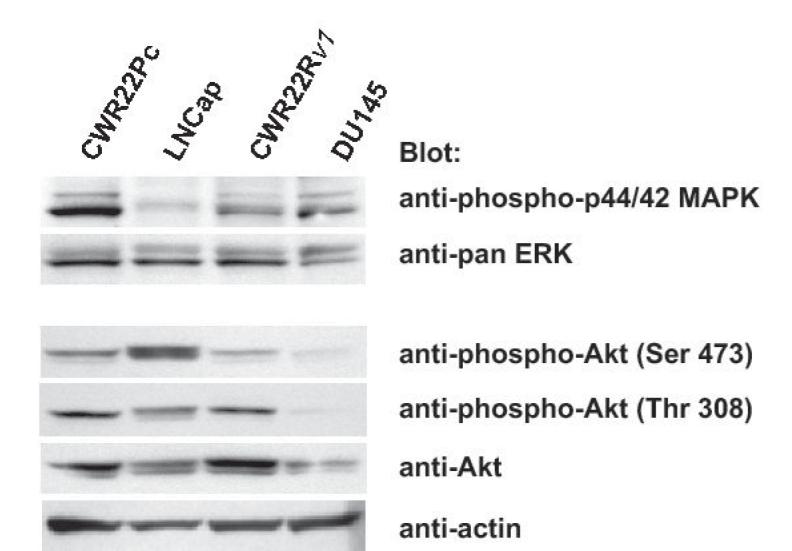
Transcription factor Stat5a/b, MAPK(p44/42) and AKT are constitutively active in CWR22Pc cells. A, Stat5ab or Stat3 were immunoprecipitated (IP) from exponentially growing CWR22Pc, CWR22Rv1, LNCaP and DU145 cells using anti-Stat5ab or anti-Stat3 pAb, respectively, and blotted with anti-phopho-Stat5a/b or anti-phospho-Stat3 antibody as indicated. Filters were stripped and re-blotted with anti-Stat5ab mAb or anti-Stat3 mAb and whole cell lysates before IP were immunoblotted with anti-actin pAb. B, Whole cell lysates of exponentially growing CWR22Pc, LNCaP, CWR22Rv1 and DU145 cells were immunoblotted with antibodies to phosphorylated MAPK (anti-phospho-p44/42 MAPK), ERK (antipanERK), serine-phosphorylated AKT (anti-phospho-AKTSer473), threonine-phosphorylated AKT (antiphospho-AKTThr308), AKT (anti-AKT) or actin (anti-actin). To investigate cytokine activation of Stat5a/b and Stat3 in CWR22Pc cells, all four cell lines (CWR22Pc, LNCaP, CWR22Rv1 and DU145) were serum-starved for 16 h and stimulated for 15 min with 10 nM human prolactin (hPrl) (C) or 4 nM interleukin-6 (IL-6) (D) and harvested. Stat5a, Stat5b and Stat3 were immunoprecipated and blotted with anti-phospho-Stat5ab or anti-phospho-Stat3 antibodies as indicated. The filters were stripped and re-blotted with anti-Stat5ab mAb or anti-Stat3 mAb to demonstrate equal loading.
In the next set of experiments, we wanted to investigate the inducibility of Stat5a/b and Stat3 activation by cytokines prolactin (Prl) and interleukin-6 (IL-6) in CWR22Pc cells in comparison to LNCaP, CWR22Rv1 and DU145 cells. The cells were first grown in a regular growth medium containing 10% FBS, serum-starved over-night and stimulated with human Prl (10 nM) or IL-6 (4 nM) for 15 min. Stat5a, Stat5b (Fig. 4C) or Stat3 (Fig. 4D) were immunoprecipitated and blotted with antibodies recognizing phosphorylated Stat5a/b or Stat3, respectively. The filters were stripped and re-blotted with anti-Stat5ab or anti-Stat3 mAbs to demonstrate equal loading. Prl stimulation of CWR22Pc cells induced predominantly activation of Stat5b while the level of Stat5a expression in CWR22Pc cells was generally low. In CWR22Rv1 cell line, Stat5a and Stat5b were expressed at equal levels and Prl induced phosphorylation of both Stat5a and Stat5b in CWR22Rv1 cells, whereas Prl did not activate Stat5a/b in DU145 or LNCaP cells (Fig. 4C). IL-6 stimulated phosphorylation of Stat3 in CWR22Pc cells. In addition, IL-6 induced activation of Stat3 in LNCaP and DU145 cells, but not in CWR22Rv1 cells. In conclusion, Stat5a/b and Stat3 in CWR22Pc cells are activated by Prl and IL-6, respectively.
The CWR22Pc cell line provides a useful research tool to address a number of key questions pertinent to the basic biology of prostate cancer as well as clinical management of the disease. First, since growth of CWR22Pc cells in culture is strictly regulated by androgens, CWR22Pc cells will be able to provide critical information about androgen-regulated growth mechanisms in a prostate-specific cell context. Second, the CWR22Pc cell line provides a model system where significance of different protein kinase signaling pathway activation can be tested for their ability to replace androgens for growth promotion and maintenance of prostate cancer cell viability in vitro and in vivo. Third, CWR22Pc cells have the H874Y mutation in the AR gene without additional genetic changes and will therefore enable studies of AR (H874Y) transcriptional activity in a biological setting where prostate-specific co-activators and co-repressors are expressed and present. Importantly, the re-growth of CWR22Pc tumors in vivo in nude mice after androgen deprivation mimics the course of development of hormone-refractory prostate cancer in humans. Future studies need to characterize genetically and epigenetically the cell clones that recur when CWR22Pc cells are grown as xenograft tumors under the biological pressure of androgen deprivation. Finally, it will be important to establish whether CWR22Pc cells metastasize when inoculated orthotopically in nude mice.
Acknowledgements
Shared Resources of Kimmel Cancer Center are partially supported by National Institute of Health Grant CA56036-08 (Cancer Center Support Grant, to Kimmel Cancer Center). The Cell cycle and ploidy analysis were performed at the Flow Cytometry Shared resource (TCSR) of the Lombardi Comprehensive Cancer Center (LCCC). This resource is partially supported by National Institutes of Health Grant 1P30-CA-51008 (Cancer Center Support Grant (CCSG) to the LCCC). This work was supported by American Cancer Society (RSG-04-196-01-MGO), DOD Department of Defense Prostate Cancer Grant (W81XWH-05-01-0062) and NIH NCI (1RO1CA113580-01A1). We thank Ms. Jacqueline Lutz for help in tumor growth experiments and critical reading of the manuscript. We also thank Dr. Karen Creswell for help in the cell cycle and ploidy analysis.
Abbreviations
- AR
androgen receptor
- DHT
dihydrotestosterone
- Prl
prolactin
- PSA
prostate specific antigen
- Stat5
signal transducer and activator of transcription 5
Footnotes
Clinical relevance Currently, one of the major obstacles in understanding the molecular mechanisms underlying the transition of prostate cancer growth from androgen dependency to hormone-refractory state is the lack of well-characterized androgen-regulated and tumorigenic human prostate cancer cell lines. We have established and characterized a new human prostate cancer cell line, CWR22Pc, derived from the primary CWR22 human prostate xenograft tumors. We demonstrate that the CWR22Pc cell line genetically originates from the primary CWR22 tumors. The growth of CWR22Pc cells is strictly regulated by androgens, and CWR22Pc cells express high levels of androgen receptor and prostate specific antigen. Importantly, when androgens were withdrawn from the established CWR22Pc tumors grown in nude mice, the tumors initially shrank but re-grew back as androgen-independent tumors. The androgen-regulated CWR22Pc cell line provides a new much-needed experimental model system for studies on androgen-regulated growth of prostate cancer cells and for research on genetic changes and kinase signaling pathways involved in growth promotion and dedifferentiation of androgen-deprived prostate cancer cells. Furthermore, CWR22Pc cells provide a model system for studies on the regulation of transcriptional activity of mutated H874YAR in a prostate cancer cell context. Since CWR22Pc cells produce prostate specific antigen (PSA) in an androgen-regulated manner, and the serum PSA levels correlate with the CWR22Pc xenograft tumor volumes in mice, CWR22Pc cells/ nude mouse tumor system will provide a valuable model for development of new diagnostics and therapeutics for prostate cancer.
References
- 1.Trachtenberg J, Walsh PC. Correlation of prostatic nuclear androgen receptor content with duration of response and survival following hormonal therapy in advanced prostatic cancer. J Urol. 1982;127:466–71. doi: 10.1016/s0022-5347(17)53868-9. [DOI] [PubMed] [Google Scholar]
- 2.DeMarzo AM, Nelson WG, Isaacs WB, Epstein JI. Pathological and molecular aspects of prostate cancer. Lancet. 2003;361:955–64. doi: 10.1016/S0140-6736(03)12779-1. [DOI] [PubMed] [Google Scholar]
- 3.Horoszewicz JS, Leong SS, Kawinski E, et al. LNCaP model of human prostatic carcinoma. Cancer Res. 1983;43:1809–18. [PubMed] [Google Scholar]
- 4.Igawa T, Lin FF, Lee MS, et al. Establishment and characterization of androgen-independent human prostate cancer LNCaP cell model. Prostate. 2002;50:222–35. doi: 10.1002/pros.10054. [DOI] [PubMed] [Google Scholar]
- 5.Pretlow TG, Wolman SR, Micale MA, et al. Xenografts of primary human prostatic carcinoma. J Natl Cancer Inst. 1993;85:394–8. doi: 10.1093/jnci/85.5.394. [DOI] [PubMed] [Google Scholar]
- 6.Wainstein MA, He F, Robinson D, et al. CWR22: androgen-dependent xenograft model derived from a primary human prostatic carcinoma. Cancer Res. 1994;54:6049–52. [PubMed] [Google Scholar]
- 7.Nagabhushan M, Miller CM, Pretlow TP, et al. CWR22: the first human prostate cancer xenograft with strongly androgen- dependent and relapsed strains both in vivo and in soft agar. Cancer Res. 1996;56:3042–6. [PubMed] [Google Scholar]
- 8.Sramkoski R, Pretlow Tn, Giaconia J, et al. A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell Dev Biol Anim. 1999;35:403–09. doi: 10.1007/s11626-999-0115-4. [DOI] [PubMed] [Google Scholar]
- 9.Helms SR, Pretlow TG, 2nd, Bueschen AJ, Lloyd KL, Murad TM. Separation of cells with histochemically demonstrable acid phosphatase activity from suspensions of cells from human prostatic carcinomas in an isokinetic gradient of Ficoll in tissue culture medium. Cancer Res. 1976;36:481–6. [PubMed] [Google Scholar]
- 10.Yamashita H, Nishio M, Fujii Y, Iwase H. Dominant-negative Stat5 inhibits growth and induces apoptosis in T47D-derived tumors in nude mice. Cancer Sci. 2004;95:662–5. doi: 10.1111/j.1349-7006.2004.tb03326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The AGT Cytogenetics Laboratory Manual. Lippincott-Raven; 1997. [Google Scholar]
- 12.An international system for human cytogenetic nomenclature 2005: ISCN; Basel: [DOI] [PubMed] [Google Scholar]
- 13.Figueiredo BC, Stratakis CA, Sandrini R, et al. Comparative genomic hybridization analysis of adrenocortical tumors of childhood. J Clin Endocrinol Metab. 1999;84:1116–21. doi: 10.1210/jcem.84.3.5526. [DOI] [PubMed] [Google Scholar]
- 14.Kochera M, Depinet TW, Pretlow TP, et al. Molecular cytogenetic studies of a serially transplanted primary prostatic carcinoma xenograft (CWR22) and four relapsed tumors. Prostate. 1999;41:7–11. doi: 10.1002/(sici)1097-0045(19990915)41:1<7::aid-pros2>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 15.van Bokhoven A, Varella-Garcia M, Korch C, et al. Molecular characterization of human prostate carcinoma cell lines. Prostate. 2003;57:205–25. doi: 10.1002/pros.10290. [DOI] [PubMed] [Google Scholar]
- 16.Tepper CG, Boucher DL, Ryan PE, et al. Characterization of a novel androgen receptor mutation in a relapsed CWR22 prostate cancer xenograft and cell line. Cancer Res. 2002;62:6606–14. [PubMed] [Google Scholar]
- 17.Tan J, Sharief Y, Hamil KG, et al. Dehydroepiandrosterone activates mutant androgen receptors expressed in the androgen-dependent human prostate cancer xenograft CWR22 and LNCaP cells. Mol Endorinol. 1997;11:450–59. doi: 10.1210/mend.11.4.9906. [DOI] [PubMed] [Google Scholar]
- 18.Craft N, Chhor C, Tran C, et al. Evidence for clonal outgrowth of androgen-independent prostate cancer cells from androgen-dependent tumors through a two-step process. Cancer Res. 1999;59:5030–6. [PubMed] [Google Scholar]
- 19.Navone NM, Olive M, Ozen M, et al. Establishment of two human prostate cancer cell lines derived from a single bone metastasis. Clin Cancer Res. 1997;3:2493–500. [PubMed] [Google Scholar]
- 20.Kaighn ME, Narayan K Shankar, Ohnuki Y, Lechner JF, Jones LW. Establishment and characterization of a human prostatic carcinoma cell line (PC-3) Invest Urol. 1979;17:16–23. [PubMed] [Google Scholar]
- 21.Stone KR, Mickey DD, Wunderli H, Mickey GH, Paulson DF. Isolation of a human prostate carcinoma cell line (DU 145) Int J Cancer. 1978;21:274–81. doi: 10.1002/ijc.2910210305. [DOI] [PubMed] [Google Scholar]
- 22.Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20:3001–15. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Linja MJ, Visakorpi T. Alterations of androgen receptor in prostate cancer. J Steroid Biochem Mol Biol. 2004;92:255–64. doi: 10.1016/j.jsbmb.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Veldscholte J, Ris-Stalpers C, Kuiper GGJM, et al. A mutation in the ligand binding domain of the androgen receptor of human LNCaP cells affects steroid binding characteristics and response to anti-androgens. Biochem Biophys Res Commun. 1990;17:534–40. doi: 10.1016/s0006-291x(05)80067-1. [DOI] [PubMed] [Google Scholar]
- 25.Zhao XY, Boyle B, Krishman AV, et al. Two mutations identified in the androgen receptor of the new human prostate cancer cell line MDA PCa 2a. J Urol. 1999;162:2192–99. doi: 10.1016/S0022-5347(05)68158-X. [DOI] [PubMed] [Google Scholar]
- 26.Zhao XY, Malloy PJ, Krishman AV, et al. Glucocorticoids can promote androgen-independent growth of prostate cancer cells through a mutated androgen receptor. Nat Med. 2000;6:703–06. doi: 10.1038/76287. [DOI] [PubMed] [Google Scholar]
- 27.Lin D-L, Keller ET. Mechanism of interleukin-6 promotion of androgen receptor function in prostate cancer cells. International Conference on Prostate Cancer Research; Iowa City. 1999. [Google Scholar]
- 28.Abreu-Martin MT, Chari A, Palladino AA, Craft NA, Sawyers CL. Mitogen-activated protein kinase kinase kinase 1 activates androgen receptor-dependent transcription and apoptosis in prostate cancer. Mol Cell Biol. 1999;19:5143–54. doi: 10.1128/mcb.19.7.5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeh S, Lin HK, Kang HY, et al. From HER2/Neu signal cascade to androgen receptor and its coactivators: a novel pathway by induction of androgen target genes through MAP kinase in prostate cancer cells. Proc Natl Acad Sci U S A. 1999;96:5458–63. doi: 10.1073/pnas.96.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ueda T, Bruchovsky N, Sadar MD. Activation of the androgen receptor N-terminal domain by interleukin-6 via MAPK and STAT3 signal transduction pathways. J Biol Chem. 2002;277:7076–85. doi: 10.1074/jbc.M108255200. [DOI] [PubMed] [Google Scholar]
- 31.Chen WY, Ramamoorthy P, Chen N, Sticca R, Wagner TE.A human prolactin antagonist, hPRLG129R, inhibits breast cancer cell proliferation through induction of apoptosis [published erratum appears in Clin Cancer Res 2000 May;6(5):2120]. Clin Cancer Res 199953583–93. [PubMed] [Google Scholar]
- 32.Tan SH, Dagvadorj A, Shen F, et al. Transcription factor Stat5 synergizes with androgen receptor in prostate cancer cells. Cancer Res. 2008;68:236–48. doi: 10.1158/0008-5472.CAN-07-2972. [DOI] [PubMed] [Google Scholar]
- 33.Ahonen TJ, Xie J, LeBaron MJ, et al. Inhibition of transcription factor Stat5 induces cell death of human prostate cancer cells. J Biol Chem. 2003;278:27287–92. doi: 10.1074/jbc.M304307200. [DOI] [PubMed] [Google Scholar]
- 34.Dagvadorj A, Kirken RA, Leiby B, Karras J, Nevalainen MT. Transcription factor signal transducer and activator of transcription 5 promotes growth of human prostate cancer cells in vivo. Clin Cancer Res. 2008;14:1317–24. doi: 10.1158/1078-0432.CCR-07-2024. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Ahonen TJ, Alanen K, et al. Activation of signal transducer and activator of transcription 5 in human prostate cancer is associated with high histological grade. Cancer Res. 2004;64:4774–82. doi: 10.1158/0008-5472.CAN-03-3499. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Zhang Y, Glass A, et al. Activation of signal transducer and activator of transcription-5 in prostate cancer predicts early recurrence. Clin Cancer Res. 2005;11:5863–8. doi: 10.1158/1078-0432.CCR-05-0562. [DOI] [PubMed] [Google Scholar]