Abstract
Background
Feeding patterns of the vector are important in the epidemiology of Chagas disease, the leading cause of heart disease in Latin America. Chagas disease is caused by the parasite, Trypanasoma cruzi, which is transmitted by blood feeding insects. Historically, feeding behaviours of haematophagous insects have been investigated using serological reactions, which have detection limits in terms of both taxonomic resolution, and quantity and quality of the blood meal. They are labor intensive, require technical expertise, need fresh or frozen samples and antibodies often are either not available commercially or the resources for synthesis and purification are not available. We describe an assay to identify vertebrate blood meal sources, and the parasite T. cruzi using species-specific PCR assays from insect vectors and use the method to provide information regarding three questions: (1) Do domestic and peri-domestic (chicken coop and animal corral) habitats vary in the blood meals detected in the vectors? (2) What is the pattern of multiple blood meals? (3) Does the rate of T. cruzi infection vary among habitats and is it associated with specific blood meal types?
Methodology/Principal Findings
Assays based on the polymerase chain reaction were evaluated for identification of the blood meal source in the heamatophagous Chagas disease vector Triatoma infestans. We evaluate a technique to identify 11 potential vertebrate food sources from the complex mixture extracted from the vector's abdomen. We tested the assay on 81 T. infestans specimens collected from the Andean highlands in the department of Chuquisaca, located in central Bolivia, one of the regions in South America where sylvatic T. infestans have been reported. This area is suggested to be the geographic origin of T. infestans and has very high human infection rates that may be related to sylvatic vector populations.
Conclusion/Significance
The results of the assays revealed that a high percentage of insects collected in human dwellings had fed on peri-domestic animals. In contrast, one insect from a chicken coop but no bugs from corrals tested positive for human blood. Forty-eight percent of insects tested positive for more than one vertebrate species. T. cruzi infection was detected in 42% of the specimens. From the epidemiological point of view, the results reveal an overall pattern of movement from peri-domestic structures to human habitations for T. infestans in this region of Bolivia as well as the important role of pigs, dogs, chickens and guinea pigs in the dynamics of T. cruzi infection.
Introduction
Triatoma infestans (Hemiptera:Reduviidae) is the main vector of Chagas disease in seven countries of South America [1]. The vector species is believed to have evolved in and be endemic to the Andean highlands of Bolivia [2]–[4] and its current geographic range includes 55% of Bolivia [5], spanning domestic, peri-domestic and sylvatic habitats [6]–[8]. In 1991 the World Health Organization launched the Southern Cone Initiative targeting vector-borne and blood-transfusion transmission in the “cone” region of South America. This initiative has shown achievement in Argentina, Brazil, Chile and Uruguay as reflected in low rates of house infestation and a decline in human T. cruzi infection [9]. Bolivian control activities, started in the mid 1980s, have not been as successful. One possible reason for the disparity is the existence of sylvatic populations of T. infestans that disperse from the natural ecotope and colonize recently insecticide treated peri-domestic structures and homes. Additional sources of reinfestation include recrudescence (hatching of eggs laid prior to spraying) [10], [11], survivors of the insecticide treatment [2], [12] and migrants from peri-domestic structures where spraying is likely not as effective. However, regions vary in their success with vector control and local vector ecology may predispose some areas to the persistence of bugs in domestic and peri-domestic habitats. For example, in the southern part of the Bolivian department of Potosí great success was observed in the elimination of T. infestans during a one-year trial [5]. However, other endemic zones in Bolivia continue to have active T. cruzi transmission and high rates of house infestation despite similar control efforts [13], [14]. Results of studies from Argentina show re-colonizers come from nearby untreated structures in the same homestead, as well as insects dispersing from neighboring localities [15].
Understanding vector movement and feeding preferences are important to the success of insecticide spraying programs against Chagas disease vectors. Infestation of insecticide-treated habitats from recrudescence, colonizers from nearby untreated areas, or sylvatic populations jeopardize the success of control program initiatives. Consequently, the strategy implemented during the surveillance phase of these programs relies on the ability to identify sources of reinvasion. Monitoring methods include investigating the population genetic structure of the vector population and analyzing the spatial association between the capture location of vectors and their feeding sources. Population genetic approaches use a variety of methods including estimates of gene flow among populations based on allozyme variability, [2], [16], [17] and more recently estimates of migration using microsatellites [18], [19].
Historically, feeding patterns of haematophagous insects have been investigated based on serological reactions [20]–[26]. Studies of feeding patters revealed that domestic insects often feed on multiple hosts [24], [25]. Feeding of Rhodnius prolixus and T. dimidiata collected from houses in Guatemala, examined with ELISA, showed that more had fed on human than other species, both species fed on opossum, and, for the insects examined, only R. prolixus fed on chicken and only T. dimidiata on cow [24]. In Argentina, dogs, chickens, and humans are the most common blood sources of domestic T. infestans [20]. The results of studies of vectors collected from and around houses using a dot-blot assay showed that Rhodnius pallescenes, a sylviatic species, collected in houses are more likely to have fed on humans than those collected from palm trees [23]. Insect density and spatial and temporal occurrence of hosts and insects contribute to host choice [20]. The number, identity and proximity of domestic and peri-domestic animals influence feeding patterns [27].
Feeding preference studies have shown that availability can be more important than preference [22]. An experiment examining T. infestans, T. dimidiata, and R. prolixus feeding on opposum, dog, chicken and toad, revealed a preference for homeothermic hosts, T. infestans showed a slight preference for dogs and only T. dimidiata, the least aggressive species, fed on toads [21]. T.infestans from houses in Argentina also showed seasonal preference. Bugs seemed to prefer dogs in the summer and chickens in winter [20].
To understand vector movement, host feeding in different ecotopes has been studied. The number, identity and proximity of domestic and peri-domestic animals influence the dispersal conduct of T. infestans. In the peri-domicile the effectiveness of insecticide treatment is reduced due to size, abundant refuge sites and physical conditions that hasten the degradation of residual insecticides [28]. Consequently, in Southern Bolivia where sylvatic populations of T. infestans occur, the peri-domicile may represent not only the major source of insects that reinvade treated domiciles and peri-domicile complexes but also may act as a corridor between sylvatic and domestic populations. Vector movement is suggested by finding that T. dimidiata collected in and around houses had fed on opossum [26].
These methods have detection limits in terms of both taxonomic resolution, and quantity and quality of the blood meal. They are labor intensive [29], require considerable technical expertise, samples need to be fresh or frozen [30] and antibodies often are either not available commercially or the resources for synthesis and purification are not available. In a previous study we described an assay based on the polymerase chain reaction (PCR) amplification of a species-specific repetitive element of nuclear DNA to identify guinea pig DNA in the complex mixture present in the abdomen of T. infestans. A PCR based assay has advantages in that samples do not need to be fresh or frozen. The assay detects DNA quite soon after feeding, we detected guinea pig DNA in 100% of the vectors up to 40 hours after bugs had fed on guinea pigs under controlled conditions. In addition, starvation does not seem to have much impact on the ability to detect prior blood meals, in vectors collected from houses and peri-domestic areas of communities in the Department of Chuquisaca and maintained for two months without feeding, we were able to amplify guinea pig DNA in 8 of 34 samples demonstrating that at least 23% of the vectors had fed on guinea pigs [31].
In the present study we extend the assay to the identification of 11 vertebrate species, and the parasite T. cruzi using species-specific PCR assays. For cow, pig, chicken, dog, guinea pig, cat, mouse and donkey the assay targets short interspersed nuclear DNA elements (SINEs) [32]–[34]. The human assay detects the Alu element-based subfamily Yb6, a long interspersed element (LINE) [35]. Subunit 8 of the mitochondrial ATP synthase gene, atp8 was amplified for goat and sheep [36]. We investigate three questions:
Are there differences among habitats (domestic vs peri-domestic) in the identity of the blood meals detected in the vectors? This information will give insight into vectors movement among habitats. For example, if domestic and peri-domestic vectors do not differ in the types of blood meals detected, that would indicate significant vector movement between habitats. At the other extreme, if human DNA is only detected in vectors collected in houses there is likely little movement from domestic to peri-domestic areas.
What is the frequency and pattern of single vs multiple blood meals? If vectors tend to feed on a single type of host, i.e., vectors feed on either dogs or humans but not both, then the likelihood of transmission among mammal species is reduced.
Does the rate of Trypanosoma cruzi infection vary among habitats and is it associated with specific blood meal types? Rates of T. cruzi infection in vectors will determine the probability of transmission to humans. High rates of T. cruzi infection in peri-domestic hosts enhance the risk for humans when insects migrate from peri-domestic to domestic habitats [15]. Because only mammals can sustain infection, T. cruzi infection should be low in bugs collected from chicken coops [27] if the bugs do not migrate and feed in other habitats. Finally, the likelihood of transmission varies among vertebrate hosts. Experiments have shown that bugs that feed on infected dogs are more likely to become infected than bugs feeding on infected humans [37].
Methods
Collection of vectors and description of study area
Eighty-one T. infestans collected from May to July of 2005 by Vector Control personnel at the Servicio Departamental de Salud (SEDES) of Chuquisaca, Bolivia were included in the present study. The insects were taken from a total sample of over one thousand insects collected from the department of Chuquisaca. The sample was stratified by habitat and insects were randomly taken from selected houses from each strata. Most insects were from domestic and peri- domestic sites in six northern localities in the department of Chuquisaca: 26 insects were collected in Capilla Llave (18°58′46S, 64°42′21″W), nine came from Carbajal (19°12′40″ S, 65°18′14″ W), 11 from Cueva Uyuni (19°25′0″S, 64°54′0″W), 13 from Serrano (19°06′0″S, 64°22′0″W) and four came from Yotala (19°9′31″S, 65°15′51″W). Eighteen insects from the southern locality of Huacareta (20°40′0″S, 63°45′0″W) were also examined.
Domiciles in this area have one or two adjacent bedrooms and a kitchen. A few have an additional room devoted to storing agricultural products; usually, part of a bedroom or a region of the kitchen is consigned to this purpose. The peri-domestic area includes structures such as pigpens; goat, sheep and cow corrals; and chicken coops. The animals, raised for subsistence, are usually at low density. Houses and fences of corrals are generally of adobe, combined with roofs of earth covered with split cane or ceramic tile. Thorn scrub branches are often also used in corrals atop adobe blocks to limit livestock movement and protect the adobe from rain. In the region under study, chickens live in a combined adobe-wall and wired enclosures with a thatched roof and they are not allowed to sleep within a room of the house, even during winter. All insects were collected from roof and walls of the enclosures.
Of the eighty-one T. infestans examined, 29 came from human habitations, 11 from chicken coops and 41 from corrals. There were 34 fifth, 12 fourth, and 11 third instar nymphs, one second and one first instars, 13 adult females and 10 adult males.
Upon collection, live insects were placed in plastic containers, transported to Sucre for species identification [38], placed in 96% ethanol, transported to Vermont, USA and stored at −20°C. No vertebrate animals were directly used in this study.
Methods of molecular genetic analysis
DNA was extracted from ∼25 mg of tissue (cut from the posterior of the abdomen using a new razor blade for each individual to prevent contamination) using the DNeasy kit (Qiagen, Inc., Valencia, CA). DNA concentration was measured using a Nanodrop 1000 spectrophotometer (Nanodrop, Bethesda, MD).
Control DNA was extracted using the same kit. Positive controls for cow, pig, sheep and chicken, were from commercially purchased meat. Donkey, cat, dog and goat DNA were extracted from hair. Human DNA was from dried blood spots on filter paper. The blood was collected from Chuquisaca with the approval of the Ethics Board, University of San Francisco Xavier, Sucre, Bolivia. Mouse and guinea pig DNA were extracted from liver. The T. cruzi positive control was from previously sequenced samples that tested positive by PCR for T. cruzi.
Extracted DNA was amplified in 25 µl reactions using 1 µl of DNA template, 0.2 µM each of forward and reverse primers, 1× PCR Master Mix (0.2 mM dNTPs, 1.5 mM MgCl2, HotStart-IT Taq DNA polymerase and reaction buffer, USB Corporation, Cleveland, Ohio).
The PCR parameters for dog, guinea pig, cow, chicken, cat, mouse and pig DNA [33], [34] were: initial denaturation of 1 min at 95°C, followed by 30 cycles of 95°C for 30 s, annealing as shown in Table 1 for 30 s, and 30 s of extension at 72°C, except for the porcine assay where 1 minute to anneal and extend at 63°C was used.
Table 1. Primer sequence and PCR product size for species-specific assays.
| Species | Primer A | Primer B | °C | size (bp) | |
| Human1 | Homo sapiens | tttgagacggagtctcgtt | gagatcgagaccacggtgaaa | 61 | 200 |
| Dog2 | Canis familiaris | agggcgcgatcctggagac | agacacaggcagagggagaa | 55 | 83 |
| Guinea pig2 | Cavia porcellus | gggatttagctcagtggcataag | attggtaccggggattgaact | 60 | 71 |
| Cow2 | Bos taurus | tttcttgttatagcccaccacac | tttctctaaaggtggttggtcag | 60 | 98 |
| Chicken2 | Gallus gallus | ctgggttgaaaaggaccacagt | gtgacgcactgaacaggttg | 60 | 169 |
| Cat2 | Felis catus | agtcggttaagcgtctgacttt | ctccaggctctgagctgtca | 55 | 98 |
| Mouse2 | Mus musculus | agatggctcagtgggtaaagg | gtggaggtcagaggacaaactt | 55 | 118 |
| Pig2 | Sus scrofa | gactaggaaccatgaggttgcg | agcctacaccacagccacag | 63 | 134 |
| Donkey2 | Equus asinus | ccaaagccccccagtacatag | gtggccaagtggttaagttcg | 60 | 152 |
| Sheep3 | Ovis aries | ctcaaggagtattttgtttc | aattctatcaatattttttagt | 48 | 117 |
| Goat3 | Capra aegagrus | tctcaaggggtgttatgc | gccacaactagacacatct | 48 | 150 |
| parasite4 | T. cruzi | cgagctcttgcccacacgggtgct | cctccaagcagcggatagttcag | 57 | 188 |
For goat and sheep [36], the protocol was: 5 min at 95°C; 35 cycles of 92°C for 1 min, followed by annealing for 2 min at 48°C, extension for 2 min at 72°C; and final extension of 5 min at 72°C. For human [35] we used: 12 min at 95°C; followed by 40 cycles at 95°C for 15 s and 1 min at 61°C to anneal and extend. For donkey [32] the protocol was: initial denaturation for 5 min at 94°C; followed by 30 cycles of 94°C for 20 s, annealing for 30 s at 60°C, extension for 40 s at 72°C; and final extension of 6 min at 72°C. For T. cruzi [39], 10 min at 94°C was followed by 30 cycles of 94°C for 20 s, annealing for 10 s at 57°C, extension for 30 s at 72°C and a final extension of 7 min at 72°C. Amplicons were visualized by electrophoresis using 2% agarose, ethidium bromide staning and UV light. Positive and negative controls were always included.
Walker et al. [33], [34] describe an elegant series of assays demonstrating the specificity of the dog, pig, guinea pig, cow, chicken, cat and mouse primers for both single species and mixed species DNA samples. They also demonstrated the primers amplify DNA in quantities from 0.01 pg to 100 ng DNA/reaction depending on the species of target template. In a previous experimental study, we showed that the assay for guinea pig DNA was successful for 100% of the vectors up to 40 hours after bugs had fed. We were also able to detect guinea pig DNA in 8 of 34 field collected specimens maintained for two months without feeding prior to testing [31]. This is our first assay using the primers to detect multiple blood meals from vectors.
To rule out PCR inhibition by unknown components in the DNA extraction, we spiked the 12 samples in which no vertebrate or parasite DNA was detected with 0.2 ng of a mixture of T. cruzi and guinea pig DNAs and ran a second PCR. All of these showed amplification.
Statistical methods
For most of the statistical analyses, the independent variables were categorical (habitat or species of vertebrate) and the dependent variable was ordinal (presence = 1 or absence = 0 for each vertebrate species or the T. cruzi parasite). For these analyses, we used contingency analysis combined with a likelihood ratio test (JMP 5.0.1.2, 2003). For statistical analysis that required another test, we describe that analysis below.
Variation among habitats in the types of blood meals detected
For each vertebrate host, we determined if each blood source was preferentially found in certain habitats. For example, was human DNA detected more often in domestic vectors than in peri-domestic vectors?
Frequency and pattern of multiple feeding
We tested whether insects that fed on a given host type were more or less likely to fed on other hosts. For example, did vectors in which we detected human DNA tend to feed exclusively on humans or did they test positive for more than one type of vertebrate host? We also determined if some pairs of DNA types co-occur in a single insect more or less often than predicted by random chance. Finally, the number of blood sources detected in a single vector was compared for domestic vs peri-domestic habitats using a nonparametric median test.
Trypanosoma cruzi association with habitats, vertebrate hosts and vector life stage
We examined if: (1) insects collected from domestic vs peri-domestic habitats differ in their likelihood of infection, (2) insects that had fed on different types of vertebrates varied in their likelihood of being infected with T. cruzi and (3) insects of different developmental stages differ in their likelihood of being infected.
Results
We were able to identify a blood meal of at least one vertebrate species in 85% (69/81) of the vectors examined. Of the 12 samples in which no blood meal was detected, only one tested positive for T. cruzi DNA. A second aliquot of the negative samples spiked with a mixture of guinea pig and T. cruzi DNA showed the characteristic 188 bp and 71 bp bands for T. cruzi and guinea pig respectively, suggesting PCR inhibition was not a problem. None of the insects tested were positive for sheep, donkey, mouse or cat DNA. Pig blood was the most common type of blood meal, found in 34 of the 69 insects that tested positive for at least one type of vertebrate DNA (49%). Dog DNA was present in 24 cases (35%) and chicken in 18 (26%). All the other species were identified at a lower frequency (Table 2).
Table 2. Percent of vectors from each habitat that tested positive by PCR for 11 different types of vertebrate DNA.
| Vertebrate host | N | Domestic | Peri-domestic | Likelihood ratio | p-value* |
| % | % | Chi Square | |||
| Human | 13 | 41 | 2 | 22.14 | <0.0001 |
| Cow | 9 | 17 | 8 | 1.65 | n.s. |
| Chicken | 18 | 7 | 31 | 7.06 | <0.01 |
| Pig | 34 | 34 | 46 | 1.05 | n.s. |
| Guinea pig | 7 | 10 | 7 | 0.16 | n.s. |
| Goat | 11 | 7 | 17 | 1.89 | n.s. |
| Dog | 24 | 41 | 23 | 2.93 | n.s. |
| Donkey, cat, sheep, or mouse | 0 | 0 | 0 | ||
| No blood sources detected | 12 | 10 | 17 | 0.75 | n.s. |
Probability a blood source is randomly distributed between domestic and peri-domestic habitat types.
Variation among habitats in the types of blood meals detected
Seven blood types were detected in insects collected from domestic habitats. Human and dog DNA were the most abundant in this ecotope, followed by pig cow, guinea pig, chicken and goat (Table 2). Of the 19 nymphs found in houses, DNA from non-domestic hosts was found in 14, while human, dog and/or guinea pig DNA was found in 15. Of the 7 adults collected from houses, 6 had DNA from non-domestic sources and 6 tested positive for human, dog and/or guinea pig DNA. In three domestic insects, no vertebrate blood was detected.
All species found in houses were also found in peri-domestic habitats. From the 42 insects collected from peri-domestic habitats, pig DNA was the most common, followed by chicken, dog, goat, guinea pig and human (Table 2). For nine peri-domestic, no vertebrate DNA was detected.
Human DNA is more prevalent in insects from houses than insects from peri-domestic habitats (Likelihood Ratio χ2 = 22.14, p<0.0001) and chicken DNA was more common in vectors from peri-domestic vectors than vectors from houses (Likelihood Ratio χ2 = 7.06, p<0.01), DNA of all the other vertebrate species is equally distributed between habitats (Figure 1 and Table 2). Those insects collected in houses that had fed on cow chicken pig or goat were defined as migrants, as were insects that had fed on humans and were found in corrals or chicken coops. Vectors are significantly more likely to migrate from houses to peri-domestic habitats than vise versa (Table 3).
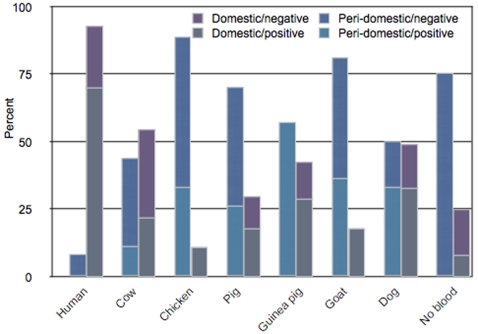
Figure 1. For domestic and peri-domestic habitats, we show the percentage of each type of blood meal found in that habitat and the T. cruzi prevalence in insect vectors that had fed on each blood source.
Vectors from domestic vs. peri-domestic habitats vary in their blood meal source. Almost all the insects that had fed on humans were collected from houses. Detection of cow, chicken pig or goat DNA in vectors collected from houses indicates a migrant from a peri-domestic to domestic habitat. Detection of human DNA in a vector collected from a peri-domestic habitat indicates migration from domestic to peri-domestic habitat.
Table 3. Vector movement between domestic and peri-domestic structures.
Number of vectors positive for human DNA collected from peri-domestic habitats.
Number of vectors positive for cow, chicken, pig or goat DNA collected in houses.
Likelihood Chi-square = 32.86, p<0.0001).
Frequency and pattern of multiple feeding
From the 69 samples that tested positive for at least one type of vertebrate or T. cruzi DNA, we detected one blood meal in 36 (52%) insects. For 22 (32%) specimens we were able to detect two different vertebrate species, in 8 we detected three different types of vertebrate DNA and finally in 3 insects we were able to identify the presence of four different types of vertebrate DNA (Table 4). The mean number of distinct blood sources detected in a single insect was significantly different among chicken coops (1.64), bedrooms (1.59), and corrals (1.27) (Median Test: Chi-square = 9.26, d.f. = 2, p<0.01). The likelihood of finding a particular blood meal mixed with at least one other type of blood (Table 4). In contrast, it was more likely that there was only one source when bugs fed on humans or cows. Pairwise co-occurrence and significant associations between species when two types of DNA were detected in a single bug are shown in Table 5. When insects were found to have fed on pigs, the frequency of human blood was significantly less than expected by chance (p<0.05). This was also the case between pig and goat DNA when both types of DNA were found in the same sample. When dog DNA was detected the probability of guinea pig DNA in the same insect was higher than expected (p<0.05).
Table 4. Frequency of single vs multiple (2, 3 or 4) vertebrate blood types identified by PCR in T. infestans from Chuquisaca, Bolivia.
| Single | 2 | 3 | 4 | Likelihood ratio Chi Square | p-value* | |
| Human | 5 | 5 | 2 | 1 | 2.73 | n.s. |
| Cow | 2 | 5 | 2 | 0 | 5.78 | <0.05 |
| Chicken | 4 | 10 | 3 | 1 | 13.29 | <0.001 |
| Pig | 16 | 9 | 6 | 3 | 3.61 | <0.05 |
| Guinea Pig | 1 | 1 | 3 | 2 | 6.64 | <0.05 |
| Goat | 5 | 3 | 1 | 2 | 0.99 | n.s. |
| Dog | 3 | 11 | 7 | 3 | 32.74 | <0.0001 |
| Total** | 36 | 22 | 8 | 3 |
Probability that vectors that fed on the specified host fed on more than one host (one-tailed test).
Total number is less than the column sum because of multiple infections.
Table 5. Pairwise probabilities of occurrence of two types of DNA in the same T. infestans specimen from Chuquisaca.
| Human | Cow | Chicken | Pig | Guinea Pig | Goat | |
| Cow | 0.27* | |||||
| n.s. | ||||||
| Chicken | 0.45 | 0.00 | ||||
| n.s. | n.s. | |||||
| Pig | 5.00 | 0.32 | 0.09 | |||
| (<0.05) ⇓ | n.s. | n.s. | ||||
| Guinea Pig | 2.57 | 1.73 | 3.70 | 0.71 | ||
| n.s. | n.s. | n.s. | n.s. | |||
| Goat | 0.52 | 0.05 | 1.50 | 3.23 | 4.10 | |
| n.s. | n.s. | n.s. | n.s. | (<0.05) ⇑ | ||
| Dog | 0.56 | 1.96 | 0.92 | 0.90 | 10.60 | 0.03 |
| n.s. | n.s. | n.s. | n.s. | (<0.01) ⇑ | n.s. |
Likelihood ratio Chi square (P-values) for pair co-occurring less than (⇓) or greater than (⇑) expected by chance.
Trypanosoma cruzi association with habitats, vertebrate hosts and vector life stage
Infection with T. cruzi was detected in 34 out of 81 bugs (42%). The prevalence of infection was higher in insects from houses (55%) compared to that observed in chicken coops (27%) and corrals (37%); however, these differences are not statistically significant (Table 6). Insects that had fed on humans, dogs or guinea pigs were more likely to be infected by T. cruzi (p<0.05) (Table 7, Figure 1). No significant difference in the frequency of T. cruzi infection was observed by developmental stage of the insect (X2 = 10.44, p>0.05) data not shown.
Table 6. Variation among habitats in T. cruzi prevalence in T. infestans from Chuquisaca.
| PCR result | Sample size | Domicile | Chicken coop | Corral |
| Negative | 47 | 13 | 8 | 26 |
| Positive | 34 | 16 | 3 | 15 |
| % Positive* | 42% | 55% | 27% | 37% |
T. cruzi infection prevalence not significantly different among habitats (Likelihood ratio Chi square = 3.56, P>0.05).
Table 7. T. cruzi association with vertebrate hosts of T. infestans from Chuquisaca.
| Vertebrate host | Likelihood ratio Chi square* | P value |
| Human | 4.70 | <0.05⇑ |
| Cow | 0.32 | n.s. |
| Chicken | 0.06 | n.s. |
| Pig | 0.11 | n.s. |
| Guinea pig | 6.29 | <0.05⇑ |
| Goat | 0.82 | n.s. |
Likelihood ratio Chi square (P-values) for pair co-occurring less than (⇓) or greater than (⇑) expected by chance.
Discussion
Reduction of the incidence of Chagas disease requires knowledge of how vectors move among habitats and especially the source of vectors that colonize houses after insecticide application. Our results show that this method can be used to show movement from peri-domestic structures to human habitations for T. infestans as well as the important role of pigs, dogs, chickens and guinea pigs in the dynamics of T. cruzi infection. Spatial analysis of variation among habitats demonstrates: (1) significantly more movement from peri-domestic to domestic structures than vice versa and (2) that vectors testing positive for human and chicken DNA are more likely to be collected from domestic structures and peri-domestic structures, respectively. Vectors testing positive for the other types of vertebrate DNA appear to be randomly distributed among habitats. Almost half of the vectors tested positive for more than one vertebrate species, T. cruzi infection was detected in 42% of the specimens and there was a significant difference in T. cruzi infection between domestic and peri-domestic habitats.
The method was used to show a dispersal from peri-domestic habitats toward the domicile. Vectors that had fed on humans were predominantly collected in human habitations, and only one insect that had fed on humans was collected outside of houses. In contrast, blood of six species of peri-domestic hosts was found in domestic vectors. Dogs and pigs provided the majority of the blood meals detected in the peri-domicile; and in vectors collected in houses, these hosts were the most common after humans. The likelihood of hosts spatially co-occurring affects the likelihood of finding vectors that have fed on both: pig blood was negatively associated with human blood, and guinea pig blood was positively associated with dog and goat blood.
The results of a study in rural villages of north-west Argentina showed T. infestans feeding patterns are non-random, the likelihood of feeding on humans decreased when dogs and chickens were present in bedroom areas [20]. The role of pig corrals as reservoirs has also been reported from studies in Argentina [28], [40]. Our insect collection methods did not include counting potential vertebrate hosts in the domicile or peri-domicile; however we do note that cow DNA was found more frequently than goat DNA in insects collected in houses. Host abundance and proximity are often considered the main determinants of host choice [20]. In this region, cows are usually kept in open corrals adjacent to human dwellings while goats are often enclosed in more distant corrals constructed with adobe blocks. The number of cows per family is usually lower than the number of goats.
We have found a high rate of T. cruzi infection in insects collected in houses (55%) and a significantly higher rate of T. cruzi infection in bugs that had fed on humans, dogs and guinea pigs. These data contrast markedly with the 4.6% of infected insects found during the surveillance phase in northwestern Argentina [42] and is consistent with the rate of 58% reported for sylvatic T. infestans in the Andean Valleys of Bolivia [43], [44] and the current high levels of transmission in this part of Bolivia. The high prevalence of T. cruzi in insects collected from houses could partially be due to the close proximity of dogs and guinea pigs, which also maintain a high parasite burden. Dogs are often employed as animal guards, watching livestock during the day and returning to the house in the evening. In this movement, they may transport T. cruzi from one habitat to another. Although, dogs in this area are not usually allowed to enter houses, they often sleep outside against the bedroom wall. Rural families in this part of Bolivia maintain guinea pigs for food in corrals located close to the house or in a particular compartment in the kitchen. We found four insects in corrals that had fed on guinea pigs. As wild guinea pigs occur in burrows in stone fences of goat and pig corrals (personal observation) in this area, we cannot ascertain whether these blood meals were from domestic or sylvatic animals.
The use of forensic DNA analysis to examine feeding in disease vectors [45]–[50], including vectors of Chagas disease is relatively new [51]. Our use of primers based on SINEs and LINEs that have high copy number and small size is advantageous for degraded DNA. For nine of the eleven vertebrate taxa we investigate, the assay is based on highly repetitive, relatively small (∼70–200 bp) sequences of Large or Small Interspersed Nuclear Element (LINE or SINE) transposable DNA, which can be an advantage when using degraded DNA. A negative PCR result for blood meal, observed in 12 specimens, could be because the insect had not fed, failure of PCR, or the blood meal source was a species not included in our assay. In 11 of these 12 samples we did not find evidence of T. cruzi infection. The T. cruzi found in the blood meal negative bug could have also been a result of co-prophagy. One specimen that tested negative for all 11 vertebrate species was a first instar nymph and could be unfed. Experimental work showed our assay detects DNA in 100% of specimens (N = 36) in as little as 1 hour and up to 40 hours after controlled feeding and identified guinea pig DNA in 23% (9 of 34) of T. infestans collected in the wild and maintained for two months, without feeding, under controlled conditions in the laboratory [31]. Estimates of mean feeding intervals in field-collected T. infestans from Argentina ranged from 3 to 7 days [41]. Thus, it is unlikely that our negative results for the other specimens were because insects were unfed. Insufficient DNA in the extracted sample or degraded DNA may offer an alternative explanation for the negative results since some specimens were analyzed even eight months after collection. In a previous study using these primers, the detection limits in complex (mixed) DNA samples using 2% agarose gels and ethidium bromide are: 5 pg in a 10 ng mixture (0.05%) for chicken, 0.005% for cow and 0.0005% for pork [33]. Non-reactive results ranged from 7% to 14% in a study on feeding patterns in T. infestans determined by double-diffusion using five genus-specific antisera [41] and 14% of Triatoma sp.specimens showed no amplification when tested for vertebrate cytB blood feedling [51]. PCR inhibition in our assay can be ruled out since negative samples amplified the two species DNA after “spiking” an aliquot of each sample with a mixture T. cruzi and guinea pig DNA's. We looked for 11 different domestic and peri-domestic species; however, we cannot eliminate the possibility that insects had fed on other species.
Although we detected guinea pig DNA in 100% of Triatoma infestans up to 40 hours after experimental feeding, a study of blood-feeding in two species of mosquitoes (Anopheles stephensi and Culex quinquefasiatus) using PCR amplification of a 358 bp region of the mtDNA cytB gene reported that all mosquitoes tested positive 1 and 6 hours after feeding, but at starting at 12 hours, there was a negative relationship between time since feeding and success of PCR amplification. There was no difference in the ability to detect blood meals between the two mosquito species, or between mosquitoes stored at +4 or −20°C up to 30 h after meal ingestion [49]. The results of an experiment of in Triatoma pallidipennis feeding on BALB/c mice amplified a 420 bp region of cytB DNA [51]. Host DNA could be detected at least until 10 weeks after the blood meal; however not all bugs tested positive for mouse DNA. Bugs that had fed for <5 min produced less intense bands or no amplification. Mota et al. [51] also reported that DNA obtained from insects collected and preserved for as long as 6 years in 70% ethanol could be amplified.
In conclusion, we have evaluated PCR-based assays for detection of the blood meal source in T. infestans. These assays tested for 11 species and were able to detect up to four species of DNA from a complex mixture in the abdominal content of bugs. Most of the species are detected based on small sized amplicons of nuclear DNA sequences with a high copy number, making them more likely to work with small amounts or degraded DNA. In addition, the expertise and equipment to perform these assays are amenable to medium equipped laboratories that may be available in many Chagas disease endemic regions.
From the epidemiological point of view, this study has disclosed the important role that pigs, dogs, chickens and guinea pigs exercise in the dynamics of the T. cruzi infection as well as in the regulation of dispersal patterns of T. infestans in this region of Bolivia.
Acknowledgments
We are grateful to Dr. Sandra Ballón, from the Laboratorio de Control Vectorial del Servicio Departamental de Salud (SEDES) de Chuquisaca for assistance with insect collection. We also thank Lindsay Christensen and Stephanie Onyekaba for technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: We gratefully acknowledge financial support from a Fulbright Fellowship (JCP). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schofield CJ. Population dynamics and control of Triatoma infestans. Ann Soc Belg Med Trop. 1985;65(Suppl 1):149–164. [PubMed] [Google Scholar]
- 2.Dujardin JP, Schofield CJ, Tibayrenc M. Population structure of Andean Triatoma infestans: allozyme frequencies and their epidemiological relevance. Med Vet Entomol. 1998;12:20–29. doi: 10.1046/j.1365-2915.1998.00076.x. [DOI] [PubMed] [Google Scholar]
- 3.Panzera F, Dujardin JP, Nicolini P, Caraccio MN, Rose V, et al. Genomic changes of Chagas disease vector, South America. Emerg Infect Dis. 2004;10:438–446. doi: 10.3201/eid1003.020812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giordano R, Cortez JC, Paulk S, Stevens L. Genetic diversity of Triatoma infestans (Hemiptera: Reduviidae) in Chuquisaca, Bolivia based on the mitochondrial cytochrome b gene. Mem Inst Oswaldo Cruz. 2005;100:753–760. doi: 10.1590/s0074-02762005000700014. [DOI] [PubMed] [Google Scholar]
- 5.Guillén G, Diaz R, Jemio A, Cassab JA, Pinto CT, et al. Chagas disease vector control in Tupiza, southern Bolivia. Mem Inst Oswaldo Cruz. 1997;92:1–8. doi: 10.1590/s0074-02761997000100001. [DOI] [PubMed] [Google Scholar]
- 6.Torrico RA. Hallazgo de Eratyrus mucronatus, infestación natural de “vinchucas” de cerro y Eutriatoma sordida en Cochabamba. An Lab Central. 1946;1:19–23. [Google Scholar]
- 7.Noireau F, Carbajal-de-la-Fuente AL, Lopes CM, Diotaiuti L. Some considerations about the ecology of Triatominae. An Acad Bras Cienc. 2005;77:431–436. doi: 10.1590/s0001-37652005000300006. [DOI] [PubMed] [Google Scholar]
- 8.Noireau F, Cortez MG, Monteiro FA, Jansen AM, Torrico F. Can wild Triatoma infestans foci in Bolivia jeopardize Chagas disease control efforts? Trends Parasitol. 2005;21:7–10. doi: 10.1016/j.pt.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Schofield CJ, Dias JC. The Southern Cone Initiative against Chagas disease. Adv Parasitol. 1999;42:1–27. doi: 10.1016/s0065-308x(08)60147-5. [DOI] [PubMed] [Google Scholar]
- 10.Salud SDd., editor. SEDES-Chuquisaca. Control vectorial del Servicio Departamental de Salud de Chuquisaca. 2006.
- 11.Ceceré MC, Vazquez-Prokopec GM, Gürtler RE, Kitron U. Reinfestation sources for Chagas disease vector, Triatoma infestans, Argentina. Emerging Infectious Diseases. 1997;12:1096–1102. doi: 10.3201/eid1207.051445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenière SF, Bosseno MF, Vargas F, Yaksic N, Noireau F, et al. Smallness of the panmictic unit of Triatoma infestans (Hemiptera: Reduviidae). J Med Entomol. 1998;35:911–917. doi: 10.1093/jmedent/35.6.911. [DOI] [PubMed] [Google Scholar]
- 13.Brenière SF, Bosseno MF, Noireau F, Yacsik N, Liegeard P, et al. Integrate study of a Bolivian population infected by Trypanosoma cruzi, the agent of Chagas disease. Mem Inst Oswaldo Cruz. 2002;97:289–295. doi: 10.1590/s0074-02762002000300002. [DOI] [PubMed] [Google Scholar]
- 14.Pizarro J, Ribera W, Aguirre A, De Muynck A. Estudio parasitológico-entomológico de traiatomíneos procedentes de la provincia Zudáñez del Departamento de Chuquisaca. Archivos Bolivianos de Medicina Vol III. 1996;52:29–33. [Google Scholar]
- 15.Ceceré MC, Gürtler RE, Chuit R, Cohen JE. Effects of chickens on the prevalence of infestation and population density of Triatoma infestans in rural houses of north-west Argentina. Med Vet Entomol. 1997;11:383–388. doi: 10.1111/j.1365-2915.1997.tb00426.x. [DOI] [PubMed] [Google Scholar]
- 16.Dujardin JP, Cardozo L, Schofield C. Genetic analysis of Triatoma infestans following insecticidal control interventions in central Bolivia. Acta Trop. 1996;61:263–266. doi: 10.1016/0001-706x(96)00008-3. [DOI] [PubMed] [Google Scholar]
- 17.Dujardin JP, Tibayrenc M, Venegas E, Maldonado L, Desjeux P, et al. Isozyme evidence of lack of speciation between wild and domestic Triatoma infestans (Heteroptera: Reduviidae) in Bolivia. J Med Entomol. 1987;24:40–45. doi: 10.1093/jmedent/24.1.40. [DOI] [PubMed] [Google Scholar]
- 18.Richer W, Kengne P, Cortez MR, Perrineau MM, Cohuet A, et al. Active dispersal by wild Triatoma infestans in the Bolivian Andes. Trop Med Int Health. 2007;12:759–764. doi: 10.1111/j.1365-3156.2007.01846.x. [DOI] [PubMed] [Google Scholar]
- 19.Pizarro JC, Gilligan LM, Stevens L. Microsatellites Reveal a High Population Structure in Triatoma infestans from Chuquisaca, Bolivia. PLoS Negl Trop Dis. 2008;2:e202. doi: 10.1371/journal.pntd.0000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gürtler RE, Cohen JE, Ceceré MC, Chuit R. Shifting Host Choices of the Vector of Chagas Disease, Triatoma Infestans, in Relation to the Availability of Host in Houses in North-West Argentina. The Journal of Applied Ecology. 1997;34:699–715. [Google Scholar]
- 21.Jiron LF, Zeledon R. [Feeding preferences in 3 species of Triatominae (Hemiptera: Reduviidae) in experimental conditions]. Rev Biol Trop. 1982;30:151–159. [PubMed] [Google Scholar]
- 22.Minter DM. Effects on transmission to man of the presence of domestic animals in infested households. New Approaches in American Trypanosomiasis Research. Washington, DC: Pan American Health Organization. Scientific Publication No. 318; 1976. pp. 330–337. [Google Scholar]
- 23.Pineda V, Montalvo E, Alvarez D, Santamaria AM, Calzada JE, et al. Feeding sources and trypanosome infection index of Rhodnius pallescens in a Chagas disease endemic area of Amador County, Panama. Rev Inst Med Trop Sao Paulo. 2008;50:113–116. doi: 10.1590/s0036-46652008000200009. [DOI] [PubMed] [Google Scholar]
- 24.Sasaki H, Rosales R, Tabaru Y. Host feeding profiles of Rhodnius prolixus and Triatoma dimidiata in Guatemala (Hemiptera: Reduviidae: Triatominae). Med Entomol Zool. 2002;51:283–289. [Google Scholar]
- 25.Wisnivesky-Colli C. Feeding patterns of Triatominae in relation to transmission of American Trypanosomiasis. In: Brener RR, Stoka AM, editors. Chagas Disease Vectors. Boca Raton, FL, USA: CRC Press; 1987. pp. 99–117. [Google Scholar]
- 26.Zeledon R, Solano G, Zuniga A, Swartzwelder JC. Biology and ethology of Triatoma dimidiata (Latreille, 1811). 3. Habitat and blood sources. J Med Entomol. 1973;10:363–370. doi: 10.1093/jmedent/10.4.363. [DOI] [PubMed] [Google Scholar]
- 27.Ceceré MC, Gürtler RE, Canale D, Chuit R, Cohen JE. The role of the peridomiciliary area in the elimination of Triatoma infestans from rural Argentine communities. Rev Panam Salud Pública. 1997;1:273–279. doi: 10.1590/s1020-49891997000400003. [DOI] [PubMed] [Google Scholar]
- 28.Ceceré MC, Vazquez-Prokopec GM, Gürtler RE, Kitron U. Spatio-temporal analysis of reinfestation by Triatoma infestans (Hemiptera: Reduviidae) following insecticide spraying in a rural community in northwestern Argentina. Am J Trop Med Hyg. 2004;71:803–810. [PMC free article] [PubMed] [Google Scholar]
- 29.Bosseno MF, García LS, Baunaure F, Gastelúm EM, Gutierrez MS, et al. Identification in triatomine vectors of feeding sources and Trypanosoma cruzi variants by heteroduplex assay and a multiplex miniexon polymerase chain reaction. Am J Trop Med Hyg. 2006;74:303–305. [PubMed] [Google Scholar]
- 30.Abad-Franch F, Monteiro FA. Molecular research and the control of Chagas disease vectors. An Acad Bras Cienc. 2005;77:437–454. doi: 10.1590/s0001-37652005000300007. [DOI] [PubMed] [Google Scholar]
- 31.Pizarro JC, Lucero D, Stevens L. A method for the identification of guinea pig blood meal in the Chagas disease vector, Triatoma infestans. Kinetoplastid Biol Dis. 2007;6:1. doi: 10.1186/1475-9292-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakagami M, Hiromura K, Chemnick LG, Ryder OA. Distribution of the ERE-1 family in Perissodactyla. Mamm Genome. 1999;10:930–933. doi: 10.1007/s003359901117. [DOI] [PubMed] [Google Scholar]
- 33.Walker JA, Hughes DA, Anders BA, Shewale J, Sinha SK, et al. Quantitative intra-short interspersed element PCR for species-specific DNA identification. Anal Biochem. 2003;316:259–269. doi: 10.1016/s0003-2697(03)00095-2. [DOI] [PubMed] [Google Scholar]
- 34.Walker JA, Hughes DA, Hedges DJ, Anders BA, Laborde ME, et al. Quantitative PCR for DNA identification based on genome-specific interspersed repetitive elements. Genomics. 2004;83:518–527. doi: 10.1016/j.ygeno.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 35.Walker JA, Kilroy GE, Xing J, Shewale J, Sinha SK, et al. Human DNA quantitation using Alu element-based polymerase chain reaction. Anal Biochem. 2003;315:122–128. doi: 10.1016/s0003-2697(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 36.Kusama T, Nomura T, Kadowaki K. Development of primers for detection of meat and bone meal in ruminant feed and identification of the animal of origin. J Food Prot. 2004;67:1289–1292. doi: 10.4315/0362-028x-67.6.1289. [DOI] [PubMed] [Google Scholar]
- 37.Gürtler RE, Ceceré MC, Castañera MB, Canale D, Lauricella MA, et al. Probability of infection with Trypanosoma cruzi of the vector Triatoma infestans fed on infected humans and dogs in northwest Argentina. Am J Trop Med Hyg. 1996;55:24–31. [PubMed] [Google Scholar]
- 38.Lent H, Wygodzinsky P. Revision of the Triatominae (Hemiptera, Reduviidae), and their significance as vectors of Chagas' disease. Bulletin of the AMNH. 1979;163:127–520. [Google Scholar]
- 39.Moser DR, Kirchhoff LV, Donelson JE. Detection of Trypanosoma cruzi by DNA amplification using the polymerase chain reaction. J Clin Microbiol. 1989;27:1477–1482. doi: 10.1128/jcm.27.7.1477-1482.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ceballos LA, Vazquez-Prokopec GM, Ceceré MC, Marcet PL, Gürtler RE. Feeding rates, nutritional status and flight dispersal potential of peridomestic populations of Triatomainfestans in rural northwestern Argentina. Acta Trop. 2005;95:149–159. doi: 10.1016/j.actatropica.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 41.Gürtler RE, Ceceré MC, Vazquez DP, Chuit R, Cohen JE. Host-feeding patterns of domiciliary Triatoma infestans (Hemiptera: Reduviidae) in Northwest Argentina: seasonal and instar variation. J Med Entomol. 1996;33:15–26. doi: 10.1093/jmedent/33.1.15. [DOI] [PubMed] [Google Scholar]
- 42.Ceceré MC, Castañera MB, Canale DM, Chuit R, Gürtler RE. Trypanosoma cruzi infection in Triatoma infestans and other triatomines: long-term effects of a control program in rural northwestern Argentina. Rev Panam Salud Pública. 1999;5:392–399. doi: 10.1590/s1020-49891999000500003. [DOI] [PubMed] [Google Scholar]
- 43.Cortez MR, Emperaire L, Piccinali RV, Gurtler RE, Torrico F, et al. Sylvatic Triatoma infestans (Reduviidae, Triatominae) in the Andean valleys of Bolivia. Acta Trop. 2007;102:47–54. doi: 10.1016/j.actatropica.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 44.Cortez MR, Pinho AP, Cuervo P, Alfaro F, Solano M, et al. Trypanosoma cruzi (Kinetoplastida Trypanosomatidae): ecology of the transmission cycle in the wild environment of the Andean valley of Cochabamba, Bolivia. Exp Parasitol. 2006;114:305–313. doi: 10.1016/j.exppara.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 45.Mukabana WR, Takken W, Knols BG. Analysis of arthropod bloodmeals using molecular genetic markers. Trends Parasitol. 2002;18:505–509. doi: 10.1016/s1471-4922(02)02364-4. [DOI] [PubMed] [Google Scholar]
- 46.Krzywinski J, Nusskern DR, Kern MK, Besansky NJ. Isolation and characterization of Y chromosome sequences from the African malaria mosquito Anopheles gambiae. Genetics. 2004;166:1291–1302. doi: 10.1534/genetics.166.3.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cook S, Diallo M, Sall AA, Cooper A, Holmes EC. Mitochondrial markers for molecular identification of Aedes mosquitoes (Diptera: Culicidae) involved in transmission of arboviral disease in West Africa. J Med Entomol. 2005;42:19–28. doi: 10.1093/jmedent/42.1.19. [DOI] [PubMed] [Google Scholar]
- 48.Oshaghi MA, Chavshin AR, Vatandoost H. Analysis of mosquito bloodmeals using RFLP markers. Exp Parasitol. 2006;114:259–264. doi: 10.1016/j.exppara.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 49.Oshaghi MA, Chavshin AR, Vatandoost H, Yaaghoobi F, Mohtarami F, et al. Effects of post-ingestion and physical conditions on PCR amplification of host blood meal DNA in mosquitoes. Exp Parasitol. 2006;112:232–236. doi: 10.1016/j.exppara.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 50.Ngo KA, Kramer LD. Identification of mosquito bloodmeals using polymerase chain reaction (PCR) with order-specific primers. J Med Entomol. 2003;40:215–222. doi: 10.1603/0022-2585-40.2.215. [DOI] [PubMed] [Google Scholar]
- 51.Mota J, Chacon JC, Gutierrez-Cabrera AE, Sanchez-Cordero V, Wirtz RA, et al. Identification of blood meal source and infection with Trypanosoma cruzi of Chagas disease vectors using a multiplex cytochrome b polymerase chain reaction assay. Vector Borne Zoonotic Dis. 2007;7:617–627. doi: 10.1089/vbz.2007.0106. [DOI] [PubMed] [Google Scholar]