Abstract
Δ9-Desaturase is a key enzyme in the synthesis of desaturated fatty acyl-CoAs. Desaturase is an integral membrane protein induced in the endoplasmic reticulum by dietary manipulations and then rapidly degraded. The proteolytic machinery that specifically degrades desaturase and other short-lived proteins in the endoplasmic reticulum has not been identified. As the first step in identifying cellular factors involved in the degradation of desaturase, liver subcellular fractions of rats that had undergone induction of this enzyme were examined. In livers from induced animals, desaturase was present in the microsomal, nuclear (P-1), and subcellular fractions (P-2). Incubation of desaturase containing fractions at physiological pH and temperature led to the complete disappearance of the enzyme. Washing microsomes with a buffer containing high salt decreased desaturase degradation activity. N-terminal sequence analysis of desaturase freshly isolated from the P-1 fraction without incubation indicated the absence of three residues from the N terminus, but the mobility of this desaturase preparation on SDS-PAGE was identical to the microsomal desaturase, which contains a masked N terminus under similar purification procedures. Addition of concentrated cytosol or the high-salt wash fraction did not enhance the desaturase degradation in the washed microsomes. Extensive degradation of desaturase in the high-salt washed microsomes could be restored by supplementation of the membranes with the lipid and protein components essential for the reconstituted desaturase catalytic activity. Lysosomotrophic agents leupeptin and pepstatin A were ineffective in inhibiting desaturase degradation. The calpain inhibitor, N-acetyl-leucyl-leucyl-methional, or the proteosome inhibitor, Streptomyces metabolite, lactacystin, did not inhibit the degradation of desaturase in the microsomal or the P-1 and P-2 fractions. These results show that the selective degradation of desaturase is likely to be independent of the lysosomal and the proteosome systems. The reconstitution of complete degradation of desaturase in the high-salt–washed microsomes by the components essential for its catalytic activity reflects that the degradation of this enzyme may depend on a specific orientation of desaturase and intramembranous interactions between desaturase and the responsible protease.
INTRODUCTION
The formation of monounsaturated fatty acids is catalyzed by Δ9 desaturase (EC1.14.99.5) in a reaction requiring acyl-CoA, NADH, NADH-reductase, cytochrome b5, phospholipid, and oxygen (Strittmatter et al., 1974). The desaturase, a 40-kDa intrinsic membrane protein, can be induced more than 50-fold in the endoplasmic reticulum by the administration of fat-free diet, insulin, or certain carbohydrate metabolites to animals (Oshino and Sato, 1972). When the dietary regimen is stopped, the desaturase activity decreases to undetectable levels with a half-life of a few hours (Oshino and Sato, 1972). The proteolytic system responsible for the rapid and selective degradation of desaturase is unknown. Eukaryotic cells contain multiple proteolytic systems including the lysosomal proteases, ATP-ubiquitin–dependent and ubiquitin-independent ATP-proteosome pathway (Hershko and Ciechanover, 1992; Inoue and Simoni, 1992). Some of these proteolytic events presumably occur on the cytoplasmic side (Gardner et al., 1993), whereas others occur within the lumen of endoplasmic reticulum (Wikstrom and Lodish, 1992). Several substrate-related peptidyl aldehydes such as N-acetyl-leucyl-leucyl norleucinal and N-acetyl-leucyl-leucyl-methional (ALLM) have been identified to inhibit the calpain, and the ubiquitin-proteosome–dependent proteolytic pathways (Jensen et al., 1995). The Streptomyces metabolite, lactacystin, is a specific inhibitor of the proteosome (Fenteany et al., 1995). It covalently modifies the highly conserved N-terminal threonine of the mammalian proteosome subunit X, a close homologue of the LMP7 proteosome subunit encoded by the major histocompatibility complex (Fenteany et al., 1995). Notwithstanding, membrane proteins are selectively degraded. A number of short-lived membrane proteins, incorrectly synthesized proteins, and partially oligomerized complexes are rapidly degraded in the endoplasmic reticulum, but it is not understood how this degradation is achieved and regulated (Klausner and Sitia, 1990; Bonifacino and Klausner, 1994).
We previously derived the cDNA sequence of the desaturase (Thiede et al., 1986) and constructed an expression vector for the production of active desaturase in Escherichia coli (Strittmatter et al., 1988). DNA constructs expressing desaturase in mice (Kaestner et al., 1989), Saccharomyces cerevisiae (Stukey et al., 1990), and a soluble form of desaturase in plants (Thompson et al., 1991; Shanklin and Somerville, 1991) have also been reported. A form of desaturase up-regulated in response to cold has been described in fish (Tiku et al., 1996).
To study the degradation of this membrane protein, desaturase was induced in rat liver membranes and isolated, and its degradation in subcellular fractions was investigated. In the present study, I report that selective degradation of desaturase can be readily monitored in microsomes and that complete degradation of this enzyme may be partially altered by a high-salt wash of the microsomes. Moreover, the inhibitors of known proteolytic systems such as lysosomes, cathepsins, or the nonlysosomal 26S proteosome complex failed to inhibit the specific degradation of desaturase. These findings should permit the design of experiments to identify the specific desaturase-degrading activity among the many previously characterized cellular proteolytic systems.
MATERIALS AND METHODS
Detergents, enzyme substrates, cofactors, and chromatographic media were obtained from Sigma (St. Louis, MO). Cytochrome b5 and cytochrome b5 reductase were prepared from rabbit liver microsomes as described previously (Ozols, 1974, 1989; Strittmatter et al., 1993). ALLM, pepstatin, leupeptin, and phenylmethylsulfonyl fluoride (PMSF) were from Sigma. Porcine erythrocyte calpain was obtained from Calbiochem (San Diego, CA). Lactacystin was obtained from Professor E.J. Corey, Department of Chemistry, Harvard University (Boston, MA).
Preparation of Desaturase Containing Subcellular Fractions
Male Sprague Dawley rats weighing 200–250 g were fasted for 48 h, fed regular diet for 48 h, fasted for second 48-h period, and refed for 20 h with Nutritional Biochemical (Cleveland, OH) “Fat Free” test diet on a schedule that permitted the animals to be killed at the beginning of a day. Control animals were refed regular diet. Subcellular fractionation of livers from induced and control animals was performed according to published procedure (Ozols, 1990). The perfused livers were homogenized in a buffer containing 0.25 M sucrose, 10 mM Tris-acetate, pH 8.1, 1 mM EDTA (6 ml/g of liver) in a glass homogenizer. Pellet P-1 was obtained by centrifugation of the homogenate at 800 × g for 10 min. The resulting supernatant was then spun at 10,000 × g for 35 min yielding pellet P-2. Centrifugation of the P-2 supernatant at 130,000 × g for 1.5 h gave pellet P-3 and the supernatant (cytosol fraction). Pelleted microsomes (P-3) were suspended in 20 volumes of 0.1 M sodium pyrophosphate, pH 7.4, and recentrifuged at 130,000 × g. High-salt washed microsomes were prepared by suspending the pellet in 20 volumes of buffer containing 0.1 M Tris-acetate, 0.5 M NaCl, 10 mM EDTA and sedimenting at 130,000 × g for 1 h to obtain high-salt washed microsomes and the high-salt supernatant. The nuclear pellet was refractionated by the method of Fleisher and Krevina (1974). Concentration of the high-salt and cytosol fractions was accomplished on a Centricon-30 concentrator (Amicon, Danvers, MA). Subcellular fractions were stored at −70°C until use. Protein concentration in the samples was determined using the Coomassie dye binding reagent (Pierce, Rockford, IL), using bovine serum albumin as a standard.
Triton X-114 Fractionation of Pellet P-2
Precondensed 4% Triton X-114 in Tris-buffered saline, pH 7.5 (Oxford Glycosystems, Abingdon Oxon, United Kingdom) was added to 120 μg of P-2 to a final concentration of 0.8%. The reaction mixture was spun at 100,000 × g for 15 min at 4°C. The supernatant was layered over a cushion of 0.25 M sucrose, and the centrifuge tube was incubated for 5 min at 37°C. Centrifugation of the reaction mixture at 12,000 × g for 5 min at 37°C yielded detergent-containing lower phase and detergent-depleted aqueous upper phase.
Isolation of Desaturase
Desaturase from the P-1 and P-2 fractions was purified in the presence of sodium deoxycholate and Triton X-100 as described previously for the purification of microsomal desaturase (Strittmatter et al., 1974, 1988). Desaturase in the P-1 or P-2 fractions for sequencing purposes was subjected to 12% SDS-PAGE and electroblotted onto the Immobilon-P membrane. Sequence analysis on the electroblotted material was performed on an Applied Biosystems (Foster City, CA) model 470A sequenator on line with a model 120A phenylhydantoin analyzer.
Reconstitution of the Desaturase System
The reconstitution and desaturase assay was performed as described previously (Strittmatter et al., 1988). Typically, 20 μl of 600 μM cytochrome b5, 6 μl of 100 μM cytochrome b5 reductase, and 20–40 μl of 30 mM egg phosphatidylcholine were added to 115 μl of desaturase sample in 2% Triton X-100 and 0.4% sodium deoxycholate. After incubation for 1.5 h at 4°C, enzyme activity was measured at 25°C by the rate of NADH oxidation in the presence and absence of stearyl-CoA.
Preparation of Antibody against Rat Liver Microsomal Desaturase
To a solution of desaturase, 20 volumes of cold acetone containing 0.2% (y/y) HCl were added. After several hours at −20°C, the protein was collected by low-speed centrifugation. About 0.5 mg of desaturase protein was mixed with 0.5 ml of Freund’s complete adjuvant and injected into rabbits. Rabbit immunoglobulin G (IgG) was prepared from serum by precipitation with ammonium sulfate as described (Hardy, 1986). IgG fractions were diluted with an equal volume of 0.1 M potassium phosphate, pH 7, and the IgG was partially purified by affinity chromatography using a column of Protein A Sepharose CL-4B equilibrated in 0.1 M potassium phosphate, pH 7, and stored in 50% glycerol at −70°C.
Immunoblotting
Complete degradation of desaturase in microsomes and nuclear fractions was determined by Western Blotting (Toubin et al., 1979). After incubation at 37°C the samples were subjected to SDS-PAGE using 12- or 10% acrylamide gel under reducing conditions in duplicate. One gel was stained with Coomassie blue, and an identical gel was electrotransferred to an Immobilon-P transfer membrane. The membrane was reacted with rabbit antidesaturase antibody, which was then complexed with antirabbit IgG-alkaline phosphatase (Sigma product A-3687). Immunoreactive desaturase bands were visualized using phosphatase substrate system detection kit (Kirkegaard & Perry Laboratories, Gaithersburg, MD). Immunoblots were quantified by densitometry using a Kodak DCS 200 digital camera with the Image software (Center for Biomedical Imaging Technology).
Assay of Desaturase Degradation
The reaction mixtures contained 10–15 μg of protein in 50 μl of 50 mM Tris-acetate, pH 7.9, and 50 mM KCl. ATP, ATP-Mg2+, and various protease inhibitors were added where indicated in the figure legend. Cytosol or high-salt fractions were concentrated on a Centricon-30 membrane (Amicon) and added to the reaction samples where indicated. The protease inhibitors were dissolved in dimethylsulfoxide and added at the concentration indicated in the figure legend. Final dimethylsulfoxide concentrations in the incubation samples were 1.5% or less. Model reactions for the protease inhibitors in microsomes were as follows: leupeptin, pepstatin, and PMSF at the concentration indicated in the figure legend was used to inhibit pepsin, papain, or endoproteases Lys-C and Asp-N. Porcine erythrocyte calpain was used as the substrate for the ALLM inhibitor. The samples were incubated 4–18 h at 37°C. Control samples were prepared at 4°C and stored at −20°C. Desaturase degradation in high-salt washed microsomes was restored by supplementation of the reaction mixture with 2 μl of 30 mM egg lecithin liposomes, 2 μl of 50 μM cytochrome b5 reductase, 2 μl of 300 μM cytochrome b5, and 5 μl of stearyl-CoA. Before gel electrophoresis of the digests, 75–150 μl of the loading buffer, containing 60 mM Tris-acetate, pH 6.8, 3% SDS, 5% β-mercaptoethanol, 10% glycerol, and 0.25% bromophenol blue were added. Ten-microliter aliquots of the reaction mixtures were subjected to electrophoresis in duplicate for Coomassie blue staining and immunoblot analysis.
RESULTS
Localization of Desaturase in Microsomes
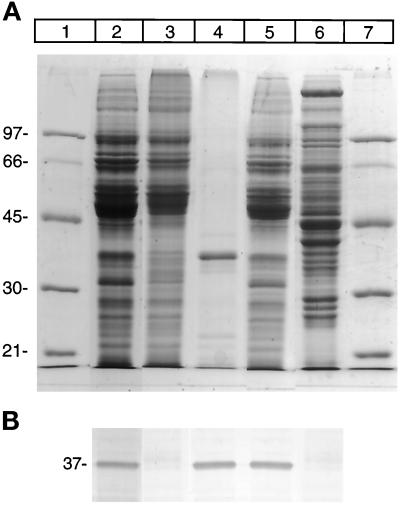
As the first step in examining the desaturase degradation of desaturase, the enzyme was induced in liver by feeding fasted rats a fat-free, high-carbohydrate diet. As seen in SDS-PAGE Coomassie blue and immunoblot analysis (Figure 1, lanes 2 and 5), a 37-kDa band reacting with desaturase antibody is evident in microsomes from rat livers that had undergone dietary manipulation. In contrast, this band was not detected in liver microsomes from control animals (Figure 1, lane 3). Purified liver microsomal desaturase migrates on SDS-PAGE as a 37-kDa band, which is lower than the predicted molecular size of 41-kDa (Thiede et al., 1986). The cytosolic fraction obtained from desaturase-induced livers is shown in Figure 1, lane 6, revealing the absence of desaturase in the cytosol fraction.
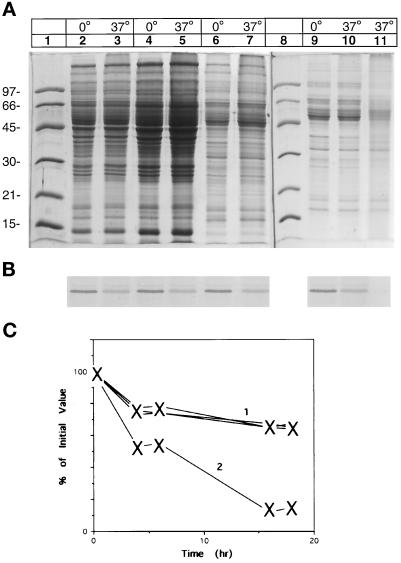
Figure 1.
Identification of desaturase in liver microsomes from rats that have undergone dietary alteration. (A) SDS-PAGE, Coomassie blue-stained gel. Lanes 1 and 7 denote molecular weight standards with the mass marked on the left of the panel. Lane 2, pyrophosphate-washed and lane 5, high-salt–washed microsomes from desaturase induced animals. Lane 3 corresponds to microsomes from control animals; lane 4, purified desaturase; lane 6, postmicrosomal supernatant from desaturase-induced liver. (B) Immunoblot analysis of desaturase in the above fractions. Samples of identical amounts to those shown in Figure 1A were electrophoresed on SDS-PAGE, transferred onto polyvinylidene difluoride Immobilon-P membrane, and incubated with antibody against desaturase, followed by incubation with secondary antibody (anti-rabbit IgG-alkaline phosphatase). The bands were visualized using phosphatase substrate detection kit as described in MATERIALS AND METHODS.
Association of Desaturase with the Particulate Cellular Fractions
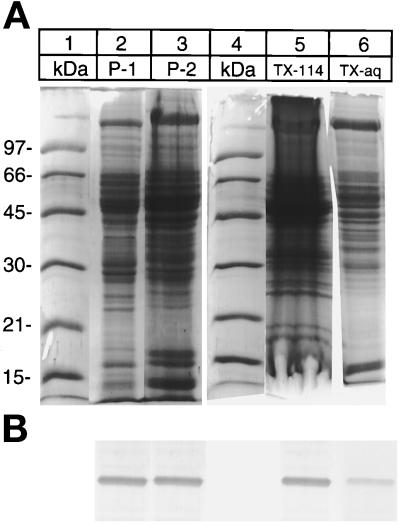
Differential centrifugation of the liver homogenate showed that desaturase is present in the subcellular fractions P-1 and P-2 as well as microsomes. Desaturase activity measurements in the P-1 and P-2 fractions indicated the presence of substantial amounts of the enzyme (Table 1). The total activity of desaturase in these fractions was estimated to be about 25% that of the microsomes. Figure 2 shows the immunoblot analysis of the desaturase in the P-1 fraction. When subcellular fractionation was repeated on the P-1 and P-2 fractions, the desaturase was again present in both the low- and the high-speed sedimenting pellet, but was absent in all of the high-speed supernatants. Partitioning of proteins in Triton X-114 phase has been used to resolve membrane proteins from the soluble proteins (Barrett, 1981). Triton X-114 is a nonionic detergent that forms small micelles at low temperature and large micelles above the cloud point temperature. Phase separation of P-2 fraction proteins in Triton X-114 is shown in Figure 2, lanes 5 and 6. Most of the desaturase partitioned in the detergent phase, although some was present in the aqueous supernatant phase.
Table 1.
Desaturase activities of hepatic subcellular fractions from animals induced for the enzyme
| Fraction | Specific Activity (nmoles/min/mg) | % |
|---|---|---|
| Microsomes | 2.5 to 2.8 | 100 |
| P-1 | 0.3 to 0.5 | 12 |
| P-2 | 0.2 to 0.8 | 15 |
| Microsomes (high-salt washed) | 2.5 to 2.8 | 100 |
| Cytosol | NDa | |
| High-salt wash | NDa |
Cell fractionation and desaturase assay were performed as described in MATERIALS AND METHODS.
ND, not detected.
Figure 2.
Association of desaturase with the components of subcellular fractions. Lane 2, P-1 (800 × g); lane 3, P-2 (10,000 × g) fractions. Lanes 5 and 6, Triton X-114 extract of P-2 fraction: lane 5, detergent-rich phase; lane 6, detergent-depleted phase. (A) Coomassie Blue-stained gel. (B) The above gel immunoblotted with antidesaturase antibody as in Figure 1B.
Structure of Desaturase in the P-1 and P-2 Material
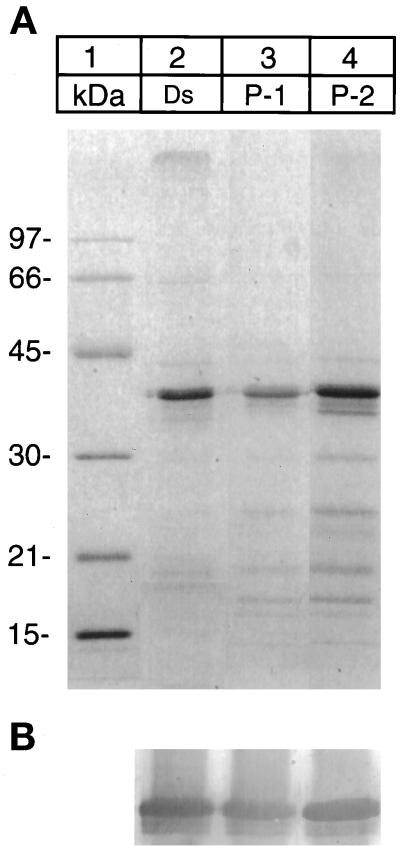
To define the relationship between the microsomal and the desaturase present in the nuclear fraction, isolation, SDS-PAGE, and the N-terminal sequence analysis of the two desaturase preparations were performed. The SDS-PAGE of the two preparations were indistinguishable (Figure 3). The N terminus of the enzyme from microsomes was blocked, but sequence analysis of the desaturase in the nuclear fractions P-1 and P-2 indicated the N terminus to be open and identical to residues 3–10 of the microsomal enzyme (Table 2).
Figure 3.
SDS-PAGE of desaturase preparations isolated from microsomes, P-1, and P-2 fractions. (A) Coomassie Blue-stained gel: lane 2, desaturase (Ds) isolated from microsomes; lane 3, desaturase isolated from P-1 fraction; and lane 4, from P-2 fraction. (B) Immunoblot of the above gel with the antidesaturase antibody as in Figure 1B.
Table 2.
N-terminal sequence analysis of desaturase in microsomes and fractions P-1/P-2
| Cycle/ position | Expected from cDNA | Microsomal | Fraction P1/P2
|
|
|---|---|---|---|---|
| Cycle | Residue (pmol) | |||
| 1 | Met | Blocked | ||
| 2 | Pro | |||
| 3 | Ala | |||
| 4 | His | 1 His | 95 | |
| 5 | Met | 2 Met | 300 | |
| 6 | Leu | 3 Leu | 230 | |
| 7 | Gln | 4 Gln | 250 | |
| 8 | Glu | 5 Glu | 250 | |
| 9 | Ile | 6 Ile | 200 | |
| 10 | Ser | 7 Ser | 50 | |
| 11 | Ser | 8 Ser | 60 | |
Selective Degradation of Desaturase Occurs in Microsomes and in P-1, P-2 Fractions
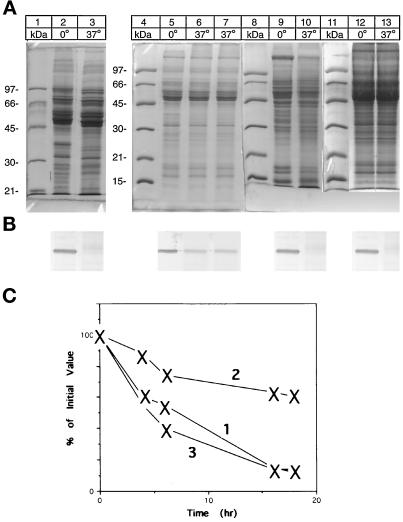
Incubation of microsomes at 37°C led to the disappearance of the desaturase as evidenced by the Coomassie blue staining and by the immunoblot (Figure 4, lanes 2 and 3). Incubation of high-salt–washed microsomes at 37°C led to an incomplete degradation of the desaturase band upon the SDS-PAGE, confirmed on Western blot (Figure 4, lanes 5 and 6). A major pathway for the intracellular degradation of proteins is an ATP-dependent reaction (Hershko and Ciechanover, 1992). Addition of ATP and Mg2+ (5 mM) to the incubation reaction did not enhance the desaturase degradation in high-salt–washed microsomes (Figure 4, lane 7). The degradation of desaturase in the P-1 or P-2 fractions was complete without supplementation, but fractions solubilized with Triton X-114 failed to be degraded under the standard incubation conditions.
Figure 4.
Selective degradation of desaturase in subcellular fractions. Aliquots of the various subcellular fractions (10–15 μg) were incubated overnight at 0 and 37°C in 50 μl of 50 mM Tris-acetate, pH 7.9 containing 50 mM KCl. Before SDS-PAGE, 75 μl of gel loading buffer were added, and a 10-μl aliquot was loaded to each lane of duplicate gels. (A) Coomassie Blue-stained gels; (B) Western Blot of the above gels with antidesaturase antibody. Lanes 2 and 3, pyrophosphate-washed microsomes; lanes 5 and 6, high-salt–washed microsomes; lane 7, high-salt–washed microsomes supplemented with ATP (2 mM) and Mg2+ (5 mM). Lanes 9 and 10, P-1 fraction incubated at 0 and 37°C; lanes 12 and 13, high-salt–washed microsomes supplemented with P-1 fraction and incubated at 0 and 37°C. (C) Estimated time course of the degradation of the desaturase. Aliquots of the various subcellular fractions were incubated at 37°C and control fractions at −20°C for the indicated time and interacted with desaturase antibody. Curve 1, desaturase-induced microsomes. Curve 2, high-salt–washed microsomes or high-salt–washed microsomes supplemented with ATP. Curve 3, high-salt– washed microsomes supplemented with P-1 fraction. The amount of desaturase was determined by densitometry and is expressed as a percentage of the amount present at the start of the experiment, which is set to 100%.
To further explore the incomplete desaturase degradation in the high-salt–washed microsomes, an aliquot of P-1 material was added to the salt-washed microsomal preparation before the incubation. As seen in Figure 4, lanes 12 and 13, when the quantity of desaturase in Figure 4, lane 5, was coincubated with the amount of P-1 fraction represented by lane 9, a complete desaturase degradation was observed. By comparison, the cytosolic fraction was unable to enhance the degradation in high-salt–washed microsomes, even at levels of 10 fold concentration (Figure 5, lanes 4 and 5). Addition of a 10-fold concentrated high-salt wash supernatant also failed to affect the desaturase degradation (Figure 5, lanes 6 and 7). Complete degradation of desaturase in the high-salt–washed microsomes could be restored by the addition of liposomes, cytochrome b5, and its reductase, components essential for the desaturase reaction (Figure 5, lanes 9, 10, and 11). The time course of the degradation of the desaturase under various conditions is presented in Figures 4C and 5C. The time course of desaturase degradation in high-salt–washed microsomes supplemented with P-1 fraction is similar to that observed in intact microsomes (Figure 4C, curves 1 and 3). In contrast, in high-salt– washed microsomes only some 30–40% of the desaturase is degraded (Figure 4C, curve 2). Supplementation of the high-salt–washed microsomes with cytosol or the high-salt wash fraction does not increase the extent of the degradation, (Figure 5C, curve 1). Whereas addition of the components of the desaturase system to the high-salt–washed microsomes yields a degradation rate similar to that observed with the intact microsomes (Figure 5C, curve 2 and Figure 4C, curve 1).
Figure 5.
Proteolysis of desaturase in the high-salt–washed microsomes cannot be reconstituted by the addition of cytosol or high-salt wash supernatant but can be restored by supplementation with lipids and cytochrome b5 reductase. Lanes 2 and 3, high-salt–washed microsomes (5 μg) were supplemented with an aliquot of cytosol (10 μg). Lanes 4 and 5, high-salt–washed microsomes supplemented with an aliquot of concentrated cytosol (20 μg) or concentrated high-salt wash (7 μg), lanes 6 and 7. Lane 9, high-salt–washed microsomes; lane 10, high-salt–washed microsomes with added lipids; lane 11, high-salt–washed microsomes with added lipids and cytochrome b5 reductase. Samples of the incubation mixtures were run on duplicate gels. (A) Coomassie Blue-stained gel. (B) Immunoblot of the above gel with antidesaturase antibody. (C) Estimated time course of the degradation of the desaturase. Aliquots of the various fractions were incubated and quantitated as described in the legend of Figure 4C. Curve 1, high-salt–washed microsomes supplemented with cytosol, or aliquot containing high-salt wash. Curve 2, high-salt–washed microsomes with added components essential for the reconstitution of the desaturase reaction. The amount of desaturase was determined by densitometry and is expressed as a percentage of the amount present at the start of the experiment, which is set to 100%.
Selective Degradation of Desaturase Is Not Inhibited by Lysosomal or Calpain Inhibitors
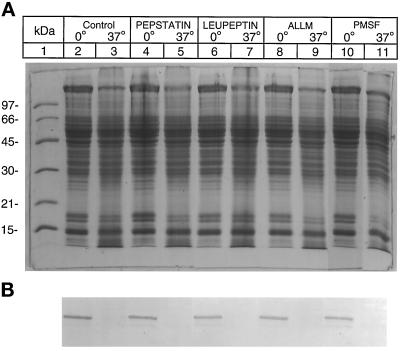
To test whether lysosomal enzymes are involved in the desaturase degradation, several lysosomal protease inhibitors were examined. No inhibition of desaturase degradation was observed with chlorquine, leupeptin, pepstatin, ALLM, or PMSF. Leupeptin and pepstatin inhibits hepatic cathepsin B and cathepsin D, respectively (Barrett, 1971). ALLM is a synthetic peptide inhibiting the activity of cathepsins B and L and calpains (Rock et al., 1994). Figure 6 shows the lack of inhibition of microsomal desaturase degradation by leupeptin and pepstatin. A similar lack of inhibition of desaturase degradation was also observed in our unpublished results when the high-salt–washed microsomes were incubated with the above inhibitors. To explore whether lysosomal proteases were responsible for desaturase degradation in the nuclear associated membranes, the above inhibitors were incubated with P-1 or P-2 fraction. As seen in Figure 7, pepstatin (180 μg/ml), leupeptin (200 μg/ml), ALLM (170 μg/ml), or PMSF (1 mM) also failed to block the degradation of desaturase in the P-2 fraction.
Figure 6.
Microsomal degradation of desaturase is not inhibited by pepstatin or leupeptin. Experimental conditions were as in Figure 4 except that pepstatin (240 μM) was present in reaction mixtures represented by lanes 4 and 5 and leupeptin (400 μM) by lanes 6 and 7. (A) Coomassie Blue-stained gel. (B) Immunoblot of the above gel with antidesaturase antibody as in Figure 1B.
Figure 7.
Degradation of desaturase in P-2 fraction is not blocked by inhibitors of lysosomal enzymes or serine proteases. Experimental conditions were as in Figure 4, except for supplementation with the protease inhibitors. Lanes 4 and 5 represent incubation mixtures containing 240 μM pepstatin; lanes 5 and 7, 400 μM leupeptin; lanes 8 and 9, 420 μM ALLM; and lanes 10 and 11, supplemented with 1 mM PMSF. Control incubation mixtures contained 1.5% dimethylsulfoxide. (A) Coomassie Blue-stained gel. (B) Immunoblot of the above gel with desaturase antibody.
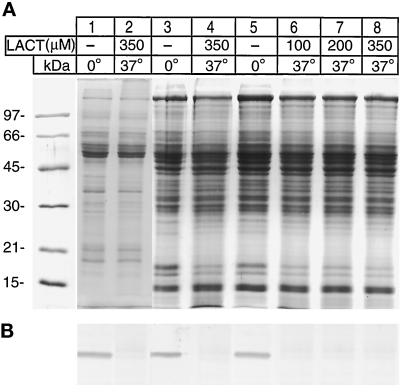
Lactacystin Does Not Block the Degradation of Desaturase in Microsomes or P-1, P-2 Fractions
Currently, the degradation of rapid turnover of cellular proteins is thought to involve the proteosome system, which is the major proteolytic activity in both the cytosol and the nucleus (Tanaka et al., 1986). Very recently, lactacystin, a Streptomyces metabolite, was demonstrated to be a highly specific inhibitor of multiple proteosome activities (Fenteany et al, 1995). In view of the profound effect of lactacystin on protein processing, the influence of this reagent on the desaturase degradation in microsomes and the nuclear fractions was examined. The results of such an experiment on the desaturase degradation in microsomes and P-2 fractions are shown in Figure 8, lanes 2, 4, 6, 7, and 8. Clearly, lactacystin caused no inhibition of the desaturase degradation.
Figure 8.
Proteosome inhibitor lactacystin does not inhibit desaturase degradation. Experimental conditions were as in Figure 4 except for addition of lactacystin in the indicated sample lanes. The incubation temperature and the final concentration of lactacystin in the samples are shown at the top of the panel. Lanes 1 and 2 represent incubation mixtures containing 10–15 μg of microsomes. Lanes 3 and 4, microsomes and P-2 fraction; lanes 5–8, samples containing 10–15 μg of P-2 fraction. Before the SDS-PAGE, 125 μl of the loading buffer containing SDS, β-mercaptoethanol, glycerol, and tracking dye were added, and 10-μl samples were run on duplicate gels. (A) Coomassie Blue-stained gel. (B) Immunoblot of the above gel with desaturase antibody.
DISCUSSION
It is now recognized that the endoplasmic reticulum is an important site for intracellular protein breakdown (for review, see Klausner and Sitia, 1990; Bonifacino and Klausner, 1994). Short-lived proteins, unassembled components of oligomeric complexes such as the T-cell receptor subunits (Wileman et al., 1993), and asialoglycoprotein receptors (Wikstrom and Lodish, 1993) are degraded in the endoplasmic reticulum. Apolipoprotein B-like proteins (Furukawa et al., 1992) and the protein product of the cystic fibrosis-associated gene (CFTR) are degraded in the endoplasmic reticulum (Jensen et al., 1995; Ward et al., 1995). Although the list of proteins degraded in this organelle tends to grow, the proteolytic machinery of endoplasmic reticulum is poorly understood, and the responsible enzymes have not been identified.
Previously, we describe the isolation, cDNA sequence, and bacterial expression of rat liver desaturase (Thiede et al., 1986; Strittmatter et al., 1988). The results described in the current report provide a glimpse of the proteolytic processing of this short-lived membrane protein. As seen in Figure 1, fasting and refeeding a fat-free, high-carbohydrate diet induced high levels of desaturase in the liver microsomal membranes. When the dietary regimen was stopped, desaturase levels rapidly decreased to levels not detectable by immunoblots (Figure 1, lane 3). An in vivo half-life of about 2 h has been estimated for the desaturase (Oshino and Sato, 1972).
Surprisingly, a considerable amount of desaturase was present in the subcellular fractions P-1 and P-2, in addition to being present in the microsomes (Table 1). The postmicrosomal supernatant or the high-salt wash of the microsomes did not contain desaturase (Figure 1, lane 6). While the P-1 fraction consisted essentially of nuclear material, P-2 subcellular fraction was heterogeneous organelle preparation. The outer nuclear membrane of hepatocytes is continuous with the endoplasmic reticulum, implying that some of the desaturase-containing membranes may traffick to the nuclear membrane. The amount of desaturase associated with P-1 and P-2 subcellular material was significant (Table 1) and unanticipated.
To determine the relationship between the microsomal and putative nuclear enzyme present in the P-1, P-2 subcellular fractions, desaturase from the latter fractions was purified for sequence analysis. N-terminal sequence analysis of the P-1, P-2 preparations showed an absence of three residues present in the microsomal protein, whereas the microsomal enzyme has a blocked N terminus (Table 2). Thus, the desaturase present in the nuclear fraction does not represent microsomal enzyme contamination, but appears to represent a specifically processed form of the enzyme. The relationship between the N-terminal processing and the nuclear localization remains to be elucidated, as discussed below.
Incubation of the microsomal membranes at 37°C resulted in the complete degradation of the desaturase, whereas in microsomes washed with a high-salt buffer, the degradation was incomplete as seen in Figure 4. The complete desaturase degradation, however, could not be restored by the addition of concentrated cytosol or high-salt wash fractions to the microsomes. Complete degradation of desaturase in the high-salt–washed microsomes could be restored by the addition of lipids, cytochrome b5 and its reductase, which constitutes functional desaturase activity. In these experiments, omission of lipid or any of the protein components limited proteolysis. It appears that in high-salt–washed microsomes, only 30–40% of the desaturase is degraded (Figure 4C). Although the possible effects of the salt wash on the desaturase degradation could be the result of many factors, one explanation may involve the formation of high-salt wash–induced conformations in the desaturase population that are resistant to the protease action. The data in Figure 5C suggest that supplementation of salt-washed microsomes with the lipid, cytochrome b5 reductase, and cytochrome b5 renders the resistant form of the desaturase to further proteolysis. The formation of insoluble desaturase aggregates have been observed during centrifugation on glycerol gradient detergent-solubilized microsomal preparations in the presence of high salt. Some of these forms retain enzymatic activity whereas others do not. Notwithstanding, the degradation reconstitution experiments imply that the procedure used to reconstitute an enzymatically active desaturase system may also yield to protein conformations that are susceptible to the proteolysis of the enzyme.
Desaturase in P-1 or P-2 fractions was degraded completely. Premixing of the salt-washed microsomes with the P-1, P-2 fraction also resulted in a complete degradation of the desaturase. Desaturase antigen bands of lower molecular mass than the desaturase could not be detected in the degradation mixtures, although the desaturase antibody can recognize the bacterial synthesis product lacking some 30 residues from the N terminus, corresponding to a decrease of 3000–5000 Da (Strittmatter et al., 1988).
Hepatic lysosomal or endosomal proteases or their precursors are ubiquitous enzymes, and their presence in microsomes and in P-1 and P-2 fractions would not be surprising, since proteases such as the procathepsin B and L may exist in the microsomal membranes as latent precursors. To determine whether lysosomal proteases are involved in the desaturase degradation, several types of protease inhibitors were examined. Leupeptin and pepstatin, inhibitors of lysosomal and endosomal proteases, had no effect on the microsomal or the P-1, P-2 desaturase degradation (Figures 6 and 7). The cysteine protease inhibitor ALLM and the serine protease inhibitor PSMF were also ineffective in blocking the desaturase degradation (Figure 7). ALLM has been shown to inhibit the regulated degradation of microsomal HMG-CoA reductase (Inoue et al., 1991), and a serine protease has been implicated in the rapid degradation of unassembled Ig light chains in endoplasmic reticulum (Gardner et al., 1993). One nonlysosomal pathway present in the cytoplasm and nuclear components that mediates rapid elimination of proteins is the proteosome pathway. Multiple types of evidence suggest that the proteosome plays a key role in the processing of antigens for the major histocompatibility complex class I presentation (Chiechanover, 1994) and is involved in generating the active forms of molecules such as the production of the 50-kDa subunit of the transcription factor NF-κB from the 105-kDa precursor (Palombella et al., 1994). The proteosome is also thought to be responsible for the degradation of the HMG-CoA reductase (McGee et al., 1996) and of the cystic fibrosis gene product (CFTR) in the endoplasmic reticulum (Rock et al., 1994; Ward et al., 1995). The proteosome is a 26S (2000-kDa) complex, containing the 20S proteosome as a key proteolytic component (Rechsteiner et al., 1993; Jentsch and Schlenker, 1995; Lowe et al., 1995). The 20S (700 kDa) complex consists of seven different α-subunits and seven unrelated β-subunits with masses ranging from 24 to 32 kDa comprising about 1% of the protein in mammalian cells (Jentsch and Schlenker, 1995). None of the individual subunits of the proteosome have proteolytic activity or show relationship to any known proteases. Recently, a highly specific, irreversible inhibitor of the proteosome, a Streptomyces metabolite–lactacystin has been identified (Fenteany et al., 1995). Lactacystin modifies covalently the highly conserved N-terminal threonine of the mammalian proteosome subunit X, a close homologue of the LMP7 proteosome subunit encoded by the major histocompatibility complex (Fenteany et al., 1995). Lactacystin has not been found to inhibit any other known protease (Fenteany et al., 1995). In view of such a remarkable housekeeping proteolytic function of the proteosome, it was of interest to determine whether the proteosome is involved in the degradation of desaturase. Lactacystin had no effect on the microsomal desaturase degradation (Figure 8). The experiment of Figure 8 also shows that lactacystin (100–350 μM) also failed to inhibit the desaturase degradation in the P-1 and P-2 subcellular fractions.
The studies reported here show that degradation of desaturase occurs in several subcellular fractions isolated by differential centrifugation. The degradation of desaturase, however, was insensitive to the lysosomal and proteosome inhibitors. If the lysosomal proteases or proteosome do not play a significant role in the desaturase degradation, what alternatives do we have to explain the desaturase degradation? Proteolytic activities such as the ER-60 protease have been detected in detergent-solubilized microsomal preparations (Otsu et al., 1995). The proteolytic activity of ER-60, however, is inhibited by leupeptin and ALLM (Otsu et al., 1995).
The observation that desaturase is present and readily degraded in subcellular fractions other than the microsomes implies that degradation of native desaturase may also involve targeting of the enzyme to compartments containing specific proteolytic machinery, which constitute a sorting pathway or the reverse process of protein targeting to the membranes. Of interest is that the amino acid sequence of desaturase has two segments that contain a potential nuclear localization sequence (NLS). In residues 33–36, Lys-Met-Lys-Lys and Arg-Lys-Lys-Val-Ser-Lys, residues 335–340 constitute potential consensus sequences for the import of proteins to the nucleus. Import of proteins to the nuclear pore complex is specified by short stretches of amino acids known as the NLSs (see review in Melchior and Gerace, 1995; Gorlich and Mattaj, 1996). Site-directed mutagenesis of desaturase in the two putative NLS segments should clarify the significance of this finding. Are posttranslation modifications involved in this process? Structure analysis of desaturase in the P-1 and P-2 fractions showed that it lacked three residues at the N terminus (Figure 2 and Table 2). The cDNA sequence predicts a Met-Pro-Ala sequence at the N terminus of the microsomal desaturase (Strittmatter et al., 1988). The N terminus of the enzyme present in microsomes is blocked, and the nature of the blocking group remains to be determined. The N-terminal–blocking groups of the two upstream essential components of the desaturase pathway, cytochrome b5 and its reductase, are an acetyl and myristoyl residue, respectively (Ozols et al., 1984; Ozols, 1989). The presence of a myristoylated residue at the N terminus of desaturase is unlikely because of the absence of a consensus Gly residue in the proximity of its N terminus. The removal of an N-blocked terminus and ProAla segment from the native desaturase is of interest because we are not aware of any reports on hepatic aminopeptidases capable of cleaving residues from N-acetylated proteins. The hepatic acylpeptide hydrolase (E.C.3.4.19.1) acts only on N-acetylated peptides that are shorter than 10 to 15 residues (Tsunasawa et al., 1983). Cathepsins that function as aminopeptidases act only on proteins with a free N terminus.
The complete degradation of desaturase in microsomes can be inhibited by a high-salt wash of the microsomes. This inhibition cannot be restored by the addition of the proteins present in the high-salt wash fraction. The partial degradation of desaturase in high-salt–washed microsomes, however, could be restored by the addition of the components essential for the in vitro catalytic activity of the desaturase. This finding implies that desaturase degradation system may necessitate a specific membrane protein assembly, similar to that observed in reconstitution of the desaturase catalytic activity in vitro. In summary, degradation of the microsomal membrane desaturase was demonstrated in this study. This specific degradation may involve several degradation pathways including removal of the N-terminal residues and the targeting of the modified desaturase to cellular components such as the nuclear material. The possibility that a short-lived protein can be degraded according to different pathways, however, would be unprecedented. Whether the removal of the N-terminal residues from the desaturase results in the formation of a specific determinant that acts as a mediator for the observed trafficking remains to be investigated.
ACKNOWLEDGMENTS
I am grateful to George Korza for his outstanding technical assistance. I thank Professor E.J. Corey, of Harvard University, for providing lactacystin. This work was supported by grant R01-Gm-26351 from The National Institutes of Health.
REFERENCES
- Barrett AJ. Cystatin, the egg white inhibitor of cysteine proteinases. Methods Enzymol. 1981;80:771–778. [Google Scholar]
- Bonifacino JS, Klausner RD. Degradation of proteins retained in the endoplasmic reticulum. In: Ciechanover AJ, Schwartz AL, editors. Cellular Proteolytic Systems. New York: Wiley-Liss; 1994. [Google Scholar]
- Chiechanover The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- Fleischer S, Kervina M. Subcellular fractionation of rat liver. Methods Enzymol. 1974;31:6–41. doi: 10.1016/0076-6879(74)31005-1. [DOI] [PubMed] [Google Scholar]
- Furukawa S, Sakata N, Ginsbergh N, Dixon JL. Studies of the sites of intracellular degradation of apolipoprotein B in Hep G2 cells. J Biol Chem. 1992;267:22630–22638. [PubMed] [Google Scholar]
- Gardner AM, Aviel S, Argon Y. Rapid degradation of an unassembled immunoglobulin light chain is mediated by a serine protease and occurs in a pre-Golgi compartment. J Biol Chem. 1993;268:25940–25947. [PubMed] [Google Scholar]
- Gorlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Hardy RR. Purification and characterization of monoclonal antibodies. In: Weir DM, editor. Handbook of Experimental Immunology, vol. 1: Immunochemistry. Oxford, UK: Blackwell Scientific; 1986. pp. 13.1–13.13. [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Inoue S, Simoni RD. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase and T cell receptor alpha subunit are differentially degraded in the endoplasmic reticulum. J Biol Chem. 1992;267:9080–9086. [PubMed] [Google Scholar]
- Inoue S, Bar-Nun S, Roitelman J, Simoni RD. Inhibition of degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase in vivo by cysteine protease inhibitors. J Biol Chem. 1991;266:13311–13317. [PubMed] [Google Scholar]
- Jensen TJ, Loo MA, Pind S, Williams DB, Goldberg AL, Riordan JR. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- Jentsch S, Schlenker S. Selective protein degradation: a journey’s end within the proteasome. Cell. 1995;82:881–884. doi: 10.1016/0092-8674(95)90021-7. [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Ntambi JM, Kelly TJ, Jr, Lane MD. Differentiation-induced gene expression in 3T3–L1 preadipocytes. A second differentially expressed gene encoding stearyl-CoA desaturase. J Biol Chem. 1989;264:14755–14761. [PubMed] [Google Scholar]
- Klausner DR, Sitia R. Protein degradation in the endoplasmic reticulum. Cell. 1990;61:611–614. doi: 10.1016/0092-8674(90)90104-m. [DOI] [PubMed] [Google Scholar]
- Lowe J, Stock D, Jap B, Swickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- McGee TP, Cheng HH, Kumagai H, Omura S, Simoni RD. Degradation of 3-hydroxy-3-methylglutaryl-CoA reductase in endoplasmic reticulum membranes is accelerated as a result of increased susceptibility to proteolysis. J Biol Chem. 1996;271:25630–25638. doi: 10.1074/jbc.271.41.25630. [DOI] [PubMed] [Google Scholar]
- Melchior F, Gerace L. Mechanisms of nuclear protein import. Curr Biol. 1995;7:310–318. doi: 10.1016/0955-0674(95)80084-0. [DOI] [PubMed] [Google Scholar]
- Oshino N, Sato R. The dietary control of the microsomal stearyl CoA desaturation enzyme system in rat liver. Arch Biochem Biophys. 1972;149:369–377. doi: 10.1016/0003-9861(72)90335-9. [DOI] [PubMed] [Google Scholar]
- Otsu M, Urade R, Kito M, Omura F, Kikuchi M. A possible role of ER-60 protease in the degradation of misfolded proteins in the endoplasmic reticulum. J Biol Chem. 1995;270:14958–14961. doi: 10.1074/jbc.270.25.14958. [DOI] [PubMed] [Google Scholar]
- Ozols J. Cytochrome b5 from microsomal membranes of equine, bovine, and porcine livers. Isolation and properties of preparations containing the membranous segment. Biochemistry. 1974;13:426–434. doi: 10.1021/bi00700a005. [DOI] [PubMed] [Google Scholar]
- Ozols J. Structure of cytochrome b5 and its topology in the microsomal membrane. Biochim Biophys Acta. 1989;997:121–130. doi: 10.1016/0167-4838(89)90143-x. [DOI] [PubMed] [Google Scholar]
- Ozols J. Subcellular fractionation of rat liver. Methods Enzymol. 1990;182:225–235. [Google Scholar]
- Ozols J, Carr SA, Strittmatter P. Identification of the NH2-terminal blocking group of NADH-cytochrome b5 reductase as myristic acid and the complete amino acid sequence of the membrane-binding domain. J Biol Chem. 1984;259:13349–13354. [PubMed] [Google Scholar]
- Palombella VJ, Rando AL, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Hoffmann L, Dubiel W. The multicatalytic and 26 S proteases. J Biol Chem. 1993;268:6065–6068. [PubMed] [Google Scholar]
- Rock LK, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Shanklin J, Somerville C. Stearoyl-acyl-carrier-protein desaturase from higher plants is structurally unrelated to the animal and fungal homologs. Proc Natl Acad Sci USA. 1991;88:2510–2514. doi: 10.1073/pnas.88.6.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter P, Kittler JM, Coghill JE, Ozols J. Interaction of non-myristoylated NADH-cytochrome b5 reductase with cytochrome b5-dimyristoylphosphatidylcholine vesicles. J Biol Chem. 1993;268:23168–13171. [PubMed] [Google Scholar]
- Strittmatter P, Spatz L, Corcoran D, Rogers MJ, Setlow B, Redline R. Purification and properties of rat liver microsomal stearyl coenzyme A desaturase. Proc Natl Acad Sci USA. 1974;71:4565–4569. doi: 10.1073/pnas.71.11.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter P, Thiede MA, Hackett CS, Ozols J. Bacterial synthesis of active rat stearyl-CoA desaturase lacking the 26-residue amino-terminal amino acid sequence. J Biol Chem. 1988;263:2532–2535. [PubMed] [Google Scholar]
- Stukey JE, McDonough VM, Martin CE. The OLE1 gene of Saccharomyces cerevisiae encodes the delta 9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J Biol Chem. 1990;265:20144–20149. [PubMed] [Google Scholar]
- Tanaka K, Ii K, Ichihara A, Waxman L, Goldberg AL. A high molecular weight protease in the cytosol of rat liver. I. Purification, enzymological properties, and tissue distribution. J Biol Chem. 1986;261:15197–15203. [PubMed] [Google Scholar]
- Thiede MA, Ozols J, Strittmatter P. Construction and sequence of cDNA for rat liver stearyl coenzyme A desaturase. J Biol Chem. 1986;261:13230–13235. [PubMed] [Google Scholar]
- Thompson GA, Scherer DE, Foxall-Van Aken S, Kenny JW, Young HL, Shintani DK, Kridl JC, Knauf VC. Primary structures of the precursor and mature forms of stearoyl-acyl carrier protein desaturase from safflower embryos and requirement of ferredoxin for enzyme activity. Proc Natl Acad Sci USA. 1991;88:2578–2582. doi: 10.1073/pnas.88.6.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiku PE, Gracey AY, Macartney AI, Beynon RJ, Cossins AR. Cold-induced expression of delta 9-desaturase in carp by transcriptional and posttranscriptional mechanisms. Science. 1996;271:815–818. doi: 10.1126/science.271.5250.815. [DOI] [PubMed] [Google Scholar]
- Toubin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polylacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunasawa S, Imanaka T, Nakazawa T. Apparent dipeptidyl peptidase activities of acylamino acid-releasing enzymes. J Biochem. 1983;93:1217–1220. doi: 10.1093/oxfordjournals.jbchem.a134248. [DOI] [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Wikstrom L, Lodish HF. Endoplasmic reticulum degradation of a subunit of the asialoglycoprotein receptor in vitro. Vesicular transport from endoplasmic reticulum is unnecessary. J Biol Chem. 1992;267:5–8. [PubMed] [Google Scholar]
- Wikstrom L, Lodish HF. Unfolded H2b asialoglycoprotein receptor subunit polypeptides are selectively degraded within the endoplasmic reticulum. J Biol Chem. 1993;268:14412–14416. [PubMed] [Google Scholar]
- Wileman T, Kane LP, Young J, Carson GR, Terhorst C. Associations between subunit ectodomains promote T cell antigen receptor assembly and protect against degradation in the ER. J Cell Biol. 1993;122:67–78. doi: 10.1083/jcb.122.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]