Abstract
Pulmonary gas exchange relies on a rich capillary network, which, together with alveolar epithelial type I and II cells, form alveolar septa, the functional units in the lung. Alveolar capillary endothelial cells are critical in maintaining alveolar structure, because disruption of endothelial cell integrity underlies several lung diseases. Here we show that targeted ablation of lung capillary endothelial cells recapitulates the cellular events involved in cigarette smoke-induced emphysema, one of the most prevalent nonneoplastic lung diseases. Based on phage library screening on an immortalized lung endothelial cell line, we identified a lung endothelial cell-binding peptide, which preferentially homes to lung blood vessels. This peptide fused to a proapoptotic motif specifically induced programmed cell death of lung endothelial cells in vitro as well as targeted apoptosis of the lung microcirculation in vivo. As early as 4 days following peptide administration, mice developed air space enlargement associated with enhanced oxidative stress, influx of macrophages, and up-regulation of ceramide. Given that these are all critical elements of the corresponding human emphysema caused by cigarette smoke, these data provide evidence for a central role for the alveolar endothelial cells in the maintenance of lung structure and of endothelial cell apoptosis in the pathogenesis of emphysema-like changes. Thus, our data enable the generation of a convenient mouse model of human emphysema. Finally, combinatorial screenings on immortalized cells followed by in vivo targeting establishes an experimental framework for discovery and validation of additional ligand-directed pharmacodelivery systems.
The alveolar septum is the critical lung functional unit involved in gas exchange. This unit is composed by a virtual cellular syncytium comprised by types I and II epithelial cells (1), which form the interface between air and blood flowing within an extensive capillary network (roughly the size of a tennis court) lined by endothelial cells. Disruption of alveolar integrity occurs in aging and in several lung diseases, particularly in emphysema, a highly prevalent pulmonary disease. Pulmonary emphysema is a chronic and incurable disease in which the air spaces distal to the terminal bronchiole are abnormally and permanently enlarged due to the destruction of their walls (2), leading to airflow limitation and impaired blood oxygenation. Historically, emphysema has been linked to an excessive lung inflammation caused by the chronic inhalation of cigarette smoke and the ensuing protease/anti-protease imbalance (3).
A “vascular hypothesis” for emphysema originated ∼40 years ago based on the observation of scarcity of pulmonary capillaries in remaining alveolar septa in emphysematous human lungs (4). More recently, the reports of emphysema caused by vascular endothelial growth factor (VEGF)5 receptor blockade and Cre-recombinase-mediated lung VEGF deletion uncovered the novel role of alveolar cell apoptosis, including endothelial cells, in the pathogenesis of the disease (5-8). This unique role of VEGF in lung homeostasis relies on the trophic effect of VEGF on alveolar endothelial cells (5) and its differentiation-promoting role on type II alveolar cells (9). We have shown that disruption of VEGF receptor signaling alters the cellular and molecular maintenance program in the lung and sets up a destructive cycle involving the mutual interaction of oxidative stress and apoptosis of alveolar cells (6), under the control of the proapoptotic lipid ceramide (10). These observations were further confirmed with the report of emphysema in mice in which the VEGF gene has been deleted by lung expression of CRE recombinase (8) and our finding of emphysema in mice treated with the combination of VEGF-R1 and -R2 blocked with neutralizing MF1 and DC101 antibodies, respectively (11). Furthermore, clinical studies supported several elements of this novel paradigm, notably that, in comparison with normal lungs, human emphysema lungs have decreased expression of VEGF and VEGF receptors, enhanced alveolar cell apoptosis, and increased levels of proapoptotic lipid ceramide (10-16).
Despite the growing recognition of alveolar cell apoptosis in the pathogenesis of emphysema, the specific contributions of individual alveolar cells (endothelial, epithelial, or myofibroblastic cells) in triggering disruption of alveolar integrity and ultimately governing the process of alveolar cell destruction in emphysema remains unknown. We previously noted that endothelial cell apoptosis predominated in the accelerated emphysema caused by cigarette smoke exposure of mice deficient in the master antioxidant transcription factor NRF-2 (17). However, other reports emphasized the potential role of epithelial cell apoptosis in cigarette smoke-induced emphysema (18). Experimental approaches aimed at identifying the structural and functional roles of the different types of alveolar cells in alveolar maintenance are crucial for understanding the mechanisms involved in lung homeostasis and for designing studies aimed at therapeutic interventions for a wide range of lung diseases.
Emphysema is characterized by a progressive decrease in alveolar gas exchange, possibly as the result of a loss of alveolar capillaries. The potential link between alveolar cell apoptosis and disruption of molecular and cellular signaling involved in alveolar structural maintenance and repair has broader implications in that cigarette smoke-induced alveolar injury may share pathogenetic features in common with alveolar enlargement due to aging (19).
There is growing evidence that endothelial cells have unique biochemical fingerprints that account for tissue- and/or blood vessel-specific biochemical heterogeneity of the vascular endothelium (20, 21). Recent proteomic approaches have provided substantial evidence that such premises apply to lung endothelial cells (22). The introduction of combinatorial approaches has enabled unbiased probing of cell surfaces for differentially expressed membrane-associated receptors and their homing ligands in functional screenings (21, 23-25). Given the phenotypic attributes of lung vasculature, we hypothesized that the expression of unique lung endothelial cell-specific peptide ligands will serve as molecular tools to interrogate the role of lung capillary endothelial cells in maintaining in vivo alveolar integrity.
Here we used ImmortoMouse-derived lung endothelial cells to screen a phage display random peptide library, since these immortalized cells retain several important molecular features corresponding to the tissue of origin (26, 27). These endothelial cells are part of a panel of tissue-specific microvascular endothelial cell lines from the H-2Kb-tsA58 mouse (termed ImmortoMouse) (27); such endothelial cells harbor a temperature sensitive SV40 large T antigen under the major histocompatibility complex H-2Kb promoter and are conditionally immortal when cultured under permissive temperatures (26, 27).
In the present work, we postulated that lung endothelial cells have unique cell surface molecular characteristics, distinct from other vascular beds, that allows for selection of specific peptides using a phage display library. Furthermore, this unique peptide would permit organ-specific in vivo targeting with a proapoptotic agent to address whether selective alveolar capillary endothelial cell apoptosis triggers the seminal features seen in cigarette smoke-induced emphysema. The combination of in vitro screening and in vivo validation presented here not only enables one to study the specific role of lung endothelial cell apoptosis in the pathogenesis of emphysema but also provides an experimental blueprint to development of a ligand-directed targeted pharmacology in other systems.
MATERIALS AND METHODS
Animals—Institutional animal care and utilization committees of the University of Texas M. D. Anderson Cancer Center and at The Johns Hopkins University approved all animal experiments. BALB/c and C57Bl6/J mice were purchased from Jackson Laboratory, and H-2Kb-tsA58 mice were purchased from Charles River Breeding Laboratories.
Cell Culture and Synthetic Peptides—H-2Kb-tsA58 mouse microvascular endothelial cell lines derived from bone marrow, brain, kidney, lung, and prostate (27) were cultured in 10% fetal bovine serum (FBS) in Dulbecco's modified Eagle's medium (DMEM) at 33 °C. Cells were cultured at 37 °C for at least 48 h to induce a quiescent state. The synthetic peptides CGSPGWVRC, CGSPGWVRC-GG-D(KLAKLAK)2, and D(KLAKLAK)2 were obtained commercially (AnaSpec; San Jose, CA) to our specifications. In some binding experiments, as indicated, a synthetic cyclic peptide (sequence CARAC) served as an unrelated negative control. For in vivo peptide homing validation, biotin-conjugated CGSPGWVRC and CARAC were also obtained.
Cell Surface Binding Assay—We used the BRASIL method (24) as described. Briefly, for the screening of the phage display random peptide library for lung endothelial cell binding peptides, 109 transducing units (TU) of CX7C phage library (23) was first incubated with 106 brain endothelial cells in 1% bovine serum albumin (BSA) in DMEM for 1 h on ice and centrifuged through organic phase for 10 min at 10,000 × g. The upper aqueous phase was then moved to a new tube with 106 lung endothelial cells and incubated on ice for 2 h. The phage/cell admixture was then centrifuged through the organic phase for 10 min at 10,000 × g. Eppendorf tubes were frozen at -80 °C for at least 10 min, and the bottom of the tube containing the cell pellet was sliced off to a new tube. Cell-bound phage were recovered by Escherichia coli k91kan bacterial infection for 30 min at room temperature, and phage recovery was determined from serial dilution on Luria-Bertani (LB) plates containing tetracycline (40 μg/ml) and kanamycin (100 μg/ml). Recovered phage were amplified in bacterial liquid culture overnight before purification for the subsequent rounds of selection. For the phage binding assays, 109 TU of phage were incubated with 2.5-5 × 105 cells in 1% BSA in DMEM for 2 h on ice and centrifuged through the organic phase for 10 min at 10,000 × g (24).
Cell Internalization Assay—Lung and prostate endothelial cells were grown in tissue chamber slides (Lab-Tek II Chamber Slide System; Nalgene Nunc International Corp., Naperville, IL), washed twice with phosphate-buffered saline (PBS), incubated with 109 TU of CGSPGWVRC phage or control phage in 1% BSA in DMEM at 37 °C, and washed five times with PBS and four times with 150 mm NaCl, 20 mm glycine, pH 2.3, to remove cell surface binding phage. Cells were washed with PBS, fixed with paraformaldehyde in PBS for 15 min, washed with PBS, permeabilized with 0.2% Triton X-100, washed with PBS, and blocked with 1% BSA in PBS. Cells were then incubated with a 1:200 dilution of the anti-M13 bacteriophage antibody (Amersham Biosciences) in 1% BSA in PBS at room temperature for 2 h, washed with PBS, and incubated with a 1:200 dilution of a Cy3-conjugated anti-rabbit secondary antibody in 1% BSA in PBS for 1 h at room temperature. Finally, cells were washed with PBS, fixed with 4% PFA in PBS, mounted, and visualized under a fluorescence microscope.
In Vitro Cell Viability and Cell Apoptosis Assays—Cells (2 × 104 cells/well) were seeded in 96-well plates for 24 h in 10% FBS in DMEM, incubated with increasing concentrations of the peptides CGSPGWVRC-GG-D(KLAKLAK)2 or CGSPGWVRC and D(KLAKLAK)2 in 80 μl of 1% FBS in DMEM for 6 hours at 37 °C. After 6 h, cell viability was measured with a cell proliferation detection reagent according to the manufacturer's instructions (WST-1; Roche Applied Science). All samples were assayed in triplicate. For the detection of cell apoptosis, cells were incubated with 100 μm CGSPGWVRC-GG-D(KLAKLAK)2 or CGSPGWVRC and D(KLAKLAK)2 peptides for 2 h. Cell apoptosis was detected by Annexin-V binding (ApoAlert; BD Biosciences) and terminal deoxynucleotidyl-transferase biotin-dUTP nick end labeling (TUNEL) (DeadEnd fluorometric TUNEL system; Promega, Madison, WI) according to the manufacturer's instructions.
Phage Binding Experiments in Vivo and ex Vivo—Avertin®-anesthetized BALB/c mice (0.015 ml/g) were injected intravenously with either 109 TU of CGSPGWVRC-displaying phage or insertless control fd-tet phage (n = 3 mice for each phage clone). Phage were allowed to circulate for 10 min, and the animals were perfused through the left ventricle of the heart with DMEM. The lung and control organs were dissected and homogenized. Homogenates were washed with ice-cold DMEM containing a protease inhibitor mixture (Sigma) and 1% BSA. Tissue-bound phage were recovered by infecting log phase k91kan bacteria with the tissue homogenates for 1 h at room temperature. Serial dilutions of the infected bacteria were plated on LB plates containing tetracycline (40 μg/ml) and kanamycin (100 μg/ml) to determine the recovered phage titer. Alternatively, anesthetized BALB/c mice were injected intravenously with 1011 TU of CGSPGWVRC-displaying phage or insertless fd-tet phage (n = 3 mice for each phage clone). After 24 h, anesthetized mice were perfused through the left ventricle of the heart with DMEM. Phage were recovered from the tissues as described above. For ex vivo phage binding experiments, lungs or spleen of a PBS-perfused BALB/c mouse were minced and digested with collagenase A (Sigma) for 3 h at 37 °C. More than 50% of the cells in the lung cell suspension were CD31-positive by flow cytometry (data not shown). The cell suspension was washed twice with 10% FBS in DMEM and filtered through a 70-μm cell strainer, resulting in single cell suspension. Lung-derived or spleen-derived cells (∼106) were incubated with CGSPGWVRC-displaying phage or control phage (∼109 TU) for 2 h on ice in 1% FBS in DMEM. The cell/phage admixtures were then centrifuged through the organic phase, and cell-bound phage were recovered and titered as described (24). For in vivo peptide homing validation, anesthetized BALB/c mice were injected intravenously with 0.2 μmol of biotin-conjugated CGSPGWVRC or CARAC peptides (n = 2 mice for each peptide-biotin). After 2 h, anesthetized mice were perfused through the left ventricle of the heart with 4% paraformaldehyde in PBS. Peptide-biotin was detected by streptavidinfluorescein (Amersham Biosciences). For blood vessel localization, tissues were co-stained with an anti-CD31 antibody (MEC 13.3) (BD Pharmingen).
Treatment of Mice with CGSPGWVRC-GG-D(KLAKLAK)2—Adult (3 months old) C57Bl6/6J mice received intraperitoneally normal saline solution, 240 μg of CGSPGWVRC-GG-D(KLAKLAK)2 peptide, or 240 μg of CGSPGWVRC and D(KLAKLAK)2 (separately or a 1:1 mixture as indicated) peptides every other day. Treatment was terminated 4 days (n = 2 mice/treatment group, treated with CGSPGWVRC-GG-D(KLAKLAK)2 or D(KLAKLAK)2), 7 days (for ceramide measurements), or 21 days (n = 5 mice/treatment group, treated with CGSPGWVRC-GG-D(KLAKLAK)2, peptide controls, or saline) after the first peptide administration.
Morphologic and Morphometric Analysis—Mice were anesthetized with halothane (Halocarbon Laboratories; North Augusta, SC), and lungs were inflated with 0.5% low melting agarose at a constant pressure of 25 cm of H2O as described (5). Lungs were then fixed in 10% formalin and paraffin-embedded. Tissue sections (5 μm) were stained with hematoxylin and eosin. Mean linear intercepts were measured by computer-assisted morphometry with Image Pro Plus (Media Cybernetics, Silver Spring, MD; utilized for the day 4 experiment) or Metamorph software (Universal Imagining, Frederick, MD). Lung sections were randomly coded, and representative images (15 per lung section) were acquired by an observer masked for the identity of the lungs, at ×20 lens magnification (6).
TUNEL Assay with Alveolar Epithelial and Endothelial Cell Colocalization—Lung sections (5 μm) from the agarose-inflated lungs were treated by the fluorescein-FragEL DNA fragmentation detection kit (EMD Biosciences, San Diego, CA) for the detection of apoptotic cells. Co-localization for type II epithelial cells (surfactant protein C; SpC) or alveolar endothelial cells (CD34) was performed as described (17). Co-localization of TUNEL within alveolar myofibroblasts was performed with anti-smooth muscle cell α-actin immunofluorescence. In brief, slides were prepared as described above and incubated with the mouse monoclonal anti-smooth muscle cell α-actin antibody (Sigma) at a 1:25 dilution and detected with the Immunogenex (La Jolla, CA) Mouse-on-Mouse kit with Texas Red as the fluorochrome. Quantification of TUNEL-positive cells co-expressing the selective alveolar cell marker was performed in selected images by an observer masked for the identity of the experimental groups. Total TUNEL-positive/4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI)-positive cells were determined using a macro operation run by the Metamorph imaging system.
Detection of Active Caspase-3—Immunohistochemical staining of the active caspase-3 was performed with the active caspase-3-specific antibody (Abcam, Cambridge, MA) on 5-μm lung sections, followed by application of DAPI (Molecular Probes, Inc., Eugene, OR) 1:10,000 from stock in PBS as described (13). Quantification of active caspase-3-positive cells was done on images (n = 15 per lung) captured by an observer masked to the identity of the slides. Specifically, quantification was done with the Metamorph software, by measurements of gray color and total intensity of positive staining, after conversion of the original red-green-blue settings to monochrome images. Reduction of background inequalities was done by using an averaging software algorithm, followed by segmentation of the positive signal and measurements based on a relative intensity scale in which the background (with isotype-matched antibody staining) is set to zero and the most intense immunohistochemical signal is set to 256 (256-gray scale). The results were normalized by the total number of cells present in the same microscopic field detected by subsequent image acquisition with the blue filter for DAPI-positive cells. Western blot analysis was performed for whole lung lysates as described (17). Densitometry data were acquired and analyzed by using ImagePro morphometric software as described.
Detection of Ceramide Levels—Paraffin-embedded lung sections (5 μm) were incubated with monoclonal anti-ceramide antibody (MID 15B4, Alexis; Axxora LLC, San Diego, CA) by using the DAKO Animal Research Kit (DAKO, Carpinteria, CA) immunohistochemistry protocol. Isotype control IgG served as a negative control. Quantification of ceramide expression was performed on coded images as described for active caspase-3 expression. We also used mass spectroscopy to measure ceramide levels, relying on the concomitant use of ceramide standards followed by normalization per total lipid concentration, as described (10). Briefly, tissue lipids were extracted by using the Bligh and Dyer method (28), and the total lipid content was quantified by measuring the total lipid phosphorus (Pi) as described (29). Seven major ceramide species as well as three major molecular species of dihydroceramide were detected via positive multiple reaction monitoring analysis of transitions of the [M + H]+ ion, as described (10).
Immunohistochemical Localization of 8-Oxo-7,8-dihydro-2′-deoxyguanosine (8-Oxo-dG)—Immunohistochemical localization of 8-oxo-dG was performed with an anti-8-oxo-dG monoclonal antibody (Oxis Research, Portland, OR) or with IgG isotype-matched negative control and detected with the Vector Mouse-on-Mouse immunodetection kit (6). Quantification and normalization were performed as described above.
Immunohistochemical Localization of Inflammatory Cells in the Lungs—Macrophages were identified by the rat anti-mouse Mac-3 antibody (BD Pharmingen, San Jose, CA). The number of Mac-3-positive cells in the lung were then normalized by the total number of DAPI-positive alveolar cells.
Statistical Analysis—For in vitro experiments and in vivo phage targeting experiments, results are expressed as mean ± S.E., and their statistical significance was determined by Student's t tests. For the in vivo experiments with the targeted proapoptotic peptide, comparisons among the study groups were assessed by analysis of variance by using the program SigmaStat (SPSS Inc., Chicago, IL), and statistical significance was set at a p value of less than 0.05. Normally distributed data are shown as bar graphs with means ± S.E., whereas nonnormally distributed data are shown with box plots in which the boxes define the 25th and 75th percentiles, with a line at a median and error bars defining the 10th and 90th percentile.
RESULTS
Isolation of a Peptide Ligand to Immortalized Lung Endothelial Cells—We used the BRASIL (biopanning and rapid analysis of selective interactive ligands) method (24) to screen a CX7C phage display random peptide library on ImmortoMouse-derived lung endothelial cells (27). After selection, 89 individual phage clones were sequenced (Table 1). Up to 78.7% of the clones included the tetrapeptide GWVR motif, and 25.8% of the clones displayed a dominant cyclic peptide insert (sequence CGSPGWVRC). Moreover, the finding that several genetically distinct phage clones (different codons) encoded the insert CGSPGWVRC supported the specificity of our overall screening approach.
TABLE 1.
Lung endothelial cell-binding peptides recovered after three rounds of selection
Areas of identity are shown in boldface type.
| Peptide sequencesa | n | Percentage |
|---|---|---|
| % | ||
| GSPGWVR | 23 | 25.8 |
| GMPGWVR | 11 | 12.4 |
| GAPGWVR | 8 | 9.0 |
| GDLGWVR | 6 | 6.7 |
| GWPGWVR | 4 | 4.5 |
| GDPGWVR | 4 | 4.5 |
| GRPGWVR | 3 | 3.4 |
| GDRGWVR | 2 | 2.2 |
| GTPGWVR | 2 | 2.2 |
| GHPGWVR | 2 | 2.2 |
| GLPGWVR | 2 | 2.2 |
| GEPGWVR | 1 | 1.1 |
| GDPGWVS | 1 | 1.1 |
| GDQGWVR | 1 | 1.1 |
| GELGWVR | 1 | 1.1 |
| RGVGWVG | 1 | 1.1 |
| QGGGVGW | 1 | 1.1 |
| Others | 16 | 18.0 |
| GXPGWVR | 62 | 69.6 |
Cysteines are omitted. Number of inserts = 89.
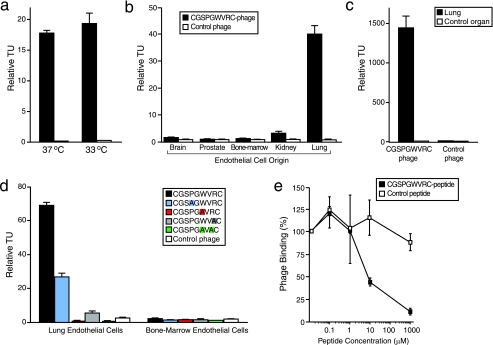
Validation of CGSPGWVRC Phage Binding to Lung Endothelial Cells—We performed a comprehensive characterization of the binding profile of the CGSPGWVRC phage relative to appropriate controls (Fig. 1). First we evaluated whether temperature (a surrogate for the level of the large T antigen in the lung endothelial cells) affected the binding of the dominant CGSPGWVRC phage clone to ImmortoMouse-derived lung endothelial cells. We performed phage-binding experiments with CGSPGWVRC phage or insertless control phage (fd-tet) on lung endothelial cells cultured either at 33 °C (permissive temperature) or 37 °C (Fig. 1a). We found no significant difference in CGSPGWVRC phage binding to lung endothelial cells cultured at either temperature (143 ± 13-fold binding at 33 °C versus 119 ± 2-fold binding at 37 °C relative to binding of negative control phage; Student's t test, p = 0.35). These data allowed us to use proliferating endothelial cells (cultured at 33 °C) for the subsequent experiments. Next, we evaluated the tissue of origin specificity of CGSPGWVRC phage binding to the lung endothelial cells. CGSPGWVRC phage or control phage were incubated with immortal endothelial cells derived from brain, prostate, bone marrow, kidney, or lung (27). We observed markedly increased binding of CGSPGWVRC phage to lung endothelial cells relative to control phage; moreover, binding of CGSPGWVRC phage to endothelial cells derived from brain, prostate, bone marrow, or kidney was detected only at background levels when compared with control phage (Fig. 1b). To rule out the possibility that the observed tissue-specific binding of CGSPGWVRC phage was related to adaptation to cell culture conditions, we also tested the CGSPGWVRC phage binding ex vivo to single-cell suspensions prepared from mouse lung or a negative control tissue (mouse spleen was used unless otherwise specified). Consistently, we observed strong binding (118 ± 12-fold) of CGSPGWVRC phage to lung single-cell suspension relative to control phage binding to the same lung single-cell suspension; in contrast, no binding or only minimal binding (1.7 ± 0.1-fold) of CGSPGWVRC phage to spleen single-cell suspension was observed when CGSPGWVRC phage binding was compared with the control phage binding to spleen-derived single-cell suspension (Fig. 1c). In order to characterize the role of individual amino acid residues in CGSPGWVRC peptide binding to lung endothelial cells, we used site-directed mutagenesis. We generated different alanine-scanning versions of the displayed peptide by cloning mutant phage inserts (CGSAGWVRC, CGSPGAVRC, CGSPGWVAC, and CGSPGAVAC) and comparing binding of each mutant insert to insertless control phage. Mutation of the tryptophan residue to alanine abolished phage binding to lung endothelial cells, and mutation of the arginine residue to alanine reduced the phage binding to lung endothelial cells by 92%. Mutation of both tryptophan and arginine residues to alanine abolished phage binding to lung endothelial cells. Although the proline to alanine mutation decreased phage binding to lung endothelial cells by 61%, binding of CGSAGWVRC peptide-displaying phage to lung endothelial cells was still significant relative to control phage binding to lung endothelial cells (p < 0.001). These results suggest critical roles for the tryptophan and arginine residues within the ligand insert CGSPGWVRC. None of the mutant phage clones showed binding to a control endothelial cell line derived from bone marrow (Fig. 1d). Finally, we found that CGSPGWVRC phage binding to lung endothelial cells is mediated specifically by the peptide, since phage binding was inhibited by the cognate synthetic CGSPGWVRC peptide; an unrelated negative control cyclic peptide at the equivalent molar concentrations had no inhibitory effect (Fig. 1e). Together, these results show that the peptide CGSPGWVRC is a potent and specific ligand to lung-derived endothelial cells.
FIGURE 1.
CGSPGWVRC phage binds specifically to lung endothelial cells. a, CGSPGWVRC phage binding to primary mouse lung microvascular endothelial cells cultured at either 33 or 37 °C detected by the BRASIL method. b, CGSPGWVRC phage binds specifically to lung endothelial cells but not to endothelial cells derived from control organs (brain, prostate, bone marrow, or kidney). c, CGSPGWVRC phage binding ex vivo to single-cell suspensions prepared from mouse lungs. CGSPGWVRC phage did not bind to single-cell suspensions prepared from the control organ (spleen). d, mutation of certain residues within CGSPGWVRC (to alanine) abolishes phage binding to lung endothelial cells. Bone marrow endothelial cells served as the negative control. Mutant residues are color-coded. e, CGSPGWVRC phage binding to lung endothelial cells is inhibited in a dose-dependent manner by the cognate synthetic peptide. An unrelated cyclic control peptide did not affect the phage binding. Shown are means ± S.E. from triplicate samples. Insertless control phage served as a negative control phage in a-d. Fd-tet phage values were set to 1 in a-c.
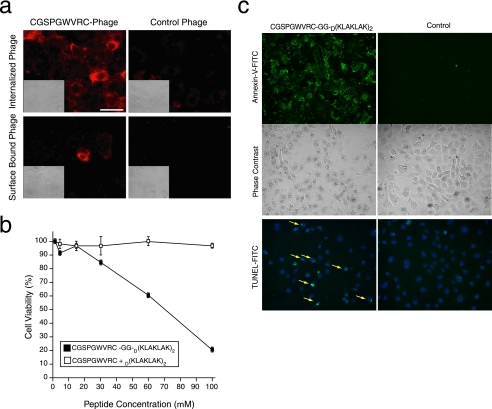
Lung Endothelial Cell Binding Peptides Mediate Cell Internalization—We next set out to evaluate whether the ligand CGSPGWVRC peptide would mediate phage internalization into lung endothelial cells. CGSPGWVRC phage or control phage were incubated with either lung-derived or prostate-derived endothelial cells. Cells were then washed to remove noninternalized phage, permeabilized, and stained with an anti-bacteriophage antibody. A Cy3-conjugated secondary antibody was used to detect the presence and localization of phage particles. After 4 h at 37 °C, CGSPGWVRC phage particles were internalized into lung endothelial cells (Fig. 2a); only background staining was observed when control phage or nonpermeabilized cells were used as negative controls. In contrast, CGSPGWVRC phage was not internalized into prostate endothelial cells (data not shown). Internalization detection of CGSPGWVRC phage is time-dependent (weak at 30 min, moderate at 2-4 h, and strong at 8 h; data not shown). These data suggest that the CGSPGWVRC peptide can mediate the internalization of ligands such as phage into lung endothelial cells. In order to evaluate whether internalization of CGSPGWVRC-directed ligands can also occur outside of the phage context (i.e. the targeted delivery of pathobiologically relevant peptide), we generated and tested a synthetic chimeric peptide of the CGSPGWVRC ligand fused to the D(KLAKLAK)2 proapoptotic motif, an amphipathic α-helix-forming antimicrobial peptide that disrupts preferentially eukaryotic mitochondrial membranes upon ligand-directed internalization (25, 30-32). Increasing concentrations of the lung endothelial cell targeted CGSPGWVRC-GG-D(KLAKLAK)2 peptide or equimolar amounts of CGSPGWVRC plus D(KLAKLAK)2 were incubated with lung endothelial cells at 37 °C, and cell viability was assessed after 6 h. Treatment of lung endothelial cells resulted in a dose-dependent decrease in cell viability with CGSPGWVRC-GG-D(KLAKLAK)2 peptide, whereas the equimolar combination of CGSPGWVRC plus D(KLAKLAK)2 peptides did not have any detectable effect on cell viability (Fig. 2b). The induction of endothelial cell apoptosis by CGSPGWVRC-GG-D(KLAKLAK)2 was further confirmed by the presence of two apoptotic markers (Annexin-V binding and TUNEL) (Fig. 2c). Taken together, these results indicate that the displayed CGSPGWVRC insert or the corresponding synthetic peptide can mediate ligand-directed internalization into lung endothelial cells both inside and outside of the phage particle context in a time- and dose-dependent manner and that internalization of the CGSPGWVRC-GG-D(KLAKLAK)2 proapoptotic peptide induces programmed cell death of the lung endothelial cells in vitro.
FIGURE 2.
CGSPGWVRC peptide mediates internalization of ligands into lung endothelial cells and cell apoptosis. a, CGSPGWVRC peptide mediates phage internalization into lung endothelial cells. CGSPGWVRC phage or control phage were incubated with lung endothelial cells at 37 °C to allow for phage internalization. Cells were stained with an anti-bacteriophage antibody after removal of the membrane-bound phage. The upper panels show staining of permeabilized cells revealing the internalized phage. The lower panels show staining of nonpermeabilized cells, demonstrating the successful removal of phage from the cell surface. Scale bar, 100 μm. b, lung endothelial cells were incubated with increasing concentrations (up to 100 μm) of proapoptotic peptide synthesized in conjunction with the targeting CGSPGWVRC peptide (CGSPGWVRC-GG-D(KLAKLAK)2) or negative control peptides (an equimolar mixture of CGSPGWVRC and D(KLAKLAK)2). Cell viability was determined by optical absorbance using a cell proliferation detection reagent. Shown are means ± S.E. from triplicate wells. c, lung endothelial cells were incubated with 100 μm proapoptotic peptide CGSPGWVRC-GG-D(KLAKLAK)2 or negative control peptides (equimolar mixture of CGSPGWVRC and D(KLAKLAK)2) for 2 h. Induction of cell apoptosis was detected by Annexin-V-fluorescein isothiocyanate (FITC) binding (top and middle panels) or TUNEL (bottom, yellow arrows).
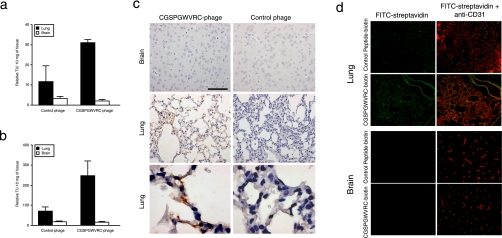
Validation of CGSPGWVRC Homing to Lung Endothelial Cells in Vivo—Having characterized ligand-directed binding and internalization in vitro, we assessed the ability of the CGSPGWVRC phage to target normal mouse lung vasculature in vivo. Circulation time is an important variant to consider while conducting in vivo phage targeting experiments, since the time frame in which phage are allowed to circulate after intravenous administration influences their biodistribution. Thus, experiments were designed toward detection of phage binding to the vascular endothelium (5-10 min of circulation before phage recovery) or toward detection of phage that were internalized by the endothelium or other nonendothelial tissue layers (circulation 24 h before phage recovery). After intravenous administration of CGSPGWVRC phage or control phage, homing was quantified by phage recovery from tissue homogenates by host bacterial infection after either 10 min or 24 h of circulation. After 10 min of circulation time, the CGSPGWVRC phage were enriched in lungs compared with either control organs or control phage (Fig. 3a). Similarly, after 24 h of circulation, the CGSPGWVRC phage were enriched in lungs compared with control organs and the control phage (Fig. 3b). Homing of CGSPGWVRC phage to lung vasculature was confirmed by immunohistochemical staining of phage on tissue sections. Strong phage staining was observed in lung tissue sections but not in control tissue sections of mice that received CGSPGWVRC phage; in contrast, insertless control phage did not home to lungs above background levels (Fig. 3c).
FIGURE 3.
Targeting mouse lung vasculature in vivo. a, the ability of the CGSPGWVRC phage to home to mouse lungs was evaluated after intravenous phage administration into BALB/c mice. Phage were recovered 10 min later from lungs or control tissues after perfusion. Shown are mean ± S.E. of TU from three mice and triplicate plating. b, the ability of the CGSPGWVRC phage to home and internalize to mouse lungs was evaluated 24 h after an intravenous phage administration into BALB/c mice. Phage were recovered from lungs or control tissues without perfusion. Shown are mean ± S.E. of TU from three mice and triplicate plating. c, CGSPGWVRC phage or control phage were administered intravenously into mice. Mice were perfused 10 min after the phage injections, and lungs and control organ were recovered. A bacteriophage-specific antibody was used for staining. Scale bar, 100 μm for the panels in the top and middle rows and 20 μm for the bottom row. Insertless phage served as a negative control. d, targeting of CGSPGWVRC synthetic peptide to the lung vasculature was evaluated by intravenous administration of CGSPGWVRC-biotin. After 1 h of circulation, peptide was detected using a streptavidin-fluorescein isothiocyanate conjugate (yellow) and co-stained with an endothelial marker specific antibody (CD31; red). Vascular co-localization of CGSPGWVRC-biotin is indicated by the yellow areas in the merged image. No specific homing was observed to a control organ or when a control peptide-biotin was injected into mice.
To confirm that targeting of the CGSPGWVRC peptide to the lung vasculature occurs outside of the context of the phage, we administered intravenously soluble CGSPGWVRC peptide linked to biotin at its carboxyl terminus and detected lung homing by streptavidin-fluorescein isothiocyanate on processed histological sections. We observed clear CGSPGWVRC-biotin labeling of alveolar capillaries (Fig. 3d), similar to the pattern observed by CD-31 immunofluorescence. Specificity was determined by lack of fluorescence in lung capillaries with an unrelated peptide or lack of brain capillary signal with the CGSPGWVRC-biotin. Of note, there was some evidence of CGSPGWVRC-biotin labeling in kidneys and liver, which can be accounted by elimination and excretion of the injected peptide (data not shown), as supported by the studies described below. These results show that CGSPGWVRC phage and the corresponding synthetic peptide target lung blood vessels in vivo.
Targeted CGSPGWVRC-GG-D(KLAKLAK)2 Causes Acute and Persistent Emphysematous Changes in the Lungs of Treated Mice—Because alveolar cell apoptosis has been linked to alveolar destructive enlargement, but there is no evidence that specific endothelial cell death can direct emphysematous alveolar enlargement, we sought to determine whether administration of CGSPGWVRC-GG-D(KLAKLAK)2 peptide to mice would induce morphological changes in the lungs, particularly emphysema. Mice given CGSPGWVRC-GG-D(KLAKLAK)2 peptide became very lethargic with difficult breathing, which improved in the ensuing hours. Mice treated with control peptide or vehicle did not change their normal behavior after anesthesia.
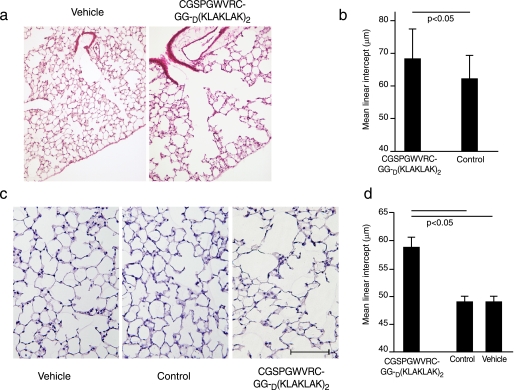
As assessed by lung morphometry, C57Bl6/6J mice that received intraperitoneal injections of 240 μg of CGSPGWVRCGG-D(KLAKLAK)2 peptide or control peptides every other day for a total of either 4 days or 21 days developed emphysema (Fig. 4). Alveolar destruction was already evident after only 4 days (two doses) of treatment (Fig. 4, a and b), and it became more prominent at 21 days (10 doses) of treatment with the peptide CGSPGWVRC-GG-D(KLAKLAK)2 (Fig. 4c). By the end of the 21-day experiment (n = 5 mice in each group), CGSPGWVRCGG-D(KLAKLAK)2-treated lungs exhibited an approximately 20% increase from a mean linear intercept of 59 ± 0.84 μmin CGSPGWVRC-GG-D(KLAKLAK)2 peptide-treated mice compared with 49.7 ± 0.9 μm in control-treated mice with CGSPGWVRC peptide, 49.5 ± 1.0 μm for untargeted D(KLAKLAK)2-treated mice and 49 ± 0.6 μm for vehicle-treated lungs (Fig. 4d). Hematoxylin and eosin-stained tissue sections of several control organs (kidney, heart, liver, and bone marrow) did not reveal any detectable histological changes in CGSPGWVRCGG-D(KLAKLAK)2-treated mice (Fig. S1).
FIGURE 4.
Induction of morphological changes in mouse lungs after administration of CGSPGWVRC-GG-D(KLAKLAK)2. a, lung sections of mice 4 days after treatment with CGSPGWVRC-GG-D(KLAKLAK)2 peptide or with control peptides (CGSPGWVRC or D(KLAKLAK)2 peptides) were stained with hematoxylin and eosin. Lung sections from the CGSPGWVRC-GG-D(KLAKLAK)2 peptide-treated mice show increased air space enlargement when compared with the lung sections from the control-treated mice. Scale bar, 400 μm. b, quantification of the mean linear intercept (μm) in CGSPGWVRC-GG-D(KLAKLAK)2 and control peptide-treated lungs after 4 days of peptide treatment. CGSPGWVRC-GG-D(KLAKLAK)2 treatment shows a significant increase in the mean linear intercept values in versus control-treated mice. c, lung sections from mice treated for 21 days with CGSPGWVRC-GG-D(KLAKLAK)2, control peptides (CGSPGWVRC or D(KLAKLAK)2), or vehicle alone were stained with hematoxylin and eosin. Lung sections from the CGSPGWVRC-GG-D(KLAKLAK)2 peptide-treated mice show increased air space enlargement when compared with the lung sections from the control-treated mice. Scale bar, 200 μm. d, the mean linear intercept (μm) was measured from vehicle, CGSPGWVRC-GG-D(KLAKLAK)2 peptide-treated, and control peptide (CGSPGWVRC or D(KLAKLAK)2)-treated animals after 21 days of treatment. CGSPGWVRC-GG-D(KLAKLAK)2 treatment shows a significant increase in mean linear intercept values compared with control peptide-treated mice.
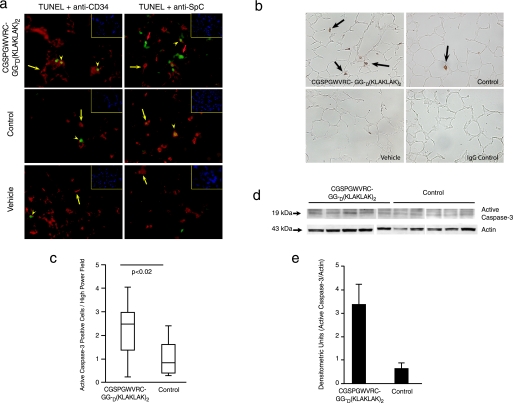
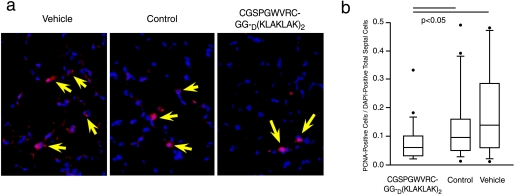
Targeted CGSPGWVRC-GG-D(KLAKLAK)2 Induces both Lung Cell Apoptosis and Decreased Lung Cell Proliferation—As shown above, CGSPGWVRC-GG-D(KLAKLAK)2 peptide treatment led to lung endothelial cell apoptosis in vitro. Given that peptide treatment with CGSPGWVRC-GG-D(KLAKLAK)2 caused alveolar destruction in mice, we determined whether this destruction was associated with directed apoptosis of alveolar endothelial cells. After 4 days of treatment with CGSPGWVRC-GG-D(KLAKLAK)2 peptide, staining of lung sections with DAPI and TUNEL showed abundant TUNEL-positive cells compared with staining of lung sections from mice receiving control peptides (supplemental Fig. S2a). TUNEL-positive cells in lung sections were abundant after 21 days of CGSPGWVRC-GG-D(KLAKLAK)2 peptide treatment (Fig. 5a) (CGSPGWVRC-GG-D(KLAKLAK)2 = 0.0288 TUNEL+/DAPI+ versus peptide = 0.0146 TUNEL+/DAPI+, p < 0.03; Mann-Whitney rank sum test). Alveolar endothelial, type II, and myofibroblastic cells were equally affected at day 4 (Fig. S2a). A similar proportion of endothelial and type II cell death was also identified on day 21 after treatment with CGSPGWVRC-GG-D(KLAKLAK)2 peptide (Fig. 5a), as detected by staining of the TUNEL-labeled lung sections with either a CD34-specific antibody (an endothelial cell marker) or an SpC-specific antibody (a type II alveolar epithelial cell marker), or smooth muscle cell α-actin-antibody (for myofibroblasts). The TUNEL findings were also confirmed by immunohistochemical staining for active caspase-3, which revealed an increased number of apoptotic cells in the alveolar septa of the CGSPGWVRC-GG-D(KLAKLAK)2 peptide-treated mice; in contrast, lungs from mice treated with control peptide or vehicle showed much less active caspase-3-positive cells (Fig. 5b). Increased active caspase-3-positive cells in the alveolar septa of the CGSPGWVRC-GG-D(KLAKLAK)2 peptidetreated mice were evident after only 4 days of treatment (Fig. S2, b and c). Enhanced activation of caspase-3 in lungs of mice after 4 days (Fig. S2, b-e) and after 21 days (Fig. 5, c and d) of treatment with CGSPGWVRC-GG-D(KLAKLAK)2 peptide was further confirmed by the detection of increased levels of the 19-kDa active caspase-3 peptide in whole-lung lysates by Western blot analysis. Densitometric assessment of the Western blot reaction at day 21 (Figs. 5, d and e, and S3) showed 3.36 ± 0.89 densitometric units of active caspase-3 in lungs of mice receiving CGSPGWVRC-GG-D(KLAKLAK)2 peptide (n = 5) compared with 0.66 ± 0.19 densitometric units in the lungs of mice receiving control peptide (n = 6). In addition to increased apoptosis of alveolar cells, we also detected decreased cell proliferation in the lungs of mice treated with CGSPGWVRC-GG-D(KLAKLAK)2 peptide for 21 days when compared with the lungs of mice treated with vehicle or control peptide (Fig. 6, a and b). In sum, these data indicate that systemic administration of CGSPGWVRC-GG-D(KLAKLAK)2 peptide caused apoptosis of alveolar cells.
FIGURE 5.
CGSPGWVRC-GG-D(KLAKLAK)2 peptide causes lung cell apoptosis after 21 days of treatment. a, identification of apoptotic (detected by TUNEL; green) endothelial cells (detected by anti-CD34 antibody; red) and type II epithelial cells (detected by anti-SpC antibody; red) in the lungs of CGSPGWVRC-GG-D(KLAKLAK)2-, control peptide-, and vehicle-treated mice after 21 days of treatment. Shown are merged images, with co-localization of cell-specific markers and apoptosis: cytoplasmic marker alone (CD34 or SpC) (red cells) (yellow arrows), TUNEL-positive cells in CD34 or SpC-positive cells (yellow arrowheads), and TUNEL-positive cells without a cytoplasmic positive markers (large orange arrows). Cell nuclei were stained with DAPI (blue) (inset, top right). b, active caspase-3 expression in lung sections of mice 21 days after treatment with CGSPGWVRC-GG-D(KLAKLAK)2, control peptides (CGSPGWVRC and D(KLAKLAK)2), or vehicle alone. CGSPGWVRC-GG-D(KLAKLAK)2 peptide-treated lungs show abundant active caspase-3-positive cells in the alveolar septa in contrast to control-treated lungs. Isotype control antibody on CGSPGWVRC-GG-D(KLAKLAK)2 peptide-treated lungs was used as a negative control for the active caspase-3 staining. c, quantification of number of alveolar septal cells positive for active caspase-3. d, increased levels of active caspase-3 were detected in the lungs of CGSPGWVRC-GG-D(KLAKLAK)2 peptide-treated mice versus control-treated mice by Western blot analysis of lung tissue lysates with an active caspase-3-specific antibody. e, densitometric quantification of active caspase-3 expression levels obtained in Western blot analysis normalized to actin levels.
FIGURE 6.
CGSPGWVRC-GG-D(KLAKLAK)2 treatment leads to decreased lung cell proliferation. a, lung sections from mice treated for 21 days with CGSPGWVRC-GG-D(KLAKLAK)2 peptide, control peptides (CGSPGWVRC and D(KLAKLAK)2 peptides), or vehicle alone were double-stained with anti-proliferating cell nuclear antigen (PCNA) antibody, and DAPI. Proliferating cell nuclear antigen- and DAPI-positive cells are indicated (arrowheads). A markedly decreased number of proliferating cells (quantification in b) is seen in CGSPGWVRC-GG-D(KLAKLAK)2-treated animals in comparison with control animals. b, quantification of proliferating cell nuclear antigen-positive cells in the lungs from CGSPGWVRC-GG-D(KLAKLAK)2-treated mice shows a decrease in proliferating cell numbers when compared with lungs from control-treated mice.
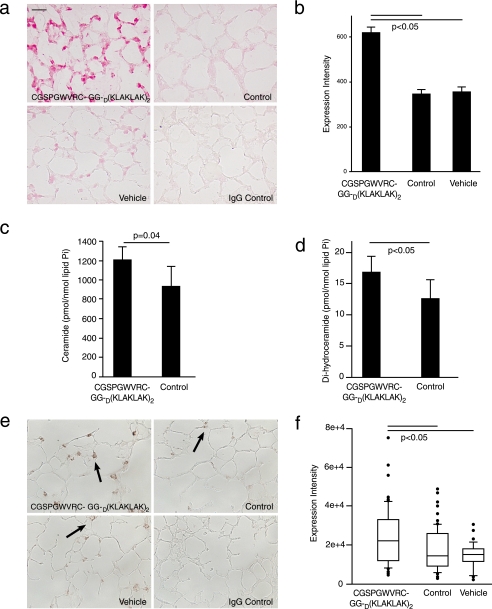
Targeted CGSPGWVRC-GG-D(KLAKLAK)2 Increases Pulmonary Oxidative Stress—We have shown that oxidative stress is critically involved in the pathogenesis of experimental cigarette smoke-induced emphysema (17). Furthermore, we demonstrated that emphysema due to VEGF receptor blockade depends on the mutual interaction between apoptosis and oxidative stress, since either apoptosis blockade or inhibition of superoxide anion prevents emphysema development (6). Immunohistochemical detection of 8-oxo-dG was used to evaluate oxidative stress in the lungs of mice receiving CGSPGWVRC-GG-D(KLAKLAK)2 for 4 and for 21 days. After 4 days of treatment, lung sections from CGSPGWVRC-GG-D(KLAKLAK)2-treated mice showed increased 8-oxo-dG expression when compared with the lung sections from the control peptide-treated mice (Fig. S4). After 21 days of treatment with vehicle or control peptides, a small number of alveolar septal cells showed staining for 8-oxo-dG (355 ± 20 and 346 ± 17 expression intensity units, respectively), whereas significantly more cells (617 ± 28 expression intensity units; p < 0.05) were stained in CGSPGWVRC-GG-D(KLAKLAK)2-treated lungs (Fig. 7, a and b). These results show that emphysema and alveolar cell apoptosis due to systemic administration of CGSPGWVRC-GG-D(KLAKLAK)2 are associated with enhanced oxidative damage to the lungs.
FIGURE 7.
Levels of 8-oxo-7,8-dihydro-2′-deoxyguanosine and ceramide are increased in CGSPGWVRC-GG-D(KLAKLAK)2-treated lungs (21 days of treatment). a, CGSPGWVRC-GG-D(KLAKLAK)2 peptide induces oxidative damage to mouse lungs, as indicated by increasing 8-oxo-dG expression. Immunohistochemical staining of 8-oxo-dG in lung sections from mice treated for 21 days with CGSPGWVRC-GG-D(KLAKLAK)2 peptide, control peptides (CGSPGWVRC and D(KLAKLAK)2), or vehicle alone. Isotype control antibody on CGSPGWVRC-GG-D(KLAKLAK)2 peptide-treated lungs served as a negative control for anti-8-oxo-dG staining. Lung sections from the CGSPGWVRC-GG-D(KLAKLAK)2-treated mice show elevated 8-oxo-dG expression compared with control-treated mice. Scale bar (a and c), 25 μm. b, quantification of the 8-oxo-dG intensity in the lung tissues from CGSPGWVRC-GG-D(KLAKLAK)2-treated mice. c and d, analysis of ceramide species by mass spectrometry shows that lungs of mice treated with the CGSPGWVRC-GG-D(KLAKLAK)2 peptide for 7 days have increased levels of ceramides (c) as well as dihydroceramides, ceramide precursors in the de novo pathway of ceramide synthesis (d), compared with the lungs from control peptide (CGSPGWVRC or D(KLAKLAK)2)-treated mice. e, CGSPGWVRC-GG-D(KLAKLAK)2 peptide induces elevation of ceramide levels in mouse lungs. Lung sections from mice after 21 days of treatment with CGSPGWVRC-GG-D(KLAKLAK)2 peptide, control peptides (CGSPGWVRC and D(KLAKLAK)2), or vehicle alone were subjected to immunohistochemical staining for ceramide. Ceramide staining in CGSPGWVRC-GG-D(KLAKLAK)2 peptide-treated lungs show numerous ceramide-positive alveolar cells, whereas lungs treated with control peptides or vehicle show only sporadic ceramide-positive cells. f, quantification of ceramide expression detected by immunohistochemistry is described under “Materials and Methods.”
Targeted CGSPGWVRC-GG-D(KLAKLAK)2 Leads to Increased Lung Ceramide Levels—Ceramide, a second messenger sphingolipid, triggers apoptosis when up-regulated in conditions associated with cellular stresses, such as during oxidative stress. We have recently shown that (i) ceramide is a critical mediator of lung tissue destruction during development of emphysema due to VEGF receptor blockade, and (ii) ceramide levels are increased in lungs of patients with cigarette smoke-induced emphysema (10). We quantified total lung ceramide levels by mass spectroscopic analysis of ceramide and its immediate precursor in the de novo synthesis pathway, dihydro-ceramide and by immunohistochemistry with a specific ceramide antibody in the lungs of mice receiving vehicle, control peptide, or CGSPGWVRC-GG-D(KLAKLAK)2 peptide after 7 or 21 days of treatment, respectively. Mass spectroscopic analysis of ceramide and its immediate precursor, dihydroceramide, showed an early and significant activation of this apoptotic signaling pathway (from 930.1 pmol/nmol lipid Pi to 1203.6 pmol/nmol lipid Pi (p = 0.04) for ceramide (Fig. 7c) and from 12.6 pmol/nmol lipid Pi to 16.8 pmol/nmol lipid Pi (p = 0.04) for dihydroceramide (Fig. 7d)) of ceramide levels in CGSPGWVRC-GG-D(KLAKLAK)2-treated lungs in comparison with control peptide after 7 days of treatment. Ceramide levels in CGSPGWVRC-GG-D(KLAKLAK)2-treated lungs were sustained after 21 days of treatment, as assessed by immunohistochemistry, which correlates closely with standard biochemical assays of ceramide in the lung (10) (p < 0.05) (Fig. 7, e and f).
Targeted CGSPGWVRC-GG-D(KLAKLAK)2 Increases the Number of Inflammatory Cells in the Lungs—Influx of inflammatory cells, particularly macrophages, is a constant finding in smokers, and it has been shown to mediate elastin fiber destruction via production of metalloprotease-12 (33). Rather than a proximal event in the course of events leading to emphysema, recent studies have suggested that macrophage influx follows alveolar lung injury and enhanced oxidative stress. Consistent with this hypothesis, we detected increased numbers of alveolar and septal macrophages in CGSPGWVRC-GG-D(KLAKLAK)2-treated lungs as compared with control peptide-treated or vehicle-treated lungs (Fig. S5).
DISCUSSION
In addition to the classical roles of endothelial cells in inflammation and coagulation, there is a clear realization that the vascular endothelium contributes to organ embryogenesis, differentiation, and acquisition of specialized functions. Furthermore, the role of endothelial cells in organ structural integrity, as initially identified in the retina (34), also pertained to a wide range of organs within the circulatory (35), nervous (36), and respiratory (37) systems. Furthermore, disruption of VEGF signaling in lung endothelial cells results in poorly developed lungs (9, 37). However, since these studies relied on blockade of VEGF signaling critical for endothelial cell survival, growth, and differentiation, they also interrupted similar effects of VEGF on nonendothelial cells, such as neuronal or type II epithelial cells, also known to express VEGF receptor 2 (38). Development of ligand-directed lung endothelial cell targeting based on the hypothesis of tissue-specific vascular ZIP codes (21-25, 39-44) allowed us to interrogate the specific role of endothelial cells in lung structural maintenance and the pathological organ consequences of its disruption. The results presented here indicate that (i) endothelial cell viability plays a central role in lung maintenance, and (ii) endothelial cell apoptosis suffices to recreate the pathophysiological processes involved in emphysematous lung destruction.
Recognition of the functional and phenotypic diversity of endothelial cells has led to a more detailed understanding of how endothelial cells provide differentiating cues to surrounding specialized cells (45). Not only do lung endothelial cells interact with branching epithelial structures during development (46), but there is evidence that pulmonary artery endothelial cells are phenotypic diverse and segregate as large pulmonary artery and lung microvascular endothelial cells (46, 47). In line with the unique molecular signatures of endothelial cells of multiple vascular beds, we have identified several peptide ligands that target specific parts of vasculature after intravenous administration from a large combinatorial random peptide library (21, 23, 40-45, 48, 49). In this study, we selected and isolated a peptide (CGSPGWVRC) that binds to immortalized lung microvascular endothelial cells (27) in vitro. We tested CGSPGWVRC phage binding to several different tissue-specific endothelial cell lines and found the CGSPGWVRC peptide-mediated phage binding to be specific to lung endothelial cells. This result was consistent with our screening strategy designed to minimize or eliminate the recovery of ubiquitous endothelial cell-binding peptides. Tryptophan and arginine residues appear to play key roles in the motif binding to lung endothelial cells, since the mutation of either residue abolishes the ability of the peptide-displaying phage to bind to lung endothelial cells. Although it is conceivable that the ImmortoMouse-derived endothelial cells do not express the full complement of proteins found in vivo, the CGSPGWVRC phage consistently showed significant binding enrichment to single-cell suspension freshly prepared from mouse lungs and homed to lung septa after intravenous administration.
We have previously utilized endothelial cell-specific peptides to deliver pharmacological compounds, such as chemotherapy drugs (49), viruses carrying toxic genes (50, 51), or a proapoptotic peptide aimed at disrupting mitochondrial integrity causing an organ-specific or tumor-specific endothelial cell ablation (25, 30-32). CGSPGWVRC peptide-mediated internalization of phage particles into cultured immortalized lung microvascular endothelial cells enabled internalization of the proapoptotic moiety D(KLAKLAK)2, which then induced dose-dependent apoptotic in vitro cell death. We confirmed several of these processes in vivo, where CGSPGWVRC peptide-displaying phage was markedly enriched in mouse lungs 24 h after intravenous administration, reflecting the biodistribution and uptake of the targeted phage particles into the cell population. Of note, the in vitro effects of the internalized CGSPGWVRC peptide fused to D(KLAKLAK)2 were validated by the pathophysiological consequences of in vivo lung endothelial cell ablation. Endothelial cell targeting and apoptosis was supported by our in vitro and in vivo localization data. These data suggest that lung endothelial cells were the primary targets of CGSPGWVRC-GG-D(KLAKLAK)2, yet a similar amount of endothelial, epithelial, and myofibroblastic cell death was occurring as early as day 4 after CGSPGWVRC-GG-D(KLAKLAK)2 administration. These findings are not surprising, since the net loss of alveolar structures has to involve increased death of all structural cells with reduced capacity of cell proliferation. Alveolar cell apoptosis caused by CGSPGWVRC-GG-D(KLAKLAK)2 led to rapid emphysema development, in which alveolar air space enlargement was detected as early as 4 days of the targeted peptide administration, further evolving during the 3-week course of peptide treatment. Characteristic pathogenetic elements present in both human and experimental emphysema were detected in the targeted mouse lung, particularly those related to alveolar cell apoptosis, oxidative stress, and macrophage infiltration, suggesting that lung microvascular apoptosis precedes (and may even initiate) these processes. Consistently, alveolar septal cell death also caused up-regulation of ceramide, a second messenger lipid present in increased levels in human emphysema that mediates and amplifies alveolar tissue destruction induced by the VEGF receptor blockade in mice and rats (10).
The finding of experimental emphysema produced by intratracheal instillation of active caspase-3 (33) or ceramide (10, 12) revealed that alveolar cell apoptosis suffices to produce destructive alveolar enlargement. Although these interventions might have affected initially alveolar epithelial cells, profound endothelial cell apoptosis was observed after ceramide instillation (10). The increase in ceramide levels in CGSPGWVRC peptide-treated lungs suggests that endothelial cell stress might underlie the enzymatic activation of ceramide synthesis, as shown after lipopolysaccharide instillation or radiation-induced endothelial cell injury (52, 53). The ensuing destructive processes and increased concentrations of ceramide (10) present in the alveolar septum may also attract macrophages in response to an excess of apoptotic cells or endothelial cell injury (54). This increase in alveolar macrophages may enhance lung destruction via activation of metalloproteases and cathepsins (7). Finally, in contrast to the self-limited injury present in most animal models of emphysema, our endothelial cell-targeted model displayed marked decreases in alveolar cell proliferation in vivo. Since the lung endothelial cell peptide does not alter endothelial cell proliferation in vitro, disruption of alveolar cell homeostasis in vivo due to endothelial cell targeting may underlie the decrease in cell proliferation. This finding may not only support a model of emphysema, which may recapitulate the irreversible process of alveolar destruction and lack of repair seen in the human disease, but may uncover an important role for the alveolar endothelial cell in lung repair mechanisms.
In summary, we provide evidence that lung endothelial cells have unique phenotypic characteristics and that lung endothelial cell-specific peptides can be identified based on these properties. We show that lung endothelial cell targeting and alveolar cell apoptotic ablation through a targeted proapoptotic homing peptide to lung vasculature is possible. We also demonstrate that endothelial cell targeting with ensuing programmed alveolar cell death sets off pathogenetic processes that ultimately result in emphysematous lung destruction. These results support the critical structural role of lung endothelial cells in maintaining lung septal integrity and may offer novel means to investigate fundamental pathobiological processes in emphysema pathogenesis. Other combinatorial screenings in vitro on immortalized cells in tandem with in vivo validation strategies may provide an experimental blueprint for development of a ligand-directed pharmacology.
Supplementary Material
Acknowledgments
We thank Emile Brown for technical support and Dr. Walter Hubbard for mass spectrometric measurements of ceramides.
This work was supported, in whole or in part, by National Institutes of Health Grants HL 066554 (to R. M. T.) and HL 077328 (to I. P.) and Grant CA 122568 (to W. A. and R. P.). This work was also supported by Department of Defense IMPACT Grant W91ZSQ-4348-N602 (to W. A. and R. P.) and a Department of Defense Grant DAMD17-01-1-0644 (to R. J. G.); an American Thoracic Society/Alpha One Foundation Research Grant (to R. M. T.); a grant from the American Lung Association (to I. P.); and awards from AngelWorks, the Gillson-Longenbaugh Foundation, the V Foundation (to W. A. and R. P.). Robert Black fellow of the Damon Runyon Cancer Research Foundation (J. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S5.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: VEGF, vascular endothelial growth factor; FBS, fetal bovine serum; DMEM, Dulbecco's modified Eagle's medium; BSA, bovine serum albumin; LB, Luria broth; PBS, phosphate-buffered saline; DAPI, 4′,6-diamidino-2-phenylindole, dihydrochloride; TUNEL, terminal deoxynucleotidyltransferase biotin-dUTP nick end labeling; 8-oxo-dG, 8-oxo-7,8-dihydro-2′-deoxyguanosine; SpC, surfactant protein C; TU, transducing units.
References
- 1.Sirianni, F. E., Chu, F. S., and Walker, D. C. (2003) Am. J. Respir. Crit. Care Med. 168 1532-1537 [DOI] [PubMed] [Google Scholar]
- 2.Snider, G. L., Kleinerman, J. L., Thurlbeck, W. M., and Bengali, Z. H. (1985) Am. Rev. Respir. Dis. 132 182-1854014865 [Google Scholar]
- 3.Shapiro, S. D. (1995) Proc. Assoc. Am. Physicians 107 346-352 [PubMed] [Google Scholar]
- 4.Liebow, A. A. (1959) Am. Rev. Respir. Dis. 80 67-93 [DOI] [PubMed] [Google Scholar]
- 5.Kasahara, Y., Tuder, R. M., Taraseviciene-Stewart, L., Le Cras, T. D., Abman, S., Hirth, P. K., Waltenberger, J., and Voelkel, N. F. (2000) J. Clin. Invest. 106 1311-1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuder, R. M., Zhen, L., Cho, C. Y., Taraseviciene-Stewart, L., Kasahara, Y., Salvemini, D., Voelkel, N. F., and Flores, S. C. (2003) Am. J. Respir. Cell Mol. Biol. 29 88-97 [DOI] [PubMed] [Google Scholar]
- 7.Tuder, R. M., Petrache, I., Elias, J. A., Voelkel, N. F., and Henson, P. M. (2003) Am. J. Respir. Cell Mol. Biol. 28 551-554 [DOI] [PubMed] [Google Scholar]
- 8.Tang, K., Rossiter, H. B., Wagner, P. D., and Breen, E. C. (2004) J. Appl. Physiol. 97 1559-1566; Discussion, 1549 [DOI] [PubMed] [Google Scholar]
- 9.Compernolle, V., Brusselmans, K., Acker, T., Hoet, P., Tjwa, M., Beck, H., Plaisance, S., Dor, Y., Keshet, E., Lupu, F., Nemery, B., Dewerchin, M., Van Veldhoven, P., Plate, K., Moons, L., Collen, D., and Carmeliet, P. (2002) Nat. Med. 8 702-710 [DOI] [PubMed] [Google Scholar]
- 10.Petrache, I., Natarajan, V., Zhen, L., Medler, T. R., Richter, A. T., Cho, C., Hubbard, W. C., Berdyshev, E. V., and Tuder, R. M. (2005) Nat. Med. 11 491-498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrache, I., Fijalkowska, I., Medler, T. R., Skirball, J., Cruz, P., Zhen, L., Petrache, H. I., Flotte, T., and Tuder, R. M. (2006) Am. J. Pathol. 169 1155-1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petrache, I., Medler, T. R., Richter, A. T., Kamocki, K., Chukwueke, U., Zhen, L., Gu, Y., Adamowicz, J., Schweitzer, K. S., Hubbard, W. C., Berdyshev, E. V., Lungarella, G., and Tuder, R. M. (2008) Am. J. Physiol. 295 L44-L53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasahara, Y., Tuder, R. M., Cool, C. D., Lynch, D. A., Flores, S. C., and Voelkel, N. F. (2001) Am. J. Respir. Crit. Care Med. 163 737-744 [DOI] [PubMed] [Google Scholar]
- 14.Koyama, S., Sato, E., Haniuda, M., Numanami, H., Nagai, S., and Izumi, T. (2002) Am. J. Respir. Crit. Care Med. 166 382-385 [DOI] [PubMed] [Google Scholar]
- 15.Kanazawa, H., Asai, K., Hirata, K., and Yoshikawa, J. (2003) Am. J. Med. 114 354-358 [DOI] [PubMed] [Google Scholar]
- 16.Kanazawa, H., Hirata, K., and Yoshikawa, J. (2003) Eur. Respir. J. 22 609-612 [DOI] [PubMed] [Google Scholar]
- 17.Rangasamy, T., Cho, C. Y., Thimmulappa, R. K., Zhen, L., Srisuma, S. S., Kensler, T. W., Yamamoto, M., Petrache, I., Tuder, R. M., and Biswal, S. (2004) J. Clin. Invest. 114 1248-1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng, T., Kang, M. J., Crothers, K., Zhu, Z., Liu, W., Lee, C. G., Rabach, L. A., Chapman, H. A., Homer, R. J., Aldous, D., DeSanctis, G., Underwood, S., Graupe, M., Flavell, R. A., Schmidt, J. A., and Elias, J. A. (2005) J. Immunol. 174 8106-8115 [DOI] [PubMed] [Google Scholar]
- 19.Tuder, R. M., Yoshida, T., Arap, W., Pasqualini, R., and Petrache, I. (2006) Proc. Am. Thorac. Soc. 3 503-510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato, M., Arap, W., and Pasqualini, R. (2007) Oncology 21 1346-1352 [PubMed] [Google Scholar]
- 21.Pasqualini, R., and Ruoslahti, E. (1996) Nature 380 364-366 [DOI] [PubMed] [Google Scholar]
- 22.Oh, Y., Mohiuddin, I., Sun, Y., Putnam, J. B., Jr., Hong, W. K., Arap, W., and Pasqualini, R. (2005) Chest 128 596S-600S [DOI] [PubMed] [Google Scholar]
- 23.Arap, W., Kolonin, M. G., Trepel, M., Lahdenranta, J., Cardó-Vila, M., Giordano, R. J., Mintz, P. J., Ardelt, P. U., Yao, V. J., Vidal, C. I., Chen, L., Flamm, A., Valtanen, H., Weavind, L. M., Hicks, M. E., Pollock, R. E., Botz, G. H., Bucana, C. D., Koivunen, E., Cahill, D., Troncoso, P., Baggerly, K. A., Pentz, R. D., Do, K. A., Logothetis, C. J., and Pasqualini, R. (2002) Nat. Med. 8 121-127 [DOI] [PubMed] [Google Scholar]
- 24.Giordano, R. J., Cardó-Vila, M., Lahdenranta, J., Pasqualini, R., and Arap, W. (2001) Nat. Med. 7 1249-1253 [DOI] [PubMed] [Google Scholar]
- 25.Kolonin, M. G., Saha, P. K., Chan, L., Pasqualini, R., and Arap, W. (2004) Nat. Med. 10 625-632 [DOI] [PubMed] [Google Scholar]
- 26.Jat, P. S., Noble, M. D., Ataliotis, P., Tanaka, Y., Yannoutsos, N., Larsen, L., and Kioussis, D. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 5096-5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langley, R. R., Ramirez, K. M., Tsan, R. Z., Van Arsdall, M., Nilsson, M. B., and Fidler, I. J. (2003) Cancer Res. 63 2971-2976 [PubMed] [Google Scholar]
- 28.Bligh, E. G., and Dyer, W. J. (1959) Can. J. Biochem. Physiol. 37 911-917 [DOI] [PubMed] [Google Scholar]
- 29.Dobrowsky, R. T., and Kolesnick, R. N. (2001) Methods Cell Biol. 66 135-165 [DOI] [PubMed] [Google Scholar]
- 30.Arap, M. A., Lahdenranta, J., Mintz, P. J., Hajitou, A., Sarkis, A. S., Arap, W., and Pasqualini, R. (2004) Cancer Cell 6 275-284 [DOI] [PubMed] [Google Scholar]
- 31.Ellerby, H. M., Arap, W., Ellerby, L. M., Kain, R., Andrusiak, R., Rio, G. D., Krajewski, S., Lombardo, C. R., Rao, R., Ruoslahti, E., Bredesen, D. E., and Pasqualini, R. (1999) Nat. Med. 5 1032-1038 [DOI] [PubMed] [Google Scholar]
- 32.Lahdenranta, J., Sidman, R. L., Pasqualini, R., and Arap, W. (2008) FASEB J. 21 3272-3278 [DOI] [PubMed] [Google Scholar]
- 33.Aoshiba, K., Yokohori, N., and Nagai, A. (2003) Am. J. Respir. Cell Mol. Biol. 28 555-562 [DOI] [PubMed] [Google Scholar]
- 34.Alon, T., Hemo, I., Itin, A., Pe'er, J., Stone, J., and Keshet, E. (1995) Nat. Med. 1 1024-1028 [DOI] [PubMed] [Google Scholar]
- 35.Carmeliet, P. (2005) Nature 438 932-936 [DOI] [PubMed] [Google Scholar]
- 36.Storkebaum, E., Lambrechts, D., and Carmeliet, P. (2004) BioEssays 26 943-954 [DOI] [PubMed] [Google Scholar]
- 37.Le Cras, T. D., Markham, N. E., Tuder, R. M., Voelkel, N. F., and Abman, S. H. (2002) Am. J. Physiol. 283 L555-L562 [DOI] [PubMed] [Google Scholar]
- 38.Brown, K. R., England, K. M., Goss, K. L., Snyder, J. M., and Acarregui, M. J. (2001) Am. J. Physiol. 281 L1001-L1010 [DOI] [PubMed] [Google Scholar]
- 39.Aird, W. C. (2003) Crit. Care Med. 31 S221-S230 [DOI] [PubMed] [Google Scholar]
- 40.Kolonin, M. G., Pasqualini, R., and Arap, W. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13055-13060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchiò, S., Lahdenranta, J., Schlingemann, R. O., Valdembri, D., Wesseling, P., Arap, M. A., Hajitou, A., Ozawa, M. G., Trepel, M., Giordano, R. J., Nanus, D. M., Dijkman, H. B., Oosterwijk, E., Sidman, R. L., Cooper, M. D., Bussolino, F., Pasqualini, R., and Arap, W. (2004) Cancer Cell 5 151-162 [DOI] [PubMed] [Google Scholar]
- 42.Pasqualini, R., Koivunen, E., and Ruoslahti, E. (1997) Nat. Biotechnol. 15 542-546 [DOI] [PubMed] [Google Scholar]
- 43.Rajotte, D., Arap, W., Hagedorn, M., Koivunen, E., Pasqualini, R., and Ruoslahti, E. (1998) J. Clin. Invest. 102 430-437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao, V. J., Ozawa, M. G., Trepel, M., Arap, W., McDonald, D. M., and Pasqualini, R. (2005) Am. J. Pathol. 166 625-636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevens, T., Rosenberg, R., Aird, W., Quertermous, T., Johnson, F. L., Garcia, J. G., Hebbel, R. P., Tuder, R. M., and Garfinkel, S. (2001) Am. J. Physiol. 281 C1422-C1433 [DOI] [PubMed] [Google Scholar]
- 46.Healy, A. M., Morgenthau, L., Zhu, X., Farber, H. W., and Cardoso, W. V. (2000) Dev. Dyn. 219 341-352 [DOI] [PubMed] [Google Scholar]
- 47.Kelly, J. J., Moore, T. M., Babal, P., Diwan, A. H., Stevens, T., and Thompson, W. J. (1998) Am. J. Physiol. 274 L810-L819 [DOI] [PubMed] [Google Scholar]
- 48.Arap, M. A., Lahdenranta, J., Hajitou, A., Marini, F. C., III, Wood, C. G., Wright, K. C., Fueyo, J., Arap, W., and Pasqualini, R. (2004) Mol. Ther. 9 305-310 [DOI] [PubMed] [Google Scholar]
- 49.Arap, W., Pasqualini, R., and Ruoslahti, E. (1998) Science 279 377-380 [DOI] [PubMed] [Google Scholar]
- 50.Hajitou, A., Trepel, M., Lilley, C. E., Soghomonyan, S., Alauddin, M. M., Marini, F. C., III, Restel, B. H., Ozawa, M. G., Moya, C. A., Rangel, R., Sun, Y., Zaoui, K., Schmidt, M., von Kalle, C., Weitzman, M. D., Gelovani, J. G., Pasqualini, R., and Arap, W. (2006) Cell 125 385-398 [DOI] [PubMed] [Google Scholar]
- 51.Hajitou, A., Lev, D. C., Hannay, J. A., Korchin, B., Staquicini, F. I., Soghomonyan, S., Alauddin, M. M., Benjamin, R. S., Pollock, R. E., Gelovani, J. G., Pasqualini, R., and Arap, W. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 4471-4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haimovitz-Friedman, A., Cordon-Cardo, C., Bayoumy, S., Garzotto, M., McLoughlin, M., Gallily, R., Edwards, C. K., 3rd, Schuchman, E. H., Fuks, Z., and Kolesnick, R. (1997) J. Exp. Med. 186 1831-1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kolesnick, R., and Fuks, Z. (2003) Oncogene 22 5897-5906 [DOI] [PubMed] [Google Scholar]
- 54.Vandivier, R. W., Fadok, V. A., Hoffmann, P. R., Bratton, D. L., Penvari, C., Brown, K. K., Brain, J. D., Accurso, F. J., and Henson, P. M. (2002) J. Clin. Invest. 109 661-670 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.