Abstract
Mechanical loading of bone initiates an anabolic, anticatabolic pattern of response, yet the molecular events involved in mechanical signal transduction are not well understood. Wnt/β-catenin signaling has been recognized in promoting bone anabolism, and application of strain has been shown to induce β-catenin activation. In this work, we have used a preosteoblastic cell line to study the effects of dynamic mechanical strain on β-catenin signaling. We found that mechanical strain caused a rapid, transient accumulation of active β-catenin in the cytoplasm and its translocation to the nucleus. This was followed by up-regulation of the Wnt/β-catenin target genes Wisp1 and Cox2, with peak responses at 4 and 1 h of strain, respectively. The increase of β-catenin was temporally related to the activation of Akt and subsequent inactivation of GSK3β, and caveolin-1 was not required for these molecular events. Application of Dkk-1, which disrupts canonical Wnt/LRP5 signaling, did not block strain-induced nuclear translocation of β-catenin or up-regulation of Wisp1 and Cox2 expression. Conditions that increased basal β-catenin levels, such as lithium chloride treatment or repression of caveolin-1 expression, were shown to enhance the effects of strain. In summary, mechanical strain activates Akt and inactivates GSK3β to allow β-catenin translocation, and Wnt signaling through LRP5 is not required for these strain-mediated responses. Thus, β-catenin serves as both a modulator and effector of mechanical signals in bone cells.
Bone tissue undergoes remodeling throughout life to adapt to its mechanical load, resulting in a mass that is optimized for daily mechanical demands. The remodeling process is tightly regulated by a balance between the number of bone-producing osteoblasts and bone-resorbing osteoclasts. Application of mechanical load promotes bone formation, whereas the removal of load leads to bone loss (1, 2). More recently, an important role for Wnt/β-catenin signaling has been recognized in promoting bone anabolism (3), and interestingly, mice with a loss-of-function mutation in the Wnt co-receptor LRP5 were shown to be resistant to the positive effects of local bone loading (4). Information further linking mechanically induced bone formation with activation of Wnt/β-catenin processes has emerged with evidence that straining cells causes translocation of β-catenin into the cell nucleus (5, 6). Consistent with β-catenin activation, osteoblasts respond to mechanical loading with increased expression of Wnt/β-catenin target genes, including Sfrp1 (secreted frizzled-related protein 1) and cyclin D1 (5).
The loss of β-catenin has been found to disrupt both skeletal development and postnatal bone acquisition (7, 8), establishing the importance of β-catenin signaling in osteoblast differentiation and function. β-Catenin can be found in at least two different cellular pools, suggesting compartmentalized roles. The pool of β-catenin found at the plasma membrane is bound to E-cadherin and α-catenin in adherens junctions. The soluble pool of β-catenin found in the cytoplasm, upon appropriate stimulation, can be translocated to the nucleus to interact with TCF/LEF (T cell factor/lymphoid enhancer binding factor) transcription factors.
The level of β-catenin in the cytoplasm is regulated by a destruction complex consisting of adenomatous polyposis coli, axin, and GSK3β (glycogen synthase kinase 3β). Constitutive phosphorylation of β-catenin by cyclin-dependent kinase at serine 45, followed by phosphorylation of the N terminus by GSK3β, targets β-catenin for degradation by the ubiquitin/proteosome pathway, limiting levels of cytoplasmic β-catenin under resting conditions. In canonical Wnt signaling, binding of Wnt ligand to its transmembrane receptors frizzled and LRP5/6 leads to disruption of the β-catenin destruction complex, which allows accumulation of stabilized β-catenin and subsequent nuclear translocation. Alteration of β-catenin action, either by a gain-of-function mutation in LRP5 (G171V) or by GSK3β inhibition, modulated the response of osteoblasts to mechanical loading (5).
GSK3β has been implicated in many cellular processes, including proliferation, differentiation, and apoptosis (9). A subset of its target proteins require phosphorylation at a priming site for subsequent phosphorylation by GSK3β. Phosphorylation of these “primed” target proteins, including β-catenin, can be blocked by phosphorylation of GSK3β at serine 9. Insulin inactivates GSK3β in this manner through Akt/protein kinase B (10). Recent studies have described β-catenin activation independent of signaling through Wnt ligands and their co-receptors (11, 12), which was mediated by phosphorylation of GSK3β by Akt downstream of signaling through heterotrimeric G protein-coupled receptors. Cyclic strain has been shown to activate Akt in multiple cell types (13, 14). Furthermore, the scaffolding protein caveolin-1 was necessary for stretch-dependent activation of Akt/protein kinase B in vascular smooth muscle cells (15). These observations suggested to us that mechanical activation of β-catenin in osteoblasts could possibly work through both Wnt-dependent and Wnt-independent pathways and that Wnt-independent signaling could involve interactions between caveolin-1, Akt, and GSK3β.
In this work, we have used a preosteoblastic cell line to study the effect of mechanical strain via substrate deformation on β-catenin signaling. We show that strain stimulates a rapid, transient accumulation of active β-catenin in the cytoplasm and its translocation to the nucleus, with subsequent up-regulation of the Wnt/β-catenin target genes Wisp1 (Wnt-induced secreted protein 1) and Cox2 (cyclooxygenase 2). Wnt signaling through LRP5 is not required for these strain-mediated responses. Akt and GSK3β phosphorylation are temporally related to the translocation of β-catenin into the nucleus, and caveolin-1 is not required for these molecular events. Finally, we show that conditions that increase basal cytoplasmic β-catenin levels, such as lithium chloride treatment or repression of caveolin-1 expression, can enhance the effects of strain. Taken together, our results suggest that β-catenin is a node where multiple influences, including mechanical inputs, are integrated to regulate cell function.
EXPERIMENTAL PROCEDURES
Reagents—Fetal bovine serum was obtained from Hyclone (Logan, UT). Culture media, glutamine, antibiotics, Lipofectamine 2000, oligofectamine, reverse transcriptase, and Taq polymerase were purchased from Invitrogen. Lithium chloride, LY294002, and indomethacin were obtained from Sigma. Wnt3a and Dkk-1 were from R&D Systems (Minneapolis, MN). The RNA isolation kit and DNase I were from Qiagen (Valencia, CA), and random primers were from Ambion (Austin, TX).
Cell Culture—CIMC-4 cells harvested from mouse calvariae express an interferon-γ-inducible, temperature-sensitive, SV40 large T antigen transgene (16). Cells were maintained in permissive conditions at 33 °C in α-minimum Eagle's medium containing 10% fetal bovine serum and 100 units/ml interferon-γ. Medium was changed every 3–4 days, and passages above 24 were not used. Prior to experiments, cells were cultured for 1 week in nonpermissive conditions at 37 °C in MEM containing 10% fetal bovine serum, 1.25 mm glutamine, and 100 μg/ml penicillin/streptomycin. Cells were plated at a density of 10,000–20,000 cells/cm2. The following pharmacological agents were used to evaluate the potential role of various signaling pathways in strain-mediated gene changes: the LRP5 inhibitor Dkk-1 (50 ng/ml), the phosphatidylinositol 3-kinase (PI3-kinase)3 inhibitor LY294002 (20 μm), and the cyclooxygenase inhibitor indomethacin (5 μm). The pharmacological agent was added to cultures 1 h prior to strain initiation and remained in the culture medium throughout the strain experiment.
Transient Transfection with siRNA—siRNA targeting murine caveolin-1 (siCav) was purchased from Ambion (Austin, TX). CIMC-4 cells (60–70% confluence) were transfected with siCav or a control siRNA (siSCR) at a concentration of 100 nm and oligofectamine (25 μl/nmol siRNA) in the absence of serum for 5 h. An equal volume of medium with serum was then added to bring the final concentration of serum to 10%. The medium was changed 24 h after transfection. Mechanical strain or pharmacological treatment was applied 48 h after transfection.
Mechanical Strain—For strain experiments, CIMC-4 cells were plated on 6-well Bioflex collagen-I-coated plates (Flexcell International, Hillsborough, NC). Uniform biaxial strain was applied (2% magnitude, 10 cycles/min) using the Flexcell FX-4000 system.
Real Time RT-PCR—Total RNA was isolated by using the RNeasy minikit (Qiagen) and treated with DNase I to remove contaminating genomic DNA. Reverse transcription was performed with 1 μg of RNA in a total volume of 20 μl/reaction. Real time PCR was performed using the iCycler instrument (Bio-Rad). Amplification reactions were performed in 25 μl containing primers at 0.5 μm, dNTPs (0.2 mm each) in PCR buffer, and 0.03 units of Taq polymerase along with SYBR green (Molecular Probes, Inc., Eugene, OR) at 1:150,000. Aliquots of cDNA were diluted 5–5000-fold to generate relative standard curves to which sample cDNA was compared. 18S and Runx2 primers were as described previously (17, 18). For Wisp1, forward and reverse primers were 5′-CTG GAC AGA AAA GGG CAT GT-3′ and 5′-AGG AAG GAG GGG AAA TCT CA-3′, respectively. For Cox2, forward and reverse primers were 5′-AGA AGG AAA TGG CTG CAG AA-3′ and 5′-GCT CGG CTT CCA GTA TTG AG-3′, respectively. Standards and samples were run in triplicate. PCR products from all species were normalized for the amount of 18S in the same RT sample, which was also standardized on a dilution curve from RT sample.
Western Blotting—Cells were washed twice with cold phosphate-buffered saline. Whole cell lysates were prepared with lysis buffer (150 mm NaCl, 50 mm Tris-HCl, 1 mm EGTA, 0.24% sodium deoxycholate, 1% IGEPAL, pH 7.5) containing 25 mm NaF and 2 mm Na3VO4. Aprotinin, leupeptin, pepstatin, and phenylmethylsulfonyl fluoride were added fresh prior to each lysis. For isolation of cytoplasmic and nuclear fractionates, cells were trypsinized and rinsed in phosphate-buffered saline and then lysed for 15 min on ice in cytoplasmic lysis buffer (0.33 m sucrose, 1 mm MgCl2, 0.1% Triton X-100, 10 mm Hepes, pH 7.4) containing phosphatases and proteinases as above. The cytoplasmic fraction was collected following centrifugation, and then the samples were rinsed in cytoplasmic lysis buffer and centrifuged to obtain a nuclear pellet. The nuclear pellet was lysed for 15 min on ice in 0.45 m NaCl, 10 mm Hepes, pH 7.4, and the nuclear fraction was collected following centrifugation. Protein (5–20 μg) was loaded onto a 10% polyacrylamide gel for chromatography and transferred to polyvinylidene difluoride membrane. After blocking, the membrane was incubated with primary antibodies overnight at 4 °C. The primary antibodies used include anti-active β-catenin (clone 8E7; Upstate Biotechnology, Inc., Lake Placid, NY), anti-total β-catenin (BD Biosciences), anti-phospho-GSK3β (serine 9, clone 2D3; Upstate Biotechnology), anti-GSK3β (Chemicon, Billerica, MA), anti-phospho-Akt (serine 473; Cell Signaling, Danvers, MA), anti-Akt (clone 11E7; Cell Signaling), and anti-actin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The antibody used for active β-catenin was specific for β-catenin dephosphorylated at serine 37 and threonine 41 (19). After washing, the membrane was then incubated with secondary antibody conjugated with horseradish peroxidase. The proteins were detected with the ECL Plus chemiluminescence kit (Amersham Biosciences).
Immunofluorescence—Following strain application, strained and control samples were fixed in 4% paraformaldehyde and permeabilized in 0.1% Triton X-100/phosphate-buffered saline. Primary antibodies (anti-active β-catenin or anti-total β-catenin) were applied overnight at 4 °C. Following rinses, an anti-mouse fluorescein isothiocyanate-conjugated antibody (Jackson Immunoresearch, West Grove, PA) was applied for 30 min. Samples were mounted in anti-fade reagent (Molecular Probes, Inc., Eugene, OR) and viewed using a Leica SP2-AOBS confocal microscope.
Luciferase Assay—CIMC-4 cells were seeded at a density of 100,000 cells/well in 6-well plates. Twenty-four hours after seeding, cells were transfected with 2.5 μg/well TCF reporter plasmid (TopFlash; Upstate Biotechnology) and 10 μl/well Lipofectamine in the absence of serum for 5 h. Then an equal volume of medium with serum was added to bring the final concentration of serum to 10%, and medium was changed 24 h after transfection. β-Galactosidase plasmid (1 μg/well) was transfected together with TopFlash to control for transfection efficiency. TopFlash signal was assessed with the Luciferase Assay System (Promega, Madison, WI). The Galacto-Star™ system (Applied Biosystems, Bedford, MA) was used for the detection of β-galactosidase. Light emission was measured with the GloMax luminometer (Promega, Madison, WI).
Immunoprecipitation—Following strain application, strained and control plates were washed on ice with cold phosphate-buffered saline and lysed in immunoprecipitation buffer (150 mm NaCl, 50 mm Tris-HCl, 1 mm EGTA, 0.24% sodium deoxycholate, 1% IGEPAL, pH 7.5). Aprotinin, leupeptin, and pepstatin were added fresh prior to lysis. Cells were lysed and transferred to Eppendorf tubes and centrifuged. Lysates (175 μg) were combined with ExactaCruzB IP matrix and an anti-caveolin-1 polyclonal antibody (Santa Cruz Biotechnology) and then incubated overnight at 4 °C. Following rinses, beads were resuspended in 40 μl of 2× loading buffer. Immunoprecipitation samples were then analyzed by immunoblotting with anti-total-β-catenin and anti-caveolin-1 (BD Biosciences).
Statistical Analysis—Results are expressed as the mean ± S.E. Statistical significance was evaluated by one-way analysis of variance or t test (GraphPad Prism, San Diego, CA).
RESULTS
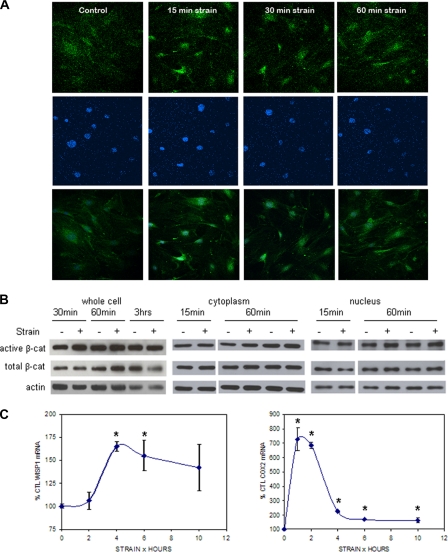
Strain Induces Nuclear Translocation of β-Catenin—Mechanical strain has been shown to induce β-catenin nuclear translocation (6) as well as to activate Wnt/β-catenin target genes (5) in osteosarcoma and osteoblastic cells. Here strain (0.17 Hz, 2%) transiently activated β-catenin in the preosteoblastic CIMC-4 cell line, as determined using an antibody to detect β-catenin that is dephosphorylated at serine 37 and threonine 41. This “active” form of β-catenin has been shown to be increased in response to Wnt stimulation and by mechanical loading (6, 19). As shown in Fig. 1A, 15 and 30 min after initiating the strain regimen, active β-catenin was strongly increased in the nuclei of strained cells as compared with unstrained control cells. The increase in active nuclear β-catenin reached a plateau by 60 min. A significant change of total β-catenin level, as assessed with an antibody against phosphorylated β-catenin, was not observed in response to strain by confocal microscopy (data not shown); however, the high level of total β-catenin present in the cytoplasm of these cells may prevent appreciation of strain-induced translocation of this species.
FIGURE 1.
Mechanical strain induces β-catenin accumulation. A, CIMC-4 cells were subjected to strain for 15–60 min and immunostained for active β-catenin (top) and DAPI (middle). Images of active β-catenin and DAPI staining were merged (bottom) to visualize active β-catenin in the nucleus. Increased nuclear active β-catenin was detected 15 min after beginning mechanical strain by confocal microscopy. B, CIMC-4 cells were strained for 15 min to 3 h. Protein from whole cell lysates and cytoplasmic and nuclear fractionates was extracted and run at 10 μg/lane on 7.5% SDS-PAGE. Western blotting showed increases in active and total β-catenin from whole cell lysates at 30 and 60 min, respectively, whereas an increase in both cytoplasmic and nuclear active β-catenin was detected 60 min after strain. Expression of actin is shown as a control for equal protein loading. C, expression of the β-catenin target genes WISP1 and COX2 in CIMC-4 cells 1–10 h after strain initiation was evaluated by real time RT-PCR. A significant increase in WISP1 expression was measured at 4 and 6 h, whereas COX2 expression was increased at all times. Two experiments grouped (mean ± S.E.) for statistical analyses are shown in the graph. *, significant effect of strain (p < 0.05). CTL, control.
Western blots of whole cell lysates showed an increase in active and total β-catenin after strain for 30 and 60 min, respectively (Fig. 1B). Three hours after strain initiation, active β-catenin had returned to base line, and total β-catenin had decreased to a level below base line. To measure changes of active and total β-catenin in the nucleus, the nuclear fraction was separated from the cytoplasmic fraction. A reproducible increase was not measurable in active or total β-catenin at 15 min of strain for either fraction. In the cytoplasmic fraction, active β-catenin was reliably increased 60 min after initiating strain. Both active and total β-catenin were increased in the nuclear fraction 60 min after strain initiation.
Consistent with the rapid activation and translocation of β-catenin, a transient increase of the Wnt/β-catenin target gene Wisp1 was measured after mechanical strain (Fig. 1C). WISP1 expression was maximally increased to 165 ± 5% of the control level at 4 h of strain and gradually returned to basal levels by 24 h (data not shown). Cox2, shown previously to respond to strain (5) and to be up-regulated by β-catenin (20, 21), was also evaluated. Cox2 expression was more rapidly and strongly enhanced by mechanical strain as compared with Wisp1, with a peak increase of 726 ± 78% at 1 h. Although peak Cox2 expression fell sharply by 4 h, the level remained significantly elevated up to 10 h following strain initiation.
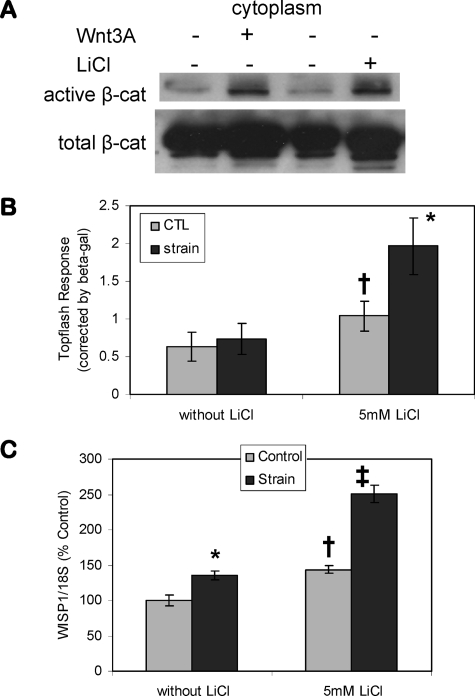
Inhibition of β-Catenin Proteolysis Augments Strain Effects—Increases in cytoplasmic β-catenin were next shown to augment the distal response to strain. LiCl was used to inactivate GSK3β; after treating cells with 20 mm LiCl for 3 h, significant increases of cytoplasmic active and total β-catenin were observed (Fig. 2A). The increase of β-catenin induced by LiCl was similar to that caused by Wnt3a treatment. The TopFlash luciferase reporter, containing TCF/LEF consensus sequences, was used to assess a functional response to nuclear β-catenin. Strain-induced nuclear accumulation of β-catenin as evidenced by confocal microscopy and Western blot, shown in Fig. 1, was not sufficient to achieve a detectable change in TopFlash luciferase activity, shown in the first set of bars in Fig. 2B. A submaximal dose of LiCl (5 mm) caused a 164 ± 28% increase in luciferase activity shown in the second set of bars. In the presence of LiCl, application of strain further increased TopFlash activity to 188 ± 33% of that of LiCl alone. Wisp1 expression was also evaluated (Fig. 2C). Consistent with the TopFlash data, 5 mm LiCl increased basal Wisp1 expression to 144 ± 6%, and strain up-regulation was significantly greater in cultures pretreated with LiCl (135 ± 6 and 174 ± 8% of control in the absence and presence of LiCl). Thus, the data suggest that increased β-catenin availability by LiCl treatment allowed the transient mechanical signal resulting in β-catenin activation to be enhanced. These results confirm that β-catenin is involved in the cellular response to strain.
FIGURE 2.
Inhibition of β-catenin proteolysis augments strain effects. A, Western blot for β-catenin in CIMC-4 cells after a 3-h treatment of 20 mm LiCl or 100 ng/ml Wnt3a showed that LiCl increased cytoplasmic active β-catenin to an extent similar to that of Wnt3a. B, CIMC-4 cells were transiently transfected with TopFlash plasmid containing six TCF/LEF binding sites driving the expression of luciferase. β-Galactosidase plasmid was transfected at the same time to control for transfection efficiency. TopFlash response in osteoblasts strained overnight was only seen when cells were also exposed to LiCl (5 mm). Six experiments grouped (mean ± S.E.) for statistical analyses are shown in the graph. *, significant effect of strain (p < 0.05); †, significant difference between unstrained basal and LiCl-treated cultures (p < 0.05). CTL, control. C, strain up-regulation of WISP1 was enhanced in the presence of LiCl. WISP1 expression at 4 h of strain was evaluated by real time RT-PCR. Cultures were treated with LiCl 1 h prior to strain initiation. The data were normalized to the expression level measured in basal control cells. Data were compiled from two experiments and are shown as mean ± S.E. *, significant effect of strain (p < 0.05); †, significant difference between basal and LiCl control cultures (p < 0.05); ‡, significant difference from all other conditions (p < 0.05).
Strain Causes Phosphorylation of GSK3β through an LRP-independent Process—Phosphorylation of GSK3β at Ser9 blocks its ability to phosphorylate β-catenin, which marks β-catenin for degradation. Whether this pathway was involved in mechanical activation of β-catenin was considered. GSK3β Ser9 phosphorylation was measurable 30 min after beginning strain exposure, with a peak response at 60 min (Fig. 3A). GSK3β phosphorylation fell to below basal level at 3 h, consistent with the decrease in total β-catenin at this time in the whole cell (Fig. 1B). Total GSK3β protein was unchanged by strain. Thus, increased β-catenin levels followed a time course consistent with strain-induced GSK3β inactivation.
FIGURE 3.
Strain inhibits GSK3β action through an LRP-independent process. A, Western blot of GSK3β phosphorylation (serine 9) showed an increase at 30 min of strain and a peak response at 1 h. Actin protein was blotted as a control for equal protein loading. B, Western blot of Akt phosphorylation (serine 473) showed a strong increase at 15 min of strain, with the response returning to base line at 1 h. C, strain up-regulation of WISP1 and COX2 expression was not disrupted by treatment with the LRP5 inhibitor Dkk-1 (50 ng/ml). WISP1 and COX2 expression at 4 h of strain were evaluated by real time RT-PCR. Cultures were treated with Dkk-1 1 h prior to strain initiation. The data were normalized to the expression level measured in basal control cells. Data were compiled from three experiments and shown as mean ± S.E. *, shows significant effect of strain, p < 0.05. D, Western blots showed that Dkk-1 did not block the increase in phospho-GSK3β (Ser9) and phospho-Akt (Ser473) levels by strain. GSK3β was evaluated at 60 min of strain, and Akt was evaluated at 30 min. E, confocal microscopy showed that Dkk-1 did not block strain-induced nuclear translocation of active β-catenin. Active β-catenin level was assessed by immunofluorescence (top) 15 min after strain initiation. Merged images of active β-catenin and nuclear DAPI staining are shown in the bottom.
Akt/protein kinase B phosphorylates and inactivates GSK3β in response to insulin stimulation (10), and strain can activate Akt in multiple cell types (13, 22). Akt activation was investigated as part of the signaling cascade in response to mechanical strain. Akt phosphorylation at Ser473 was strongly induced at 15 min of strain, with a return to nearly basal levels by 60 min (Fig. 3B), consistent with the transient mechanical effect.
The LRP5 inhibitor Dkk-1 was used to evaluate whether the canonical Wnt/LRP5 pathway was required for strain activation of β-catenin signaling. CIMC-4 cultures were strained in the presence of Dkk-1 and analyzed for Wisp1 and Cox2 gene expression. Dkk-1 did not disrupt strain up-regulation of Wisp1 (123 ± 8% of control for no treatment and 158 ± 16% of control with Dkk-1; Fig. 3C) or Cox2 (297 ± 53% of control for no treatment and 317 ± 33% of control with Dkk-1). The basal levels of Wisp1 and Cox2 expression were unchanged by Dkk-1. As well, strain increased GSK3β Ser9 phosphorylation in the presence of Dkk-1, consistent with strain-induced Akt activation (Fig. 3D). Basal levels of GSK3β phosphorylation and Akt phosphorylation were unchanged by Dkk-1. Examined by confocal microscopy, Dkk-1 did not disrupt strain-induced nuclear accumulation of active β-catenin (Fig. 3E). Cumulatively, the data suggest that Wnt signaling through LRP5 is not involved in the mechanisms by which strain activates β-catenin.
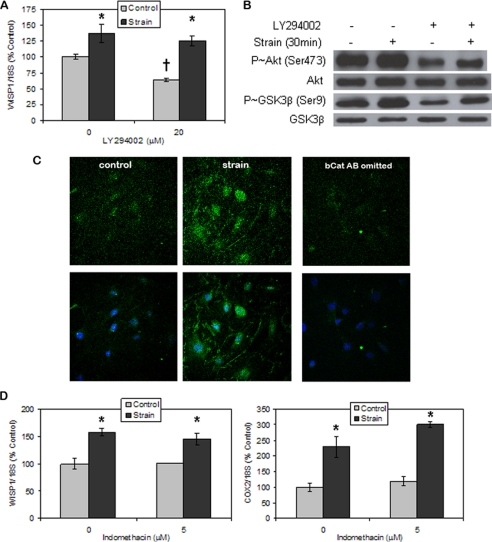
Mechanical Activation of β-Catenin Target Genes Does Not Require PI3-kinase or PGE2 Generation—The PI3-kinase inhibitor LY294002 was used to evaluate whether PI3-kinase activation by mechanical strain was proximal to Akt activation. Although basal Wisp1 gene expression was diminished by 36 ± 3% in the presence of LY294002, PI3-kinase inhibition did not block, and may have enhanced, the strain-induced increase in Wisp1 gene expression (138 ± 14% of control for no treatment and 197 ± 20% of control with LY294002; Fig. 4A). Thus, strain induction of Wisp1 expression did not require signaling through the PI3-kinase pathway.
FIGURE 4.
Mechanical activation of β-catenin targets does not require PI3-kinase or PGE2 generation. A, WISP1 expression was decreased by treatment with the PI3-kinase inhibitor LY294002 (20 μm), but strain up-regulated WISP1 expression in the presence of LY294002. WISP1 expression at 4 h of strain was evaluated by real-time RT-PCR. Cultures were treated with the inhibitor 1 h prior to strain initiation. The data were normalized to the expression level measured in basal control cells. Data were compiled from two experiments and shown as mean ± S.E. *, significant effect of strain (p < 0.05); †, significant difference between basal and LY294002 control cultures (p < 0.05). B, Western blots showed that strain-induced increases in phospho-GSK3β (P∼GSK3β) (Ser9), and phospho-Akt (P∼Akt) (Ser473) were not blocked by LY294002 treatment. C, CIMC-4 cells were subjected to strain for 15 min, and active β-catenin level was assessed by immunofluorescence (top). Merged images of active β-catenin and nuclear DAPI staining are shown in the bottom. Confocal microscopy showed that LY294002 did not block strain-induced nuclear translocation of active β-catenin. A sample in which the active β-catenin antibody was omitted was also included (right). D, strain up-regulation of WISP1 and COX2 expression was not disrupted by treatment with the cyclooxygenase inhibitor indomethacin (5 μm). WISP1 and COX2 expression at 6 h of strain were evaluated by real time RT-PCR. Cultures were treated with indomethacin 1 h prior to strain initiation. The data were normalized to the expression level measured in basal control cells. Representative data are shown as mean ± S.E., and the experiment was repeated one time. *, significant effect of strain (p < 0.05).
Indeed, despite the fact that basal Akt phosphorylation was strongly decreased by LY294002 treatment, strain significantly increased phosphorylation of Akt as compared with the unstrained condition when the PI3-kinase inhibitor was present (Fig. 4B). Basal phospho-GSK3β protein was less affected by the presence of LY294002, confirming that other kinases target the Ser9 site. Mechanical strain induction of GSK3β phosphorylation was unaffected by the presence of LY294002, shown in the figure. Total GSK3β and Akt levels were unchanged by LY294002 treatment or by strain. Consistent with a strain mechanism that does not rely on PI3-kinase, LY294002 treatment failed to block strain-induced increases in nuclear accumulation of active β-catenin assessed by confocal microscopy (Fig. 4C).
The rapid rise in COX2 mRNA upon strain application might implicate PGE2 in the subsequent gene response. To block COX2-generated PGE2 synthesis that might arise in response to strain, the cyclooxygenase inhibitor indomethacin was added. Indomethacin did not disrupt the strain-induced increase in Wisp1 expression, which rose to 159 ± 7 and 144 ± 11% of that of control in the absence and presence of indomethacin, respectively (Fig. 4D). Indomethacin treatment also did not affect the increase in Cox2 expression by strain (230 ± 33 and 255 ± 9% of control in the absence and presence of indomethacin). Basal levels of Wisp1 and Cox2 expression were unchanged by indomethacin. The persistence of Wisp1 and Cox2 up-regulation by strain in the presence of indomethacin indicates that PGE2 is not required for downstream strain effects.
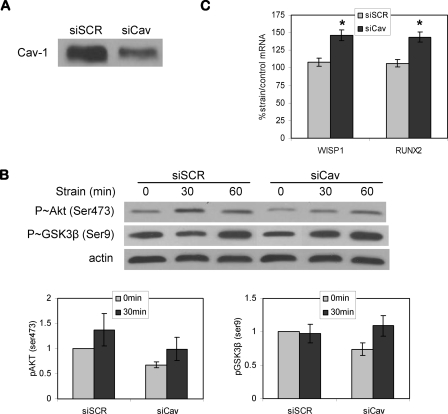
Caveolin-1 Knockdown Does Not Prevent Strain Activation of Kinase and Gene Targets—Strain-induced activation of Akt in vascular cells has previously been shown to require the presence of caveolin (15), such that disruption of caveolar structures by inhibition of caveolin-1 prevented Akt activation. We therefore evaluated whether caveolin might be part of the proximal mechanical response apparatus in bone cells. Delivery of siCav was used to knock down caveolin-1 protein by more than 50% at 48 h compared with cells transfected with a scrambled control siRNA, shown in Fig. 5A. Fig. 5B reveals that bone cells transfected with siCav had decreased basal levels of phosphorylated Akt and GSK3β. Reductions in Akt and GSK3β phosphorylation were a consistent finding over three experiments, shown by averaged densitometries for phosphorylated proteins (graphs under the representative Western blots). The activation of Akt following 30 and 60 min of strain in cells treated with siCav was similar to that in cells transfected with control siRNA. Thus, in contrast to vascular cells, mechanical strain induced activation of Akt in bone cells despite decreases in caveolin-1.
FIGURE 5.
Caveolin-1 knockdown does not prevent strain activation of kinase and gene targets. A, caveolin-1 protein levels were decreased by treatment with siRNA targeting caveolin-1 for 48 h. B, Western blot showed that caveolin-1 knockdown did not inhibit strain activation of Akt. Strain-induced inactivation of GSK3β, shown by the increase of phospho-GSK3β (Ser9), was enhanced after strain for 30 min when caveolin-1 expression was decreased. The base-line levels of phospho-Akt (Ser473) and phospho-GSK3β (Ser9) were decreased in cells transfected with siCav. Representative blotting is shown here. Averaged densitometries (mean ± S.E.) for phosphorylated proteins from three experiments are shown in the graphs. C, real time RT-PCR after 4 h of mechanical strain in cells transfected with control siRNA (siSCR) or siCav. The strain effect on WISP1 expression was masked in cells transfected with siSCR. Increases in WISP1 and RUNX2 gene expression by strain were seen in cells treated with siCav. Eight experiments grouped (mean ± S.E.) for statistical analyses are shown in the graph. *, significant effect of strain (p < 0.01).
Compared with nontransfected cells where strain induced GSK3β inactivation by 30 min, as shown in Fig. 3, delivery of control siRNA to cells dampened strain effects on GSK3β phosphorylation. There was no significant change in GSK3β Ser9 phosphorylation 30 min after beginning strain in cells transfected with control siRNA. The reasons for this are not known. In contrast, strain-induced GSK3β phosphorylation appeared to be enhanced when caveolin-1 was decreased, since inactivation of GSK3β by strain was strongly increased at 30 min in cells treated with siCav.
The absence of strain-induced changes in GSK3β phosphorylation upon delivery of control siRNA was underscored by a loss of strain-induced Wisp1 expression. As shown in Fig. 5C, in cells transfected with control siRNA, Wisp1 mRNA did not increase in response to strain at 4 h. However, in cells transfected with siRNA targeting caveolin-1, where we showed enhanced GSK3β phosphorylation by strain, up-regulation of Wisp1 mRNA was significant, increasing 147 ± 8% compared with unstrained cells. This finding further implicates strain-induced GSK3β inactivation to the downstream response. A Runx2 strain response had previously been detected only after 24 h of strain application (18) and was not affected by strain in cells transfected with control siRNA, although it is considered to be a β-catenin-responsive gene (23). Unexpectedly, however, Runx2 expression was increased 143 ± 7% after strain in cells that had been transfected with caveolin-1 siRNA. Thus, silencing caveolin-1 may have enhanced the ability of strain to induce β-catenin responses.
Caveolin-1 Knockdown Increases β-Catenin Nuclear Translocation—To inform our results implicating caveolin-1 in the mechanical responses of osteoblasts, we asked how silencing caveolin-1 would affect cytoplasmic β-catenin and its nuclear translocation in the absence of strain. It has been reported that caveolin-1 inhibits canonical Wnt/β-catenin signaling by sequestering β-catenin in the caveolar membrane (24). As shown in Fig. 6A, delivery of siCav to CIMC-4 cells significantly increased cytoplasmic β-catenin, especially total β-catenin. We were not able to demonstrate a significant change of nuclear β-catenin level by Western blot (data not shown). However, increased functional nuclear translocation of β-catenin was indicated by the TopFlash assay. The TopFlash signal was doubled in cells transfected with siCav as compared with cells transfected with a control siRNA (Fig. 6B); comparing the black bars, TopFlash was 100 ± 18 in siSCR cells and 203 ± 32 in siCav cells. Treatment with 20 mm LiCl increased TopFlash by nearly 3-fold in both conditions, indicating that the downstream effect of LiCl inactivation of GSK3β depended on the amount of β-catenin available for translocation. In the case of cells where caveolin-1 was silenced, more β-catenin was translocated when GSK3β was inactivated. As such, mechanical induction of β-catenin activation might also involve transfer of β-catenin from caveolae to cytoplasm.
FIGURE 6.
Caveolin-1 knockdown increases β-catenin nuclear translocation. A, CIMC-4 cells transfected with siRNA targeting caveolin-1 showed increased levels of cytoplasmic active and total β-catenin by Western blot. B, TopFlash response in cells treated with siCav showed increased base-line TopFlash signal. LiCl (20 mm) induced an increase of TopFlash signal, and its effect was further enhanced when caveolin-1 was decreased. Three experiments grouped (mean ± S.E.) for statistical analyses are shown in the graph. **, significant effect of LiCl (p < 0.01); ††, significant difference between control siRNA- and siCav-treated cultures (p < 0.01). C, Western blots of caveolin-1 immunoprecipitates after strain was applied for 15–60 min. The basal association of β-catenin with caveolin-1 was not disrupted by strain application. Caveolin-1 was blotted to show equal immunoprecipitate levels in samples.
To further explore a possible link between caveolin-1 and the accessibility of β-catenin, we evaluated whether the basal association of these two proteins was altered by strain. Caveolin-1 was immunoprecipitated from lysates made from unstrained control cultures and cultures subjected to 15–60 min of mechanical strain. Consistent with previous reports (24), caveolin-1 immunoprecipitates pulled down β-catenin (Fig. 6C). Comparable amounts of β-catenin were associated with caveolin-1 in control and strained cultures. These data indicated that β-catenin and caveolin-1 were associated but that short term mechanical strain did not significantly disrupt their association. This suggests that the pool of β-catenin associated with the plasma membrane is not acutely altered by mechanical strain and that increases in the cytoplasmic and nuclear levels of active β-catenin primarily result from the alteration in GSK3β activity by strain.
DISCUSSION
Maintenance of the skeleton is a sum game, balancing influences from hormones, growth factors, inflammation, and pregnancy, to name but a few factors. During health, these diverse inputs are integrated at a cellular level, where bone cells adapt skeletal structure to achieve success (i.e. load bearing without fracture). Load bearing itself has been increasingly recognized as the ultimate determinant of structure, where incoming signals not only serve as information from the functional environment but also activate pathways that lead to bone remodeling through control of osteoblast and osteoclast function. Mechanical input, importantly, promotes the addition of new bone to relieve stress during load, and it does this through increasing osteoblast numbers and activity at the sites of stress. A recent understanding that β-catenin is paramount to precursor entry into the osteoblast lineage and to an increase in markers consistent with modeling activity (3, 7) has also impacted our understanding of mechanical signaling. Mice lacking LRP5, the osteoblast receptor that responds to Wnt signaling to increase β-catenin activation, respond poorly to loading (4). As well, Wnt signaling was shown to be increased by loading, as were Wnt/β-catenin-responsive genes (5). Most recently, β-catenin was shown to translocate into the nucleus after loading in vitro (6, 25). In the work presented here, we have elucidated the proximal signals that contribute to the β-catenin response by mechanical loading of osteoblasts. Mechanical strain stimulated a rapid β-catenin response in osteoblasts, with increased cytoplasmic active β-catenin and subsequent nuclear translocation occurring within 30 min. Although the increase of nuclear active β-catenin was transient, the magnitude of change was sufficient to stimulate induction of target genes Wisp1 and Cox2.
Regulation of Cox2 by mechanical loading is well established in osteoblasts. Recent studies provide evidence that Cox2 is a downstream target of β-catenin in skeletal lineage cells. Overexpression of constitutively active β-catenin or LiCl treatment led to increased Cox2 expression in osteoblasts and articular chondrocytes (25, 26) as well as enhanced Cox2 promoter activity (21). In addition to transcriptional regulation, a recent report shows that β-catenin can stabilize Cox2 mRNA (27), a mechanism that might contribute to the steep rise in Cox2 expression we observed at 1 h of strain.
The possibility that canonical Wnt signaling might play a role in regulating bone responsiveness to mechanical loading was first suggested by observations that LRP5 gain- or loss-of-function mutations were associated with high or low bone mass, respectively, in humans and in mice. In vivo studies showed that mechanical stress up-regulated Wnt target genes and that this effect was enhanced in LRP5 gain-of-function transgenic mice (5). Our in vitro experiments indicate that mechanical loading-induced activation of β-catenin signaling in osteoblasts does not directly involve Wnt/LRP5. Application of Dkk-1, which disrupts Wnt signaling by binding to LRP5, failed to inhibit strain-induced nuclear translocation of β-catenin as well as subsequent up-regulation of Wisp1 and Cox2. This would suggest that direct mechanical stimulation of β-catenin does not require the Wnt molecule and, furthermore, that the LRP5 mutation amplifies, rather than initiates, the mechanical response, similarly to what we see in cells treated with LiCl. Interestingly, after binding to Wnt, LRP6 receptors are found as clusters in caveolar signalosomes (28), and effective downstream transmission of a Wnt signal requires caveolin (29). That we can repress caveolin-1 expression and still see mechanical activation of β-catenin, as discussed below, provides further evidence that strain's activation of β-catenin does not require Wnt binding of LRP.
GSK3β constitutively phosphorylates the N terminus of β-catenin following monophosphorylation by cyclin-dependent kinase at serine 45, which then marks β-catenin for degradation. When GSK3β is phosphorylated at serine 9, it cannot bind the priming site on its target protein, and its ability to phosphorylate the protein is blocked. LiCl treatment activates β-catenin by phosphorylating GSK3β at serine 9 (30). Similarly, we showed that mechanical strain induced a rapid but transient increase in serine 9 phosphorylation of GSK3β in parallel with the changes in β-catenin levels, indicating inactivation of GSK3β by strain.
Akt/protein kinase B is the probable effector of strain inactivation of GSK3β. Cyclic strain has been shown to activate Akt in multiple cell types, including smooth muscle cells and epithelial cells (13, 14). Akt phosphorylation was rapid in these systems, with the peak response occurring by 30 min after load initiation, and was dependent on PI3-kinase, since treatment with the inhibitor LY294002 blocked the response. Although we similarly found that Akt activation was rapidly increased by strain in osteoblasts, Akt phosphorylation was not dependent on PI3-kinase. This dissimilarity could be due to a difference in the cell type or in the strain regimen applied, since we used a more modest strain level (2%) compared with the other studies (10–25%). With regard to cell type, fluid flow-induced activation of Akt in osteoblasts was independent of PI3-kinase (31). Recent reports suggest that mTORC2 phosphorylates Akt at serine 473 and that integrin-linked kinase can contribute to Akt activation (32, 33). Another contributor to this signaling pathway may be caveolin-1, which was required for mechanical strain to cause activation of Akt in vascular smooth muscle and mesangial cells (15, 34). In our osteoblast cells, strain activation of Akt was not significantly changed when caveolin-1 was knocked down. Thus, in contrast to high magnitude strain responses, which appear to require both PI3-kinase and caveolin, low magnitude activation of Akt by strain in osteoblasts requires neither.
Although caveolin-1 was not required for the mechanical response studied here, it did modify β-catenin signaling. The increase in β-catenin levels and β-catenin directed transcriptional activity that were consequent to caveolin-1 knockdown probably resulted from a chronic disruption in sequestration of β-catenin at the plasma membrane. As well, overexpression of caveolin-1 decreases canonical β-catenin signaling (35). These findings may be important in explaining the larger and stronger bone mass observed in caveolin-1 null mice; furthermore, marrow stromal cells from caveolin-1 null mice more rapidly enter the osteoprogenitor lineage, and siRNA targeting caveolin-1 in normal murine stromal cells likewise promotes osteogenesis (36). Thus, the altered phenotype of the caveolin-1 null mouse is probably due to an amplification of β-catenin signaling.
Although mechanical strain influences β-catenin stability, alterations in the accessible pool of β-catenin also influence strain effects. Treatment with LiCl or caveolin-1 knockdown led to a chronic increase in cytoplasmic β-catenin levels and downstream events; mechanical responses were enhanced by both of these conditions. Mechanical induction of β-catenin target genes can also be increased by Wnt3a treatment (5). The increase in not only active but also total β-catenin measured after strain in our study might suggest that strain increases an accessible pool of β-catenin previously sequestered in a storage pool (e.g. caveolar membrane). The size of the accessible pool of β-catenin thus appears to be an important factor influencing the magnitude of response to mechanical loading.
In summary, we have shown that mechanical strain stimulates a rapid, transient β-catenin response in osteoblasts. Mechanical activation of Akt and inactivation of GSK3β contribute to β-catenin translocation independently of Wnt signaling through LRP5. Conditions that increase basal β-catenin levels, such as LiCl treatment or repression of caveolin-1 expression, further enhance downstream effects of strain, pointing to β-catenin-mediated processes as a common distal pathway whereby multiple factors influence osteoblast function. Our work lends mechanistic support to evidence that exercise can augment multiple strategies that increase bone.
This work was supported, in whole or in part, by National Institutes of Health Grants AR42360 and AR52014. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PI3-kinase, phosphatidylinositol 3-kinase; siRNA, small interfering RNA; siCav, siRNA targeting murine caveolin-1; siSCR, control siRNA; RT, reverse transcription; DAPI, 4′,6-diamidino-2-phenylindole.
References
- 1.Rubin, C., and Lanyon, L. (1984) J. Bone Joint Surg. 66A 397-402 [PubMed] [Google Scholar]
- 2.Rubin, J., Fan, X., Biskobing, D., Taylor, W., and Rubin, C. (1999) J. Orthop. Res. 17 639-645 [DOI] [PubMed] [Google Scholar]
- 3.Krishnan, V., Bryant, H. U., and Macdougald, O. A. (2006) J. Clin. Invest. 116 1202-1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawakami, K., Robling, A. G., Ai, M., Pitner, N. D., Liu, D., Warden, S. J., Li, J., Maye, P., Rowe, D. W., Duncan, R. L., Warman, M. L., and Turner, C. H. (2006) J. Biol. Chem. 281 23698-23711 [DOI] [PubMed] [Google Scholar]
- 5.Robinson, J. A., Chatterjee-Kishore, M., Yaworsky, P. J., Cullen, D. M., Zhao, W., Li, C., Kharode, Y., Sauter, L., Babij, P., Brown, E. L., Hill, A. A., Akhter, M. P., Johnson, M. L., Recker, R. R., Komm, B. S., and Bex, F. J. (2006) J. Biol. Chem. 281 31720-31728 [DOI] [PubMed] [Google Scholar]
- 6.Armstrong, V. J., Muzylak, M., Sunters, A., Zaman, G., Saxon, L. K., Price, J. S., and Lanyon, L. E. (2007) J. Biol. Chem. 282 20715-20727 [DOI] [PubMed] [Google Scholar]
- 7.Rodda, S. J., and McMahon, A. P. (2006) Development 133 3231-3244 [DOI] [PubMed] [Google Scholar]
- 8.Holmen, S. L., Zylstra, C. R., Mukherjee, A., Sigler, R. E., Faugere, M. C., Bouxsein, M. L., Deng, L., Clemens, T. L., and Williams, B. O. (2005) J. Biol. Chem. 280 21162-21168 [DOI] [PubMed] [Google Scholar]
- 9.Cohen, P., and Frame, S. (2001) Nat. Rev. Mol. Cell Biol. 2 769-776 [DOI] [PubMed] [Google Scholar]
- 10.Cross, D. A., Alessi, D. R., Cohen, P., Andjelkovich, M., and Hemmings, B. A. (1995) Nature 378 785-789 [DOI] [PubMed] [Google Scholar]
- 11.Castellone, M. D., Teramoto, H., Williams, B. O., Druey, K. M., and Gutkind, J. S. (2005) Science 310 1504-1510 [DOI] [PubMed] [Google Scholar]
- 12.Haq, S., Michael, A., Andreucci, M., Bhattacharya, K., Dotto, P., Walters, B., Woodgett, J., Kilter, H., and Force, T. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 4610-4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasaneen, N. A., Zucker, S., Lin, R. Z., Vaday, G. G., Panettieri, R. A., and Foda, H. D. (2007) Am. J. Physiol. 293 L1059-L1068 [DOI] [PubMed] [Google Scholar]
- 14.Kippenberger, S., Loitsch, S., Guschel, M., Muller, J., Knies, Y., Kaufmann, R., and Bernd, A. (2005) J. Biol. Chem. 280 3060-3067 [DOI] [PubMed] [Google Scholar]
- 15.Sedding, D. G., Hermsen, J., Seay, U., Eickelberg, O., Kummer, W., Schwencke, C., Strasser, R. H., Tillmanns, H., and Braun-Dullaeus, R. C. (2005) Circ. Res. 96 635-642 [DOI] [PubMed] [Google Scholar]
- 16.Ragab, A. A., Nalepka, J. L., Bi, Y., and Greenfield, E. M. (2002) Am. J. Physiol. 283 C679-C687 [DOI] [PubMed] [Google Scholar]
- 17.Rubin, J., Murphy, T. C., Zhu, L., Roy, E., Nanes, M. S., and Fan, X. (2003) J. Biol. Chem. 278 34018-34025 [DOI] [PubMed] [Google Scholar]
- 18.Fan, X., Rahnert, J. A., Murphy, T. C., Nanes, M. S., Greenfield, E. M., and Rubin, J. (2006) J. Cell Physiol. 207 454-460 [DOI] [PubMed] [Google Scholar]
- 19.van Noort, M., Meeldijk, J., van der Zee, R., Destree, O., and Clevers, H. (2002) J. Biol. Chem. 277 17901-17905 [DOI] [PubMed] [Google Scholar]
- 20.Araki, Y., Okamura, S., Hussain, S. P., Nagashima, M., He, P., Shiseki, M., Miura, K., and Harris, C. C. (2003) Cancer Res. 63 728-734 [PubMed] [Google Scholar]
- 21.Yun, K., and Im, S. H. (2007) Biochem. Biophys. Res. Commun. 364 270-275 [DOI] [PubMed] [Google Scholar]
- 22.Rice, K. M., Desai, D. H., Preston, D. L., Wehner, P. S., and Blough, E. R. (2007) Exp. Physiol. 92 963-970 [DOI] [PubMed] [Google Scholar]
- 23.Gaur, T., Lengner, C. J., Hovhannisyan, H., Bhat, R. A., Bodine, P. V., Komm, B. S., Javed, A., van Wijnen, A. J., Stein, J. L., Stein, G. S., and Lian, J. B. (2005) J. Biol. Chem. 280 33132-33140 [DOI] [PubMed] [Google Scholar]
- 24.Galbiati, F., Volonte, D., Engelman, J. A., Watanabe, G., Burk, R., Pestell, R. G., and Lisanti, M. P. (1998) EMBO J. 17 6633-6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norvell, S. M., Alvarez, M., Bidwell, J. P., and Pavalko, F. M. (2004) Calcif. Tissue Int. 75 396-404 [DOI] [PubMed] [Google Scholar]
- 26.Kim, S. J., Im, D. S., Kim, S. H., Ryu, J. H., Hwang, S. G., Seong, J. K., Chun, C. H., and Chun, J. S. (2002) Biochem. Biophys. Res. Commun. 296 221-226 [DOI] [PubMed] [Google Scholar]
- 27.Lee, H. K., and Jeong, S. (2006) Nucleic Acids Res. 34 5705-5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bilic, J., Huang, Y. L., Davidson, G., Zimmermann, T., Cruciat, C. M., Bienz, M., and Niehrs, C. (2007) Science 316 1619-1622 [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto, H., Komekado, H., and Kikuchi, A. (2006) Dev. Cell 11 213-223 [DOI] [PubMed] [Google Scholar]
- 30.Hedgepeth, C. M., Conrad, L. J., Zhang, J., Huang, H. C., Lee, V. M., and Klein, P. S. (1997) Dev. Biol. 185 82-91 [DOI] [PubMed] [Google Scholar]
- 31.Pavalko, F. M., Gerard, R. L., Ponik, S. M., Gallagher, P. J., Jin, Y., and Norvell, S. M. (2003) J. Cell Physiol. 194 194-205 [DOI] [PubMed] [Google Scholar]
- 32.Troussard, A. A., Mawji, N. M., Ong, C., Mui, A., St-Arnaud, R., and Dedhar, S. (2003) J. Biol. Chem. 278 22374-22378 [DOI] [PubMed] [Google Scholar]
- 33.Sarbassov, D. D., Guertin, D. A., Ali, S. M., and Sabatini, D. M. (2005) Science 307 1098-1101 [DOI] [PubMed] [Google Scholar]
- 34.Zhang, B., Peng, F., Wu, D., Ingram, A. J., Gao, B., and Krepinsky, J. C. (2007) Cell. Signal. 19 1690-1700 [DOI] [PubMed] [Google Scholar]
- 35.Torres, V. A., Tapia, J. C., Rodriguez, D. A., Lladser, A., Arredondo, C., Leyton, L., and Quest, A. F. (2007) Mol. Cell Biol. 27 7703-7717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubin, J., Schwartz, Z., Boyan, B. D., Fan, X., Case, N., Sen, B., Drab, M., Smith, D., Aleman, M., Wong, K. L., Yao, H., Jo, H., and Gross, T. S. (2007) J. Bone Miner. Res. 22 1408-1418 [DOI] [PubMed] [Google Scholar]