Abstract
Septins are filamentous GTPases that associate with cell membranes and the cytoskeleton and play essential roles in cell division and cellular morphogenesis. Septins are implicated in many human diseases including cancer and neuropathies. Small molecules that reversibly perturb septin organization and function would be valuable tools for dissecting septin functions and could be used for therapeutic treatment of septin-related diseases. Forchlorfenuron (FCF) is a plant cytokinin previously shown to disrupt septin localization in budding yeast. However, it is unknown whether FCF directly targets septins and whether it affects septin organization and functions in mammalian cells. Here, we show that FCF alters septin assembly in vitro without affecting either actin or tubulin polymerization. In live mammalian cells, FCF dampens septin dynamics and induces the assembly of abnormally large septin structures. FCF has a low level of cytotoxicity, and these effects are reversed upon FCF washout. Significantly, FCF treatment induces mitotic and cell migration defects that phenocopy the effects of septin depletion by small interfering RNA. We conclude that FCF is a promising tool to study mammalian septin organization and functions.
Septins were originally identified in screens for genes that regulate the cell cycle in budding yeast Saccharomyces cerevisiae (1). In this organism, septins function as scaffolds and diffusion barriers to localize many proteins required for bud site selection, cell cycle progression, and cell division (2-6). Septins have since been identified in many other eukaryotes ranging from fungi to animals (7, 8). In mammalian cells, septins regulate cytoskeleton organization (9-14) and membrane traffic and fusion (15-17), and during mitosis, they are required for proper chromosome alignment, segregation (18, 19), and cytokinesis (13, 20, 21).
Septins are guanine nucleotide-binding proteins that form filaments associated with membranes and the cytoskeleton (22, 23). Mammalian septin filaments assemble from heteromeric complexes, whose composition varies depending on cell type and level of expression (22, 24). Mammalian septin complexes have been purified from mouse brain and tissue culture cells (9, 16), and the ubiquitously expressed Septin 2 (SEPT2) has been reconstituted in vitro into filamentous structures by itself and in a complex with SEPT6 and SEPT7 (9, 25-27). Recently, x-ray crystallography and electron microscopy revealed that SEPT2, SEPT6, and SEPT7 assemble into filaments through their GTP binding domains (27).
An increasing body of evidence implicates the mammalian septin family in the pathogenesis of diverse disease states including neoplasia, neurodegenerative conditions, and infections (28, 29). Many septin isoforms are abnormally expressed in carcinomas (30-34), and altered levels of septin expression strongly correlate with tumorigenic phenotypes such as increased cell growth, motility and invasiveness, and resistance to microtubule-disrupting reagents (35, 36). In neurodegenerative disorders (e.g. Alzheimer disease), septins are present in cytosplasmic inclusions and neurofibrillary tangles, and a septin isoform (SEPT4) was recently shown to suppress α-synuclein neurotoxicity in Parkinson disease (37-39). Finally, some septins have been identified as host proteins involved in viral and bacterial infections (40, 41).
Various tools have been developed to study septin organization and functions, including function-blocking antibodies and RNA interference. However, these reagents are not readily reversible, and we have, therefore, turned to small molecule compounds, which could also be of therapeutic value in translational research. A recent study showed that the plant cytokinin forchlorfenuron (FCF;2 1-(2-chloro-4-pyridyl)-3-phenylurea, 4PU300) disrupted septin localization and caused cytokinesis defects in budding yeast (42). However, it was unclear whether FCF directly and specifically affected septins, and if so, how FCF affected septin organization and functions. Here, we analyzed the effects of FCF on mammalian septins using recombinant SEPT2/6/7 as an in vitro septin assembly system and mammalian cells to study effects in vivo. Our studies show that FCF directly alters septin assembly in vitro and affects septin organization in mammalian cells by stabilizing septin filaments; these effects of FCF in cells are reversible upon FCF washout. Cells treated with FCF display mitosis defects and decreased cell migration that phenocopy the effects of septin knockdown by siRNAs.
EXPERIMENTAL PROCEDURES
Cell Culture, Cell Synchronization, and siRNA Transfections—Madin-Darby canine kidney (MDCK) clone II and HeLa (ATCC CCL-2) cells were maintained as described previously (18). HeLa S3 cells were maintained in high glucose DMEM supplemented with 10% fetal bovine serum (FBS). MDCK-SEPT2-YFP (18), MDCK-actin-GFP (43), and HeLa-α-tubulin-GFP (a gift from L. Wordeman, University of Washington) cells were maintained in DMEM (Sigma-Aldrich) supplemented with 10% FBS and 0.5 mg/ml G418. HeLa S3 cells were synchronized in mitosis by blocking with 2 mm thymidine (Sigma) for 18 h followed by a 4-h release into regular media, and a 12-h incubation with nocodazole (Sigma; 40 ng/ml). HeLa cells were transfected with siCONTROL non-targeting siRNA and SEPT2 siRNAs as described previously (18).
Assembly of Recombinant SEPT2/6/7 in Vitro—Recombinant His-SEPT2/6/7 was purified from insect cells and assembled into filamentous structures as described previously (9). Briefly, His-SEPT2/6/7 was clarified by centrifugation at 100,000 × g, diluted to ∼0.2 mg/ml, and dialyzed against low salt buffer (5 mm potassium phosphate buffer (pH 7.2), 50 mm KCl, 10% glycerol, 2 mm MgCl2, and 1 mm dithiothreitol) in the presence of FCF (Sigma) or DMSO (Sigma) at 4 °C. Assembly of higher-order SEPT2/6/7 filaments was reversed by dialysis in high salt buffer (5 mm potassium phosphate buffer (pH 7.2), 1 m KCl, 10% glycerol, 2 mm MgCl2, and 1 mm dithiothreitol) for 12 h. Septin structures were transferred to poly-l-lysine-coated glass coverslips and fixed with 3% paraformaldehyde (EM Sciences) in phosphate-buffered saline for 10 min, blocked with 2% bovine serum albumin, and stained with anti-SEPT2 (N5N; rabbit polyclonal antibody). The surface area of septin structures was quantified using the ImageJ software. Negative stain electron microscopy was performed by adsorbing His-SEPT2/6/7 complexes onto glow-discharged carbon-coated copper grids (200 mesh copper hexagonal) for 2 min. Samples were washed twice with water and stained with 1% uranyl acetate for 2 min. Following staining, samples were dried and examined with a CM-12 Philips transmission electron microscope. In pelleting assays of recombinant septin assembly, higher-ordered filamentous structures were centrifuged at 16,000 × g for 10 min. Supernatants and pellets were then analyzed by SDS-PAGE and Coomassie Brilliant Blue staining.
Actin and Microtubule Polymerization Assays—Pyrene actin (Cytoskeleton, Inc.) polymerization assay was performed according to the manufacturer's instructions in the presence of different concentrations of FCF or DMSO. Fluorescence intensity was measured using a fluorescence plate reader (Tecan Group Ltd.; excitation 360 nm, emission 405 nm). A bovine tubulin (Cytoskeleton, Inc.) polymerization assay was performed according to the manufacturer's instructions in the presence of various concentrations of FCF or DMSO. Turbidity was measured at 340 nm using a plate reader (Tecan Group Ltd.).
Immunofluorescence Microscopy—Subconfluent cells were fixed with warm PHEM (60 mm Pipes-KOH, pH 6.9, 25 mm Hepes, 10 mm EGTA, and 1 mm MgCl2) containing 3% paraformaldehyde (EM Sciences) and 0.1% Triton X-100 and stained with antisera to SEPT2 (N5N; rabbit polyclonal antibody), SEPT7 (anti-SEPT7; rabbit polyclonal antibody), α-tubulin (DM1α; Sigma-Aldrich), and secondary fluorescein isothiocyanate- or rhodamine red X-conjugated F(ab′)2 goat anti-mouse or anti-rabbit IgGs (Jackson ImmunoResearch Laboratories). Microfilaments were stained with Alexa Fluor-355-conjugated phalloidin (Invitrogen). Samples were mounted in VECTASHIELD mounting medium (Vector Laboratories) and imaged with an inverted Zeiss microscope (Axiovert 200) or the Marianas system (Intelligent Imaging Innovations). Cell morphology (axis length ratio) was quantified using the Statistics module in the Slidebook 4.2 software. Staining of MDCK cells for centromere-associated protein E (CENP-E) and CREST serum was performed as described previously (18). Images were captured and deconvolved with an Applied Precision Delta Vision wide field deconvolution system (×63 oil objective; ×1.5 optovar). Quantification of CENP-E-containing kinetochores (n =∼100 per cell) was performed in every fourth optical section from a deconvolved z-stack.
Cytotoxicity Assays—HeLa or MDCK cells were plated overnight on collagen-coated 35-mm tissue culture dishes and then treated with DMSO or FCF for 4 h. Cells were trypsinized, resuspended in DMEM supplemented with 10% FBS, and mixed in a 1:1 ratio with trypan blue (Invitrogen) at room temperature for 1 min. Blue and non-blue cells were counted using a hemocytometer. For alamarBlue assays, HeLa cells (1 × 104/well) were plated onto a 96-well plate (Costar) and cultured overnight. Cells were then treated with FCF or DMSO, and 50 μl of medium was removed and mixed with 5.5 μl of alamarBlue reagent (AbD Serotec) for 2 h. Fluorescence intensities were measured using a plate reader (Tecan Group Ltd.; excitation 560 nm, emission 590 nm). Each measurement was performed in quadruplicates.
Live Cell Imaging and FRAP—Cells were grown on collagen-coated coverslips for 24 h at subconfluent density and imaged in phenol red-free DMEM supplemented with 10% FBS, 25 mm Hepes (Invitrogen) using the Marianas system (Intelligent Imaging Innovations) equipped with the MicroPoint fluorescence recovery after photobleaching (FRAP) laser system (Photonic Instruments). Photobleaching experiments were carried out using the FRAP module of the Marianas system. Equivalent laser intensity, repetition, and exposure time were used for FRAP measurements in FCF- or DMSO-treated samples. Bleached areas were accurately positioned along septin filaments, actin stress fibers, or microtubule bundles. Images were taken 5 s before photobleaching and 5 min after photobleaching at a rate of one frame per second. Fluorescence intensities were quantified after background subtraction and normalization against unbleached areas using the Slidebook 4.2 software as described previously (43). Fluorescence intensities (FI) were plotted against time (t) and fitted to an exponential recovery curve: FI = FIst - (FIst - FIblch)exp(-In2 × T/t½) in which FIblch is the fluorescence intensity at the time of bleaching, and FIst is the fluorescence intensity at the final steady status. From this equation, half-times of fluorescence recovery were derived. The koff was calculated as In2/t½, and recovery percentage was calculated as: ((FIst - FIblch)/(FIprebleach - FIblch)) × 100 in which FIprebleach is the fluorescence intensity before photobleaching (44, 45). Plots and curve fittings were performed with the Origin 6.1 software.
Cell Motility and Wound-healing Assays—A modified cell motility assay was used (46). Fluorescent beads (1 μm Fluo-Spheres; Invitrogen) were vortexed, sonicated, and plated onto 18-mm coverslips precoated with collagen and incubated at 4 °C overnight. Excess beads were washed off, and HeLa cells (3 × 103) were plated for 16 h at 37 °C in DMEM and 10% FBS. Cells were fixed, random images were taken, and the surface areas covered by single cells were quantified using the ImageJ software. For wound-healing assays, HeLa cells (2 × 106) were grown on 35-mm dishes overnight in DMEM and 10% FBS. MDCK cells were first plated in low calcium DMEM (5 μm Ca2+) and 10% dialyzed FBS for 75 min, and then the medium was exchanged for DMEM with 10% FBS. Confluent monolayers were scratched with a pipette tip. HeLa and MDCK cells were imaged for 8 and 14 h, respectively. Displacement surface areas (ΔArea) were calculated as Si - St in which Si is the initial wounded surface area and St is the wounded surface area at time point t. The surface areas were quantified using ImageJ.
RESULTS
FCF Alters SEPT2/6/7 Assembly in Vitro—To determine whether FCF (Fig. 1A) has a direct effect on mammalian septin assembly, we used recombinant SEPT2/6/7 complex purified from insect cells (9). SEPT2/6/7 complexes were dialyzed in the presence of FCF or DMSO (the vehicle for FCF) in low salt buffer to induce septin self-assembly into higher-order structures, which were analyzed by immunofluorescence microscopy (Fig. 1B) or transmission electron microscopy (Fig. 1F). Under control conditions (DMSO), SEPT2/6/7 complexes assembled into filamentous structures as reported previously (9). In the presence of 20 μm FCF, SEPT2/6/7 complexes formed structures that were larger and denser than those formed in DMSO (Fig. 1B). These structures were highly inter-twined (Fig. 1B, see inset) and had significantly larger surface area than control filaments (Fig. 1, C and D), giving rise to a higher percentage (30.2% versus 6.2%) of structures with a surface area greater than 500 μm2 (Fig. 1D). The effect of FCF on SEPT2/6/7 assembly was concentration-dependent, with a significant effect at a concentration as low as 5 μm (Fig. 1E), and was reversible upon dialysis of filaments in high salt buffer with-out FCF (supplemental Fig. 1).
FIGURE 1.
FCF alters SEPT2/6/7 assembly in vitro. A, the chemical structure of FCF. B, SEPT2/6/7 complexes (0.2 μg/μl) were dialyzed for 2 h in low salt buffer containing FCF (20 μm) or DMSO to induce septin filament assembly. Immunofluorescence images show filamentous structures stained for SEPT2. Insets show magnified images of selected regions; scale bars, 2 μm. C, the bar graph shows the surface areas (mean ± S.E.) of septin filaments after dialysis in DMSO (red, n = 132) or FCF (green, n = 129); ****, p < 0.0001. D, the surface area histogram of septin filaments formed in DMSO (red) or FCF (green). E, the bar graph shows the surface areas (mean ± S.E.) of septin filaments after 1 h dialysis of SEPT2/6/7 (0.1 μg/μl) in low salt buffer containing various concentrations of DMSO (red) or FCF (green, 1 μm; blue, 5 μm; cyan, 20 μm); ****, p < 0.0001. F, representative images of negative stained septin filaments after dialysis in DMSO (left panel) or 20 μm FCF (right panel). G, septin filaments formed in DMSO or FCF were spun at 16,000 × g. Supernatant (Sup) and pellet (Pellet) fractions were resuspended in SDS-sample buffer, analyzed by SDS-PAGE, and stained with Coomassie Blue. H and I, pyrene actin (H) and tubulin (I) polymerization assays in the presence of FCF or DMSO.
The ultrastructure of septin filaments assembled in vitro in the presence or absence of FCF was examined by transmission electron microscopy. In DMSO, SEPT2/6/7 formed extended fibers with an average diameter of 10.9 ± 0.3 nm (n = 40) that appeared to be composed of two parallel filaments (Fig. 1F, left panel, see inset). The morphology of these septin structures is similar to that formed by recombinant septins and septin complexes purified from mammalian cells (9, 25, 26). In FCF, SEPT2/6/7 formed much thicker filaments with an average diameter of 21.0 ± 1.1 nm (n = 40) that appear to comprise many parallel filaments (Fig. 1F, right panel, see inset). This change in septin assembly was confirmed by analyzing the ratio of septins in the supernatant and pellet after centrifugation at 16,000 × g. After dialysis in the presence of FCF, more SEPT2/6/7 was pelleted (∼90% versus ∼50%) (Fig. 1G).
We also examined whether FCF has similar effects on the assembly of other filamentous cytoskeleton polymers such as actin and tubulin. In vitro actin and tubulin polymerization assays were performed in the presence of a range of FCF or DMSO concentrations. FCF did not affect the rate or steady state amount of actin or tubulin polymerization (Fig. 1, H and I).
Taken together, these results show that FCF alters SEPT2/6/7 assembly and formation of higher-order structures in a dose-dependent manner. FCF did not affect actin or tubulin polymerization in vitro under the same conditions.
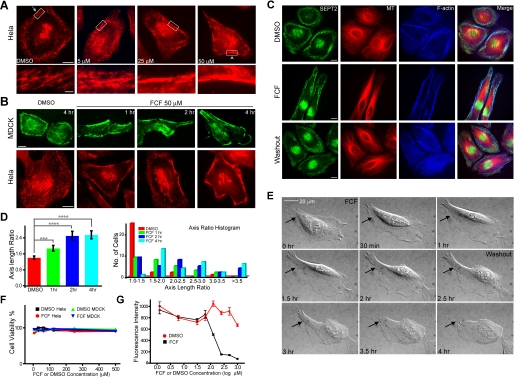
FCF Alters Septin Organization in HeLa and MDCK Cells Reversibly—We next tested whether FCF affected septin organization in mammalian cells. HeLa cells were treated with increasing concentrations of FCF for 2 h and stained for SEPT2 (Fig. 2A). SEPT2 organization changed from thin, short fibrillar structures (Fig. 2A, arrow, see magnified image of the boxed region in the lower panel) to thick, long filamentous structures localized at the cell periphery (Fig. 2A, arrowhead, see magnified image of the boxed region in the lower panel). These effects were dose-dependent and pronounced at 50 μm FCF (Fig. 2A). To examine the time course of FCF effects, HeLa and MDCK cells were treated with 50 μm FCF and fixed after 1, 2, or 4 h. FCF began to induce the reorganization of septins after 1 h of treatment (Fig. 2B) with a concomitant increase in SEPT2 filament length and width (supplemental Fig. 2, A and B). Significantly, FCF-induced changes in septin organization were reversed upon FCF washout (2 h; Fig. 2C), demonstrating that the effects of FCF were not permanent and that septins were dynamic and could reorganize within 2 h. We also examined whether septins other than SEPT2 were affected by FCF and found that organization of SEPT7 was also affected during FCF treatment similar to that of SEPT2 (supplemental Fig. 2C).
FIGURE 2.
FCF reversibly alters septin organization in HeLa and MDCK cells. A, immunofluorescence images of HeLa cells stained for SEPT2 after treatment with 5, 25, or 50 μm FCF or DMSO for 2 h; scale bars, 10 μm. Lower panels show the boxed regions at higher magnification; scale bar, 2 μm. B, MDCK and HeLa cells were treated with DMSO for 4 h or FCF for 1,2, or 4 h and stained for SEPT2; scale bars, 10 μm. C, immunofluorescence images of HeLa cells stained for SEPT2 (green),α-tubulin (red), and F-actin (blue) after treatment with DMSO for 4 h (top panel) or 50 μm FCF for 4 h (middle panel) followed by a 2-h washout with regular cell medium (bottom panel); scale bars, 10 μm. D, the bar graph (left panel) shows ratios (mean ± S.E.) of lengths of major cell axes to lengths of minor cell axes for MDCK cells treated with DMSO (red) or 50 μm FCF for 1 (green), 2 (blue), and 4 h (cyan); ***, p = 0.0049; ****, p < 0.0001. The histogram (right panel) shows the distribution of axis ratio values after DMSO and FCF treatments. E, differential interference contrast images from time-lapse microscopy (supplemental movies 1 and 2) of HeLa cells. FCF was added at 0 h and washed out at 2.5 h. Arrows point to a cell region that becomes elongated during FCF treatment and retracts upon FCF washout. F, a trypan blue cytotoxicity assay of HeLa or MDCK cells treated with various concentrations of DMSO or FCF (6.25-500 μm) for 4 h. Trypsinized cells were stained with trypan blue reagent and counted as the percentage of total cells (n =∼200). G, an alamarBlue cytotoxicity assay of HeLa cells treated with various concentrations of DMSO (red circles) or FCF (1.25 μm-1 mm; black squares) for 24 h. Cell medium was incubated with alamarBlue reagent, and fluorescence intensities (mean ± S.E.) were plotted as a function of time. All measurements were performed in quadruplicates.
We observed that FCF-treated cells had elongated morphology (Fig. 2, A-C). We used the axis-length ratio (major axis length/minor axis length) to measure cell morphology (Fig. 2D). Quantification of the axis-length ratio of MDCK cells treated with FCF or DMSO revealed a time-dependent effect of FCF on cell morphology (Fig. 2D); this time course of changes in cell shape paralleled changes in septin organization observed by immunofluorescence microscopy (Fig. 2B). Live cell imaging of HeLa cells treated with FCF also showed gradual changes in cell morphology, which were reversed upon FCF washout (Fig. 2E, arrows, and see supplemental movies 1 and 2).
Although the effects of FCF were reversible, indicating that cells were viable, we evaluated the cytotoxicity of FCF using trypan blue and alamarBlue assays. The trypan blue assay shows that the viability of HeLa and MDCK cells was unaffected even at FCF concentrations as high as 500 μm for 4 h (Fig. 2F). The alamarBlue assay indicates that after 24 h of treatment, FCF-treated HeLa cells were as viable as DMSO-treated cells at a concentration of FCF ≤ 62.5 μm. However, HeLa cells began to show decreasing viability at higher FCF concentrations (Fig. 2G). The results show that FCF is not toxic over short time periods even at high concentrations, and for long treatments, FCF is not toxic at a concentration of FCF ≤ 62.5 μm.
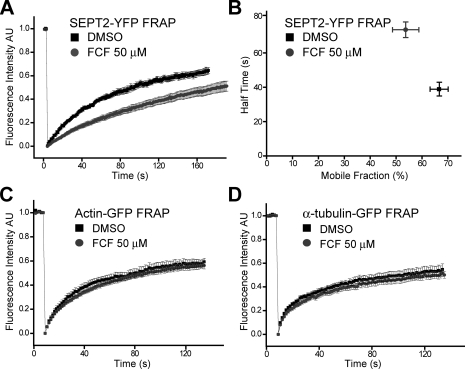
FCF Dampens SEPT2 Dynamics—To explore how FCF acts on septins, we measured septin filament dynamics using FRAP in MDCK cells stably expressing SEPT2-YFP (18). From the profile of fluorescence recovery, we calculated the mobile fraction and half-time of fluorescence recovery (t½) of SEPT2-YFP (see “Experimental Procedures”). The mobile fraction of SEPT2-YFP filaments was 66.6 ± 3.5% in the presence of DMSO when compared with 53.7 ± 5.1% in the presence of FCF (Fig. 3, A and B, and Table 1). The difference in the mobile fraction indicates that FCF increased the stable, immobile fraction of septin filaments. FCF treatment also resulted in a doubling of the half-time of fluorescence recovery (73.5 ± 4.5 s versus 38.9 ± 4.0 s, Fig. 3, A and B, Table 1, and supplemental movies 3 and 4). Since SEPT2-YFP dynamics reflect mainly the association and dissociation of septin complexes into filamentous structures (the recovery is much slower than free cytoplasmic diffusion), we derived the dissociation rate constant koff (koff = In2/t½) of SEPT2-YFP as described previously for FRAP analysis of cytoskeleton-binding proteins (44, 45). SEPT2-YFP in FCF-treated cells had a koff of 9.8 ± 0.6 × 10-3 s-1 when compared with 14.8 ± 1.1 × 10-3 s-1 in DMSO-treated cells. This further suggests that FCF stabilizes septin complexes into higher-order, less dynamic filamentous structures.
FIGURE 3.
FCF dampens SEPT2 dynamics in MDCK cells. A, normalized FRAP curves for SEPT2-YFP in MDCK cells treated with DMSO (n = 10; supplemental movie 3) or 50 μm FCF (n = 15; supplemental movie 4). AU, arbitrary units. B, the plot shows half-times of SEPT2-YFP fluorescence recovery (mean ± S.E.; y axis) and the percentage of SEPT2-YFP mobile fraction (mean ± S.E.; x axis) in MDCK cells treated with DMSO (n = 10; black squares) or 50 μm FCF (n = 15; gray circles). C, normalized FRAP curves for actin-GFP in MDCK cells treated with DMSO (n = 13) or FCF 50 μm (n = 15). D, normalized FRAP curves for α-tubulin-GFP in HeLa cells treated with DMSO (n = 14) or FCF 50 μm (n = 14). All error bars represent S.E.
TABLE 1.
Half time of recovery (t½) and percentage recovery of SEPT2 filaments, actin microfilaments, and microtubules
All values for t½ and % recovery represent mean ± S.E.
| Subject | Treatment | t½ | Recovery | Number | p value |
|---|---|---|---|---|---|
| s | % | ||||
| SEPT2-YFP filaments | DMSO | 38.9 ± 4.0 | 66.6 ± 3.5 | 10 | t½ < 0.001 |
| FCF 50 μm | 73.5 ± 4.5 | 53.7 ± 5.1 | 15 | Recovery 0.0268 | |
| Actin-GFP bundles | DMSO | 20.1 ± 1.9 | 59.3 ± 3.0 | 13 | t½ 0.8112 |
| FCF 50 μm | 20.6 ± 1.1 | 56.5 ± 2.9 | 15 | Recovery 0.5060 | |
| α-tubulin-GFP microtubules | DMSO | 13.5 ± 2.0 | 54.6 ± 4.6 | 14 | t½ 0.5477 |
| FCF 50 μm | 15.1 ± 1.7 | 51.4 ± 3.0 | 14 | Recovery 0.5651 |
To test the specificity of FCF on septins, similar experiments were performed in cells stably expressing actin-GFP or α-tubulin-GFP. FCF had no effect on actin micro-filament or microtubule dynamics when compared with the DMSO control (Fig. 3, C and D, and Table 1).
These FRAP studies on septin filaments, actin, and microtubule filaments demonstrate that FCF stabilizes septin filaments by decreasing the mobile fraction and koff of septins and that neither actin microfilament nor microtubule dynamics is affected by FCF. This result is consistent with our observations that FCF caused increased assembly of septin complexes into large filamentous structures in vitro but had little effect on either actin or tubulin polymerization (Fig. 1).
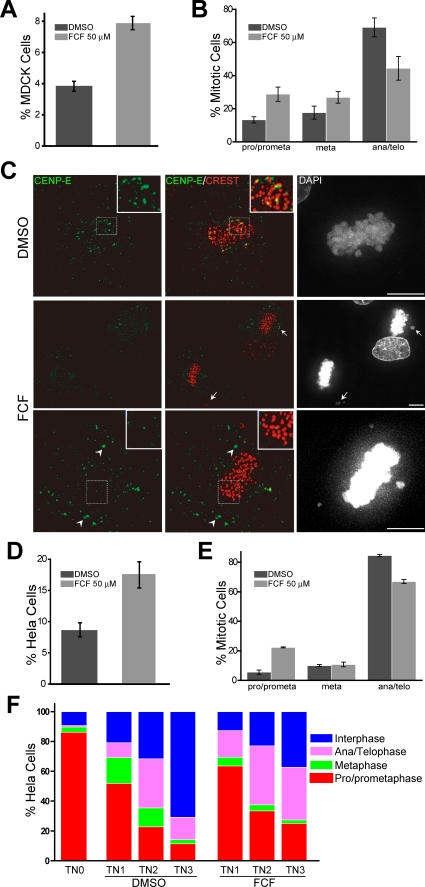
FCF Causes Mitotic Defects—Given that septins have important functions in mitosis (13, 18, 19) and FCF is able to change septin organization and dynamics, we sought to examine whether FCF caused mitotic defects. FCF treatment of MDCK and HeLa cells for 8 h caused an increase in the percentage of mitotic cells (Fig. 4, A and D). These cells appeared to be delayed in prophase/prometaphase (Fig. 4, B and E). Time-lapse microscopy of HeLa cells expressing histone 2B-GFP revealed an increase in the average duration of mitosis in FCF-treated cells (125 ± 8.7 min; n = 70) when compared with DMSO-treated cells (81.7 ± 3.9 min; n = 70). In these movies, histone 2B-GFP labeled chromosomes of the cells treated with FCF failed to properly congress and align at the metaphase plate (supplemental movies 5-7). Similar failure in chromosome congression and alignment was observed in FCF-treated MDCK cells (Fig. 4C, arrows). These data resemble the effects of SEPT2 and SEPT7 depletion in chromosome alignment (18, 19), which are caused by mislocalization of CENP-E, a mitotic motor that is required for proper kinetochore-microtubule attachment. To examine whether the localization of CENP-E is disrupted upon FCF treatment, mitotic MDCK cells were stained for CENP-E and the kinetochore marker protein CREST (Fig. 4C). Quantitative analysis of these images revealed a reduction in the percentage of kinetochores that contained CENP-E (20.8 ± 0.7% in FCF-treated cells versus 40.6 ± 1.9% in DMSO-treated cells; n = 10). In addition, CENP-E was often observed to mislocalize outside the kinetochore regions of mitotic chromosomes (Fig. 4C, arrowheads).
FIGURE 4.
FCF causes mitotic defects. A, the bar graph shows the mitotic index (mean ± S.E.) of MDCK cells after treatment with DMSO or 50 μm FCF for 8 h. Mitotic cells were identified by staining with DAPI and anti-α-tubulin and calculated as the percentage of total MDCK cells (n =∼1800; three independent experiments). B, the bar graph shows the percentage (mean ± S.E.) of DMSO-(n =∼100) and FCF-treated (n =∼150) mitotic cells in prophase/prometaphase (pro/prometa), metaphase (meta), and anaphase/telophase (ana/telo). Results are representative of three independent experiments. C, maximum intensity projections of deconvolved optical sections from FCF- and DMSO-treated (50 μm; 8 h) MDCK cells stained for CENP-E, CREST and DNA (DAPI). Arrows point to misaligned chromosomes; arrowheads point to CENP-E accumulations outside the kinetochore regions. Scale bars, 5 μm. D, the bar graph shows the mitotic index (mean ± S.E.) of HeLa cells after treatment with DMSO (n =∼800) or 50 μm FCF (n =∼500) for 8 h. Results are representative of three independent experiments. E, the bar graph shows the percentage (mean ± S.E.) of DMSO-(n =∼150) and FCF-treated (n =∼150) mitotic cells in prophase/pro-metaphase, metaphase, and anaphase/telophase. Results are representative of three independent experiments. F, after a thymidine-nocodazole block, HeLa S3 cells were released into mitosis for 1 (TN1), 2 (TN2), and 3 (TN3) h. For each time point, FCF (50 μm) or DMSO was added in the medium 30 min prior to fixation and staining with DAPI. The stack bar graphs show the percentage of HeLa cells (n =∼400) in interphase and each phase of mitosis.
Since previous studies have shown that septin disruption results in cytokinesis defects (20, 21), we tested whether FCF has similar effects downstream of chromosome alignment. Synchronized HeLa cells were arrested in prophase/prometaphase and treated with FCF at various time points after release into mitosis (Fig. 4F). Although early FCF treatments delayed the transition from prophase/prometaphase to metaphase, FCF treatments after a 2.5-h release into mitosis delayed the transition from telophase/cytokinesis to interphase. These data are consistent with a septin function in cytokinesis (13, 20, 21). In summary, our results indicate that FCF may cause mitotic defects by disrupting septin organization and dynamics during cell division.
FCF Impairs Cell Migration—Having investigated the specific effects of FCF on septin organization and dynamics, we examined whether FCF affects cell migration, which has been previously shown to be disrupted by septin mutants (47). We used a single cell motility assay (Fig. 5, A and B) and a “wound-healing” assay (Fig. 5, C and D) to evaluate the effects of FCF on cell migration. In the single cell motility assay, HeLa cells were plated on fluorescent beads and treated with 50 μm FCF or DMSO for 16 h; as the cells migrate during this time, they remove beads by endocytosis, leaving behind a cleared area devoid of beads that is a direct measure of the area covered by the cell (Fig. 5A, areas outlined by the red lines). Quantification of the surface areas covered by HeLa cells clearly indicates that FCF inhibited cell migration (Fig. 5B). In the wound-healing assay, cell monolayers were scratched to remove a line of cells, which were then filled in by cells migrating in from the edges of the monolayer. This assay also reveals that FCF decreased the rate of MDCK and HeLa cell migration (Fig. 5, C and D, and supplemental movies 8 and 9).
FIGURE 5.
FCF impairs cell migration. A, representative images from HeLa cell motility assays. HeLa cells were plated on glass coverslips coated with green fluorescent beads, incubated in medium containing DMSO or 5, 25, or 50 μm FCF, and fixed 16 h later. Red lines mark the areas traveled by HeLa cells. B, the bar graph shows the surface areas (mean ± S.E., n = 20) traveled by HeLa cells after treatment with different concentrations of DMSO or FCF; ****, p < 0.0001. C, representative differential interference contrast images from HeLa (left; supplemental movie 8) and MDCK (right; supplemental movie 9) wound-healing assays at different time points. Cell monolayers were wounded with a pipette tip and then imaged in medium containing DMSO or 50 μm FCF. D, the plots show the changes in wounded surface area (mean ± S.E.; three independent experiments for each cell type and eight areas imaged for each experiment) as a function of time in cell monolayers treated with DMSO (black squares) or 50 μm FCF (gray circles).
To test directly whether septins are involved in cell migration, we knocked down SEPT2 in HeLa cells and performed the single cell motility assay. We found that upon SEPT2 knockdown, the average surface area traveled by knockdown cells was reduced by ∼50% when compared with control cells (supplemental Fig. 3). These data are consistent with previous studies showing cell motility defects upon exogenous expression of septin mutants (47). Taken together, these results show that disruption of septin organization and dynamics by FCF treatment results in defects in cell migration that phenocopy the effects of septin depletion by siRNA.
DISCUSSION
Septins comprise a filamentous system with regulatory and structural properties for diverse cellular functions. Function-blocking antibodies and siRNAs have been used to study septin functions, but these reagents are not readily reversible, and the time scale of their use leads to cumulative effects. Therefore, small molecules that perturb septin organization quickly and reversibly would be powerful tools to dissect septin organization and functions.
An earlier study showed that the small molecule compound FCF disrupted septin organization and caused cytokinesis defects in budding yeast (42). However, that study did not show whether FCF acted directly and specifically on septins, and if so, how FCF acted on septins and whether FCF could be used as a tool to study mammalian septin organization and functions. Our results show that FCF specifically altered septin assembly in vitro but did not affect actin or tubulin polymerization. These observations are in agreement with our studies in mammalian cells showing that FCF induced changes in septin organization from short, thin fibrilla to long, thick bundle-like filaments. These effects of FCF in cells were reversible. In summary, FCF targets the filamentous assembly of divergent septin isoforms (e.g. Cdc3/10/11/12 (42), SEPT2, SEPT7) reversibly and across species.
To elucidate how FCF acts on septins, we examined SEPT2 dynamics in vivo using FRAP. The results reveal that FCF increased both the half-time of recovery and the immobile fraction, indicating an increase in the stability of septin filaments. This is consistent with the FCF-induced changes in septin organization observed both in vitro (Fig. 1) and in cells (Fig. 2). Moreover, our FRAP data indicate that the thick bundle-like septin filaments induced by FCF arise from a decrease in the koff of septin assembly (lower dissociation rate), which progressively results in accumulation of abnormally large filamentous structures.
FCF-induced changes in septin dynamics and organization resulted in cell morphology changes, mitotic defects, and decreased cell migration. These phenotypes are similar to those reported from septin depletion and overexpression studies (13, 18, 21, 47). We suggest that stabilization of septin filaments by FCF reduces the turnover rate of septin filaments and therefore the relative abundance of cytoplasmic and filamentous septins, which is critical for their function. These results are in accordance with the effects of septin depletion or overexpression, which alter overall septin organization and function.
FCF provides a high degree of temporal control over septin function, acting within 1 h (Fig. 2B), and is reversible (Fig. 2C); therefore, it may prove more useful than siRNA as a probe for septin functions. Taken together, our results show that FCF alters mammalian septin assembly in vitro and alters septin organization and dynamics in cells by stabilizing septin filaments. These results suggest that FCF will be useful as a tool to study septin biology. With low levels of cytotoxicity, anti-mitotic, and anti-migratory efficacies, FCF could be considered as a septin-based anti-tumorigenic agent.
Supplementary Material
Acknowledgments
We are indebted to Florian Wessel (Max Planck Institute, Münster, Germany) for technical help and data analysis, Dr. Makoto Kinoshita (Kyoto University, Kyoto, Japan) for recombinant proteins, and Dr. Linda Wordeman (University of Washington) for HeLa cells expressing α-tubulin-GFP. We also thank Nafisa Ghori for technical assistance with transmission electron microscopy and members of the Nelson/Frydman laboratories for help and advice.
This work was supported, in whole or in part, by National Institutes of Health Grant GM35527 (to W. J. N.). This work was also supported by a Cell and Molecular Biology Training Grant and by grants from the Department of Defense Breast Cancer Research Program Predoctoral Fellowship BC083077 (to Q. H.) and the Stanford Digestive Disease Center (N. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains three supplemental figures and nine supplemental movies.
Footnotes
The abbreviations used are: FCF, forchlorfenuron; MDCK, madin-darby canine kidney; FRAP, fluorescence recovery after photobleaching; CENP-E, centromere associated protein E; DMSO, dimethyl sulfoxide; siRNA, small interfering RNA; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; GFP, green fluorescent protein; YFP, yellow fluorescent protein; FI, fluorescence intensities; PIPES, 1,4-piperazinediethanesulfonic acid; DAPI, 4′,6-diamidino-2-phenylindole.
References
- 1.Hartwell, L. H. (1971) Exp. Cell Res. 69 265-276 [DOI] [PubMed] [Google Scholar]
- 2.Longtine, M. S., DeMarini, D. J., Valencik, M. L., Al-Awar, O. S., Fares, H., De Virgilio, C., and Pringle, J. R. (1996) Curr. Opin. Cell Biol. 8 106-119 [DOI] [PubMed] [Google Scholar]
- 3.Gladfelter, A. S., Pringle, J. R., and Lew, D. J. (2001) Curr. Opin. Microbiol. 4 681-689 [DOI] [PubMed] [Google Scholar]
- 4.Faty, M., Fink, M., and Barral, Y. (2002) Curr. Genet. 41 123-131 [DOI] [PubMed] [Google Scholar]
- 5.Longtine, M. S., and Bi, E. (2003) Trends Cell Biol. 13 403-409 [DOI] [PubMed] [Google Scholar]
- 6.Versele, M., and Thorner, J. (2005) Trends Cell Biol. 15 414-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindsey, R., and Momany, M. (2006) Curr. Opin. Microbiol. 9 559-565 [DOI] [PubMed] [Google Scholar]
- 8.Kinoshita, M. (2003) Genome Biol. 4 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinoshita, M., Field, C. M., Coughlin, M. L., Straight, A. F., and Mitchison, T. J. (2002) Dev. Cell 3 791-802 [DOI] [PubMed] [Google Scholar]
- 10.Kremer, B. E., Adang, L. A., and Macara, I. G. (2007) Cell 130 837-850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagata, K.-I., and Inagaki, M. (2004) Oncogene 24 65-76 [DOI] [PubMed] [Google Scholar]
- 12.Kremer, B. E., Haystead, T., and Macara, I. G. (2005) Mol. Biol. Cell 16 4648-4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joo, E., Surka, M. C., and Trimble, W. S. (2007) Dev. Cell 13 677-690 [DOI] [PubMed] [Google Scholar]
- 14.Nagata, K.-I., Kawajiri, A., Matsui, S., Takagishi, M., Shiromizu, T., Saitoh, N., Izawa, I., Kiyono, T., Itoh, T. J., Hotani, H., and Inagaki, M. (2003) J. Biol. Chem. 278 18538-18543 [DOI] [PubMed] [Google Scholar]
- 15.Beites, C. L., Xie, H., Bowser, R., and Trimble, W. S. (1999) Nat. Neurosci. 2 434-439 [DOI] [PubMed] [Google Scholar]
- 16.Hsu, S.-C., Hazuka, C. D., Roth, R., Foletti, D. L., Heuser, J., and Scheller, R. H. (1998) Neuron 20 1111-1122 [DOI] [PubMed] [Google Scholar]
- 17.Spiliotis, E. T., Hunt, S. J., Hu, Q., Kinoshita, M., and Nelson, W. J. (2008) J. Cell Biol. 180 295-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spiliotis, E. T., Kinoshita, M., and Nelson, W. J. (2005) Science 307 1781-1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu, M., Wang, F., Yan, F., Yao, P. Y., Du, J., Gao, X., Wang, X., Wu, Q., Ward, T., Li, J., Kioko, S., Hu, R., Xie, W., and Yao, X. (2008) J. Biol. Chem. M710591200. [DOI] [PMC free article] [PubMed]
- 20.Kinoshita, M., Kumar, S., Mizoguchi, A., Ide, C., Kinoshita, A., Haraguchi, T., Hiraoka, Y., and Noda, M. (1997) Genes Dev. 11 1535-1547 [DOI] [PubMed] [Google Scholar]
- 21.Surka, M. C., Tsang, C. W., and Trimble, W. S. (2002) Mol. Biol. Cell 13 3532-3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spiliotis, E. T., and Nelson, W. J. (2006) J. Cell Sci. 119 4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weirich, C. S., Erzberger, J. P., and Barral, Y. (2008) Nat. Rev. Mol. Cell Biol. 9 478-489 [DOI] [PubMed] [Google Scholar]
- 24.Barral, Y., and Kinoshita, M. (2008) Curr. Opin. Cell Biol. 20 12-18 [DOI] [PubMed] [Google Scholar]
- 25.Sheffield, P. J., Oliver, C. J., Kremer, B. E., Sheng, S., Shao, Z., and Macara, I. G. (2003) J. Biol. Chem. 278 3483-3488 [DOI] [PubMed] [Google Scholar]
- 26.Mendoza, M., Hyman, A. A., and Glotzer, M. (2002) Curr. Biol. 12 1858-1863 [DOI] [PubMed] [Google Scholar]
- 27.Sirajuddin, M., Farkasovsky, M., Hauer, F., Kuhlmann, D., Macara, I. G., Weyand, M., Stark, H., and Wittinghofer, A. (2007) Nature 449 311-315 [DOI] [PubMed] [Google Scholar]
- 28.Russell, S. E. H., and Hall, P. A. (2005) Br. J. Cancer 93 499-503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall, P. A., and Russell, S. E. H. (2004) J. Pathol. 204 489-505 [DOI] [PubMed] [Google Scholar]
- 30.Montagna, C., Lyu, M.-S., Hunter, K., Lukes, L., Lowther, W., Reppert, T., Hissong, B., Weaver, Z., and Ried, T. (2003) Cancer Res. 63 2179-2187 [PubMed] [Google Scholar]
- 31.Larisch, S. (2004) Cell Cycle 3 1021-1023 [PubMed] [Google Scholar]
- 32.Craven, R. A., Stanley, A. J., Hanrahan, S., Dods, J., Unwin, R., Totty, N., Harnden, P., Eardley, I., Selby, P. J., and Banks, R. E. (2006) Proteomics 6 2853-2864 [DOI] [PubMed] [Google Scholar]
- 33.Capurso, G., Crnogorac-Jurcevic, T., Milione, M., Panzuto, F., Campanini, N., Dowen, S. E., Di Florio, A., Sette, C., Bordi, C., Lemoine, N. R., and Delle Fave, G. (2005) Neuroendocrinology 81 311-321 [DOI] [PubMed] [Google Scholar]
- 34.Burrows, J. F., Chanduloy, S., McIlhatton, M. A., Nagar, H., Yeates, K., Donaghy, P., Price, J., Godwin, A. K., Johnston, P. G., and Russell, S. E. H. (2003) J. Pathol. 201 581-588 [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez, M. E., Peterson, E. A., Privette, L. M., Loffreda-Wren, J. L., Kalikin, L. M., and Petty, E. M. (2007) Cancer Res. 67 8554-8564 [DOI] [PubMed] [Google Scholar]
- 36.Amir, S., and Mabjeesh, N. J. (2007) Cancer Biol. Ther. 6 1926-1931 [DOI] [PubMed] [Google Scholar]
- 37.Kinoshita, A., Kinoshita, M., Akiyama, H., Tomimoto, H., Akiguchi, I., Kumar, S., Noda, M., and Kimura, J. (1998) Am. J. Pathol. 153 1551-1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ihara, M., Tomimoto, H., Kitayama, H., Morioka, Y., Akiguchi, I., Shi-basaki, H., Noda, M., and Kinoshita, M. (2003) J. Biol. Chem. 278 24095-24102 [DOI] [PubMed] [Google Scholar]
- 39.Ihara, M., Yamasaki, N., Hagiwara, A., Tanigaki, A., Kitano, A., Hikawa, R., Tomimoto, H., Noda, M., Takanashi, M., Mori, H., Hattori, N., Miyakawa, T., and Kinoshita, M. (2007) Neuron 53 519-533 [DOI] [PubMed] [Google Scholar]
- 40.Kim, C. S., Seol, S. K., Song, O. K., Park, J. H., and Jang, S. K. (2007) J. Virol. 81 3852-3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pizarro-Cerda, J., Jonquieres, R., Gouin, E., Vandekerckhove, J., Garin, J., and Cossart, P. (2002) Cell. Microbiol. 4 101-115 [DOI] [PubMed] [Google Scholar]
- 42.Iwase, M., Okada, S., Oguchi, T., and Toh-e, A. (2004) Genes Genet. Syst. 79 199-206 [DOI] [PubMed] [Google Scholar]
- 43.Yamada, S., Pokutta, S., Drees, F., Weis, W. I., and Nelson, W. J. (2005) Cell 123 889-901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bulinski, J. C., Odde, D. J., Howell, B. J., Salmon, T. D., and Waterman-Storer, C. M. (2001) J. Cell Sci. 114 3885-3897 [DOI] [PubMed] [Google Scholar]
- 45.Sprague, B. L., and McNally, J. G. (2005) Trends Cell Biol. 15 84-91 [DOI] [PubMed] [Google Scholar]
- 46.Turner, D. P., Moussa, O., Sauane, M., Fisher, P. B., and Watson, D. K. (2007) Cancer Res. 67 1618-1625 [DOI] [PubMed] [Google Scholar]
- 47.Chacko, A. D., Hyland, P. L., McDade, S. S., Hamilton, P. W., Russell, S. H., and Hall, P. A. (2005) J. Pathol. 206 458-465 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.