Abstract
Proteoglycans (PGs) are composed of a protein moiety and a complex glycosaminoglycan (GAG) polysaccharide moiety. GAG chains are responsible for various biological activities. GAG chains are covalently attached to serine residues of the core protein. The first step in PG biosynthesis is xylosylation of certain serine residues of the core protein. A specific linker tetrasaccharide is then assembled and serves as an acceptor for elongation of GAG chains. If the production of endogenous GAG chains is selectively inhibited, one could determine the role of these endogenous molecules in physiological and developmental functions in a spatiotemporal manner. Biosynthesis of PGs is often blocked with the aid of nonspecific agents such as chlorate, a bleaching agent, and brefeldin A, a fungal metabolite, to elucidate the biological roles of GAG chains. Unfortunately, these agents are highly lethal to model organisms. Xylosides are known to prime GAG chains. Therefore, we hypothesized that modified xylose analogs may able to inhibit the biosynthesis of PGs. To test this, we synthesized a library of novel 4-deoxy-4-fluoroxylosides with various aglycones using click chemistry and examined each for its ability to inhibit heparan sulfate and chondroitin sulfate using Chinese hamster ovary cells as a model cellular system.
Proteoglycans are composed of a core protein and one or more glycosaminoglycan (GAG)4 side chains such as chondroitin sulfate (CS) and heparan sulfate (HS). A common linkage tetrasaccharide, GlcAβ(1–3)Galβ(1–3)Galβ(1–4)Xylβ(1-O-Ser), is found between serine residues in core proteins and the GAG polysaccharide side chains (1, 2). Various biological activities of proteoglycans depend critically on interactions of the sulfated GAG side chains with a wide array of proteins, including proteases and protease inhibitors, growth factors and receptors, morphogens, cytokines, and extracellular matrix structural proteins (3, 4). These proteins differentially recognize and bind to specific sulfate groups of GAG chains. GAG chain formation involves the following events: chain initiation by the transfer of xylose residues to certain serine amino acids in the core proteins, assembly of the tetrasaccharide linkage region, elongation by alternate addition of d-glucuronic acid and N-acetyl-d-hexosamine (GlcNAc for HS and GalNAc for CS) units to the linker tetrasaccharide, and finally highly coordinated multiple sulfation/epimerization steps along the GAG chains (5–8).
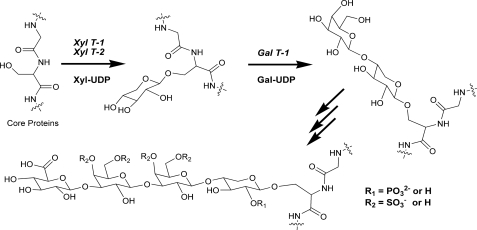
The linkage tetrasaccharide is synthesized by sequential transfer of xylose, galactose, galactose, and glucuronic acid residues from their corresponding sugar nucleotides. The following glycosyltransferases have been shown to be involved in assembly of the linkage region: xylosyltransferase-1/2, galactosyltransferase-1, galactosyltransferase-2, and glucuronyltransferase-1 (see Fig. 1) (9–14). Furthermore, various chemical modifications such as phosphorylation of xylose at C-2 or sulfation of galactose residues at C-4 and C-6 in the linkage region are predicted to play an important regulatory role in the assembly and maturation of the GAG chains (15–18).
FIGURE 1.
Assembly of the PG tetrasaccharide linkage region. XYL T1 and XYL T2, xylosyltransferase-1 and xylosyltransferase-2; GAL T1, galactosyltransferase-1.
To elucidate the biological significance of GAG chains, several chemical approaches have been employed to alter or inhibit their biosynthesis. The most important ones are sodium chlorate, a bleaching agent, and brefeldin A (BFA), a fungal isoprenoid metabolite (19–22). Sodium chlorate inhibits the formation of 3′-phosphoadenosine 5′-phosphosulfate, the sulfate donor, by competitively binding to 3′-phosphoadenosine-5′-phosphosulfate synthase at the active site and thereby affects the sulfation events in the Golgi (23). As a result, chlorate-containing medium has been widely used in various cellular systems to define the biological functions of sulfated GAG chains. However, a very high concentration of chlorate (up to 30 mm) is required to affect the sulfation. Such high concentrations are not suitable for in vivo studies in model organisms. On the other hand, BFA inhibits secretory transport of Golgi vesicles, an essential process for the functional Golgi. Various biosynthetic enzymes that are involved in the assembly and maturation of GAG chains have been shown to be localized to different regions of the Golgi apparatus. BFA blocks proteoglycan (PG) biosynthesis by preventing the transport of Golgi vesicles, an important process for the functional Golgi. It has been shown that BFA differentially affects HS and CS PG biosynthesis (19, 24, 25). In addition, BFA affects the biosynthesis of various other glycoconjugates (22, 26). Thus, it is very difficult to pinpoint the biological actions of GAG chains alone without affecting other glycosylation events using BFA. In an another approach, a series of 4-deoxyglucosamine analogs were examined for their ability to inhibit GAG biosynthesis in hepatocytes (27). GAG chains isolated from analog-treated hepatocytes were shown to be smaller in size than those isolated from control cells. Unfortunately, these analogs are not selective inhibitors of GAG biosynthesis and affect the biosynthesis of other glycoconjugates as well.
Many xylosides containing hydrophobic aglycones act as acceptors for the elongation of GAG chains. Various xylosides have been used to induce free GAG chains in various cellular systems for nearly 4 decades (28–31). As an approach to perturb GAG biosynthesis, we propose that 4-deoxy-4-fluoroxylosides cannot prime GAG chains as they do not have an acceptor hydroxyl group at C-4 for subsequent sugar attachment and elongation. Furthermore, these modified xylosides could affect galactosyltransferase-1, an enzyme involved in assembly of the linkage region, and thereby inhibit PG biosynthesis. In our efforts to develop new approaches to selectively block PG biosynthesis, we synthesized a library of 4-deoxy-4-fluoroxylosides using click chemistry and examined them for their ability to modulate GAG biosynthesis in wild-type Chinese hamster ovary (CHO) cells. We identified a number of 4-deoxy-4-fluoroxylosides that efficiently inhibit PG biosynthesis in a selective manner. We expect that these novel xyloside derivatives will become powerful chemical biology tools to define the developmental and physiological actions of PGs in model organisms by inhibiting or perturbing their biosynthesis in a spatiotemporal manner.
EXPERIMENTAL PROCEDURES
Materials—Wild-type CHO-K1 cells were obtained from American Type Culture Collection. All cell culture reagents were obtained from HyClone. The radiochemical Na 352SO4 was purchased from MP Biomedicals, and Ultima-FloAP flow scintillation mixture for flow radiometric analysis was obtained from PerkinElmer Life Sciences. All other chemicals and biochemicals were obtained from Sigma. The Pronase solution (×6) was prepared using Streptomyces griseus protease type XIV (1 mg/ml). The DEAE-Sepharose gel was purchased from Amersham Biosciences. The TSK-GEL DEAE-3SW HPLC ion-exchange chromatography column (7.5 mm × 7.5 cm, 10-μm particle size), was obtained from Tosoh Bioscience.
General Synthetic Methods—All reactions were carried out under a nitrogen atmosphere in oven-dried glassware using standard syringe and septum techniques. 1H, 13C, and 19F NMR spectra were obtained on a Bruker 400-MHz spectrometer. 1H chemical shifts are reported relative to tetramethylsilane at 0.0 ppm and referenced to the residual proton signal of the deuterated solvents. High resolution mass spectrometry of the synthetic scaffolds was performed using a Finnigan LCQ mass spectrometer in either positive or negative ion mode. TLC was carried out using 0.25-mm thick precoated Silica Gel HF254 aluminum sheets. Chromatograms were observed under short and long wavelength UV light and were visualized by heating plates that were dipped in a solution of ammonium(VI) molybdate tetrahydrate (12.5 g) and cerium(IV) sulfate tetrahydrate (5.0 g) in 10% aqueous sulfuric acid (500 ml). Flash column chromatography was performed using Silica Gel 60 (230–400 mesh) with a stepwise solvent polarity gradient, correlated with TLC mobility.
1,2,3-Tri-O-benzoyl-4-deoxy-4-fluoro-d-xylopyranoside (Compound 3)—The xyloside derivative 3 was synthesized using a previously published synthetic procedure (32). In brief, diethylaminosulfur trifluoride (9.370 mmol) was added to the solution containing compound 2 (3.244 mmol) in dry CH2Cl2 (50 ml) at –40 °C. The reaction mixture was allowed to come to room temperature slowly and was then stirred for an additional 3 h. After addition of MeOH (4 ml), the organic solvent was washed with saturated sodium bicarbonate solution, dried over MgSO4, and evaporated. The crude material was purified by flash chromatography on silica gel (9:1 hexane/EtOAc) to give 0.910 g of compound 3 (60% yield) as a colorless oil, which could not be crystallized and contained traces of the inseparable β-anomer.
2,3-Di-O-benzoyl-4-deoxy-4-fluoro-d-xylopyranosyl Azide (Compound 4)—Tetramethylsilyl azide (0.549 mmol) and SnCl4 (0.183 mmol) were added dropwise to a stirred solution of compound 3 (0.366 mmol) in dry CH2Cl2 (5 ml), and stirring was continued at room temperature. The progress of the reaction was checked by TLC, followed by evaporation in vacuo to give a residue, which was purified by column chromatography (12:1 hexane/EtOAc) to yield compound 4 (60 mg, 43% yield).
4-Deoxy-4-fluoroxlosides (Compounds 5a–5i)—Catalytic amounts of CuSO4·H2O and sodium l-ascorbate were added to a stirred solution of compound 4 (0.13 mmol) and a triple bond-containing aryl/alkyl scaffold (0.195 mmol) in 4 ml of 4:1.3 N,N-dimethylformamide/H2O. Stirring was continued overnight. The reaction mixture was concentrated and purified over a silica gel column (3:1 hexane/EtOAc) to yield the protected xylosides. NaOMe (25 mg) was added to a solution of protected xyloside (0.09 mmol) in 10 ml of 1:1 MeOH/CH2Cl2 and stirred for 3 h at room temperature. When TLC showed product formation, the reaction was neutralized by addition of Amberlite IR-120 resins, filtered, and concentrated to give a crude product, which was further purified by HPLC using a C18 column to obtain the pure final products (compounds 5a–5i). Structural data and NMR spectra are provided in the supplemental material.
Screening of 4-Deoxy-4-fluoroxylosides (Compounds 5a–5i) in CHO Cells—Inhibition of PG biosynthesis by modified xylosides (compounds 5a–5i) was examined using wild-type CHO-K1 cells as described briefly below. Cells (∼1 × 105 cells/well, containing the appropriate complete growth medium) were plated in a 24-well plate and incubated at 37 °C in a humidified incubator for 24 h to reach a confluence of ∼50%. The cells were then washed with sterile phosphate-buffered saline and replaced with 495 μl of the appropriate medium containing 10% dialyzed fetal bovine serum (HyClone). A stock solution containing various 4-deoxy-4-fluoroxylosides was prepared, and appropriate amounts of modified xylosides were added to various wells to yield final concentrations of 30 μm, 100 μm, 300 μm, and 1 mm. 50 μCi of Na 352SO4 was then added to each well to radiolabel the GAG side chains synthesized. The 24-well plates were placed in the incubator for 24 h before the addition of Pronase solution, followed by overnight incubation at 37 °C to harvest and analyze the effect of modified xylosides on PG biosynthesis.
Analysis of GAG Chains—After treating each well overnight with Pronase solution, the entire contents of the wells were transferred to a microcentrifuge tube and subjected to centrifugation at 16,000 × g for 5 min. The supernatant was transferred to a fresh tube, and 1 ml of 0.016% Triton X-100 was added. The diluted supernatant was loaded onto a DEAE-Sepharose column (0.2 ml) pre-equilibrated with 10 column volumes of wash buffer (20 mm NaOAc buffer (pH 6.0) containing 0.1 m NaCl and 0.01% Triton X-100), and the column was washed with 30 column volumes of wash buffer. The bound HS/CS chains were eluted using 6 column volumes of elution buffer (20 mm NaOAc (pH 6.0) containing 1 m NaCl). The extent of inhibition of PG biosynthesis was determined using the elution profile of GAG chains on an anion-exchange column by HPLC with an in-line radiodetector. Approximately 45,000 cpm of radioactive material isolated from each well was diluted to 1 ml with buffer containing 10 mm KH2PO4 (pH 6.0) and 0.2% CHAPS, loaded onto a HPLC anion-exchange column, and eluted with a linear NaCl gradient of 0.2–1.0 m over 80 min.
Cell Viability Assay—CHO-K1 cells were examined for viability in the presence of 4-deoxy-4-fluoroxyloside derivatives using CellTiter-Blue® reagent (Promega). Cells were seeded into duplicate wells of a 96-well plate at a density of 2.5 × 104 viable cells/well in 125 μl of Ham's F-12 complete growth medium supplemented with 10% fetal bovine serum and grown for 24 h with vehicle alone (Control 1) or xyloside derivatives. Cells were also treated with 3% H2O2 as an additional control (Control 2) for the cell viability assay. After 24 h of treatment with vehicle, H2O2, or xyloside derivative, 25 μl of CellTiter-Blue reagent was added to each well according to the manufacturer's protocol. After incubation in the humidified incubator for 2 h, the fluorescence generated in the assay was then stopped and stabilized by addition of 50 μl of 3% SDS solution. The fluorescence was immediately measured using a Spectra-Max M5 microplate reader (Molecular Devices, Sunnyvale, CA) at an excitation wavelength of 560 nm and an emission wavelength of 590 nm (33).
Estimation of Glycolipids and Glycopeptides—CHO-K1 cells were seeded onto a 24-well plate at a density of 1 × 105 cells/well (containing 50 μCi of [3H]GlcN) and grown for 24 h with vehicle alone or in the presence of xyloside inhibitors. After 24 h, 100 μl of Pronase (1 mg/ml) was added to each well to digest the proteins and to release glycopeptides. The samples were centrifuged at 21,000 × g for 20 min. The supernatant was used for the estimation of glycopeptides, whereas the pellet was used for the estimation of glycolipids (34). The pellet obtained after Pronase digestion was washed three times with 1 ml of phosphate-buffered saline. The glycolipids were then extracted from the pellet using 1:1:0.3 (v/v/v) chloroform/methanol/water, and the total radioactivity was measured by liquid scintillation. To estimate the effect of xylosides on glycosylation of proteins, the supernatants containing glycopeptides, after removal of residual GAG chains, were concentrated by lyophilization. The lyophilized samples were resuspended in distilled water and fractionated using a Bio-Gel P-2 column (with an eluant of 10 mm ammonium bicarbonate) to remove excess [3H]GlcN. Fractions were collected at 500 μl/fraction using a Gilson FC-204 fraction collector and measured using a liquid scintillation counter to estimate the level of [3H]GlcN incorporation in glycopeptides.
RESULTS AND DISCUSSION
Synthesis of 4-Deoxy-4-fluoroxylosides—Selective inhibition of PG biosynthetic pathways using modified xylosides is more desirable than nonspecific inhibition using toxic agents such as BFA and chlorate. With selective inhibition, these novel reagents assist in divulging the complex biological roles of heparan sulfate and other sulfated GAG chains at much higher molecular resolution. To accomplish our vision, we designed a library of triazolyl-4-fluoroxylosides that can be synthesized using a simple procedure, “click” chemistry, as shown in Scheme 1. Hydrophobic groups are required to transport xylosides across membranes and may also be required for recognition by enzymes involved in assembly of the linkage region. We have previously employed the same click methodology to assemble a library of xylosides and demonstrated that these xylosides are able to stimulate GAG biosynthesis (31). This approach introduced a diverse set of hydrophobic aglycones very quickly and allowed us to examine the effect of aglycone moieties on GAG biosynthesis in a rigorous manner.
SCHEME 1.
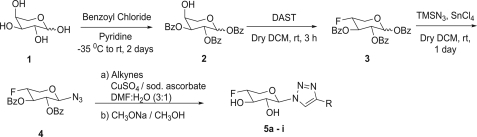
Synthesis of 4-deoxy-4-fluoroxylosides. DAST, diethylaminosulfur trifluoride; TMSN3, tetramethylsilyl azide; rt, room temperature; DMF, N,N-dimethylformamide; DCM, dichloromethane.
Our synthetic strategy employed the selective benzoylation of l-arabinopyranose (compound 1) in the first step using the previously reported procedure to obtain the partially protected 1,2,3,-tri-O-benzoyl-l-arabinopyranoside (compound 2) (35). Deoxyfluorocarbohydrate analogs, which contain a fluorine atom instead of a hydroxyl group, are extensively used as mechanistic probes or inhibitors of various enzymes involved in glycosylation. Therefore, we predicted that replacement of the C-4 hydroxyl group with a fluorine atom in xylose might block the transfer of the galactose residue from Gal-UDP catalyzed by galactosyltransferase-1, one of the key enzymes involved in assembly of the tetrasaccharide linkage region, and thereby prevent the assembly of PGs (Fig. 1). The direct reaction of an unprotected hydroxyl group with diethylaminosulfur trifluoride very often leads to nucleophilic displacement of the hydroxyl group by fluoride with concomitant inversion of configuration (36). The decisive step in our synthetic strategy is the inversion of the axial hydroxyl group at C-4 of the partially benzoylated l-arabinose derivative (compound 2) with diethylaminosulfur trifluoride to obtain the 4-deoxy-4-fluoroxylopyranoside derivative (compound 3).
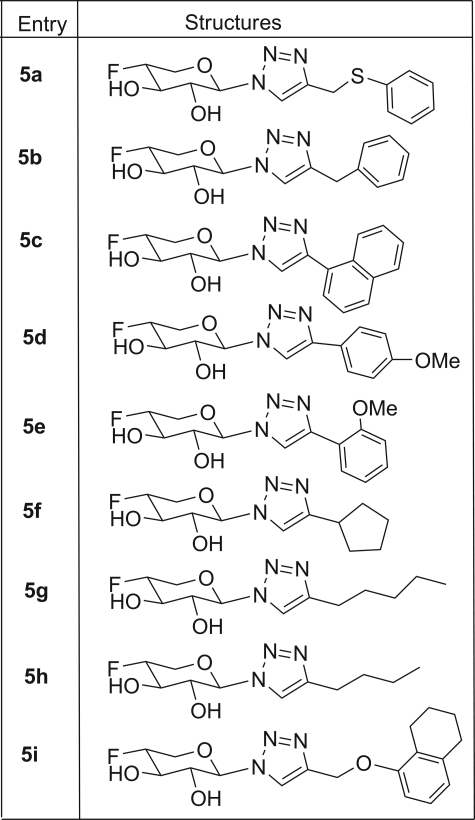
Xylosides require a hydrophobic aglycone moiety to efficiently prime GAG chains. Therefore, we predicted that 4-deoxy-4-fluoroxylosides would also require a hydrophobic aglycone moiety to efficiently inhibit PG biosynthesis. Thus, compound 3 needs to be appended with a hydrophobic group (aglycone) at the anomeric carbon. We and others have shown previously that the nature of the hydrophobic aglycone dramatically influences the priming ability of xylosides, as different hydrophobic aglycones are differentially recognized by the enzymes involved in assembly of the linkage region (30, 31). We identified a number of hydrophobic groups that were found to be important for priming GAG chains in our previous studies (31). On the basis of those findings, we have selectively chosen a set of promising hydrophobic groups to be appended at the anomeric carbon of 4-deoxy-4-fluoroxylopyranoside. We utilized this robust synthetic methodology in the present study to prepare various 4-deoxy-4-fluoroxylosides as potential biosynthetic inhibitors (Scheme 1). We synthesized the xylopyranosyl azide derivative 4 by treating compound 3 with tetramethylsilyl azide under dry solvent conditions. Compound 4 was then treated with a wide variety of azide-reactive triple bond-containing hydrophobic agents in the presence of Cu2+ salt/ascorbic acid, a step known as click chemistry. After click chemistry, benzoylated derivatives were deprotected under Zemplén conditions to obtain the final xyloside derivatives (compounds 5a–5i), which were then purified using a conventional chromatographic system. 1H, 13C, and 19F NMR analysis confirmed the structural identity of these potential PG biosynthetic inhibitors (Table 1).
TABLE 1.
Structures of 4-deoxy-4-fluoroxylosides
Screening of Potential Inhibitors of PG Biosynthesis—Our ability to manipulate the biosynthesis of the GAG chains will enhance our understanding of the functional interactions and significance of GAG chains. Various previous investigations utilized chlorate to inhibit 3′-phosphoadenosine-5′-phosphosulfate synthase, a key enzyme that is required for 3′-phosphoadenosine 5′-phosphosulfate production, which leads only to overall reduction in GAG sulfation. Chlorate does not block the elongation of GAG chains. BFA, which disrupts the Golgi apparatus, is known to differentially affect CS and HS PG assembly. Furthermore, BFA affects numerous glycosylation pathways, and therefore, BFA precludes our ability to selectively deduce the specific roles of GAG chains. The biosynthesis of the PG linkage tetrasaccharide, a common precursor for both HS and CS chain elongation, is a key step in the assembly of PGs. The completion of this specific linkage tetrasaccharide synthesis is an indispensable prerequisite for the conversion of core proteins to biologically active PGs. In this study, we investigated, for the first time, the influence of the fluorination of the xylosides at C-4 on the assembly of GAG chains by affecting the activity of biosynthetic enzymes involved in assembly of the linkage region, particularly the galactosyltransferase-1 enzyme, which catalyzes the transfer of the first Gal residue to xylosylated core proteins. We have examined various 4-deoxy-4-fluoroxylosides (compounds 5a–5i) for their ability to inhibit PG biosynthesis using wild-type CHO cells as a model cellular system. CHO cells produce both HS and CS chains, and therefore, the extent of selective inhibition of either HS or CS chains can also be determined.
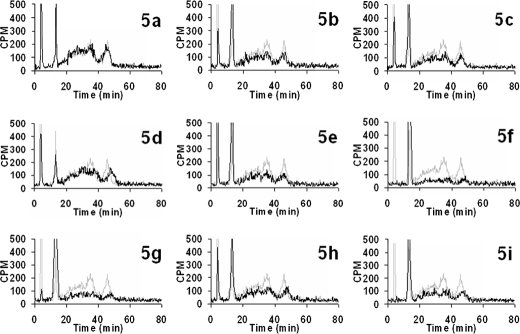
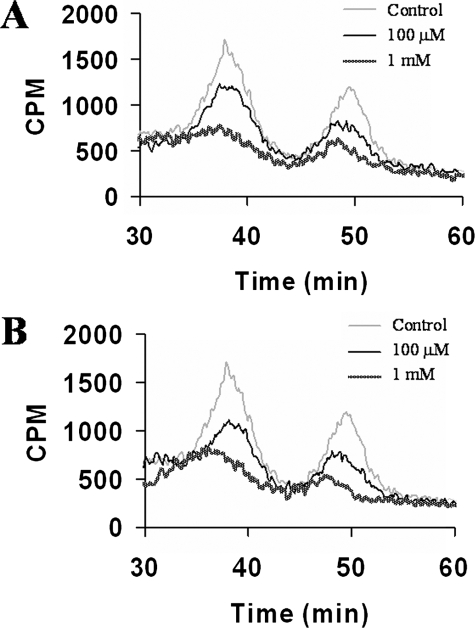
HS and CS chains could be resolved and identified using DEAE anion-exchange HPLC columns. Furthermore, elution profiles of GAG chains indicate both the extent of sulfation and the sulfation pattern. Thus, GAG chains that have less sulfation elute at lower ionic strength, whereas heavily sulfated GAG chains elute at higher ionic strength. Therefore, we utilized anion-exchange HPLC to deduce whether 4-deoxy-4-fluoroxylosides inhibit PG biosynthesis and the extent of inhibition. Wild-type CHO cells were grown in medium containing radioactive Na 352SO4 in the absence (control) or presence of various potential inhibitors at various concentrations for ∼24 h (Table 1). CHO cells were then subjected to Pronase treatment and subsequently passed through small disposable DEAE-Sepharose columns to enrich and elute the radioactive GAG chains. The resulting extract was analyzed by HPLC with an in-line radiodetector as described under “Experimental Procedures.” GAG chains composed of both HS and CS chains and extracted from cells that were not treated with the inhibitor migrated between 20 and 50 min as two peaks. The earlier peak is attributed to HS chains, whereas the later one is attributed to CS chains (37). We also observed two very sharp peaks eluting at ∼5 and 17 min. The structural identities of these peaks are currently under investigation. The chromatogram served as a control and was utilized to deduce the extent of inhibition of biosynthesis by 4-deoxy-4-fluoroxylosides. We then analyzed extracts from CHO cells that were grown in the presence of various inhibitors and compared them with the elution profile of GAG chains extracted from control CHO cells. GAG chains from xyloside 5a-treated cells were shown to have an elution profile that was very identical to that of wild-type cells and eluted between 20 and 50 min, suggesting that xyloside 5a is a poor inhibitor of PG biosynthesis (Fig. 2). On the other hand, xylosides 5b–5d were able to reduce the amount of GAG chains by up to 20%, suggesting that they are weak inhibitors of PG biosynthesis. Xylosides 5e, 5h, and 5i were able to inhibit ∼50% of total GAG production. Xylosides 5f and 5g were found to be the best inhibitors, as they reduced 90% of the total GAG chains produced. Dose-response experiments were done to investigate the extent of inhibition of biosynthesis by various xyloside derivatives. Cells were treated with inhibitors at various concentrations in the presence of [3H]GlcN for 24 h. As shown in Fig. 3, the amounts of isolated GAG chains decreased with increasing concentrations of xyloside derivatives 5e and 5g. BFA was shown to have differential effects on the biosynthesis of HS and CS. However, it is interesting to note that we did not observe any differential effects of these scaffolds on the biosynthesis of HS and CS.
FIGURE 2.
Inhibition of PG biosynthesis by xyloside derivatives. 35S-Labeled GAG chains were isolated from control and 4-deoxy-4-fluoroxyloside-treated cells by protease digestion and purification as described under “Experimental Procedures.” Radiolabeled GAG chains were then fractionated on a DEAE HPLC column and analyzed for radioactivity with the aid of an in-line radiodetector. The bound GAG chains were eluted with a linear gradient of 1 m NaCl as described under “Experimental Procedures.” The elution profiles of GAG chains isolated from cells that were untreated (gray trace; control) or treated (black trace) with PG biosynthetic inhibitors (compounds 5a–5i) are shown above.
FIGURE 3.
Dose-dependent decrease in GAG biosynthesis by 4-deoxy-4-fluoroxylosides in CHO cells. Samples of 3H-labeled GAG chains from control and 4-deoxy-4-fluoroxyloside-treated (compounds 5e (A) and 5g (B)) CHO cells were applied to a DEAE HPLC column, eluted with a linear NaCl gradient starting with 0.2 m NaCl as described under “Experimental Procedures,” and analyzed for radioactivity with an in-line radiodetector.
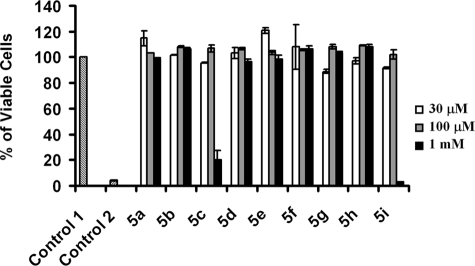
Assessment of cell viability is critical before utilizing these novel molecular scaffolds in animal models to deduce the biological roles of GAGs. Therefore, after screening xyloside derivatives for their ability to inhibit PG biosynthesis, we investigated whether these derivatives were toxic to the cells. Although several methods are available, we chose a fluorescence assay, the alamarBlue assay, which is convenient for the measurement of cell viability in a reliable and rapid manner. The fluorescent dye used in this assay is easily soluble in growth medium, is stable in solution, and is least toxic to cells. Cell viability assay confirmed that these xyloside derivatives were not toxic to cells at various concentrations. However, we observed that xylosides 5c and 5i were toxic at 1 mm and higher concentrations to CHO cells (Fig. 4).
FIGURE 4.
Cytotoxicity of 4-deoxy-4-fluoroxylosides. CHO cells were treated with different concentrations of xylosides (compounds 5a–5i) for 24 h and then incubated with CellTiter-Blue® reagent for 2 h as described under “Experimental Procedures” to estimate the percent of viable cells. CHO cells were grown in the absence of inhibitors (vehicle alone) as a control (Control 1), and the values of treated cells were compared with Control 1. Cells were also grown in the presence of 3% H2O2 as an additional control (Control 2).
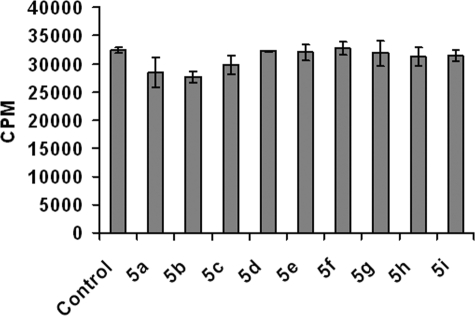
After determining that most of these scaffolds are not toxic to the cells at different concentrations, we then examined whether these fluorinated xylosides affect the biosynthesis of other glycoconjugates such as glycolipids and glycopeptides. Cells were labeled with [3H]glucosamine in the presence of xyloside derivatives to determine whether these scaffolds affect other glycosyltransferases involved in glycoprotein/glycolipid biosynthesis. We isolated radiolabeled glycolipids and glycopeptides from treated and untreated (control) cells. Quantitation of [3H]glucosamine-labeled glycolipids extracted from both treated and untreated cells suggested that these scaffolds did not affect glycolipid biosynthesis (Fig. 5). Xyloside 5f and 5g inhibited PG biosynthesis to a greater extent compared with any other scaffolds. Therefore, we examined whether these scaffolds affect glycosylation of proteins. The total radioactivity of fractions containing glycopeptides was largely the same for control cells (7.29 × 106 cpm), xyloside 5f-treated cells (7.01 × 106 cpm), and xyloside 5g-treated cells (7.91 × 106 cpm). Thus, we can conclude that these scaffolds selectively inhibit GAG biosynthesis without affecting other glycoconjugates.
FIGURE 5.
Comparison of the effects of various 4-deoxy-4-fluoroxylosides on glycolipid biosynthesis in CHO cells. Cells were incubated with vehicle alone (control) or with xylosides (compounds 5a–5i) for 24 h in the presence of [3H]GlcNH2. Glycolipids were isolated and quantified as described under “Experimental Procedures.” All values are expressed as a percent of the control sample without any xyloside derivative.
The priming ability of xylosides is greatly influenced by the nature of the hydrophobic aglycone group. We found that many of the best stimulatory aglycones of click xylosides, examined in our previous studies, seem to be essential in the design and construction of potential PG biosynthetic inhibitors (31). Thus, our observation clearly suggests that the hydrophobic nature of aglycones is as important as the fluoro atom replacing the acceptor hydroxyl group at C-4. It is interesting that these modified xylosides inhibit PG biosynthesis to different extents, thus providing novel approaches to fine-tune the extent of inhibition of PG biosynthesis in the quest to determine the biological consequences of variable inhibition in vivo.
In summary, a library of 4-deoxy-4-fluoroxylosides with various aglycone moieties was synthesized and screened for its ability to inhibit PG biosynthesis. The extent of inhibition of GAG biosynthesis depends on the hydrophobic aglycone group of modified xylosides. These modified xylosides are, as predicted, more selective in blocking the biosynthesis of PGs without challenging the Golgi machinery, and therefore, it is now possible to unveil the biological actions of GAG chains at much higher molecular resolution in animal models. We are currently utilizing these novel scaffolds to perturb the biosynthesis of PGs in zebrafish to elucidate the signaling pathways that are regulated by HS PGs during development. It is highly desirable to decipher the contribution of each GAG type by blocking selectively the production of either HS or CS only with the design of new synthetic scaffolds. Finally, it is important to note that GAG chains have been implicated in several pathological conditions, including amyloid formation, cancer cell growth, axonal degeneration, and vascular diseases. Therefore, the inhibitors of PG biosynthesis are anticipated to have a greater impact in the discovery and development of novel therapeutic agents to tackle various diseases (38, 39).
Supplementary Material
This work was supported, by National Institutes of Health Grant GM075168 (to B. K.). This work was also supported by the Human Frontiers Science Program (to B. K.) and by a grant-in-aid for science research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to M. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental material.
Footnotes
The abbreviations used are: GAG, glycosaminoglycan; CS, chondroitin sulfate; HS, heparan sulfate; BFA, brefeldin A; PG, proteoglycan; CHO, Chinese hamster ovary; HPLC, high pressure liquid chromatography; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
References
- 1.Muir, H. (1958) Biochem. J. 69 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindahl, U., and Roden, L. (1966) J. Biol. Chem. 241 2113–2119 [PubMed] [Google Scholar]
- 3.Bernfield, M., Gotte, M., Park, P. W., Reizes, O., Fitzgerald, M. L., Lincecum, J., and Zako, M. (1999) Annu. Rev. Biochem. 68 729–777 [DOI] [PubMed] [Google Scholar]
- 4.Schwartz, N. B., and Domowicz, M. (2004) Glycoconj. J. 21 329–341 [DOI] [PubMed] [Google Scholar]
- 5.Salmivirta, M., Lidholt, K., and Lindahl, U. (1996) FASEB J. 10 1270–1279 [DOI] [PubMed] [Google Scholar]
- 6.Esko, J. D., and Selleck, S. B. (2002) Annu. Rev. Biochem. 71 435–471 [DOI] [PubMed] [Google Scholar]
- 7.Sasisekharan, R., and Venkataraman, G. (2000) Curr. Opin. Chem. Biol. 4 626–631 [DOI] [PubMed] [Google Scholar]
- 8.Prydz, K., and Dalen, K. T. (2000) J. Cell Sci. 113 193–205 [DOI] [PubMed] [Google Scholar]
- 9.Gotting, C., Kuhn, J., Zahn, R., Brinkmann, T., and Kleesiek, K. (2000) J. Mol. Biol. 304 517–528 [DOI] [PubMed] [Google Scholar]
- 10.Almeida, R., Levery, S. B., Mandel, U., Kresse, H., Schwientek, T., Bennett, E. P., and Clausen, H. (1999) J. Biol. Chem. 274 26165–26171 [DOI] [PubMed] [Google Scholar]
- 11.Bai, X., Zhou, D., Brown, J. R., Crawford, B. E., Hennet, T., and Esko, J. D. (2001) J. Biol. Chem. 276 48189–48195 [DOI] [PubMed] [Google Scholar]
- 12.Kitagawa, H., Tone, Y., Tamura, J., Neumann, K. W., Ogawa, T., Oka, S., Kawasaki, T., and Sugahara, K. (1998) J. Biol. Chem. 273 6615–6618 [DOI] [PubMed] [Google Scholar]
- 13.Schon, S., Prante, C., Bahr, C., Kuhn, J., Kleesiek, K., and Gotting, C. (2006) J. Biol. Chem. 281 14224–14231 [DOI] [PubMed] [Google Scholar]
- 14.Cuellar, K., Chuong, H., Hubbell, S. M., and Hinsdale, M. E. (2007) J. Biol. Chem. 282 5195–5200 [DOI] [PubMed] [Google Scholar]
- 15.Moses, J., Oldberg, A., Cheng, F., and Fransson, L. A. (1997) Eur. J. Biochem. 248 521–526 [DOI] [PubMed] [Google Scholar]
- 16.Gulberti, S., Lattard, V., Fondeur, M., Jacquinet, J. C., Mulliert, G., Netter, P., Magdalou, J., Ouzzine, M., and Fournel-Gigleux, S. (2005) J. Biol. Chem. 280 1417–1425 [DOI] [PubMed] [Google Scholar]
- 17.Yamada, S., Okada, Y., Ueno, M., Iwata, S., Deepa, S. S., Nishimura, S., Fujita, M., Van Die, I., Hirabayashi, Y., and Sugahara, K. (2002) J. Biol. Chem. 277 31877–31886 [DOI] [PubMed] [Google Scholar]
- 18.Oegema, T. R., Jr., Kraft, E. L., Jourdian, G. W., and Van Valen, T. R. (1984) J. Biol. Chem. 259 1720–1726 [PubMed] [Google Scholar]
- 19.Calabro, A., and Hascall, V. C. (1994) J. Biol. Chem. 269 22764–22770 [PubMed] [Google Scholar]
- 20.Greve, H., Cully, Z., Blumberg, P., and Kresse, H. (1988) J. Biol. Chem. 263 12886–12892 [PubMed] [Google Scholar]
- 21.Humphries, D. E., and Silbert, J. E. (1988) Biochem. Biophys. Res. Commun. 154 365–371 [DOI] [PubMed] [Google Scholar]
- 22.Sampath, D., Varki, A., and Freeze, H. H. (1992) J. Biol. Chem. 267 4440–4455 [PubMed] [Google Scholar]
- 23.Safaiyan, F., Kolset, S. O., Prydz, K., Gottfridsson, E., Lindahl, U., and Salmivirta, M. (1999) J. Biol. Chem. 274 36267–36273 [DOI] [PubMed] [Google Scholar]
- 24.Uhlin-Hansen, L., and Yanagishita, M. (1993) J. Biol. Chem. 268 17370–17376 [PubMed] [Google Scholar]
- 25.Sugumaran, G., Katsman, M., and Silbert, J. E. (1992) J. Biol. Chem. 267 8802–8806 [PubMed] [Google Scholar]
- 26.Sherwood, A. L., and Holmes, E. H. (1992) J. Biol. Chem. 267 25328–25336 [PubMed] [Google Scholar]
- 27.Berkin, A., Szarek, W. A., and Kisilevsky, R. (2005) Glycoconj. J. 22 443–451 [DOI] [PubMed] [Google Scholar]
- 28.Okayama, M., Kimata, K., and Suzuki, S. (1973) J. Biochem. (Tokyo) 74 1069–1073 [PubMed] [Google Scholar]
- 29.Robinson, H. C., Brett, M. J., Tralaggan, P. J., Lowther, D. A., and Okayama, M. (1975) Biochem. J. 148 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fritz, T. A., Lugemwa, F. N., Sarkar, A. K., and Esko, J. D. (1994) J. Biol. Chem. 269 300–307 [PubMed] [Google Scholar]
- 31.Kuberan, B., Ethirajan, M., Victor, X. V., Tran, V., Nguyen, K., and Do, A. (2008) ChemBioChem 9 198–200 [DOI] [PubMed] [Google Scholar]
- 32.Wicki, J., Schloegl, J., Tarling, C. A., and Withers, S. G. (2007) Biochemistry 46 6996–7005 [DOI] [PubMed] [Google Scholar]
- 33.O'Brien, J., Wilson, I., Orton, T., and Pognan, F. (2000) Eur. J. Biochem. 267 5421–5426 [DOI] [PubMed] [Google Scholar]
- 34.Bailey, D. S., Burke, J., Sinclair, R., and Mukherjee, B. B. (1981) Biochem. J. 195 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Batey, J. F., Bullock, C., O'Brien, E., and Williams, J. M. (1975) Carbohydr. Res. 43 43–50 [Google Scholar]
- 36.Dax, K., Albert, M., Ortner, J., and Paul, B. J. (2000) Carbohydr. Res. 327 47–86 [DOI] [PubMed] [Google Scholar]
- 37.Zhang, L., Beeler, D. L., Lawrence, R., Lech, M., Liu, J., Davis, J. C., Shriver, Z., Sasisekharan, R., and Rosenberg, R. D. (2001) J. Biol. Chem. 276 42311–42321 [DOI] [PubMed] [Google Scholar]
- 38.Turnbull, J. E., and Field, R. A. (2007) Nat. Chem. Biol. 3 74–77 [DOI] [PubMed] [Google Scholar]
- 39.Lindahl, U. (2007) Thromb. Haemostasis 98 109–115 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.