Abstract
Hepatocyte growth factor activator inhibitors (HAI)-1 and -2 are recently identified and closely related Kunitz-type transmembrane serine protease inhibitors. Whereas HAI-1 is well established as an inhibitor of the serine proteases matriptase and hepatocyte growth factor activator, the physiological targets of HAI-2 are unknown. Here we show that HAI-2 displays potent inhibitory activity toward matriptase, forms SDS-stable complexes with the serine protease, and blocks matriptase-dependent activation of its candidate physiological substrates proprostasin and cell surface-bound pro-urokinase plasminogen activator. To further explore the potential functional relationship between HAI-2 and matriptase, we generated a transgenic mouse strain with a promoterless β-galactosidase marker gene inserted into the endogenous locus encoding HAI-2 protein and performed a global high resolution mapping of the expression of HAI-2, matriptase, and HAI-1 proteins in all adult tissues. This analysis showed striking co-localization of HAI-2 with matriptase and HAI-1 in epithelial cells of all major organ systems, thus strongly supporting a role of HAI-2 as a physiological regulator of matriptase activity, possibly acting in a redundant or partially redundant manner with HAI-1. Unlike HAI-1 and matriptase, however, HAI-2 expression was also detected in non-epithelial cells of brain and lymph nodes, suggesting that HAI-2 may also be involved in inhibition of serine proteases other than matriptase.
Recent mining of vertebrate genomes uncovered an unexpectedly large number of new membrane-associated trypsin-like serine proteases. The biochemical and physiological functions of most of these new serine proteases are undefined and the subject of active investigation. Trypsin-like serine proteases are typically synthesized as inactive zymogens that are irreversibly activated by a single endoproteolytic cleavage within a highly conserved activation site. They are subsequently inactivated by specific serine protease inhibitors that bind directly to the active site (1-3). Three functionally distinct classes of serine protease inhibitors, termed serpin-, Kazal-, and Kunitz-type inhibitors, have been identified in vertebrates. Whereas the serpin-type inhibitors have been extensively studied due to their preeminent role in regulating coagulation and fibrinolysis (3), the Kazal-and Kunitz-type serine protease inhibitors in vertebrates are comparatively less explored.
Hepatocyte growth factor activator inhibitor (HAI2)-1 and HAI-2 (also known as placental bikunin), encoded by Spint1 and Spint2 genes, respectively, are two recently discovered and closely related membrane-associated Kunitz-type serine protease inhibitors. These unusual serine protease inhibitors are type I transmembrane glycoproteins that contain two extracellular Kunitz-type inhibitory domains (4-8). HAI-1 was originally described as an endogenous inhibitor of hepatocyte growth factor activator (8). However, studies published shortly after the identification of HAI-1 show that the shed extracellular domain of HAI-1 can be isolated from tissue fluids and cell culture supernatants in a complex with the extracellular domain of the transmembrane serine protease matriptase (encoded by the ST14 gene) (9), strongly suggesting a physiological role of HAI-1 in matriptase inhibition. This was confirmed in several recent genetic studies in mice and zebrafish that revealed an essential role of matriptase inhibition by HAI-1 during vertebrate embryonic development (10-14).
The analysis of the functions of HAI-2 in vertebrate physiology has been complicated by the reported early embryonic lethality of Spint2 null mice (15). However, HAI-1 and HAI-2 are both type I transmembrane proteins and display 39-56% amino acid identity in their two Kunitz-type inhibitor domains, suggesting that HAI-2 could be a second physiological inhibitor of matriptase. Indeed, we show here that the kinetics of matriptase inhibition by HAI-2 is equipotent to that of HAI-1, and that matriptase and HAI-2 form SDS-stable complexes. Furthermore, HAI-2 efficiently blocks matriptase-mediated activation of two physiological candidate substrates, the prostasin zymogen and cell surface-bound pro-urokinase plasminogen activator (uPA). By generating a mouse strain with a promoterless β-galactosidase marker gene inserted into the endogenous Spint2 locus, we show that HAI-2 co-localizes with matriptase and HAI-1 in the epithelia of all major organ systems. Collectively, these new data strongly implicate HAI-2 as a physiologically relevant inhibitor of matriptase, possibly acting in a redundant or partially redundant manner with HAI-1 to regulate epithelial cell surface proteolysis in adult tissues.
EXPERIMENTAL PROCEDURES
Chromogenic Peptide Hydrolysis Assay—The chromogenic peptide Glu-Gly-Arg-p-nitroanilide was purchased from Bachem Bioscience (King of Prussia, PA), the soluble recombinant human HAI-1 and HAI-2 extracellular domains were from R&D Systems (Minneapolis, MN). To study the inhibition of matriptase proteolytic activity, 1 nm recombinant human matriptase serine protease domain (16) was incubated with 100 μm chromogenic peptide substrate in 50 mm Tris-HCl, pH 8.0, 1 mm CaCl2, 0.1% Tween 20 for 20 min at 37 °C in the presence of 0-500 nm HAI-1 or HAI-2 proteins. Changes in absorbance at 405 nm were monitored over time on a SAFIRE2™ Microplate Reader (Tecan, Durham, NC). All measurements were performed in triplicate.
Formation of HAI/Matriptase Inhibitory Complexes—100 ng of the recombinant active human matriptase serine protease domain in 50 mm Tris/HCl, pH 8.0, 100 mm NaCl buffer was incubated with 300 ng of human recombinant HAI-1 or HAI-2 for 30 min at room temperature. Protein complexes were analyzed by 4-12% reducing SDS-PAGE and Western blotting using a polyclonal anti-mouse HAI-2 primary antibody (R&D Systems) and anti-goat secondary antibody conjugated with alkaline phosphatase (EMD Chemicals-Calbiochem, La Jolla, CA) and a 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium detection system (Roche Applied Science). For non-immunological detection of HAI-matriptase complex formation, the assay was performed using 20 ng of active human matriptase serine protease domain and 50 ng of human recombinant HAI-1 or HAI-2, followed by 4-12% reducing SDS-PAGE and silver staining using a SilverQuest kit (Invitrogen) according to the manufacturer's instructions.
Prostasin Zymogen Activation Assay—0.1 μm human soluble prostasin zymogen prepared as described (17) was incubated with 5 nm recombinant active human matriptase serine protease domain for 1 h at 37 °C in 50 mm Tris/HCl, pH 8.0, 100 mm NaCl buffer. To evaluate the effect of HAI-1 and HAI-2 on matriptase-mediated activation of pro-prostasin, matriptase was preincubated with 100 nm human recombinant HAI-1 or HAI-2 proteins for 30 min at 37 °C prior to the pro-prostasin activation assay. Proteins were boiled and analyzed by reduced SDS-PAGE followed by Western blot as described above using a monoclonal anti-prostasin antibody (BD Biosciences, San Jose, CA), goat anti-mouse secondary antibody conjugated with alkaline phosphatase (DakoCytomation, Glostrup, Denmark).
Activation of Pro-uPA on the Surface of THP-1 Cells—Matriptase-dependent cell surface activation of pro-uPA was determined as described previously (18). Briefly, THP-1 acute monocytic leukemia cells (ATCC, Manassas, VA) were grown in RPMI 1640 medium supplemented with 10% fetal calf serum, l-glutamine, and antibiotics (all from Invitrogen). Before the experiment, the cells were washed twice in serum-free RPMI 1640 buffered with 25 mm HEPES, pH 7.4, and incubated for 5 min at room temperature in 100 mm NaCl, 50 mm glycine/HCl, pH 3.0, buffer to dissociate cell surface-associated uPA. Cells were washed twice in serum-free RPMI 1640, 25 mm HEPES, pH 7.4, resuspended in the same medium at 107 cells/ml, and incubated with 300 nm HAI-2, 250 nm plasminogen activator inhibitor-1 (R&D Systems), or in the absence of inhibitors for 15 min at room temperature. 100 nm pro-uPA (a kind gift of Dr. Stephen Leppla, NIAID, National Institutes of Health) or 100 nm high molecular weight two-chain uPA (Molecular Innovations, Novi, MI) were then added, and the mixture was incubated for 30 min at 37 °C. Cells were washed twice with serum-free RPMI 1640, 25 mm HEPES, pH 7.4, resuspended at 106 cells/ml in 50 mm Tris/HCl, pH 7.4, 100 mm NaCl, 0.01% Tween 20 with 0.5 mm SpectrozymeUK (American Diagnostica, Stamford, CT), and incubated at 37 °C for 4 h. uPA substrate hydrolysis was measured by the increase in absorbance at 405 nm using a Victor3V spectrophotometer (PerkinElmer Life Sciences).
Tissue Acquisition—All procedures involving live animals were performed in an Association for Assessment and Accreditation of Laboratory Animal Care International-accredited vivarium following Institutional Guidelines and standard operating procedures. Spint2 knock-in mice containing a promoterless β-galactosidase gene trap inserted into intron 1 of the mouse Spint2 gene were generated from the embryonic stem cell line KST272 obtained from Bay Genomics (San Francisco, CA) (15). The generation of the Spint1 null mice and β-galactosidase-tagged ST14 knock-in mice have previously been described (12, 19, 20). The Spint2 knock-in mice were genotyped for the presence of the β-galactosidase gene trap by PCR using HAI-2-geo51 (5′-ATCTGCAACCTCAAGCTAGC-3′) and HAI-2-geo31 (5′-CAGAACCAGCAAACTGAAGG-3′) primers. The Spint1 null mice and ST14 knock-in mice were genotyped by PCR as described previously (12, 19, 20).
Whole Mount X-gal Staining—Six-month-old wild-type or heterozygous β-galactosidase-tagged Spint2 knock-in mice or β-galactosidase-tagged ST14 knock-in mice were euthanized by CO2 inhalation. Organs were excised, and slices of each tissue were placed in 4% paraformaldehyde in phosphate-buffered saline for 30 min, rinsed in phosphate-buffered saline, and stained overnight at 37 °C with a β-galactosidase staining kit (Roche Applied Science). The tissues were post-fixed for 16 h in 4% paraformaldehyde, embedded in paraffin, and sectioned. The sections were counterstained with nuclear fast red and subsequently examined for HAI-2 or matriptase expression. All microscopic images were acquired on a Zeiss AxioImager Z1 light microscope using an AxioCam HRc digital camera system (both Carl Zeiss AG, Gottingen, Germany).
Immunohistochemistry—Six-month-old wild-type or Spint1 null mice were euthanized by CO2 inhalation. Tissues were fixed overnight in 4% paraformaldehyde, embedded in paraffin, and cut into sections 5 μm thick. Antigens were retrieved by incubation for 10 min at 37 °C with 10 μg/ml proteinase K (Fermentas, Hanover, MD). The sections were blocked for 1 h at room temperature with 2% bovine serum albumin in phosphate-buffered saline and incubated overnight at 4 °C with the goat anti-mouse HAI-1 primary antibody (R&D Systems). Bound antibodies were visualized using a biotin-conjugated anti-goat secondary antibody (Vector Laboratories, Burlingame, CA) and a Vectastain ABC kit (Vector Laboratories) using 3,3′-diaminobenzidine substrate (Sigma).
In Situ Hybridization—In situ hybridization of digoxigenin-labeled probes to paraformaldehyde-fixed, paraffin-embedded sections was carried out using the Link-Label ISH Core Kit II (BioGenex, San Ramon, CA) following the manufacturer's instructions. The Spint2-specific digoxigenin-labeled RNA probes were prepared by in vitro transcription of the +40- to +347-bp fragment of the mouse Spint2 open reading frame cloned into a pCRII-TOPO vector using the DIG RNA Labeling Kit (Roche Applied Science) according to the manufacturer's instructions. 100 ng of sense or antisense RNA probe was applied on parallel tissue sections and hybridized overnight at 37 °C. After the hybridization, signals were detected using the Link-Label ISH Core Kit II (BioGenex) as described in the instruction manual, and the sections were counterstained with nuclear fast red.
RESULTS
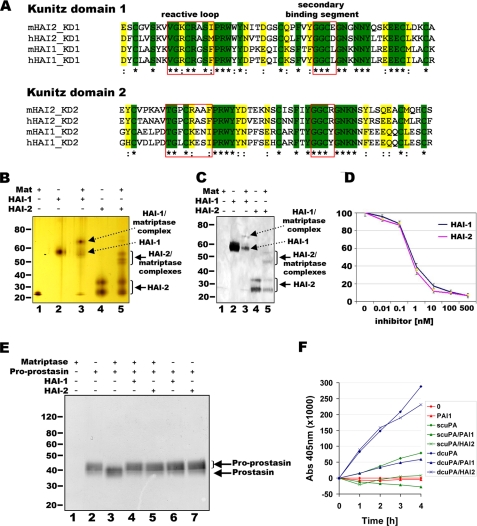
HAI-2 Is a Potent Inhibitor of Matriptase That Blocks the Proteolytic Activation of Its Candidate Physiological Targets the Prostasin Zymogen, and Cell Surface Pro-uPA—HAI-2 consists of two Kunitz-type inhibitor domains followed by a single span transmembrane domain and lacks the additional N-terminal region of unknown function found in HAI-1 (7). Although this accounts for the significant difference in the overall molecular weight between the two inhibitors (28.2 and 56.9 kDa for nascent unmodified proteins, respectively), the two proteins nevertheless exhibit a high degree of homology in their inhibitory domains. Comparing the two Kunitz-type inhibitor domains of the human and murine forms of HAI-1 and HAI-2 revealed a 39-56% overall amino acid identity and a 57-67% amino acid homology (Fig. 1A). The most highly conserved motifs included the reactive site loop and the secondary binding segment of the Kunitz domain (Fig. 1A), which were previously shown to be involved in the binding of the Kunitz domain to target proteases, including matriptase (16). Because HAI-1 is an essential inhibitor of matriptase in both mice and zebrafish (12, 14), this suggested that HAI-2 could also be a relevant inhibitor of this widely expressed membrane serine protease. HAI-1 has been shown previously to form SDS-stable complexes with matriptase that can be visualized by SDS-PAGE under non-boiling conditions (9). Therefore, to investigate the possible interaction between matriptase and HAI-2, we incubated the recombinant matriptase serine protease domain with recombinant soluble extracellular domain of either HAI-2 or HAI-1. Protease-inhibitor complex formation was analyzed by reducing SDS-PAGE followed by either silver staining or Western blotting using an HAI-2 antibody that cross-reacts with HAI-1. As reported previously, a novel protein species appeared with a molecular weight predicted for the matriptase-HAI-1 complex when matriptase was incubated with HAI-1 (Fig. 1, B and C, lane 3). This was associated with a decrease in the amount of non-complexed HAI-1 (Fig. 1, B and C, compare lanes 2 and 3). Interestingly, the formation of similar matriptase-HAI-2 complexes was readily detected when matriptase was incubated with HAI-2 (Fig. 1, B and C, lane 5), also associated with a decrease in non-complexed HAI-2 (Fig. 1, B and C, compare lanes 4 and 5). Next, we determined if HAI-2 displayed inhibitory activity against matriptase by determining the IC50 of the soluble HAI-2 toward the matriptase serine protease domain. In agreement with the high amino acid identity of the Kunitz-type inhibitor domains of the two transmembrane serine protease inhibitors, HAI-2 potently inhibited matriptase, displaying an IC50 that was indistinguishable from that of HAI-1 (0.5 nm and 0.63 nm, respectively) (Fig. 1D).
FIGURE 1.
HAI-2 is a potent inhibitor of matriptase. A, alignment of the amino acid sequences of Kunitz domains (KD) 1 (top panel) and 2 (bottom panel) of mouse and human HAI-2 and HAI-1. Identical residues are highlighted in green and indicated by an asterisk. Homologous substitutions are highlighted in yellow and indicated by a colon. Residues of the reactive site loop and the secondary binding segment that are involved in binding of the protease target are boxed in red. B and C, matriptase forms stable inhibitory complexes with HAI-2. 20 (B) or 100 (C) ng of human recombinant matriptase serine protease domain was incubated with 50 (B) or 300 (C) ng of human recombinant HAI-1 or HAI-2 for 30 min at room temperature. The formation of protease-inhibitor complexes was analyzed by silver staining (B) or Western blot using an antibody that recognizes both HAI-1 and HAI-2 (C). Positions of non-complexed HAI-1, non-complexed HAI-2, matriptase-HAI-1 complexes, and matriptase-HAI-2 complexes are indicated by arrows. The positions of molecular weight markers in kilodaltons are shown at the left. D, HAI-2 inhibits matriptase peptidolytic activity. 1 nm human recombinant matriptase serine protease domain was incubated in the presence of 0-500 nm of human recombinant HAI-1 or HAI-2 for 20 min at 37 °C. Matriptase activity toward the chromogenic peptide Glu-Gly-Arg-p-nitroanilide was measured as an increase in absorbance at 405 nm. Measurements were performed in triplicate and shown as the mean ± the S.D. E, HAI-2 inhibits activation of the prostasin zymogen by matriptase. 100 nm human recombinant soluble prostasin zymogen was incubated with 5 nm recombinant matriptase serine protease domain alone or in the presence of 500 nm HAI-1 or HAI-2 for 1 h at 37 °C. Activation of the prostasin zymogen was analyzed by Western blot using anti-prostasin antibody. Positions of the prostasin zymogen and active prostasin are indicated by arrows. The positions of molecular weight markers in kilodaltons are shown at the left. F, HAI-2 inhibits the physiological activation of pro-uPA by matriptase on the surface of THP-1 monocyte cells. THP-1 cells were stripped of endogenous uPA and incubated with 300 nm HAI-2 or 250 nm plasminogen activator inhibitor-1 for 15 min at room temperature. Pro-uPA (scuPA) or active high molecular weight two-chain uPA (dcuPA) was added to a final concentration of 100 nm for 30 min at 37 °C. The cells were washed, and cell surface-associated uPA activity was measured by the increase in the absorbance at 405 nm in the presence of 0.5 mm SpectrozymeUK.
HAI-2 Blocks Matriptase-mediated Activation of Its Candidates Substrates Pro-prostasin and Cell Surface-bound Pro-uPA—Matriptase likely promotes oral epithelial and epidermal differentiation by proteolytically activating the prostasin (PRSS8/CAP1) zymogen, because ST14- and PRSS8-ablated epidermis are phenocopies of each other and active prostasin is absent in ST14 null mice (17, 21). To determine if HAI-2 can block prostasin activation by matriptase, recombinant prostasin zymogen was expressed in HEK293T cells and released from the cell surface by phosphatidylinositol-specific phospholipase C. As reported previously (17), incubation of soluble prostasin zymogen with low amounts of matriptase lead to the formation of active two-chain prostasin, which displayed a slightly increased electrophoretic mobility in high percentage SDS-PAGE after reduction of the single disulfide bridge that links the two chains (Fig. 1E, compare lanes 2 and 3). This activation of prostasin by matriptase was completely blocked when matriptase was preincubated with either HAI-1 (Fig. 1E, compare lanes 3 and 4) or HAI-2 (Fig. 1E, compare lanes 3 and 5).
We next determined if HAI-2 was capable of inhibiting matriptase activity in a physiologically relevant setting. Recent studies have shown that matriptase is required for the conversion of pro-uPA to active two-chain uPA on the surface of the acute monocytic leukemia cell line THP-1, ultimately leading to plasminogen activation on the cell surface (18). To investigate if HAI-2 could regulate matriptase activity under physiologically relevant conditions, we therefore measured uPA activity on the surface of THP-1 cells that were preincubated with either pro-uPA or active high molecular weight uPA in the presence or absence of soluble HAI-2 protein. HAI-2 displayed no inhibitory activity toward active two-chain uPA when assayed in vitro (data not shown). In agreement with this, the presence of HAI-2 had no effect on cell surface uPA activity when THP-1 cells were preincubated with active uPA (Fig. 1F). However, HAI-2 almost completely abolished uPA activity on the surface of cells that were preincubated with pro-uPA (Fig. 1F). In contrast, when the known uPA inhibitor plasminogen activator inhibitor-1 was added to THP-1 cells instead of HAI-2, a strong reduction of cell surface uPA activity was evident irrespectively of whether the cells were incubated with pro-uPA or active uPA (Fig. 1F). These data indicate that, unlike plasminogen activator inhibitor-1, HAI-2 does not target uPA protease activity per se, but rather the activation of the uPA zymogen by endogenous matriptase.
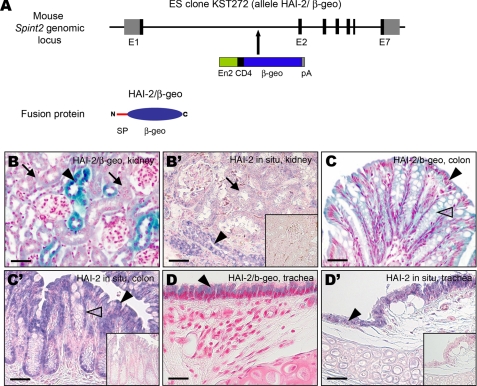
Generation of β-Galactosidase-tagged Spint2 Mice for Delineation of HAI-2 Matriptase Co-localization Studies in Vivo—The data presented above potentially implicated HAI-2 in the regulation of matriptase proteolytic activity. However, for a protease inhibitor to serve as a physiological inhibitor of a cognate protease requires the physical proximity of the two proteins in tissues. HAI-2 and matriptase are both membrane-anchored proteins, making it likely that HAI-2 must be expressed by the same cells that express matriptase to inhibit the protease. Therefore, we next set out to delineate the potential co-localization of HAI-2 and matriptase in cell lineages that form adult murine tissues. Furthermore, we mapped the expression of HAI-2 relative to HAI-1 to determine the possible functional overlap between the two membrane serine protease inhibitors.
An initial test of several commercially available anti-mouse HAI-2 antibodies revealed prohibitively high nonspecific immunohistochemical background using a variety of staining conditions, as well as cross reactivity with HAI-1 (data not shown). A similar lack of specificity was observed with all commercially available and in-house-generated matriptase antibodies. To analyze the expression of HAI-2 in mouse tissues, we therefore employed the technique of enzymatic gene trapping. A search of ES cell clones available through the International Gene Trap Consortium (www.genetrap.org) revealed one ES cell clone, KST272, with a promoterless β-galactosidase-neomycin phosphotransferase fusion gene (β-geo) inserted into intron 1 of the mouse Spint2 gene, which encodes HAI-2 (15) (HAI-2/β-geo allele, Fig. 2A). The insertion results in the expression of a β-geo reporter protein from the endogenous promoter of the Spint2 gene, thus allowing identification of HAI-2-expressing cells in situ by X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining. This technique has previously been shown to be a reliable alternative to immunohistochemistry in the absence of suitable antibodies or when proteins are expressed below the level of immunological detection (20, 22, 23).
FIGURE 2.
HAI-2 generation of a β-galactosidase-tagged spint2 mouse strain. A, schematic representation of the mouse HAI-2 gene locus targeted with a β-geo gene trap vector. ES cell clone KST272 has a gene trap consisting of the engrailed-2 (En2) splice acceptor site/CD4 transmembrane domain/β-galactosidase-neomycin phosphotransferase fusion gene (β-geo), and a bovine growth hormone polyadenylation site (pA) inserted between exon 1 and 2 (E1 and E2) of mouse Spint2 gene. Clone KST272 gives rise to a fusion protein that consists of an N-terminal signal peptide (SP) from the endogenous mouse HAI-2 protein fused to the β-galactosidase-neomycin phosphotransferase (β-geo) fusion protein (HAI-2/β-geo). B-D, representative histological sections showing high expression of HAI-2-β-galactosidase protein (blue) in epithelium of distal tubules in kidney (B, arrowhead), surface epithelium (C, arrowhead), and goblet cells (C, open arrowhead) in colon, epithelium in trachea (D, arrowhead), and low expression in proximal tubules in kidney (B, arrows). B′-D, expression of endogenous HAI-2 mRNA detected by in situ hybridization in distal and proximal tubules in kidney (B′, arrowhead and arrow, respectively), surface epithelium and goblet cells (C′, arrowhead and open arrowhead, respectively) in colon, and in tracheal epithelium (D′, arrowhead) in adult wild-type mouse. Insets: In situ hybridization of mouse HAI-2 sense probe in wild-type kidney (B′), colon (C′), and trachea (D′). Scale bars:25 μm (B, B′, D, and D′), 50 μm (C and C′).
To validate the use of Spint2 β-geo-targeted mice to delineate HAI-2 expression, staining with X-gal for detection of β-galactosidase activity and in situ hybridization of Spint2 mRNA was performed on parallel sections. In several tissues, including kidney, colon, trachea, and gallbladder, the pattern of β-geo protein expression closely matched that of Spint2 mRNA (Fig. 2, B-D′, and data not shown). This preliminary analysis established that the expression pattern of the β-geo protein is a faithful representation of the expression of the endogenous HAI-2 in mouse tissues.
For the detection of HAI-1, we performed immunohistochemistry using a previously validated polyclonal antibody (12). Sections of the corresponding tissues from adult HAI-1-deficient mice3 were used as negative controls in all experiments. To locate the expression of matriptase, we used a knock-in mouse strain with an insertion of a promoterless β-geo marker between exons 16 and 17 of the mouse ST14 gene that was previously validated for detecting matriptase expression in adult tissues (20, 24).
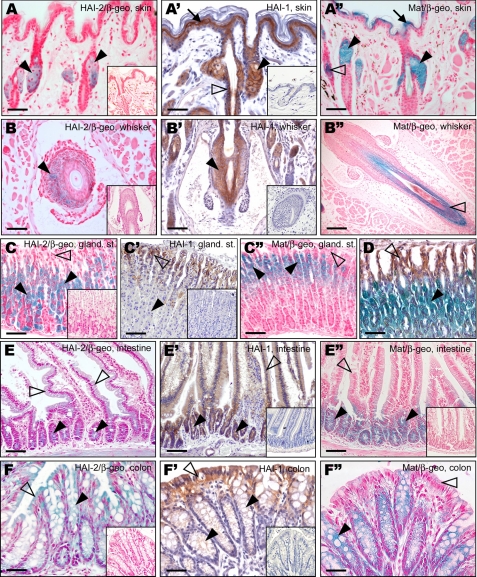
HAI-2 Co-localizes with Matriptase and HAI-1 in Sebaceous Glands and Vibrissal Inner Root Sheath Cells of the Epidermis—The expression of HAI-2 in the integumentary system appeared to be confined to the cells of sebaceous glands and the inner root sheath of vibrissal, but not pelage, hair follicles (Table 1, Fig. 3 (A and B), and data not shown). In contrast, examination of adult skin confirmed the previously reported widespread and overlapping expression of HAI-1 and matriptase in the keratinocytes of the suprabasal layer of the interfollicular epidermis, and the inner root sheath and matrix of pelage and vibrissal hair follicles, as well as the sebaceous glands (Fig. 3, A′, A″, B′, and B″). None of the three proteins were expressed in the basal layer of the epidermis or in any of the cell types in the dermis. Similar patterns of expression were found in keratinized stratified epithelia of several other organs, including the oral cavity, tongue, esophagus, and forestomach. Although all these tissues showed expression of both HAI-1 and matriptase in the keratinocytes of their respective suprabasal, but not basal layers, none of them showed any detectable expression of HAI-2 (Table 1 and supplemental Fig. S1).
TABLE 1.
Expression of HAI-2, HAI-1, and matriptase in integumentary and digestive systems
| Tissue | Cell population | HAI-2 | HAI-1 | Matriptase |
|---|---|---|---|---|
| Skin | Interfollicular epidermis | |||
| Basal layer | − | +/− | − | |
| Spinous layer | − | + | − | |
| Granular layer | − | + | + | |
| Hair follicles | ||||
| Outer root sheath | − | − | − | |
| Inner root sheath | + | + | + | |
| Medulla | − | + | + | |
| Cortex | − | + | + | |
| Cuticle | − | − | − | |
| Sebaceous gland | + | + | + | |
| Oral cavity | Tongue epithelium | − | + | + |
| Oral mucosal epithelium | − | + | + | |
| Lip epithelium | − | + | + | |
| Digestive tract | ||||
| Esophagus | Epithelium, suprabasal layer | − | + | + |
| Forestomach | Epithelium, suprabasal layer | − | + | + |
| Glandular stomach | Surface epithelium | − | + | − |
| Parietal cells | + | − | + | |
| Zymogenic cells | − | − | − | |
| Jejunum/ileum | Crypts | + | + | + |
| Villous epithelium | +/− | + | − | |
| Colon | Crypts | + | +/− | + |
| Goblet cells | + | + | + | |
| Surface epithelium | + | + | +/− | |
| Liver | Hepatocytes | − | − | − |
| Bile duct epithelium | − | + | +/− | |
| Gallbladder | Epithelium | + | + | − |
| Pancreas | Acinar cells | − | − | − |
| Islets of Langerhans | + | + | + |
FIGURE 3.
HAI-2 co-localizes with HAI-1 and matriptase in skin and epithelia of gastrointestinal tract. A-F, X-gal staining of HAI-2 expression (blue). In adult skin, HAI-2 was detected in sebaceous glands (A, arrowheads) and in cells of inner root sheath in vibrissal follicles (B, arrowhead). In gastrointestinal tissues, HAI-2 staining was localized to parietal cells of gastric glands in glandular stomach (C and D, arrowheads), in the epithelium of crypts (E, arrowheads) and intestinal villi (E, open arrowheads) within small intestine, and in goblet cells (F, arrowhead) and surface epithelium (F, open arrowhead) in colon. A′-F′ and A″-F″, immunohistochemical staining of HAI-1 expression (A′-F′, brown), and X-gal staining of matriptase expression (A″-F″, blue). In skin, HAI-1 and matriptase co-localize in the suprabasal keratinocytes in epidermis (A′ and A″, arrows), and in sebaceous glands (A′ and A″, arrowheads). HAI-1 also shows a widespread expression in keratinocytes of pelage (A′, open arrowhead) and vibrissal (B′, arrowhead) follicles, whereas matriptase expression is restricted to cells of hair matrix, medulla, and cortex (A″ and B″, open arrowheads and data not shown). In glandular stomach, HAI-1 was detected predominantly in surface epithelium (C′ and D, open arrowheads) and did not co-localize with HAI-2 (C and D, arrowheads) or matriptase (C″, open arrowhead), both of which were expressed in parietal cells, but not in surface mucosal cells (C and C″, open arrowheads). In small intestine, matriptase was found predominantly in crypt epithelium (E″, arrowheads), whereas HAI-1 showed a more widespread expression in both crypt and villous epithelium (E′, arrowheads and open arrowhead, respectively). In colon, HAI-1 and matriptase co-localized with HAI-2 in both surface mucosal cells (F′ and F″, open arrowhead) and in goblet cells (F′ and F″, arrowheads). Insets: X-gal staining of wild-type tissues (A, B, C, E″, and F), or HAI-1 immunohistochemical staining of tissues from HAI-1-deficient mice (A′-C′, E′, and F′) used as negative controls. Scale bars:50 μm (A, A″, B, D, F, and F″), 75 μm (C, C″, E, and E″), and 100 μm (B′ and B″).
Widespread and Coordinated Expression of HAI-2 with Matriptase and HAI-1 in the Digestive Tract—Unlike the stratified epithelium of the upper digestive tract, the simple epithelium of the glandular stomach and the entire intestinal tract showed a high level of HAI-2 expression (Table 1). In the glandular stomach, HAI-2 was detected in the parietal cells, whereas no expression was observed in the surface mucous cells or the chief cells (Fig. 3C). Matriptase was also found exclusively in the parietal cells, especially in the region proximal to the surface of the gastric gland (Fig. 3C″). On the other hand, the expression of HAI-1 was highly restricted to the surface mucous cells, with only very low levels of HAI-1 detectable in the parietal cells (Fig. 3C′), thus restricting the expression of the two inhibitors to non-overlapping populations of gastric epithelial cells (Fig. 3D). In the small intestine, both HAI-2 and HAI-1 showed widespread expression in the surface epithelium of both the intestinal villi and the crypts of Lieberkuhn (Fig. 3, E and E′). Matriptase, on the other hand, was expressed predominantly in the stem cells and proliferating regions of the crypt epithelium, with progressively decreased expression toward the mature absorptive cells of the villous epithelium (Fig. 3E″). Similarly, the two inhibitors were strongly expressed in both goblet cells and the surface epithelial cells of the colon, whereas matriptase showed high expression in the cells at the base of the colonic crypts and in the goblet cells and only much lower expression in the surface mucosal cells (Fig. 3, F-F″).
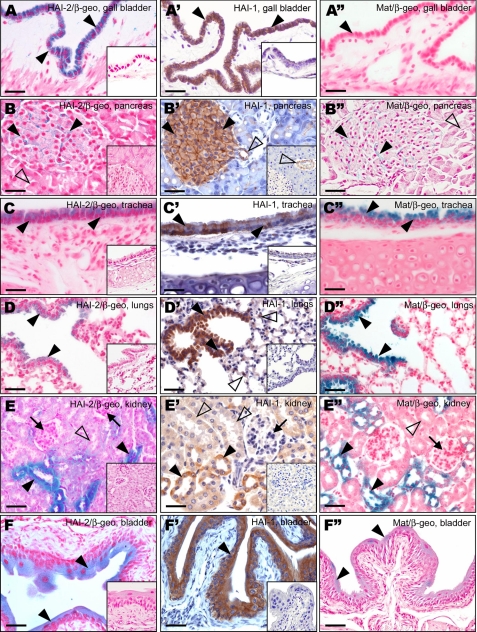
No HAI-2 was detected in the liver, whereas HAI-1 and matriptase showed specific expression in the bile ducts (supplemental Fig. S2). HAI-1 was also detected in a subpopulation of periportal cells, but the presence of an equally strong signal in tissues from HAI-1-deficient animals indicates that this is a result of a cross-reactivity of anti-HAI-1 antibody with a different protein target (supplemental Fig. S2). Columnar epithelium of the gallbladder stained positively for both HAI-2 and HAI-1, but not for matriptase (Fig. 4, A-A″). In the pancreas, the expression of all three proteins was restricted to the islets of Langerhans that showed significant expression of HAI-1 and low, but clearly identifiable expression of HAI-2 and matriptase (Fig. 4, B-B″). Neither the protease nor the inhibitors were present in the acinar or ductal cells of the exocrine portion of the pancreas (Fig. 4, B and B″, and data not shown).
FIGURE 4.
Expression profiles of HAI-2, HAI-1, and matriptase in gallbladder, pancreas, respiratory, and urinary tissues. A-F, X-gal staining of HAI-2 expression (blue). High levels of HAI-2 were observed in epithelium of gallbladder (A, arrowheads), respiratory epithelium of the trachea (C, arrowheads), and bronchioles (D, arrowheads), distal and collecting tubules in kidney (E, arrowheads), and in epithelium of urinary bladder (F, arrowheads). Low expression was detected in pancreas in islets of Langerhans (B, arrowheads) and in proximal tubules in kidney (E, open arrowhead), whereas no HAI-2 expression was observed in exocrine pancreas (B, open arrowhead) or in glomeruli (E, arrows). A′-F′, immunohistochemical staining of HAI-1 expression (brown). A″-F″, X-gal staining of matriptase expression (blue). Expression of HAI-1 (A′, arrowheads), but not matriptase (A″, arrowhead) was detected in epithelium of gallbladder. Both proteins co-localized in islets of Langerhans in pancreas (B′ and B″, arrowheads), respiratory epithelium of the trachea (C′ and C″, arrowheads) and bronchioles (D′ and D″, arrowheads), distal and collecting tubules in kidney (E′ and E″, arrowheads), and to epithelia of urinary bladder (F′ and F″, arrowheads). Low expression of HAI-1 (E′, open arrowheads) but not matriptase (E″, open arrowhead) was also observed in kidney proximal tubules, and neither HAI-1 nor matriptase were expressed in glomeruli (E′ and E″, arrows). The immunostaining seen in some endothelial cells within pancreas (B′, open arrowhead) is nonspecific, because the same signal was present in tissues of animals expressing no HAI-1 protein (inset of B′, open arrowhead). Scale bars:30 μm (A-A″), 50 μm (B-F″).
Coordinated Expression of HAI-2 with Matriptase and HAI-1 in the Respiratory System—HAI-2 and HAI-1, as well as matriptase, showed a widespread expression throughout the epithelia of respiratory tissues (Table 2). In the trachea, the proteins were expressed in the ciliated columnar epithelial cells (Fig. 4, C-C″), and the same pattern of expression was maintained also in bronchi and bronchioles (Fig. 4, D-D″, and supplemental Fig. S2). In addition, only HAI-1 was detected in lung alveoli, where it localized specifically to type II alveolar cells (Fig. 4D′). None of the proteins was observed in supporting cartilage, fibroblast stroma, or smooth muscle cells or capillary endothelium of the lungs.
TABLE 2.
Expression of HAI-2, HAI-1, and matriptase in respiratory and urogenital systems
| Tissue | Cell population | HAI-2 | HAI-1 | Matriptase |
|---|---|---|---|---|
| Respiratory system | ||||
| Trachea | Epithelium | + | + | + |
| Bronchi/bronchioles | Epithelium | + | + | + |
| Lungs | Alveolar type II cells | − | + | − |
| Urinary system | ||||
| Kidney | Glomeruli | − | − | − |
| Proximal tubules | +/− | +/− | + | |
| Distal/collecting tubules | + | + | + | |
| Bladder | Epithelium | + | + | + |
| Reproductive system | ||||
| Mammary gland | Ductal epithelium | − | + | + |
| Uterus | Surface epithelium | +/− | + | + |
| Uterine glands | + | + | +/− | |
| Ovary | Granulosa cells, antral follicles | + | − | + |
| Corpus luteum | + | − | − | |
| Testis | Undifferentiated spermatogonia | − | + | − |
| Seminal vesicle | Epithelium | + | + | − |
HAI-2, HAI-1, and Matriptase Co-localize in the Urogenital System—High levels of HAI-2 expression were detected in the kidney (Table 2). In this organ, HAI-2 specifically localized to the epithelium of the distal convoluting ducts and collecting ducts (Fig. 4E). Proximal tubules displayed much weaker staining, and no signal was detected in the glomeruli. This pattern of expression was identical to that of HAI-1, whereas matriptase expression appeared to be restricted to the distal and collecting ducts (Fig. 4, E′ and E″). All three proteins were also expressed in the epithelium of the urinary bladder. Whereas the expression of HAI-2 and matriptase was restricted to the surface (suprabasal) layer, HAI-1 displayed a more general distribution and was detected in both basal and suprabasal cells of this transitional epithelium (Fig. 4, F-F″).
Although most of the mouse reproductive tissues showed expression of either HAI-1, HAI-2, or matriptase, each of the proteins exhibited a unique pattern of expression (Table 2). In females, HAI-2 was detected in the surface epithelium of the uterus and in the uterine glands, whereas no expression was observed in adult virgin mammary gland (supplemental Fig. S2). In ovary, HAI-2 expression was detected both in the granulosa cells of the antral follicles and in the progesterone-producing cells of the corpora lutea (Fig. 5A). Matriptase was also present in the antral follicles (Fig. 5A″), whereas HAI-1 did not show detectable levels of expression in this organ (Fig. 5A′). In the male reproductive system, HAI-2 was present in the columnar epithelium of the seminal vesicle, but not in the testis (Fig. 5B and supplemental Fig. S2). The expression of the HAI-2/β-geo fusion protein in prostate could not be evaluated due to prominent endogenous β-galactosidase activity in this tissue, which resulted in a strong X-gal signal even in tissues from control wild-type mice (data not shown). Similar to HAI-2, HAI-1 was detected in the epithelium of the seminal vesicle (Fig. 5B′) but was also expressed in seminiferous tubules of the adult testis, where it localized specifically to undifferentiated spermatogonia that are in contact with the basement membrane (supplemental Fig. S2). Unlike HAI-1 or HAI-2, no expression of matriptase was detected in testis or in seminal vesicle (Fig. 5B″ and supplemental Fig. S2).
FIGURE 5.
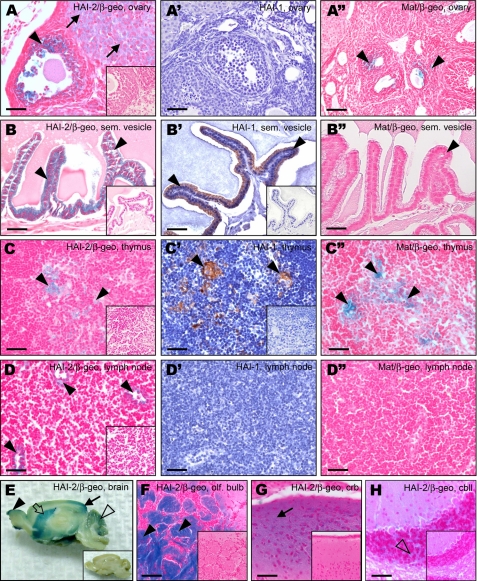
Expression profiles of HAI-2, HAI-1, and matriptase in male and female reproductive tissues, thymus, lymph node, and brain. A-H, X-gal staining of HAI-2 expression (blue). HAI-2 was detected in ovary in corpus luteus (A, arrows) and in granulosa cells of the follicles (A, arrowhead), in epithelium of seminal vesicle (B, arrowheads), medullar epithelial cells in thymus (C, arrowheads) and in endothelial cells of lymphatic vessels within lymph nodes (D, arrowheads). Within brain, high levels of HAI-2 were found in glomerular layers of olfactory bulb (E and F, arrowheads), neurons of cerebral cortex (E and G, arrows), and in striatum (E, open arrow), and lower levels of HAI-2 were detected also in inner granular layer of cerebellum (E and H, open arrowheads). A′-D′, immunohistochemical staining of HAI-1 expression (brown). HAI-1 was detected in epithelial cells of seminal vesicle (B′, arrowheads) and in epithelial cells in thymus (C′, arrowheads). No HAI-1 immunostaining was observed in ovary (A) or in lymph node (D). A″-D″, X-gal staining of matriptase expression (blue). Matriptase was detected in granulosa cells of antral follicles in ovary (A″, arrowheads), as well as in medullar epithelium in thymus (C″, arrowheads). No matriptase staining was observed in epithelium in seminal vesicle (B″, arrowhead) or in lymph node. Insets: X-gal staining of wild-type tissues (A-H), or HAI-1 immunohistochemical staining of tissues from HAI-1-deficient mice (B′ and C′) used as negative controls. Scale bars:25 μm (B-B″), 50 μm (A-A″, C-C″, D-D″, G, and H), and 100 μm (F).
HAI-2, HAI-1, and Matriptase Co-localization in Thymic Epithelium—Thymic epithelial matriptase plays a critical role in thymocyte maturation (19). HAI-2 expression was detected in the medullary epithelial cells of the thymus, where it precisely co-localized with the expression of HAI-1 and matriptase (Table 3 and Fig. 5 (C-C″)). No expression of HAI-2 was found in any of the salivary glands from adult male or female mice (supplemental Fig. S3, and data not shown). This was in contrast to HAI-1 and matriptase that showed high levels of expression in the ductal epithelium of the submandibular, sublingual, and parotid glands (supplemental Fig. S3, and data not shown). None of the three proteins was detected in the acinar cells of any of the glands. In hematopoietic/lymphatic tissues, HAI-2, but not matriptase or HAI-1, was detected in the endothelial cells of lymphatic vessels within the lymph nodes (Fig. 5, D and D″). No detectable levels of any of the three proteins were found in the spleen (data not shown). In the brain, HAI-2 was highly expressed in the glomerular layer of the olfactory bulb, in the cerebral cortex, and in the striatum (Fig. 5, E-G, and data not shown), and to a lesser extent in the inner granular layer in the cerebellum (Fig. 5, E and H). Consistent with their reported epithelium-specific expression, neither HAI-1 nor matriptase was found in the central nervous system (data not shown). Examination of other mouse tissues did not reveal any detectable expression of HAI-2, HAI-1, or matriptase in cells of the circulatory system, including the arteries, veins, and capillaries, in muscle cells, including smooth, striated and heart muscle tissue, in the cells of the connective or adipose tissues (Table 3 and data not shown).
TABLE 3.
Expression of HAI-2, HAI-1, and matriptase in other organs
| Tissue | Cell population | HAI-2 | HAI-I | Matriptase |
|---|---|---|---|---|
| Salivary glands | ||||
| Submandibular gland | Ductal epithelium | − | + | + |
| Mucous/serous acini | − | − | − | |
| Sublingual gland | Ductal epithelium | − | + | + |
| Mucous acini | − | +/− | − | |
| Parotid gland | Ductal epithelium | − | + | + |
| Serous acini | − | − | − | |
| Thymus | Medullary epithelium | + | + | + |
| Lymph nodes | Lymphatic endothelium | + | − | − |
| Brain | ||||
| Olfactory bulb | Glomerular layer | + | − | − |
| Cerebrum | Cortex | + | − | − |
| Striatum | + | − | − | |
| Cerebellum | Inner granular layer | + | − | − |
| Spleen | Lymphoid/endothelial cells | − | − | − |
| Skeletal muscle | Myocytes | − | − | − |
| Heart | Cardiomyocytes | − | − | − |
| Vascular system | Arteries, veins, and capillaries | − | − | − |
DISCUSSION
HAI-2 (also known as placental bikunin) is a relatively little studied Kunitz-type transmembrane serine protease inhibitor. Spurred by the strong structural similarity of HAI-2 and HAI-1, we examined the potential of HAI-2 for serving as a physiological inhibitor of matriptase, which is a critical in vivo inhibitory target of HAI-1. Interestingly, we found that HAI-2 was a highly efficient inhibitor of matriptase that displayed in vitro inhibitory properties toward matriptase that were indistinguishable from those of HAI-1, as evidenced by near identical IC50 values, the formation of SDS-stable complexes with recombinant matriptase serine protease domain, and the abrogation of matriptase activation of its candidate substrate prostasin. Furthermore, HAI-2 was capable of blocking endogenous matriptase activity within the physiologically relevant context of cell surface pro-uPA activation (18).
The Kunitz-type inhibitor domains of HAI-1 and HAI-2 have been reported to have broad inhibitor specificity toward trypsin-like serine proteases (25-31), and the efficient inhibition of matriptase by HAI-2 in vitro would be of little significance unless the membrane serine protease and protease inhibitor pair were in physical proximity in tissues. To address this issue in the absence of suitable antibodies, we created a mouse strain that expressed a β-galactosidase marker gene under the control of the endogenous Spint2 regulatory sequences and performed a global high resolution mapping of the co-localization of HAI-2, HAI-1, and matriptase. Overall, the HAI-2 expression data generated this way were in good agreement with previous reports of HAI-2 mRNA and protein expression in individual human and mouse tissues (5, 6, 32-36). This analysis identified multiple epithelial cell lineages that expressed both HAI-2 and matriptase. These included the sebaceous glands and inner root sheath cells of the epidermis, the parietal cells of the glandular stomach, the transitional region between crypt and surface epithelium of the small intestine, the goblet cells of the colon, the ciliated columnar cells of the trachea, bronchi, and bronchioles, the collecting ducts of the kidney, the suprabasal urinary bladder epithelium, the antral follicles of the ovary, and the thymic epithelium (Tables 1, 2, 3). This widespread co-localization of matriptase indicated that HAI-2 would have the capacity to inhibit matriptase in multiple tissues.
Most of the cell lineages expressing HAI-2 and matriptase also expressed HAI-1. Exceptions included the parietal cells of the glandular stomach and the antral follicle cells of the ovary. This suggests that HAI-2 regulation of matriptase may be mostly redundant or partially redundant with HAI-1, but in a few cell lineages, HAI-2 would be the principal matriptase inhibitor. Finally, a few epithelial tissues, as well as the lymphatic and central nervous systems, displayed prominent expression of HAI-2 in the absence of matriptase, suggesting that HAI-2 may have additional matriptase-independent substrates.
In summary, our biochemical analysis and high resolution expression analysis strongly suggests that HAI-2 is a physiologically relevant inhibitor of matriptase in multiple adult epithelial tissues.
Supplementary Material
Acknowledgments
We thank Drs. J. Silvio Gutkind and Mary Jo Danton for critically reviewing this manuscript.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Program. This work was also supported by a Grant DAMD-17-02-1-0693 from the Department of Defense (to T. H. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S3.
Footnotes
The abbreviations used are: HAI, hepatocyte growth factor activator inhibitor; β-geo, fusion of β-galactosidase and neomycin phosphotransferase; IC50, half-maximal (50%) inhibitory concentration; PAI, plasminogen activator inhibitor; uPA, urokinase plasminogen activator.
R. Szabo, and T. H. Bugge, unpublished data.
References
- 1.Puente, X. S., Sanchez, L. M., Overall, C. M., and Lopez-Otin, C. (2003) Nat. Rev. Genet. 4 544-558 [DOI] [PubMed] [Google Scholar]
- 2.Rawlings, N. D., and Barrett, A. J. (1994) Methods Enzymol. 244 19-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rau, J. C., Beaulieu, L. M., Huntington, J. A., and Church, F. C. (2007) J. Thromb. Haemost. 5 Suppl. 1, 102-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marlor, C. W., Delaria, K. A., Davis, G., Muller, D. K., Greve, J. M., and Tamburini, P. P. (1997) J. Biol. Chem. 272 12202-12208 [DOI] [PubMed] [Google Scholar]
- 5.Itoh, H., Kataoka, H., Hamasuna, R., Kitamura, N., and Koono, M. (1999) Biochem. Biophys. Res. Commun. 255 740-748 [DOI] [PubMed] [Google Scholar]
- 6.Muller-Pillasch, F., Wallrapp, C., Bartels, K., Varga, G., Friess, H., Buchler, M., Adler, G., and Gress, T. M. (1998) Biochim. Biophys. Acta 1395 88-95 [DOI] [PubMed] [Google Scholar]
- 7.Kawaguchi, T., Qin, L., Shimomura, T., Kondo, J., Matsumoto, K., Denda, K., and Kitamura, N. (1997) J. Biol. Chem. 272 27558-27564 [DOI] [PubMed] [Google Scholar]
- 8.Shimomura, T., Denda, K., Kitamura, A., Kawaguchi, T., Kito, M., Kondo, J., Kagaya, S., Qin, L., Takata, H., Miyazawa, K., and Kitamura, N. (1997) J. Biol. Chem. 272 6370-6376 [DOI] [PubMed] [Google Scholar]
- 9.Lin, C. Y., Anders, J., Johnson, M., and Dickson, R. B. (1999) J. Biol. Chem. 274 18237-18242 [DOI] [PubMed] [Google Scholar]
- 10.Tanaka, H., Nagaike, K., Takeda, N., Itoh, H., Kohama, K., Fukushima, T., Miyata, S., Uchiyama, S., Uchinokura, S., Shimomura, T., Miyazawa, K., Kitamura, N., Yamada, G., and Kataoka, H. (2005) Mol. Cell Biol. 25 5687-5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan, B., Brennan, J., Grant, D., Peale, F., Rangell, L., and Kirchhofer, D. (2007) Dev. Biol. 303 222-230 [DOI] [PubMed] [Google Scholar]
- 12.Szabo, R., Molinolo, A., List, K., and Bugge, T. H. (2007) Oncogene 26 1546-1556 [DOI] [PubMed] [Google Scholar]
- 13.Mathias, J. R., Dodd, M. E., Walters, K. B., Rhodes, J., Kanki, J. P., Look, A. T., and Huttenlocher, A. (2007) J. Cell Sci. 120 3372-3383 [DOI] [PubMed] [Google Scholar]
- 14.Carney, T. J., von der Hardt, S., Sonntag, C., Amsterdam, A., Topczewski, J., Hopkins, N., and Hammerschmidt, M. (2007) Development 134 3461-3471 [DOI] [PubMed] [Google Scholar]
- 15.Mitchell, K. J., Pinson, K. I., Kelly, O. G., Brennan, J., Zupicich, J., Scherz, P., Leighton, P. A., Goodrich, L. V., Lu, X., Avery, B. J., Tate, P., Dill, K., Pangilinan, E., Wakenight, P., Tessier-Lavigne, M., and Skarnes, W. C. (2001) Nat. Genet. 28 241-249 [DOI] [PubMed] [Google Scholar]
- 16.Friedrich, R., Fuentes-Prior, P., Ong, E., Coombs, G., Hunter, M., Oehler, R., Pierson, D., Gonzalez, R., Huber, R., Bode, W., and Madison, E. L. (2002) J. Biol. Chem. 277 2160-2168 [DOI] [PubMed] [Google Scholar]
- 17.Netzel-Arnett, S., Currie, B. M., Szabo, R., Lin, C. Y., Chen, L. M., Chai, K. X., Antalis, T. M., Bugge, T. H., and List, K. (2006) J. Biol. Chem. 281 32941-32945 [DOI] [PubMed] [Google Scholar]
- 18.Kilpatrick, L. M., Harris, R. L., Owen, K. A., Bass, R., Ghorayeb, C., Bar-Or, A., and Ellis, V. (2006) Blood 108 2616-2623 [DOI] [PubMed] [Google Scholar]
- 19.List, K., Haudenschild, C. C., Szabo, R., Chen, W., Wahl, S. M., Swaim, W., Engelholm, L. H., Behrendt, N., and Bugge, T. H. (2002) Oncogene 21 3765-3779 [DOI] [PubMed] [Google Scholar]
- 20.List, K., Szabo, R., Molinolo, A., Nielsen, B. S., and Bugge, T. H. (2006) Am. J. Pathol. 168 1513-1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leyvraz, C., Charles, R. P., Rubera, I., Guitard, M., Rotman, S., Breiden, B., Sandhoff, K., and Hummler, E. (2005) J. Cell Biol. 170 487-496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voss, A. K., Thomas, T., and Gruss, P. (1998) Dev. Dyn. 212 171-180 [DOI] [PubMed] [Google Scholar]
- 23.Wurst, W., and Gossler, A. (2000) in Gene Targeting: A Practical Approach, 2nd Ed. (Joyner, A., ed) pp. 207-254, Oxford University Press, New York
- 24.List, K., Hobson, J. P., Molinolo, A., and Bugge, T. H. (2007) J. Cell. Physiol. 213 237-245 [DOI] [PubMed] [Google Scholar]
- 25.Denda, K., Shimomura, T., Kawaguchi, T., Miyazawa, K., and Kitamura, N. (2002) J. Biol. Chem. 277 14053-14059 [DOI] [PubMed] [Google Scholar]
- 26.Kirchhofer, D., Peek, M., Lipari, M. T., Billeci, K., Fan, B., and Moran, P. (2005) FEBS Lett. 579 1945-1950 [DOI] [PubMed] [Google Scholar]
- 27.Kirchhofer, D., Peek, M., Li, W., Stamos, J., Eigenbrot, C., Kadkhodayan, S., Elliott, J. M., Corpuz, R. T., Lazarus, R. A., and Moran, P. (2003) J. Biol. Chem. 278 36341-36349 [DOI] [PubMed] [Google Scholar]
- 28.Herter, S., Piper, D. E., Aaron, W., Gabriele, T., Cutler, G., Cao, P., Bhatt, A. S., Choe, Y., Craik, C. S., Walker, N., Meininger, D., Hoey, T., and Austin, R. J. (2005) Biochem. J. 390 125-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delaria, K. A., Muller, D. K., Marlor, C. W., Brown, J. E., Das, R. C., Roczniak, S. O., and Tamburini, P. P. (1997) J. Biol. Chem. 272 12209-12214 [DOI] [PubMed] [Google Scholar]
- 30.Fan, B., Wu, T. D., Li, W., and Kirchhofer, D. (2005) J. Biol. Chem. 280 34513-34520 [DOI] [PubMed] [Google Scholar]
- 31.Qin, L., Denda, K., Shimomura, T., Kawaguchi, T., and Kitamura, N. (1998) FEBS Lett. 436 111-114 [DOI] [PubMed] [Google Scholar]
- 32.Yamauchi, M., Kataoka, H., Itoh, H., Seguchi, T., Hasui, Y., and Osada, Y. (2004) J. Urol. 171 890-896 [DOI] [PubMed] [Google Scholar]
- 33.Yamauchi, M., Itoh, H., Naganuma, S., Koono, M., Hasui, Y., Osada, Y., and Kataoka, H. (2002) Biol. Chem. 383 1953-1957 [DOI] [PubMed] [Google Scholar]
- 34.Kataoka, H., Itoh, H., Uchino, H., Hamasuna, R., Kitamura, N., Nabeshima, K., and Koono, M. (2000) Cancer Lett. 148 127-134 [DOI] [PubMed] [Google Scholar]
- 35.Parr, C., Watkins, G., Mansel, R. E., and Jiang, W. G. (2004) Clin. Cancer Res. 10 202-211 [DOI] [PubMed] [Google Scholar]
- 36.Itoh, H., Kataoka, H., Tomita, M., Hamasuna, R., Nawa, Y., Kitamura, N., and Koono, M. (2000) Am. J. Physiol. 278 G635-G643 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.