Abstract
The thermophilic filamentous fungus Talaromyces emersonii secretes a variety of hydrolytic enzymes that are of interest for processing of biomass into fuel. Many carbohydrases have been isolated and characterized from this fungus, but no studies had been performed on peptidases. In this study, two acid-acting endopeptidases were isolated and characterized from the culture filtrate of T. emersonii. One of these enzymes was identified as a member of the recently classified glutamic peptidase family and was subsequently named T. emersonii glutamic peptidase 1 (TGP1). The second enzyme was identified as an aspartyl peptidase (PEP1). TGP1 was cloned and sequenced and shown to exhibit 64 and 47% protein identity to peptidases from Aspergillus niger and Scytalidium lignocolum, respectively. Substrate profiling of 16 peptides determined that TGP1 has broad specificity with a preference for large residues in the P1 site, particularly Met, Gln, Phe, Lys, Glu, and small amino acids at P1′ such as Ala, Gly, Ser, or Thr. This enzyme efficiently cleaves an internally quenched fluorescent substrate containing the zymogen activation sequence (kcat/Km = 2 × 105 m-1 s-1). Maximum hydrolysis occurs at pH 3.4 and 50 °C. The reaction is strongly inhibited by a transition state peptide analog, TA1 (Ki = 1.5 nm), as well as a portion of the propeptide sequence, PT1 (Ki = 32 nm). Ex vivo studies show that hyphal extension of T. emersonii in complex media is unaffected by the aspartyl peptidase inhibitor pepstatin but is inhibited by TA1 and PT1. This study provides insight into the functional role of the glutamic peptidase TGP1 for growth of T. emersonii.
As chemoheterotrophs, filamentous fungi secrete a variety of polymer-degrading hydrolases such as peptidases and carbohydrases that degrade organic material in the local environment to provide essential nutrients for the growing hyphae. Many fungi release organic acids to acidify the environment while secreting enzymes that are optimally active under these conditions. Traditionally, acid-acting fungal peptidases were assigned to the aspartic protease family and include aspergillopepsin from various Aspergillus species (1) and penicillopepsin from Penicillium species (2). These enzymes have two active-site aspartic acid residues and are strongly inhibited by pepstatin. Recently, two distinct groups of acid peptidases were identified that are insensitive to pepstatin and lack the catalytic motif observed in pepsin-type peptidases. One group, “sedolisins” possess a fold similar to members of the subtilisin family but contain an active site triad of serine (S), glutamic acid (E), and aspartic acid (D) (3). They are sensitive to leupeptin and have homologs in bacteria (4), fungi (5), slime mold (6), and mammals (7). The second group of pepstatin-insensitive acid peptidases share a distinct set of structural, enzymatic, and physicochemical properties, and to date have only been identified in a select group of fungi (8). Members of this novel family possess a catalytic dyad consisting of a glutamate (E) and a glutamine (Q) and have been classified as a new family of peptidases called “eqolisins” (9, 10). The EQ dyad is unique in nature and differs from other known mechanistic classes. Consequently, these enzymes have also been termed “glutamic peptidases” and have been reclassified in the MEROPS data base as “Family G1” (11). The substrate specificity and catalytic mechanism of these peptidases have been reported using peptide substrates (12) and transition state analogs (13), and Kubota et al. (14) have reported the role of the propeptide sequence in stability and activation of the zymogen.
Biochemical and functional data have been obtained for few eqolisins to date. Consequently, there is little known about the regulation of these novel peptidases in their fungal hosts, and the in vivo role has proven elusive. Studies on eqolisin expression in Aspergillus niger (15) and Sclerotinia sclerotiorum (16) suggest that these enzymes are regulated by extracellular levels of carbon, nitrogen, and pH. Poussereau et al. (16) proposed that eqolisins are an adaptation to host invasion or pathogenicity; however, they are only found in a few select pathogenic species and are absent in many others.
In this study we have identified, isolated, and characterized a glutamic peptidase (TGP1)4 from the secretome of Talaromyces emersonii. This thermophile secretes commercially valuable biopolymer-degrading enzymes, such as cellulases (17) and xylanases (18); however, no characterization of peptidases has previously been performed. We show that fungal growth under select conditions is absolutely dependent on TGP1 activity but not the co-secreted aspartyl peptidase PEP1.
EXPERIMENTAL PROCEDURES
Culture and Growth Conditions—T. emersonii was obtained from laboratory stocks and maintained on Sabouraud dextrose agar (SDA) by transfer of mycelia plugs to the center of fresh plates. Mycelia plugs were used to inoculate starter liquid cultures containing 4% w/v glucose and 1% w/v peptone. For protein production, mineral salt media (19) containing 2% glucose with or without yeast extract and ammonium sulfate were inoculated with mycelia from starter cultures and grown for 72 h on an orbital shaker. For gene expression analysis, T. emersonii was grown in media containing yeast nitrogen base without amino acids and ammonium sulfate (Sigma) and 2% glucose. The media were supplemented with an organic or inorganic nitrogen source (1% w/v) as outlined in Fig. 3. Bacto™ casitone and peptone were obtained from BD Biosciences. Hyphal extension assays were performed in the presence of growth agonists and antagonists (n = 4) dried onto sterile filter paper (Whatman No.2(3 × 6 mm)). Filter paper was placed in front of the growing edge of T. emersonii on SDA plates, and hyphal growth was measured after 18 h of incubation. All incubations were performed at 45 °C.
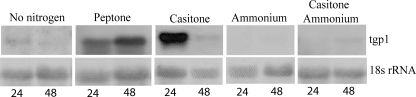
FIGURE 3.
Northern analysis of tgp1 expression in total RNA extracted from T. emersonii after 24 and 48 h growth in media containing glucose and yeast nitrogen base and supplemented with the indicated nitrogen source. An 18 S rRNA probe was used as a loading control.
Endoprotease Assay—Proteolytic activity was measured using a modification of the method described by Buroker-Kilgore and Wang (20). Briefly, 20 μl of enzyme sample was added to 80 μl of 7.5 μm bovine serum albumin (BSA) in 0.1 m citric acid/sodium phosphate, pH 3.3, and incubated at 50 °C. 20 μl was removed after 5, 15, or 45 min and added to 180 μl of Coomassie Plus™ protein assay reagent (Pierce) in a microtiter plate. Absorbance was read at 590 nm, and activity (IU) was calculated relative to BSA degradation using a standard curve. One IU is defined as degradation of 1 μmol of BSA min-1 ml-1.
Purification of Peptidases—All purification steps were performed at 4 °C. Mycelia were harvested through several layers of muslin cloth, and the spent media were centrifuged at 3,000 × g for 60 min. The supernatant was vacuum-filtered (Whatman No. 1) and then concentrated and dialyzed against 0.03 m ammonium acetate, pH 5.0 (Buffer A), in an Amicon DC2 apparatus fitted with an HIP 10-43 hollow fiber dialyzer. Further concentration was performed in a stirred ultrafiltration cell equipped with a 10-kDa cutoff membrane (Millipore Corp.). The dialyzed culture filtrate was applied to a DEAE-cellulose column (15 × 2.5 cm) pre-equilibrated with Buffer A, and protein was eluted in a linear gradient of 0–1.0 m NaCl in Buffer A. Fractions were collected, assayed for proteolytic activity, pooled, and dialyzed against Buffer A. The concentrate was reapplied to a DEAE-cellulose column (5 × 2 cm) and eluted in steps of increasing NaCl concentration. Proteolytically active fractions displaying pepstatin-sensitive (PEP1) and -insensitive (TGP1) activity were pooled separately, dialyzed, and concentrated against Buffer A containing 0.1 m NaCl. Each concentrate was applied to a Sephacryl S-100 HR column (150 × 1.5 cm), pre-equilibrated with Buffer A containing 0.1 m NaCl. Fractions with proteolytic activity were collected and pooled. PEP1 was further purified on a TEAE-cellulose column (8 ×3 cm). Aspergillopepsin I (PEP1Asp) was purchased from Sigma.
Analysis of Purity and Determination of Mr, pI, and N-terminal Sequence—Protein purity and molecular weight were assessed by Coomassie Brilliant Blue R-250 staining of a 4–20% Tris-glycine gel and MALDI-TOF mass spectrometry on a Voyager DE™ Pro BioSpectrometry work station (Applied Biosystems) following crystallization in sinapinic acid. N-terminal sequencing of TGP1 was performed on protein adsorbed to polyvinylidine fluoride. PEP1 was digested with trypsin, and peptides were sequenced by tandem mass spectrometry. Isoelectric focusing was performed using a flat bed apparatus (GE Healthcare) according to the manufacturer's instructions.
DNA Extraction and Cloning of tgp1—Mycelia from T. emersonii were harvested by filtration through sterile muslin, flash-frozen in liquid N2, and ground to a fine powder with a mortar and pestle. DNA was isolated according to the method of Raeder and Broda (21) and RNase-treated. A partial gene sequence encoding tgp1 was PCR-amplified from genomic DNA using degenerate primers designed from the N-terminal sequence (5′-AAYTGGGCNGGNGCNGT-3′) and a highly conserved region of other glutamic peptidases (5′-RAARTCYTCNACDATCCA-3′). The PCR product was subcloned into the pGEM-TEASY vector (Promega) and sequenced. A digoxigenin-labeled DNA probe (Roche Applied Science) was generated using gene-specific oligonucleotides (5′-GTGCTCATCGGCAGCGGCTAC-3′ and 5′-GGCGTTGGTCTCGCACAGCTG-3′). This probe was used to enrich a T. emersonii genomic library (22) for bacteriophage encoding TGP1, and the full-length gene was sequenced using 5′-CGAGTGGATCGTCGAGGACTTCTCCGAG-3′ and 5′-CTCGGAGAAGTCCTCGACGATCCACTCG-3′ oligonucleotides.
RNA Extraction and Northern Blot Analysis—Total RNA was isolated from T. emersonii with TRI-Reagent (Molecular Research Centre). RNA was treated with DNase and converted to cDNA using Moloney murine leukemia virus reverse transcriptase (Ambion). The complete coding sequence of tgp1 was confirmed by PCR amplification and sequencing of cDNA using gene-specific oligonucleotides (5′-CAGGTCATTCCCAAGATCA-3′ and 5′-CAAGATATTCTTCTCAACA-3′). Northern blots were performed on total RNA as outlined previously (19), using the digoxigenin-labeled tgp1 probe. Membranes were stripped in 0.2 m Tris-HCl, pH 7.5, 50% (v/v) formamide at 80 °C and hybridized with the digoxigenin-labeled 18 S rRNA probe. Images were visualized using a Fluor-S™ MultiImager (Bio-Rad).
Peptide Synthesis—Two internally quenched fluorescent substrates, AOD1 ((K-Mca)PLGKQVEY(K-Dnp) where Mca indicates 7-methoxycoumarin-4-yl) and AOD2 ((K-Mca)PPGFSAFT(K-Dnp)), were synthesized by Fmoc chemistry. A 20-residue peptide encoding Glu29 to Thr48 of the TGP1 propeptide (PT1) and a 6-residue transition state analog of glutamic peptidases (TA1) (Ac-Phe-Lys-Phe-AHPPA-Leu-Arg-NH2) containing a (3S,4S)-phenylstatinyl (AHPPA) were also synthesized. All Fmoc-amino acids were obtained from Nova-Biochem, except Fmoc-AHPPA-OH (Chem-Impex International). Synthesis was performed on a Symphony Quartet (Protein Technologies).
Enzyme Digestion of Peptides and Analysis of Cleavage Sites—Substrate specificity was evaluated by MALDI-TOF MS analysis following incubation of TGP1 with a variety of peptides purchased from AnaSpec (human ACTH-(1–24), human ACTH-(18–39), bombesin, protein kinase C substrate, [Gln4]neurotensin, somatostatin-14, and substance P), Bachem (α-Bag cell peptide and glycoprotein IIb fragment 656–667), Sigma (human angiotensin I, [Val5]angiotensin II, oxidized bovine insulin B chain, and porcine angiotensinogen-(1–14)), and in-house peptide stocks. Cleavage products were identified using the FindPept tool (ExPASy) after incubation of TGP1 (20 nm) with substrate (10 μm) for 1, 5, and 15 min in 0.1 m citrate phosphate buffer, pH 3.3. Reactions were stopped by heating to 95 °C. Digestive products were crystallized in α-cyano-4-hydroxycinnamic acid and subjected to MALDI-TOF MS as outlined previously.
Fluorimetric Peptidase Assay—Proteolytic activity of purified TGP1 (7.5 nm) and PEP1Asp (111 nm) was measured by continuous assay with of AOD1 (20 μm) and AOD2 (15 μm), respectively. Cleavage of each substrate yields an increase in fluorescence at λex 328 nm and λem 393 nm. All assays were carried out in triplicate at 30 °C in 0.1 m citrate phosphate buffer, pH 3.4, 0.01% (v/v) Brij-35, 1% (v/v) DMSO, unless otherwise stated. Kinetic constants, Km and Vmax, were calculated by nonlinear regression, following continuous assay at pH 3.4, with a substrate concentration range of 0.5–20 μm. Assays were performed in 96-well plates in a Spectra Max Gemini EM (Molecular Devices). Initial velocities (relative fluorescent units/s) were calculated using SoftMax Pro software (Molecular Devices) and data fit using KaleidaGraph (Synergy Software).
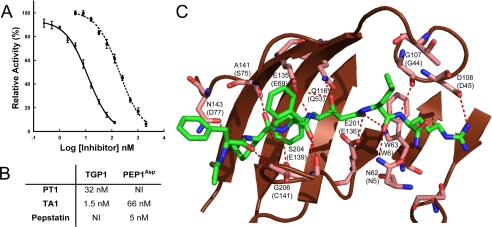
Effect of pH, Temperature, and Inhibitors on Enzyme Activity—The pH optimum of TGP1 was determined by continuous assay over a pH range of 1.95–6 in 0.1 m citrate phosphate buffer. pH stability was assessed by preincubation in 0.015 m citrate phosphate buffer (pH 3 to 7.8) for 30 min followed by dilution into assay buffer. The temperature optimum was determined from end point fluorescence after incubating the enzyme with substrate at defined temperatures for 15 min. The reaction was stopped by heat denaturation at 95 °C. Temperature stability was assessed by preincubation of TGP1 for 30 min at temperatures ranging from 4 to 90 °C. Enzyme inhibition was assessed using 10 μm of TA1, PT1, pepstatin, leupeptin, 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, trans-epoxysuccinyl-l-leucylamido-(4-guanidino)butane, and EDTA. Dose-response plots were generated for PT1 and TA1. Ki values were estimated using a modified Cheng and Prusoff equation (23) for tight binding inhibitors, Ki = (IC50 - [Et]/2)/(1 + [S]/Km). A TGP1/TA1 model was constructed from the co-crystal structure of SGP with TA1 (PDB code 2IFW) using Swiss model. The predicted structure was viewed with PyMOL.
Hyphal Extension Assay—Growth of fungal hyphae in the presence of peptidase inhibitors was assayed as outlined previously (24). Briefly, a mycelial plug was positioned in the center of an SDA plate and allowed to grow for 48 h. 100 nmol of antagonist was dried onto filter paper (18 mm2) and placed at the edge of the mycelial perimeter. Pepstatin has low solubility in water, so methanol was chosen as the vehicle for all inhibitors. Plates were incubated for a further 18 h, and growth of the hyphal front was measured over this time. Statistical differences in growth rate were assessed by Student's t test. A dose-response plot was generated for TA1 and PT1 inhibition. For rescue experiments, 0.89 nmol of TGP1 in 10 mm citrate phosphate buffer, pH 3.4, was added to filter paper containing 35 nmol of PT1 and placed at the hyphal growth front as outlined previously. Control experiments were performed using 10 mm citrate phosphate buffer, pH 3.4.
RESULTS
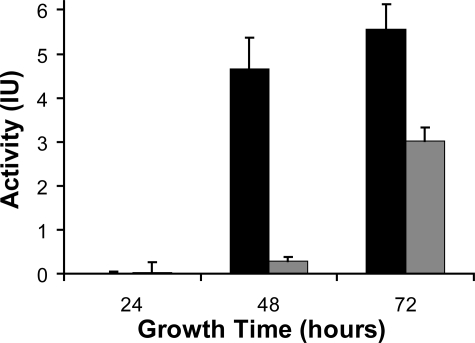
General Endopeptidase Assay—In liquid cultures, T. emersonii secretes organic acids that alter the extracellular pH from 5.5 to 3.5 in less the 24 h (results not shown). We have developed a simple, rapid assay to monitor endopeptidase activity in the media. In our assay, digestion of BSA by peptidases in the secretome of T. emersonii correlates with a reduction in absorbance after Coomassie Blue staining. Using this technique we observed that acidic endopeptidase activity in spent media is maximum after 72 h of growth, particularly in media with reduced nitrogen content (Fig. 1).
FIGURE 1.
Degradation of BSA by peptidases in spent media. T. emersonii was grown in the presence (gray) or absence (black) of both ammonium sulfate and yeast extract for 24, 48, and 72 h. Spent media were incubated with BSA for 45 min, and degradation was assessed by Coomassie staining.
Purification of Peptidases from T. emersonii Secretome—Spent media from T. emersonii liquid cultures was concentrated, dialyzed, and bound to a DEAE-cellulose column. Protein was eluted in a linear salt gradient, and all fractions with endopeptidase activity were pooled, dialyzed, and loaded on a second DEAE-cellulose column. Two protein peaks with acidic endopeptidase activity were eluted in 0.12 and 0.14 m salt (Table 1). The activity in the later peak was completely abolished by the aspartyl peptidase inhibitor pepstatin; however, this antagonist had no effect on activity in the early peak. These fractions were pooled separately and further purified by gel filtration. Analysis of the pepstatin-insensitive peptidase yielded a homogeneous sample of 27 kDa by SDS-PAGE and 21.37 ± 0.01 kDa by MALDI-TOF MS, whereas isoelectric focusing produced a single band with a pI ≤ 3.5. N-terminal sequencing (VEYSSNWAGAVLIGS) confirmed that this enzyme belonged to the glutamic peptidase family and was subsequently named T. emersonii glutamic peptidase 1 (TGP1). The pepstatin-sensitive enzyme (PEP1) was further purified on a TEAE-cellulose column and trypsin-digested and subjected to tandem mass spectrometry. Two peptides (PQTTFFDTVK and PLFAVTLK) revealed that PEP1 belonged to the pepsin-fold protein family. These peptides were 90% identical to poorly conserved regions of aspergillopepsin I isolated from Aspergillus saitoi (PEP1Asp). PEP1Asp is commercially available and was used in place of PEP1 for all assays.
TABLE 1.
Purification of extracellular TGP1 and PEP1 from T. emersonii
| Sample | Purification step | Total volume | Total protein | Total activity | Specific activity | Yield | Purification factor |
|---|---|---|---|---|---|---|---|
| ml | mg | units | units/mg | % | -fold | ||
| Crude | Spent media | 12,100 | 1077 | 28333 | 26.31 | 100 | 1 |
| Concentrate/dialyzed | 20 | 820 | 26445 | 32.25 | 93.34 | 1.23 | |
| DEAE-cellulose | 90 | 380 | 25145 | 66.17 | 88.75 | 2.52 | |
| TGP1 | DEAE-cellulose | 60 | 51.5 | 16459 | 319.59 | 58.09 | 12.15 |
| Sephacryl S-100 | 33 | 19.83 | 15000 | 756.43 | 52.94 | 28.75 | |
| PEP1 | DEAE-cellulose | 70 | 20.5 | 3625 | 176.83 | 12.79 | 6.72 |
| Sephacryl S-100 | 40 | 5.3 | 1684 | 317.74 | 5.94 | 12.07 | |
| TEAE-cellulose | 12 | 0.35 | 245.2 | 700.57 | 0.87 | 26.6 |
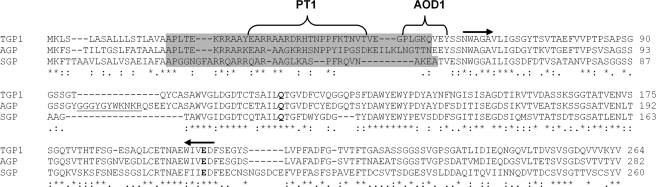
Cloning and Sequencing of the tgp1 Gene—Degenerate oligonucleotides corresponding to the N terminus of TGP1 and a conserved region found in other glutamic peptidases (AEWIVED) facilitated PCR amplification and cloning of a 425-bp fragment from T. emersonii genomic DNA. Sequencing of multiple clones revealed the presence of the tgp1 gene fragment that encodes the native N terminus. This sequence was used to design a tgp1-specific DNA probe to screen a T. emersonii genomic library. The full-length tgp1 gene sequence was obtained from the genomic library, and the coding region was confirmed by cDNA sequencing (GenBank™ accession number AF439998). The gene consists of an intronless open reading frame of 792 bases, which encodes a polypeptide of 264 amino acids. A comparison of the gene sequence with the N terminus of TGP1 revealed that the peptidase is synthesized as a zymogen consisting of a 56-residue prepro-sequence (Met1–Gln56). Signal P (25) analysis indicates that cleavage of the signal peptide occurs between Ala17–Ala18 of the prepro-sequence (p value = 0.914). The propeptide contains an abundance of positively charged residues (pI = 10.9) that may stabilize the zymogen by electrostatic interactions with the mature enzyme (pI ≤ 3.5). The calculated molecular mass of the mature form is 21.37 kDa, which is identical to the value obtained by MALDI-TOF MS. Protein alignment studies indicate that mature TGP1 shares 64 and 47% identity with glutamic peptidases from A. niger (AGP) (26) and Scytalidium lignocolum (SGP) (27), respectively, and the active site dyad is proposed by homology to be Gln116 and Glu201 (Fig. 2).
FIGURE 2.
Protein alignment of TGP1 with glutamic peptidases from A. niger and S. lignocolum. The signal peptide and mature enzyme are separated by the proregion (shaded), and the area used to design a competitive inhibitor (PT1) and fluorescent substrate (AOD1) are indicated. The active site residues are in boldface, and the regions used to design degenerate oligonucleotides are indicated with arrows. Mature AGP consists of a noncovalently linked two chain structure formed by removal of an intervening peptide (underlined) during autoactivation. Asterisks indicate complete identity between all protein, and colons and periods indicate conserved and semi-conserved substitutions.
The tgp1-specific probe was used to profile total RNA from T. emersonii following growth in a variety of media containing a source of organic or inorganic nitrogen (Fig. 3). These studies show that expression of tgp1 is up-regulated at 24 h of growth in the presence of extracellular protein such as peptone or casitone but not by inorganic nitrogen such as ammonium sulfate. In fact, addition of ammonium sulfate to casitone media represses tgp1 expression. Growth for a further 24 h results in an increase in tgp1 expression in peptone media but a decrease in casitone media. Because of the stable expression of tgp1 in media containing peptone, growth inhibition studies using peptidase inhibitors were performed in complex media containing peptone.
A gene sequence displaying 67% identity to tgp1 was also identified during the cloning procedure but did not encode any protein identified in the secretome of T. emersonii. This gene fragment was termed tgp2 and has been deposited in the GenBank™ data base (accession number AF439999).
Substrate Specificity and Enzyme Kinetics—Oxidized insulin B chain is a commonly used bioactive peptide to profile the substrate specificity of glutamic peptidases (28–30). TGP1 was incubated with this peptide, and the digestion products were analyzed by MALDI-TOF MS. This study showed that TGP1 cleaves oxidized insulin B chain at two sites that are also cleaved by other eqolisins (Table 2, part A). An apparent third cleavage site occurred between the oxidized Cys19 and Gly20 residues, but this site was later identified as a product of laser fragmentation (31). A library of diverse peptides was assayed with TGP1, and the digestive products were subjected to MALDI-TOF MS analysis. MALDI-TOF MS is extremely sensitive but largely qualitative; however, time course analysis of proteolysis reveals the subsite preference (Table 2, part B). Substrate profiling of the peptide library confirmed that TGP1 has endopeptidase but not exopeptidase activity. In general, tripeptides are the smallest unit released from either terminus; however, removal of a Ser8–Leu9 dipeptide from the C terminus of α-Bag cell peptide was detected after extended incubation with TGP1. The lack of exopeptidase activity was demonstrated by analysis of angiotensinogen-(1–14) and angiotensin II digestion. The His9–Leu10 bond of angiotensinogen-(1–14) is efficiently cleaved by TGP1 causing the release of a pentapeptide fragment from the C termini. However, in angiotensin II, this bond is located at the C termini, and cleavage is not observed even after extended incubation. In total, 16 peptides containing 32 cleavage sites were examined. These data indicate that TGP1 has broad substrate recognition; however, in 72% of the cleavage sites the residue in the P1 position is larger than the P1′. In fact, Met, Gln, Phe, Lys, Glu, and on the amino side of Ala, Gly, Ser, or Thr are the most efficient reactions observed. However, when a large amino acid is found in the P1′ position, it is almost always a charged group (Arg, Lys, or Glu).
TABLE 2.
A, comparison of insulin B chain cleavage sites by glutamic peptidases; B, hydrolysis of a diverse peptide library by TGPI A, cleavage sites are indicated by a triangle. B, time in minutes that digestion products are first observed are indicated above the triangle. Oxidized and iodoacetylated amino acids are underlined and double underlined, respectively.
To validate the substrate library data, an internally quenched fluorescent substrate (AOD1) was synthesized using sequence from the pro-TGP1 activation site encoding Pro52 to Tyr59. Cleavage was detected by MALDI-TOF MS between Gln56 and Val57 only (data not shown). Under optimal pH conditions, Km, kcat, and kcat/Km values of 4.2 μm, 0.83 s-1, and 2 × 105 m-1 s-1 respectively, were obtained.
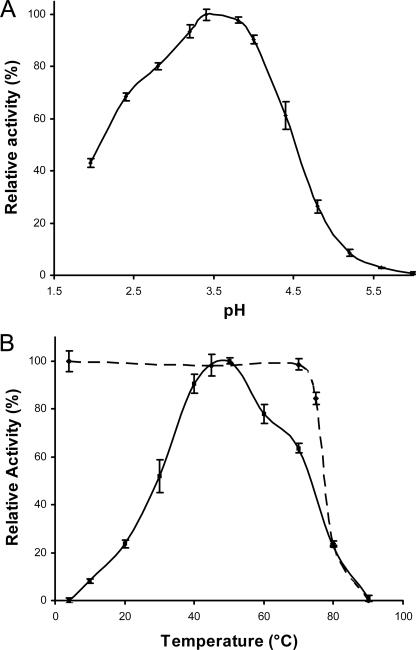
Effect of pH and Temperature on TGP1 Activity and Stability—TGP1 displays optimum activity against AOD1 at pH 3.5 with 50% remaining at pH 2.1 and 4.6 (Fig. 4A). TGP1 was irreversibly inactivated above pH 7.4 (data not shown). The activity profile of purified TGP1 in response to increasing temperature is shown in Fig. 4B. Optimum activity was observed at 50 °C with 50% activity remaining at 75 and 30 °C. The enzyme is stable at 70 °C for 3 h. No significant loss in activity was determined after 24 h of incubation under physiological conditions (45 °C, pH 3.5) or 2 years in storage (-20 °C, pH 5).
FIGURE 4.
A, effect of pH on TGP1 activity. The maximum activity obtained at pH 3.4 was taken as 100% relative activity. B, effect of temperature on TGP1 stability and activity. For stability experiments (broken line), TGP1 was preincubated at various temperatures for 30 min, cooled to 4 °C and assayed at 30 °C. Stability is measured relative to samples stored on ice. For temperature optimum experiments (solid line), end point fluorescence was monitored after 15 min of incubation of enzyme and substrate at defined temperatures. Activity was calculated relative to maximum fluorescence at 50 °C.
Inhibition of TGP1 and PEP1Asp Activity—TGP1 activity was not significantly affected by active site antagonists of serine, cysteine, and aspartyl peptidases, whereas EDTA insensitivity indicated that metal ions are unnecessary for activity (data not shown). Two distinct peptides were synthesized to probe the inhibitory characteristics of TGP1. First, a transition state mimetic (TA1) of scytalidoglutamic peptidase (13) competitively inhibits AOD1 processing by TGP1. Structural homology modeling of TGP1 with TA1 indicated that the inhibitor binds in an extended conformation to a wide groove on the concave surface of the upper β-sheet. The nonhydrolyzable bond of TA1 is located between the catalytic dyad of Glu201 and Gln116 (Fig. 5C). A pro-TGP1 fragment encompassing residues Glu29–Thr48 (PT1) was also synthesized and shown to be a specific and potent competitive inhibitor of TGP1 (Fig. 5A).
FIGURE 5.
A, determination of IC50 for PT1 (broken line) and TA1 (solid line) for TGP1. B, comparison of Ki values for inhibitors of TGP1 and PEP1Asp. NI = no inhibition at 10 μm. C, homology model of TGP1 with SGP (2IFW) containing TA1. TGP1 residues within 10 Å of TA1 (green stick model) are highlighted, and predicted hydrogen bonding interactions between the enzyme and inhibitor are indicated (broken red line). Residues that are structurally equivalent in SGP are indicated in parentheses.
AOD2 was developed as a useful substrate for characterizing metallo- and aspartyl peptidases. PEP1Asp cleaves AOD2 with Km, kcat, and kcat/Km values of 1.5 μm, 0.02 s-1, and 1.2 × 104 m-1 s-1, respectively. Pepstatin and TA1 competitively inhibit PEP1Asp cleavage of AOD2; however, there is no inhibition by PT1 at 10 μm (Fig. 5B).
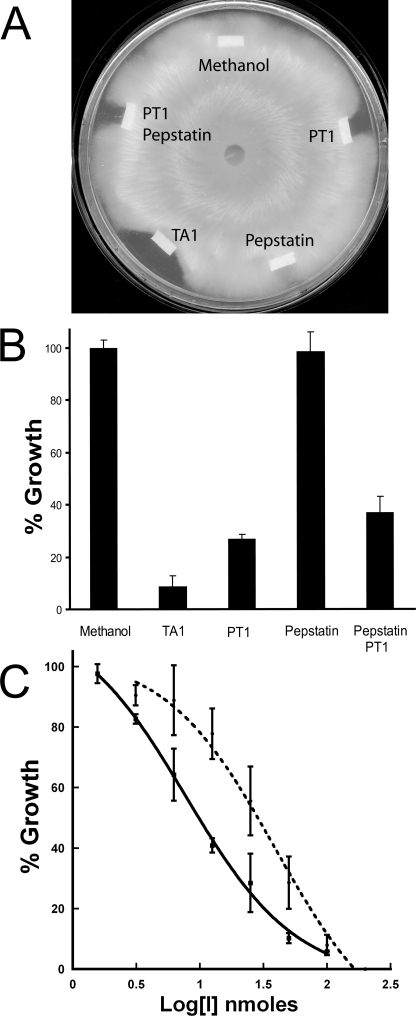
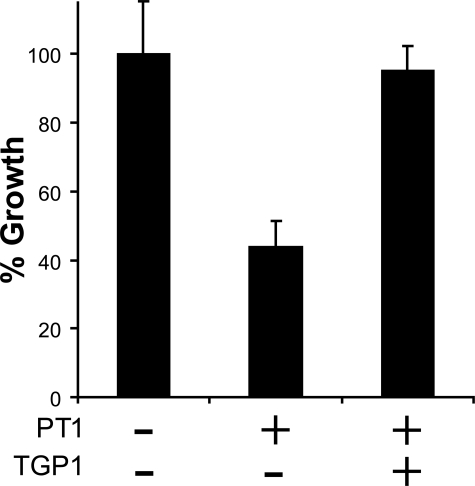
Hyphal Growth Inhibition Assay—To investigate the role of secreted acid peptidases in the growth of T. emersonii, we performed a hyphal extension assay in the presence of 100 nmol of pepstatin, PT1, and the dual inhibitor TA1 (Fig. 6, A and B). Pepstatin showed no effect, whereas PT1 caused 73% reduction in growth relative to the vehicle control (Student's t test p < 0.0001). A combination of pepstatin and PT1 failed to retard growth more than PT1 alone indicating that glutamic and not aspartyl peptidases are essential for growth of T. emersonii on complex media. This result was validated using the dual antagonist TA1, which caused a 91% reduction in hyphal extension (Student's t test p < 0.0001). The crescent shaped zone surrounding TA1 and PT1 is indicative of an antifungal agent (32), so we investigated the inhibitory properties further. T. emersonii was grown in the presence of 1.56–100 nmol of TA1 and 3.125–200 nmol of PT1. Inhibition of growth was dose-dependent, and a 50% reduction in hyphal extension was observed in the presence of 7.75 nmol of TA1 and 35 nmol of PT1 (Fig. 6C). To investigate if reduction in growth by PT1 was solely because of TGP1 inhibition, we added purified TGP1 to the growth front. Hyphal extension rate was rescued (Student's t test p = 0.001) to a level comparable with the vehicle control (Fig. 7).
FIGURE 6.
A, effect of acid peptidase inhibitors on growth of T. emersonii. A mycelial plug was positioned in the center of an SDA plate and allowed to grow for 48 h. 100 nmol of antagonist was dried onto filter paper and placed at the growing edge. Plates were incubated for a further 18 h. B, hyphal extension in the presence of each antagonist was measured, and growth was calculated relative to methanol. C, dose response of fungal growth in the presence of TA1 (solid line) and PT1 (broken line).
FIGURE 7.
Rescue from PT1 inhibition. TGP1 (0.89 nmol) in 10 mm citrate phosphate buffer, pH 3.4, was added to filter paper containing 35 nmol of PT1. Hyphal extension was measured relative to buffer alone.
DISCUSSION
The thermophilic, aerobic fungus T. emersonii secretes substantial amounts of hydrolytic enzymes that can be used to process inert, low value polymers into high value metabolites such as sugars (17, 18, 33, 34). T. emersonii also has GRAS status making it an excellent model species to investigate hydrolytic enzymes that have orthologs in plant and human pathogens. Growth of T. emersonii in complex media causes a rapid reduction in extracellular pH most likely because of the conversion of sugars into organic acids (35). In this study, we have shown that spent media contain abundant acid-acting proteolytic activity against macromolecular substrates such as BSA, particularly when T. emersonii is grown in media with reduced nitrogen content. We report the purification to homogeneity of two acid-acting endopeptidases from the secretome of T. emersonii using a rapid colorimetric assay that monitors BSA degradation. These peptidases bear no sequence similarity to each other and are easily distinguished by molecular weight and sensitivity to pepstatin. The aspartyl peptidase PEP1 was sequenced and shown to be highly similar to other well characterized fungal pepsins. Fungal pepsins are used in the food industry to induce flavor development during cheese production (36) and fermentation of traditional Japanese foods (1). Consequently, these enzymes have been well characterized so we focused our attention on TGP1, the first glutamic peptidase isolated and characterized from a thermophile and nonpathogenic fungus. TGP1 is the most abundant peptidase secreted by T. emersonii, which allowed us to isolate sufficient protein for biochemical analysis. N-terminal sequence data facilitated the cloning and sequencing of the full-length tgp1 gene sequence from genomic DNA. The deduced protein sequence revealed the presence of signal and propeptide components at the N terminus and active site residues that are located in highly conserved regions of this peptidase family (Fig. 2).
Biochemical characterization of this protein family has been limited to the plant pathogen S. lignicolum (SGP) (37) and opportunistic human pathogens A. niger (AGP) (38, 39) and Penicillium marneffei (PMAP-1) (30). These enzymes degrade oxidized insulin B chain at multiple positions; however, only the Glu13–Ala14 cleavage site is a common cleavage site among previously reported eqolisins (28–30). In this study, we utilized MALDI-TOF MS to monitor TGP1 digestion of oxidized insulin B chain over time. These data indicated that cleavage occurred between Glu13–Ala14 and Tyr26–Thr27 (Table 2, part A). These data prompted further characterization of TGP1 specificity using a diverse peptide library. MALDI-TOF MS data of cleavage products indicated that substrate interaction from P3 to P3′ is required for efficient cleavage to occur. Large residues with the exception of tryptophan are preferred in the S1 pocket, whereas the S1′ subsite preferentially binds smaller amino acids such as Gly, Ser, and Ala (Table 2, part B). These data are comparable with subsite specificity profiling of SGP, which indicates that cleavage is favored between bulky P1 residues (Phe > Tyr > His) and small P1′ residues (Ala > Gly/Thr > Ser) (40).
Kinetic analysis was performed using an internally quenched fluorescent peptide designed from the activation site of pro-TGP1. Cleavage of this substrate occurs between the predicted Gln56–Val57 site to yield a new N terminus that is identical to the mature enzyme. Using this substrate, TGP1 has maximum activity at pH 3.4 and 50 °C, which is comparable with the optimal environment for T. emersonii growth (pH 3.0–4.0; 45 °C).
Structural comparison of SGP and AGP indicated that their overall structures are almost identical, with a root mean square deviation of 0.89 Å (13). A common catalytic mechanism has been proposed for both enzymes, which can be conferred upon TGP1. This mechanism involves the active site Glu201 donating a proton to the carbonyl oxygen of the scissile bond, whereas the carbonyl carbon obtains an OH- from a water molecule to form the tetrahedral intermediate. This transition state is stabilized by hydrogen bonding with Glu201 and Gln116. The protonated Glu201 then donates a proton to the amide nitrogen atom of the scissile bond, which triggers the breakdown of the tetrahedral intermediate and subsequently the peptide bond (9, 12). A transition state analog (TA1) containing a nonhydrolysable scissile bond was synthesized and shown to be a potent inhibitor of TGP1. TA1 was developed by Pillai et al. (13) to provide structural insight into the mechanism of substrate binding in SGP. TA1 contains a hydroxyethylene group in the P1 residue, where the hydroxyl group is the functional mimic of the oxyanion, and the methylene group mimics the NH+2 of the transition state. A homology model of TGP1 was created using structural data of SGP bound to TA1 (Fig. 5C). The inhibitor binds in an extended conformation by hydrogen bonding interactions to residues that are structurally equivalent in SGP. Furthermore, with the exception of a Thr to Ser substitution, amino acids forming the S1 and S1′ subsites in SGP (Trp6, Thr37, Ile51, Gln53, Asp57, Trp67, Tyr71, Glu136, and Phe138) are found in identical positions in TGP1, which supports the similarity in substrate specificity.
In this study we show that TA1 is not only a potent antagonist of TGP1 but also the fungal aspartyl peptidase, PEP1Asp. To develop a specific inhibitor of TGP1, we synthesized a 20-mer peptide encoding a portion of the propeptide sequence. Kubota et al. (14) had previously identified the AGP propeptide and a series of propeptide fragments as potent competitive inhibitors of the enzyme. Alanine substitution of this peptide indicated that a central region (Lys18–Pro24) was associated with AGP inhibition. The homologous central region of the TGP1 propeptide (PT1) displayed comparable inhibition of TGP1. Both peptides contain a proline couplet, which may confer secondary structure to the propeptide allowing binding to the active site pocket. Mutations to either of these residues decreased inhibition of AGP. PT1 shows no inhibition of PEP1Asp or partially purified PEP1 at micromolar concentrations and was used for ex vivo studies to specifically target TGP1.
To date, genetic disruption of T. emersonii proteins has proven unsuccessful; however, knock-out studies in other fungal species have highlighted a role of extracellular peptidases in growth and virulence. Disruption of AGP and the aspartyl peptidase, PEPA in A. niger showed a 6 and 84% reduction in extracellular peptidase activity, respectively (41). Other studies have shown that mutation of EapA, an aspartyl peptidase, resulted in a loss of 99% of the milk-clotting activities in the plant pathogen Cryphonectria parasitica (42). Double mutants of EapA and a glutamic peptidase, EapC, led to a further 30% reduction of the residual activity but did not adversely affect the survival of the fungus. However, disruption of a second glutamic peptidase gene, EapB, appeared to be lethal, suggesting this enzyme may be essential for growth (43).
The dual inhibition characteristics of TA1 and the class-specific antagonists PT1 and pepstatin provided us with excellent tools to analyze the role of acid-acting peptidases in T. emersonii cultures by chemical knockdown. Hyphal extension assays were performed in the presence of equal amounts of pepstatin, PT1, and TA1. Pepstatin had little effect on hyphal extension, whereas PT1 and TA1 significantly retarded fungal growth. These data strongly indicated that hyphal extension was dependent on TGP1 but not PEP1 activity. The wide zone of inhibition around PT1 and TA1 was characteristic of an antifungal agent; therefore, we analyzed these compounds further and found that inhibition was dose-dependent. A 50% reduction in fungal growth was observed in the presence of 7.75 nmol of TA1 (7.54 μg) and 35 nmol of PT1 (82 μg) (Fig. 6C). This inhibition is comparable with antifungal agents isolated from bacteria (44, 45) and plants (46, 47) that specifically target peptidases. To investigate if growth retardation by PT1 is the result of TGP1 inhibition, we added both PT1 and purified enzyme to the hyphal extension assay and observed a rescue of growth to 95% of vehicle control (Fig. 7).
Fungi express membrane proteins to transport amino acids and small peptides (2–5 amino acids) into the cell for use as nutrients, whereas larger peptides cannot be assimilated (48). TGP1 displays broad specificity and readily digests a host of peptides and proteins into smaller peptides that can be readily utilized by the growing mycelia. Added to this, gene expression analysis indicated that tgp1 is induced in the presence of an extracellular protein source but repressed by inorganic nitrogen (Fig. 3). These data correlate with a reduction in secreted acid peptidases activity in cultures containing ammonium sulfate (Fig. 1). Casitone and peptone are enzymatic digests of protein and largely differ by the degree of hydrolysis. Casitone has a greater abundance of amino acids and short peptides that are readily utilizable by the fungus. These data correlate with a sharp induction of tgp1 expression during the first 24 h of growth. In contrast, 80% of peptides in peptone are greater than 5 amino acids and require processing by extracellular peptidases into utilizable nitrogen. These data correlate with continuous expression of TGP1 by T. emersonii over 48 h of growth. Interestingly, the major transcription factor that regulates nitrogen catabolism in fungi, AreA/Nit2, binds with high affinity to a pair of GATA elements in close proximity (49). These elements are found 163 and 205 nucleotides upstream of the tgp1 start codon, which may provide a means of regulating tgp1 expression in the presence or absence of extracellular nitrogen. Taken together, we predict that the role of TGP1 is to provide T. emersonii with a supply of nitrogen from organic matter.
A BLAST search of the NCBI data base identified 18 fungal species that possess one or more genes encoding a putative eqolisin. Of these, 14 are either human or plant pathogens. Moon et al. (30) showed that the major acid peptidase secreted by P. marneffei, the causative agent of penicilliosis, is the glutamic peptidase, PMAP-1. Penicilliosis cases were rare before the human immunodeficiency virus pandemic but now account for up to 15% of all human immunodeficiency virus-related illnesses in Southeast Asia (50). In this study, we have characterized a glutamic peptidase and demonstrated that its activity is essential for growth in T. emersonii. This fungus is nonpathogenic but serves as a model species to investigate homologous proteins in pathogenic species.
Acknowledgments
We thank Dr. Gerard Cagney for assistance with MALDI-TOF MS and Dr. Daniel R. Hostetter for critical review of this manuscript.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) AF439998 and AF439999.
This work was supported, in whole or in part, by National Institutes of Health Grant GM082250 (to C. S. C.). This work was also supported in part by Enterprise Ireland Research Innovation and Higher Education Authority pre-PRTLI, PRTLI awards (to M. G. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TGP1, T. emersonii glutamic peptidase 1; PEP1, T. emersonii pepsin-type peptidase 1; BSA, bovine serum albumin; MALDI-TOF MS, matrix-assisted laser desorption/ionization with time of flight mass spectrometer; SDA, Saboraud dextrose agar; Dnp, 2,4-dinitrophenol; Fmoc, N-(9-fluorenyl)methoxycarbonyl; ACTH, adrenocorticotropic hormone.
References
- 1.Ichishima, E. (2004) in Handbook of Proteolytic Enzymes (Barrett, A. J., Rawlings, N. D., and Woessner, J. F., eds) pp. 92-99, Elsevier Academic Press, London, UK
- 2.Hofmann, T. (2004) in Handbook of Proteolytic Enzymes (Barrett, A. J., Rawlings, N. D., and Woessner, J. F., eds) pp. 99-104, Elsevier Academic Press, London, UK
- 3.Wlodawer, A., Li, M., Gustchina, A., Oyama, H., Dunn, B. M., and Oda, K. (2003) Acta Biochim. Pol. 50 81-102 [PubMed] [Google Scholar]
- 4.Oda, K., Sugitani, M., Fukuhara, K., and Murao, S. (1987) Biochim. Biophys. Acta 923 463-469 [DOI] [PubMed] [Google Scholar]
- 5.Lee, B. R., Furukawa, M., Yamashita, K., Kanasugi, Y., Kawabata, C., Hirano, K., Ando, K., and Ichishima, E. (2003) Biochem. J. 371 541-548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishii, W., Ueki, T., Miyashita, R., Kojima, M., Kim, Y. T., Sasaki, N., Murakami-Murofushi, K., and Takahashi, K. (2003) Biochem. Biophys. Res. Commun. 301 1023-1029 [DOI] [PubMed] [Google Scholar]
- 7.Sleat, D. E., Gin, R. M., Sohar, I., Wisniewski, K., Sklower-Brooks, S., Pullarkat, R. K., Palmer, D. N., Lerner, T. J., Boustany, R. M., Uldall, P., Siakotos, A. N., Donnelly, R. J., and Lobel, P. (1999) Am. J. Hum. Genet. 64 1511-1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sims, A. H., Dunn-Coleman, N. S., Robson, G. D., and Oliver, S. G. (2004) FEMS Microbiol. Lett. 239 95-101 [DOI] [PubMed] [Google Scholar]
- 9.Fujinaga, M., Cherney, M. M., Oyama, H., Oda, K., and James, M. N. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 3364-3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yabuki, Y., Kubota, K., Kojima, M., Inoue, H., and Takahashi, K. (2004) FEBS Lett. 569 161-164 [DOI] [PubMed] [Google Scholar]
- 11.Rawlings, N. D., Morton, F. R., and Barrett, A. J. (2006) Nucleic Acids Res. 34 D270-D272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasaki, H., Kojima, M., Sawano, Y., Kubota, K., Suganuma, M., Muramatsu, T., Takahashi, K., and Tanokura, M. (2005) Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 81 441-446 [Google Scholar]
- 13.Pillai, B., Cherney, M. M., Hiraga, K., Takada, K., Oda, K., and James, M. N. (2007) J. Mol. Biol. 365 343-361 [DOI] [PubMed] [Google Scholar]
- 14.Kubota, K., Nishii, W., Kojima, M., and Takahashi, K. (2005) J. Biol. Chem. 280 999-1006 [DOI] [PubMed] [Google Scholar]
- 15.Jarai, G., van den Hombergh, J. P., and Buxton, F. P. (1994) Gene (Amst.) 145 171-178 [DOI] [PubMed] [Google Scholar]
- 16.Poussereau, N., Creton, S., Billon-Grand, G., Rascle, C., and Fevre, M. (2001) Microbiology 147 717-726 [DOI] [PubMed] [Google Scholar]
- 17.Murray, P. G., Grassick, A., Laffey, C. D., Cuffe, M. M., Higgins, T., Savage, A. V., Planas, A., and Tuohy, M. G. (2001) Enzyme Microb. Technol. 29 90-98 [DOI] [PubMed] [Google Scholar]
- 18.Tuohy, M. G., Puls, J., Claeyssens, M., Vrsanska, M., and Coughlan, M. P. (1993) Biochem. J. 290 515-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray, P. G., Collins, C. M., Grassick, A., and Tuohy, M. G. (2003) Biochem. Biophys. Res. Commun. 301 280-286 [DOI] [PubMed] [Google Scholar]
- 20.Buroker-Kilgore, M., and Wang, K. K. (1993) Anal. Biochem. 208 387-392 [DOI] [PubMed] [Google Scholar]
- 21.Raeder, U., and Broda, P. (1985) Lett. Appl. Microbiol. 1 17-20 [Google Scholar]
- 22.Grassick, A., Murray, P. G., Thompson, R., Collins, C. M., Byrnes, L., Birrane, G., Higgins, T. M., and Tuohy, M. G. (2004) Eur. J. Biochem. 271 4495-4506 [DOI] [PubMed] [Google Scholar]
- 23.Copeland, R. A., Lombardo, D., Giannaras, J., and Decicco, C. P. (1995) Bioorg. Med. Chem. Lett. 5 1947-1952 [Google Scholar]
- 24.Roberts, W. K., and Selitrennikoff, C. P. (1986) Biochim. Biophys. Acta 880 161-170 [DOI] [PubMed] [Google Scholar]
- 25.Bendtsen, J. D., Nielsen, H., von Heijne, G., and Brunak, S. (2004) J. Mol. Biol. 340 783-795 [DOI] [PubMed] [Google Scholar]
- 26.Huang, X. P., Kagami, N., Inoue, H., Kojima, M., Kimura, T., Makabe, O., Suzuki, K., and Takahashi, K. (2000) J. Biol. Chem. 275 26607-26614 [DOI] [PubMed] [Google Scholar]
- 27.Oda, N., Gotoh, Y., Oyama, H., Murao, S., Oda, K., and Tsuru, D. (1998) Biosci. Biotechnol. Biochem. 62 1637-1639 [DOI] [PubMed] [Google Scholar]
- 28.Iio, K., and Yamasaki, M. (1976) Biochim. Biophys. Acta 429 912-924 [DOI] [PubMed] [Google Scholar]
- 29.Oda, K., and Murao, S. (1976) Agric. Biol. Chem. 40 1221-1225 [Google Scholar]
- 30.Moon, J. L., Shaw, L. N., Mayo, J. A., Potempa, J., and Travis, J. (2006) Biol. Chem. 387 985-993 [DOI] [PubMed] [Google Scholar]
- 31.Brown, R. S., Feng, J., and Reiber, D. C. (1997) Int. J. Mass Spectrom. Ion Proc. 169 1-18 [Google Scholar]
- 32.Koiwa, H., Kato, H., Nakatsu, T., Oda, J., Yamada, Y., and Sato, F. (1997) Plant Cell Physiol. 38 783-791 [DOI] [PubMed] [Google Scholar]
- 33.Tuohy, M. G., Walsh, D. J., Murray, P. G., Claeyssens, M., Cuffe, M. M., Savage, A. V., and Coughlan, M. P. (2002) Biochim. Biophys. Acta 1596 366-380 [DOI] [PubMed] [Google Scholar]
- 34.Heneghan, M. N., McLoughlin, L., Murray, P. G., and Tuohy, M. G. (2007) Enzyme Microb. Technol. 41 677-682 [Google Scholar]
- 35.Magnuson, J., and Lasure, L. (2004) in Advances in Fungal Biotechnology for Industry, Agriculture, and Medicine (Tkacz, J. S., and Lange, L., eds) pp. 307-340, Springer, New York
- 36.Cooper, J. B. (2004) in Handbook of Proteolytic Enzymes (Barrett, A. J., Rawlings, N. D., and Woessner, J. F., eds) pp. 104-107, Elsevier Academic Press, London, UK
- 37.Oda, K. (2004) in Handbook of Proteolytic Enzymes (Barrett, A. J., Rawlings, N. D., and Woessner, J. F., eds) pp. 219-221, Elsevier Academic Press, London, UK
- 38.Takahashi, K. (2004) in Handbook of Proteolytic Enzymes (Barrett, A. J., Rawlings, N. D., and Woessner, J. F., eds) 2nd Ed., pp. 221-224, Elsevier Academic Press, London, UK
- 39.Fianchi, L., Picardi, M., Cudillo, L., Corvatta, L., Mele, L., Trape, G., Girmenia, C., and Pagano, L. (2004) Mycoses 47 163-167 [DOI] [PubMed] [Google Scholar]
- 40.Kataoka, Y., Takada, K., Oyama, H., Tsunemi, M., James, M. N., and Oda, K. (2005) FEBS Lett. 579 2991-2994 [DOI] [PubMed] [Google Scholar]
- 41.van den Hombergh, J. P., Sollewijn Gelpke, M. D., van de Vondervoort, P. J., Buxton, F. P., and Visser, J. (1997) Eur. J. Biochem. 247 605-613 [DOI] [PubMed] [Google Scholar]
- 42.Razanamparany, V., Jara, P., Legoux, R., Delmas, P., Msayeh, F., Kaghad, M., and Loison, G. (1992) Curr. Genet. 21 455-461 [DOI] [PubMed] [Google Scholar]
- 43.Jara, P., Gilbert, S., Delmas, P., Guillemot, J. C., Kaghad, M., Ferrara, P., and Loison, G. (1996) Mol. Gen. Genet. 250 97-105 [DOI] [PubMed] [Google Scholar]
- 44.Vernekar, J. V., Ghatge, M. S., and Deshpande, V. V. (1999) Biochem. Biophys. Res. Commun. 262 702-707 [DOI] [PubMed] [Google Scholar]
- 45.Dash, C., Ahmad, A., Nath, D., and Rao, M. (2001) Antimicrob. Agents Chemother. 45 2008-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, X., Li, J., Wang, X., Fang, W., Bidochka, M. J., She, R., Xiao, Y., and Pei, Y. (2006) Peptides 27 1726-1731 [DOI] [PubMed] [Google Scholar]
- 47.Lorito, M., Broadway, R. M., Hayes, C. K., Woo, S. L., Noviello, C., Williams, D. L., and Harman, G. E. (1994) Mol. Plant-Microbe Interact. 7 525-527 [Google Scholar]
- 48.Wiles, A. M., Cai, H., Naider, F., and Becker, J. M. (2006) Microbiology 152 3133-3145 [DOI] [PubMed] [Google Scholar]
- 49.Marzluf, G. A. (1997) Microbiol. Mol. Biol. Rev. 61 17-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ustianowski, A. P., Sieu, T. P., and Day, J. N. (2008) Curr. Opin. Infect. Dis. 21 31-36 [DOI] [PubMed] [Google Scholar]