Abstract
Mutations in the RECQL4 helicase gene have been linked to Rothmund-Thomson syndrome, which is characterized by genome instability, cancer susceptibility, and premature aging. To better define the cellular function of the RecQ4 protein, we investigated the subcellular localization of RecQ4 upon treatment of cells with different DNA-damaging agents including UV irradiation, 4-nitroquinoline 1-oxide, camptothecin, etoposide, hydroxyurea, and H2O2. We found that RecQ4 formed discrete nuclear foci specifically in response to UV irradiation and 4-nitroquinoline 1-oxide. We demonstrated that functional RecQ4 was required for the efficient removal of UV lesions and could rescue UV sensitivity of RecQ4-deficient Rothmund-Thomson syndrome cells. Furthermore, UV treatment also resulted in the colocalization of the nuclear foci formed with RecQ4 and xeroderma pigmentosum group A in human cells. Consistently, RecQ4 could directly interact with xeroderma pigmentosum group A, and this interaction was stimulated by UV irradiation. By fractionating whole cell extracts into cytoplasmic, soluble nuclear, and chromatin-bound fractions, we observed that RecQ4 protein bound more tightly to chromatin upon UV irradiation. Taken together, our findings suggest a role of RecQ4 in the repair of UV-induced DNA damages in human cells.
Genome instability is thought to play an important role in the development and progression of cancer and has also been implicated in the aging process. The RecQ family of DNA helicases maintains genomic integrity through their participation in DNA repair, replication, and recombination pathways. There are at least five distinct RecQ helicases in human: RECQL1, BLM, WRN, RECQL4, and RECQL5 (1). Mutation in WRN, BLM, or RECQL4 results in the rare autosomal recessive disorders of Werner syndrome, Bloom syndrome, and Rothmund-Thomson syndrome (RTS),2 respectively. RTS patients display clinical profiles that include growth deficiency, photosensitivity with poikiloderma, signs of premature aging including the graying and loss of hair and cataracts, and an increased cancer predisposition especially to osteosarcoma (2).
The RECQL4 gene encodes a 1208-amino-acid (133-kDa) protein (RecQ4) that contains the conserved DNA helicase domain homologous to the Escherichia coli RecQ helicase (3). RecQ4 has a DNA-dependent ATPase activity and single-stranded DNA annealing activity; however, unlike other RecQ family members, no detectable DNA unwinding ability has been observed for RecQ4 (4, 5). The cellular function of RecQ4 is obscure, although some recent studies indicate that RecQ4 may be implicated in different DNA metabolic processes. Firstly, RecQ4 has been proposed to function in the initiation of DNA replication with its N terminus required for the recruitment of DNA polymerase α (6, 7). Secondly, Petkovic et al. (8) report that after etoposide treatment, RecQ4 nuclear foci coincide with the foci formed by Rad51, a crucial protein functioning in homologous recombination of double strand break repair pathway. In addition, fibroblasts from RTS patients (RTS cells) are sensitive to ionizing radiation (9), and Kumata et al. (10) recently provide evidence that RecQ4 participates in double strand break repair in Xenopus egg extracts. Thirdly, Woo and Werner (11, 12) have reported that RecQ4 plays a role in oxidative stress. Fourthly, cells defective in RecQ4 escaped from the S-phase arrest following UV or hydroxyurea treatment (13). These data altogether suggest that RecQ4 is involved in normal DNA replication, distinct DNA repair processes, and cell cycle arrest.
Nucleotide excision repair (NER) is a highly versatile and sophisticated DNA damage removal pathway as it deals with a wide range of bulky DNA lesions, including UV-induced photoproducts, bulky chemical adduct, and certain types of DNA cross-links (14). There are two subpathways of NER: global genome NER (GG-NER) and transcription-coupled NER (TC-NER). The NER pathway consists of at least 30 proteins involved in sequential damage recognition, dual incision, repair synthesis, and ligation steps (15). Briefly, XPC-hHR23B senses NER lesions in GG-NER, whereas lesions are detected by stalled RNA polymerase II in TC-NER. Then transcript factor IIH is recruited to the lesions and unwinds DNA duplex followed by binding of xeroderma pigmentosum group A and replication protein A (RPA) to stabilize the opened DNA complex. XPG and ERCC1-XPF make the dual incision. DNA repair synthesis is carried out by polymerase δ and polymerase ε and their cofactors proliferating cell nuclear antigen and replication factor C. Finally the newly synthesized patch is sealed by DNA ligase I to restore the original DNA sequence. XPA plays an indispensable role and is required for both GG-NER and TC-NER. Given its affinity for damaged DNA and its ability to interact with many core NER factors, XPA is anticipated to verify NER lesions and to play a central role in positioning the repair machinery correctly around the injury (15).
In this study, we have examined the cellular localization of RecQ4 in response to multiple DNA damage agents. We found that RecQ4 formed nuclear foci specifically upon induction of UV and 4-nitroquinoline 1-oxide (4NQO) DNA damage, and importantly, that complementation with functional RecQ4 accelerated the removal of UV lesions and rescued the UV sensitivity in RecQ4-deficient RTS cells. Moreover, RecQ4 foci colocalized with XPA foci in nuclei after UV irradiation. Further, two proteins could interact with each other, and their interaction was increased by UV exposure. Altogether these findings suggest a role of RecQ4 in the repair of UV DNA damage.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatments—HeLa and H1299 cells were obtained from the American Type Culture Collection and maintained at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. The AG05013 fibroblasts, from a patient with RTS, and GM04429 XPA fibroblasts, from a xeroderma pigmentosum patient, were obtained from Coriell Cell Repositories. Cells were maintained in the minimum Eagle's medium supplemented with essential and nonessential amino acids, vitamins, and 20% uninactivated fetal bovine serum. For UV exposure, cells were irradiated with various doses of UV using a UV cross-linker and further incubated for 4 h at 37°C before harvesting. For drug treatments, cells were incubated with 1.25 μg/ml 4NQO for 1 h, 20 μm camptothecin for 4 h, 20 μm etoposide for 6 h, 2 mm hydroxyurea for 24 h, and 100 μm H2O2 for 3 h before cells were fixed.
Antibody Production—The RecQ4 antibody used in this study was raised against recombinant N-terminal RecQ4 (residues 239–357) fused to a glutathione S-transferase (GST) tag in rabbit. The RecQ4 antibody was affinity-purified with Amino-Link Plus immobilization kit (Pierce) following the manufacturer's instructions. Western blotting of whole cell lysates from 293T cells and HeLa cells showed that the purified antibody only recognized RecQ4 protein.
Immunofluorescence—Cells were grown on 18-mm coverslips overnight prior to treatment. After treatment, cells were fixed with 100% methanol and blocked in phosphate-buffered saline containing 15% fetal bovine serum. Primary antibody dilutions used are as follows: rabbit anti-RecQ4 1:250, mouse anti-XPA 1:100 (Kamiya). Secondary antibody dilutions are as follows: rhodamine red-X-conjugated AffiniPure goat anti-rabbit 1: 250 and fluorescein isothiocyanate-conjugated AffiniPure goat anti-mouse 1: 50 (Jackson ImmunoResearch, Inc.). Images were captured with a Nikon inverted fluorescent microscope with attached camera at ×60 magnifications and processed using PhotoShop 7.0 (Adobe) software.
Subcellular Fractionation—The cellular protein fractionation was performed essentially as described (30). Briefly, to prepare whole cell extracts, cells were lysed in solution A (50 mm Tris-HCl, pH 7.8, 420 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40, 0.34 m sucrose, 10% glycerol, 1 mm Na3VO4, 10 mm NaF, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture). Lysates were cleared by centrifugation, and protein concentration was determined by the Bradford assay (Bio-Rad). For chromatin fractionation, cells were first lysed in buffer B (10 mm HEPES at pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 10% glycerol, 0.1% Triton X-100, protease, and phosphatase inhibitors) and left on ice for 5 min. Cytoplasmic proteins were separated from nuclei by low speed centrifugation (1300 × g for 4 min). Isolated nuclei were washed once with solution B and then lysed in solution C (3 mm EDTA, 0.2 mm EGTA, 1 mm dithiothreitol). After a 10-min incubation on ice, soluble nuclear proteins were released from chromatin by low speed centrifugation (1700 × g for 4 min). Isolated chromatin was resuspended in BC100 (20 mm Tris-Cl, pH 7.9, 100 mm NaCl, 20% glycerol, 0.1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture) and sheared by sonication. Chromatin-bound proteins were obtained after centrifugation at 13,000 rpm for 20 min.
Slot Blot Analysis—Cells were harvested at 0, 2, 4, 8, and 24 h after UV irradiation. Genomic DNA was isolated using the DNeasy kit (Qiagen) according to the manufacturer's protocol. For the control, DNA was harvested from nonirradiated cells. DNA was denatured for quantification of thymine dimers as follows: DNA (100 ng in 0.32 m NaOH, 5 mm ethylenediamine tetraacetic acid) was boiled for 10 min followed by the addition of acetic acid and cold ammonium acetate to final concentrations of 1.4 and 0.6 m, respectively. To quantify the DNA lesions, denatured DNA was spotted onto nitrocellulose membrane prewetted with 6× SSC buffer using a slot blot apparatus (Schleicher & Schuell Inc.). The filter was baked at 80 °C for 2 h. Quantification of thymine dimers was carried out using anti-cyclobutane pyrimidine dimer (CPD) antibody (MBL International Corp.). Antibody binding was determined by using the ECL chemiluminescent method (Amersham Biosciences).
Western Blotting—Cell lysates and immunoprecipitates were separated on 8% SDS-polyacrylamide gels and transferred to nitrocellulose membrane. The membranes were blocked with Tris-buffered saline with Tween buffer containing 5% powered milk and probed using following primary antibodies: anti-RecQ4 and anti-XPA (Santa Cruz Biotechnology) and anti-RPA32 (Kamiya). The membranes were then incubated with horseradish peroxidase-linked secondary anti-rabbit or anti-mouse antibodies (Amersham Biosciences), and bound antibodies were visualized using the ECL chemiluminescent method.
Co-immunoprecipitation Assays—Whole cell lysates were diluted with dilution buffer (15 mm Tris-HCl, pH 7.8, 1 mm EDTA, 10% glycerol, protease, and phosphatase inhibitors) and incubated with 2 μg of rabbit anti-XPA or 20 μl of mouse anti-FLAG M2 agarose (Sigma) for 10–14 h at 4 °C with end-over-end rotation. Then protein A/G-agarose beads were added, and the reaction mixtures were further mixed for 1 h at 4°C. The immunoprecipitates were separated from supernatant by centrifugation and washed with phosphate-buffered saline containing 0.05% Nonidet P-40. Proteins were extracted from the agarose beads by boiling in 1× SDS gel loading buffer and resolved on 8% SDS-polyacrylamide gels.
Cell Viability Assay—RecQ4 cDNA fragments were cloned into pBabe-puro retroviral vector to make the pBabe-RecQ4 construct. We infected RTS cells with pBabe vector alone or pBabe-RecQ4, after which infected RTS cells were selected with 0.5 μg/ml puromycin. Stably infected RTS cells were seeded in 96-well plates (1 × 103/well), allowed to attach overnight, and then treated with increasing doses of UV irradiation and left to grow for another 48 h. Aliquots of 10 μl of MTT (5 mg/ml) were added to each well. After 4 h, the color formed was quantitated by a spectrophotometric plate reader (Berkman, Inc.) at 595-nm wavelength after solubilization in 150 μl of dimethyl sulfoxide (DMSO).
GST Pull-down Assay—35S-labeled in vitro translated XPA protein was incubated with immobilized GST or GST-RecQ4 protein at 4 °C for 2 h. The beads were washed five times with BC200 (20 mm Tris-Cl, pH 7.9, 200 mm NaCl, 20% glycerol, 0.1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture). The bound proteins were eluted by boiling in sample buffer and analyzed by SDS-PAGE and autoradiography, and GST fusion proteins were analyzed by SDS-PAGE and Coomassie Blue staining.
RESULTS
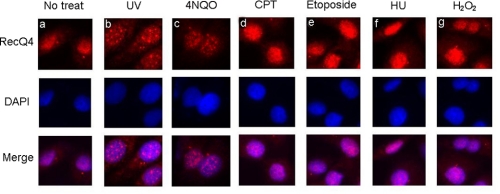
RecQ4 Forms Nuclear Foci in Response to UV Irradiation and 4NQO—To examine the function of RecQ4 protein in response to DNA damage, we used the affinity-purified RecQ4 antibody to visualize the localization of RecQ4 in cells. HeLa cells were treated with different DNA-damaging agents including UV irradiation and 4NQO, which induce NER repair; camptothecin and etoposide, which produce DNA double strand breaks; hydroxyurea, which inhibits DNA replication; and H2O2, which induces DNA damage via oxygen-derived reactive species. Indirect immunofluorescence staining analysis revealed that most RecQ4 was homogeneously localized in nucleus of mock-treated HeLa cells (Fig. 1a), consistent with the report by Kitao et al. (16). Remarkably, UV or 4NQO treatment induced a significant redistribution of RecQ4 protein in nuclei as discrete RecQ4 nuclear foci were observed upon treatment (Fig. 1, b and c). Notably, there was no significant RecQ4 foci formation in cells treated with other DNA damage agents (Fig. 1, d–g). These results suggest that RecQ4 may relocalize to the sites of DNA damage induced specifically by UV irradiation and 4NQO treatment.
FIGURE 1.
RecQ4 formed nuclear foci in response to UV DNA damage. HeLa cells were treated with different DNA damage agents as indicated. For UV treatment, cells were irradiated with 20 J/m2 UV followed by a 4-h recovery. For chemical treatment, cells were incubated with 1.25 μg/ml 4NQO for 1 h, 20μm camptothecin (CPT) for 4 h, 20 μm etoposide for 6 h, 2 mm hydroxyurea (HU) for 24 h, or 100 μm H2O2 for 3 h. After treatment, cells were rinsed with phosphate-buffered saline, and immunostaining was done as described under “Experimental Procedures.” Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI)(blue). No treat, left untreated.
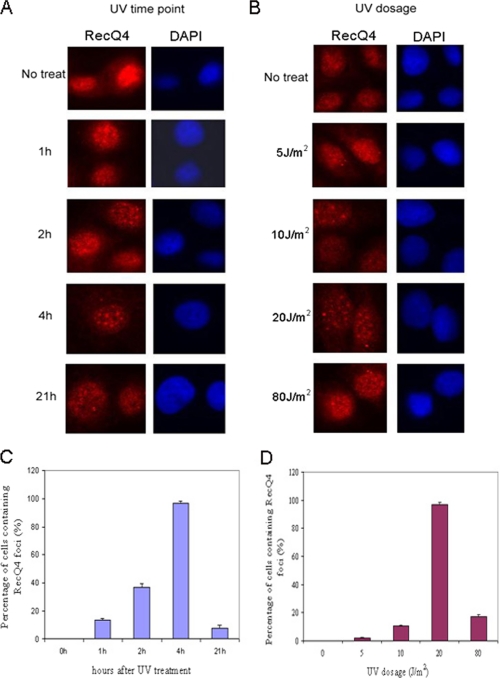
Since both UV irradiation and 4NQO treatment induce NER repair, we next examined the kinetics of RecQ4 foci formation upon UV exposure. Immunofluorescence studies were performed at 1, 2, 4, and 21 h after recovery from 20 J/m2 UV exposure in HeLa cells. As shown in Fig. 2, A and C, the formation of RecQ4 nuclear foci could be observed at 1 h after UV exposure, and the percentage of cells containing RecQ4 foci increased over recovery time. The foci formation peaked at 4 h after UV exposure (∼97% of cells contained foci) and dramatically decreased at 21 h after exposure with 7.6% cells containing RecQ4 foci. We also examined RecQ4 foci formation in response to different doses of UV irradiation and found that the percentage of cells having RecQ4 foci increased with the increment of UV doses up to 20 J/m2. After 80 J/m2 of UV irradiation, however, only 17% of cells were found to form RecQ4 foci (Fig. 2, B and D).
FIGURE 2.
The formation of RecQ4 foci peaked at 4 h after UV exposure and reached the maximum in response to 20 J/m2 UV irradiation. A, HeLa cells were irradiated with 20 J/m2 UV followed by a 1-, 2-, 4-, or 21-h recovery, and then immunostaining was done as described under “Experimental Procedures.” 4′,6-Diamidino-2-phenylindole (DAPI) staining of the nuclei is shown in blue. No treat, left untreated. B, HeLa cells were exposed to different dosages of UV irradiation (5, 10, 20 and 80 J/m2). At 4 h after UV irradiation, cells were immunostained with anti-RecQ4 antibody (red). C, quantitative results of A. The averages of three different experiments are shown. For each experiment, at least 100 cells for each time point were counted, and the percentage of cells containing RecQ4 foci was calculated. The error bar represents S.D. D, quantitative results of B. The averages of three different experiments are shown. For each experiment, at least 100 cells for each dose were counted, and the percentage of cells containing RecQ4 foci was calculated. The error bar represents S.D.
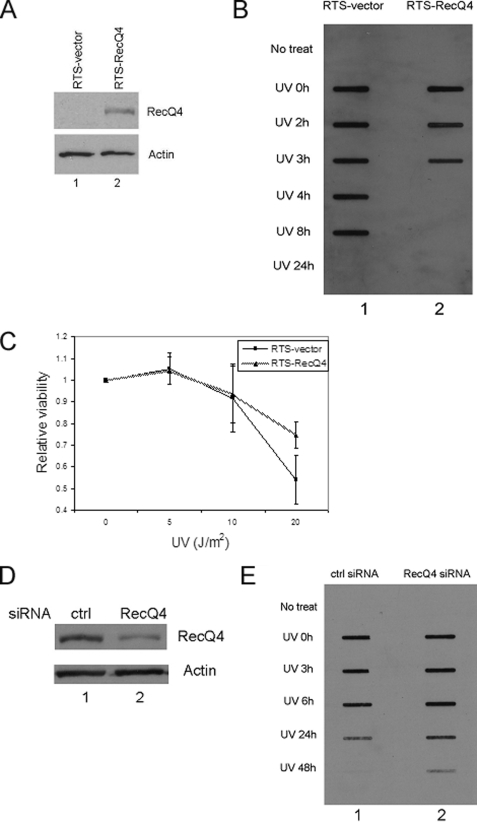
RecQ4 Has a Functional Role in NER—Since RecQ4 formed nuclear foci specifically in response to UV irradiation and 4NQO treatment, we postulated that RecQ4 might be involved in the NER pathway. To test this hypothesis, we generated pBabe-RecQ4 retroviral construct. The pBabe control virus and pBabe-RecQ4 virus were used to infect RTS cells (RecQ4-deficient fibroblasts from an RTS patient), and whole cell lysates were prepared to probe the expression level of RecQ4. As shown in Fig. 3A, RecQ4 was detected in RTS cells infected with pBabe-RecQ4 virus but not in RTS cells infected with control virus (lane 2 versus lane 1). The stable cell lines infected with control virus or pBabe-RecQ4 virus were generated. To study whether RecQ4 affects DNA repair efficiency in global genomic repair, we detected the levels of UV-induced CPDs in the two cell lines. We found RTS cells complemented with RecQ4 had a much faster removal rate of CPDs when compared with RTS control cells (Fig. 3B). At 4 h after UV irradiation, the CPD lesions were almost completely removed in RTS-RecQ4 cells, whereas no significant removal was observed at this time point in RTS control cells. This observation demonstrated that RecQ4 was required for the efficient removal of UV-induced DNA lesions. Since reduced DNA repair is often linked to the hypersensitivity to UV irradiation, we performed the MTT assay to determine the cellular viability in response to UV irradiation with these two cell lines. Cells were seeded onto 96-well plate and irradiated with increasing doses of UV. At 48 h after irradiation, cell viability was determined. Fig. 3C showed that there was no significant difference in UV sensitivity between the two cell lines upon exposure to a lower dose of UV irradiation (5 and 10 J/m2). However, in response to 20 J/m2 UV irradiation, RTS-RecQ4 cells survived significantly better than RTS control cells, indicating that RecQ4 helps cells to sustain a higher dose of UV damage. To further confirm that RecQ4 function in UV-induced DNA repair, RecQ4 was down-regulated by siRNA in HeLa cells (Fig. 3D, lane 2 versus lane 1). The cells were treated with UV irradiation, and the levels of CPDs were detected. As shown in Fig. 3E, down-regulation of RecQ4 significantly slowed the removal of UV-induced DNA lesions (lane 2 versus lane 1).
FIGURE 3.
RecQ4 has a functional role in NER. A, RTS cells were infected with pBabe vector or pBabe-RecQ4. Total cell lysates were prepared from stably transfected cells and probed with anti-RecQ4 (upper panel) and actin (lower panel) antibodies, respectively. B, RTS-RecQ4 cells and RTS control cells were seeded into a 24-well plate and harvested at different time points after 20 J/m2 UVC irradiation. The slot blot analysis was carried out, and UV-induced CPDs were detected with anti-CPDs antibody. No treat, left untreated. C, RTS-RecQ4 cells and RTS control cells were seeded in a 96-well plate and allowed to attach overnight. The cells were treated with increasing doses of UV irradiation followed by a 48-h recovery. MTT assays were performed as detailed under “Experimental Procedures.” Data were normalized to the mock treatment controls (as the value of 1). Points, mean of four different measurements; bars, S.D. D, total cell lysates were prepared from HeLa cells treated with control (ctrl) siRNA or RecQ4 siRNA and detected with anti-RecQ4 (upper panel) and actin (lower panel) antibodies, respectively. E, HeLa cells were seeded onto a 24-well plate, treated with control siRNA and RecQ4 siRNA, and harvested at different time points after 20 J/m2 UVC irradiation. The slot blot analysis was carried out, and UV-induced CPDs were detected with anti-CPDs antibody.
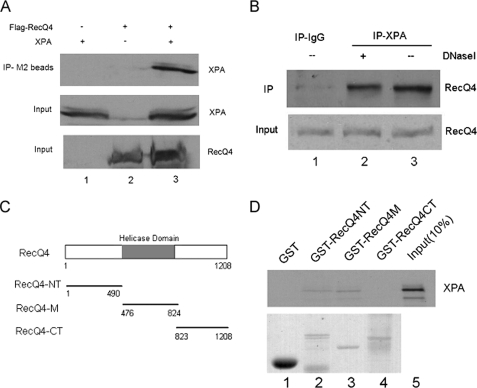
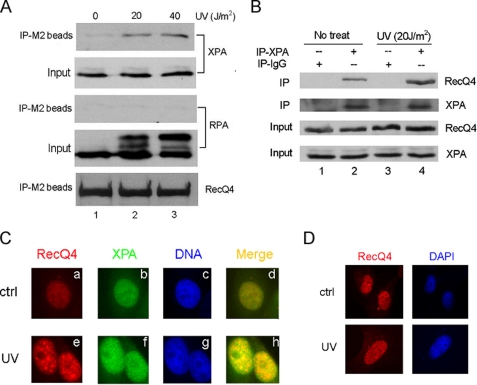
RecQ4 and XPA Interacted with Each Other—Next we set out to determine how RecQ4 plays a role in NER repair by examining whether RecQ4 interacts with any components of NER pathway. XPA is a center factor for NER and is required for both subpathways of NER; we therefore performed co-immunoprecipitation assay to examine the possible interaction of RecQ4 and XPA. FLAG-RecQ4 and XPA plasmids were transiently transfected into 293T cells, respectively, or co-transfected. Whole cell extracts were then prepared and precipitated with anti-FLAG resin (M2 beads). The immunoprecipitates were analyzed by Western blotting with anti-XPA antibody. As shown in Fig. 4A (upper panel), XPA was efficiently pulled down by the anti-FLAG resin in co-transfected cells (lane 3) but not in cells that were transfected with RecQ4 or XPA alone (lanes 1 and 2). Immunoblotting of cell lysates confirmed the expression of XPA/RecQ4 in transfected cells (Fig. 4A, middle and lower panels). Next we sought to determine whether endogenous RecQ4 and XPA could interact with one another. Whole cell lysates from HeLa cells were incubated with control IgG or anti-XPA antibody, and immunoprecipitates were detected by anti-RecQ4 antibody. Endogenous RecQ4 could be significantly precipitated by anti-XPA antibody from HeLa cell lysates but not by control IgG antibody (lane 3 versus lane 1). Furthermore, DNase I treatment of cell lysates did not abrogate the interaction of XPA and RecQ4, indicating that this interaction is not mediated by the separate binding of two proteins to the chromatin (Fig. 4B, lane 2). Furthermore, we tested whether RecQ4 directly interacted with XPA and the specific region of RecQ4 mediating this interaction. GST fusion proteins were generated for the N-terminal (GST-RecQ4-NT), central (GST-RecQ4-M), and C-terminal (GST-RecQ4-CT) regions of RecQ4 and immobilized on GST-agarose. As shown in Fig. 4D, 35S-labeled in vitro translated XPA bound to immobilized GST-RecQ4-NT and GST-RecQ4-M but not to immobilized GST-RecQ4-CT and GST alone (lanes 2 and 3 versus lanes 1 and 4). Taken together, these data demonstrated that RecQ4 and XPA could interact with each other both in vivo and in vitro.
FIGURE 4.
RecQ4 and XPA interacted with each other. A, 293T cells were transiently transfected with FLAG-RecQ4 and XPA plasmids, respectively, or co-transfected. Then whole cell lysates were prepared for co-immunoprecipitation assays with anti-FLAG resin (IP-M2 beads) as described under “Experimental Procedures.” Proteins from the immunoprecipitates were detected by Western blot using anti-XPA antibody (upper panel). 10% of the total volumes of the whole cellular lysates were used as input (middle and lower panel). B, HeLa whole cell lysates were treated with 100 μg/ml DNase I for 20 min at 37 °C or left untreated. Then co-immunoprecipitation assays were performed with anti-XPA antibody, and immunoprecipitates were detected by anti-RecQ4 antibody (upper panel). 10% of whole cell lysates were also included as input (lower panel). C, schematic representation of the RecQ4 protein and different GST fusion proteins. RecQ4-NT, N-terminal region of RecQ4; RecQ4-M, central region of RecQ4; and RecQ4-CT, C-terminal region of RecQ4. D, 35S-labeled in vitro translated XPA protein was incubated with immobilized GST or GST-RecQ4 protein at 4 °C for 2 h. The beads were washed five times with BC200, the bound proteins were eluted by boiling in sample buffer and analyzed by SDS-PAGE and autoradiography (upper panel), and GST fusion proteins were analyzed by SDS-PAGE and Coomassie Blue staining (lower panel).
UV Irradiation Stimulated the Interaction between RecQ4 and XPA and Induced Nuclear Colocalization of Two Proteins—The observations that RecQ4 formed nuclear foci in response to UV irradiation and interacted with NER factor XPA constitutively promoted us to investigate the RecQ4 interaction with XPA in cells after UV irradiation. H1299 cells that were stably transfected with FLAG-RecQ4 were irradiated with increasing doses of UV or left unirradiated. At 4 h after irradiation, whole cell lysates were prepared for co-immunoprecipitation with anti-FLAG resin (M2-beads). As shown in Fig. 5A, UV treatment significantly stimulated the interaction between two proteins as more RecQ4 was precipitated by anti-XPA antibody in UV-treated cells (lanes 2 and 3 versus lane 1). XPA forms complex with RPA to function in NER (17). However, no interaction between RecQ4 and RPA was detected under the same conditions, suggesting that the interaction of RecQ4 with XPA is intrinsic to two proteins and is not mediated by the formation of multiprotein complex in cells. To examine the interaction of endogenous RecQ4 and XPA after UV irradiation, HeLa cells were left untreated or exposed to 20 J/m2 UV, and whole cell extracts were used for immunoprecipitation with XPA antibody. The UV treatment did not change the cellular expression of RecQ4 and XPA (Fig. 5B, lower two panels); however, it did enhance the interaction between two proteins (Fig. 5B, upper panel, lane 2 versus 4). Since XPA also forms UV-induced nuclear foci, we were interested in whether the foci formed with RecQ4 and XPA could colocalize in cells after UV irradiation. Immunofluorescence microscopy was performed. As shown in Fig. 5C, in the mock-treated HeLa cells, RecQ4 and XPA proteins appeared to be homogeneously distributed throughout the nucleus (subpanels a and b). Upon exposure to UV irradiation, RecQ4 and XPA redistributed to form discrete nuclear foci, and an obvious colocalization of RecQ4 and XPA foci was observed, as indicated in Fig. 5C (subpanels e, f, and h).
FIGURE 5.
UV irradiation stimulated the interaction between RecQ4 and XPA. A, H1299 cells stably transfected with FLAG-RecQ4 were irradiated with 20 or 40 J/m2 UV or left untreated. At 4 h after UV treatment, whole cell lysates were prepared and immunoprecipitated with anti-FLAG resin (IP-M2 beads) followed by Western blotting with anti-XPA, anti-RPA, and anti-RecQ4 antibodies. 10% of whole cell lysates were used as input. B, HeLa cells were treated with 20 J/m2 UV or left untreated. 4 h after treatment, whole cell lysates were immunoprecipitated with anti-XPA antibody, and precipitated proteins were detected with anti-RecQ4 antibody. 10% of whole cell lysates were also included as input. C, HeLa cells cultured on coverslips were either untreated (upper panels) or treated with 20 J/m2 UV (lower panels). 4 h after treatment, cells were immunostained by anti-RecQ4 (red) and anti-XPA (green) antibodies. The colocalization of RecQ4 and XPA foci were shown in yellow. Nuclei were stained with 4′,6-diamidino-2-phenylindole (blue). ctrl, control. D, XPA-deficient cells cultured on coverslips were either untreated (upper panels) or treated with 20 J/m2 UV (lower panels). 4 h after treatment, cells were immunostained by anti-RecQ4 (red) antibodies. DAPI, 4′,6-diamidino-2-phenylindole.
RecQ4 Function in NER Is through Interaction with XPA—We further examined whether the XPA is necessary for RecQ4 function in NER. XPA null cells from a xeroderma pigmentosum patient (GM04429, Coriell Institute) were used for immunofluorescence staining analysis. RecQ4 was normally expressed in these cells (data not shown). After UV irradiation, XPA cells were stained by RecQ4 antibody. Remarkably, the RecQ4 foci were no longer formed (Fig. 5D), indicating that the function of RecQ4 in NER was dependent on its interaction with XPA protein.
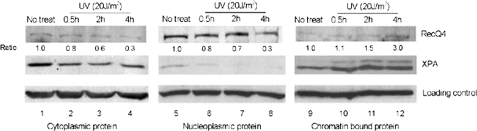
UV Irradiation Increased Chromatin-bound Form of RecQ4—It has been reported that XPA protein translocated from cytoplasm to nucleus in response to UV irradiation (18). To investigate whether RecQ4 could also increasingly associate with chromatin after UV treatment, extracts of HeLa cells were fractionated into three parts: cytoplasmic protein, soluble nuclear protein, and chromatin-bound protein. HeLa cells were treated with 20 J/m2 UV, and at 0.5, 2, and 4 h after treatment, three fractions of cell lysates were prepared for Western blotting. As shown in Fig. 6, most RecQ4 protein was located in nucleoplasm in untreated cells, consistent with our immunofluorescence results (Fig. 6, upper panel, lane 5 versus lanes 1 and 9). At 4 h after UV irradiation, the cytoplasmic and nucleoplasmic portion of RecQ4 decreased 70%, and correspondingly, the chromatin-bound form of RecQ4 increased ∼3-fold when compared with that in untreated cells (Fig. 6, upper panel, lane 12 versus lanes 4 and 8). As a positive control, we clearly detected that XPA translocated from cytoplasm to nuclei, with most of XPA bound to chromatin at 0.5 h after UV treatment (Fig. 6, middle panel).
FIGURE 6.
More RecQ4 protein bound to chromatin after UV irradiation. HeLa cells were irradiated with 20 J/m2 UV or left untreated (No treat). At 0.5, 2, and 4 h after UV treatment, cells were collected, and chromatin fractionation was carried out as described under “Experimental Procedures.” Cytoplasmic fraction, nucleoplasmic fraction, and chromatin-bound fraction were separated on 8% SDS-PAGE gels and then probed with anti-RecQ4 (upper panels) and anti-XPA (middle panels) antibodies. β-Actin (lanes 1–4), γ-tubulin (lanes 5–8) and histone H1 (lanes 9–12) were used as loading control for three fractions, respectively (lower panels). The quantities of RecQ4 protein were estimated by densitometry and then normalized to untreated controls, which were designated as 1.0.
DISCUSSION
Mutation in the RecQ4 causes Rothmund-Thomson syndromes, which have severe physiological consequences in humans, the most prominent of which are cancer susceptibility and premature aging. At the cellular level, defects in RecQ4 cause genome instability, particularly trisomy, aneuploidy, deletions, translocations, and high frequencies of chromosomal rearrangements (19). Several other members of the RecQ family, such as BLM (20) and WRN (21), have been shown to play an important role in DNA damage repair pathways. However, there are limited and conflicting data regarding the potential role of RecQ4 in the DNA damage response (8, 10–12). Therefore, we investigated RecQ4 localization in response to several forms of DNA damage. We have shown that, for the first time, endogenous RecQ4 formed discrete nuclear foci in response to UV irradiation and 4NQO, a UV mimetic agent (Fig. 1, b and c). In addition, the foci were specific to UV DNA damage and 4NQO treatment since no such foci formed upon treatment with other DNA-damaging agents including camptothecin, etoposide, hydroxyurea, and H2O2. In addition, RecQ4 nuclear foci colocalized with XPA foci after UV irradiation (Fig. 5C, subpanel h), suggesting that the formation of RecQ4 nuclear foci is relevant to the NER of UV-induced DNA damage. There are two studies about RecQ4 foci published recently. Petkovic et al. (8) reported that endogenous RecQ4 formed nuclear foci in various human cell lines in the absence of DNA damage agent treatment. Werner et al. (12) reported that RecQ4 formed nuclear foci after H2O2 treatment, whereas Woo et al. (11) reported that the RecQ4 localization pattern did not change in response to a number of DNA-damaging agents, including UV irradiation. The discrepancy in these reports is hard to interpret, but the different cell fixation approaches and the different sources of antibodies used for immunostaining might be the reason, as discussed previously (8).
We have found that the formation of RecQ4 nuclear foci reached the maximum at 4 h after 20 J/m2 UV irradiation (Fig. 2C), and fewer cells were observed to form RecQ4 foci in the early post-UV exposure hours (<4 h), during which the cellular NER function is believed to be very active (22). The RecQ4 nuclear foci greatly decreased 21 h after UV irradiation, and at this time point, either DNA lesions are removed or cells die from apoptosis. Biochemical studies have shown that almost 100% pyrimidine 6-4 pyrimidone photoproducts and 50% of CPDs are removed from the whole genome within 4 h (23). Therefore, our observations suggest that RecQ4 may not participate in NER in the early cellular response to UV irradiation and that it may play a role in the late stage of NER. We have also observed that RecQ4 nuclear foci augmented with the increasing dose of UV irradiation up to 20 J/m2 (Fig. 2D). However, the formation of RecQ4 foci dramatically reduced following 80 J/m2 UV irradiation. The lower doses of UV irradiation mainly lead to the induction of protection mechanisms in the cells, including active DNA repair processes and cell cycle arrest (24). In contrast, the high doses of UV irradiation usually trigger acute cell death through apoptosis or necrosis as the majority of cells were found to be killed after 80 J/m2 UV irradiation (data not shown). Such an UV dose-dependent formation of RecQ4 foci demonstrated that RecQ4 is involved in the repair of UV-induced DNA damage and not a nonspecific response to UV irradiation.
Our results have shown that RecQ4 was required for the optimal removal of UV-induced DNA lesions (Fig. 3, B and E) and that RecQ4 could also rescue the UV sensitivity of RTS cells (Fig. 3C), indicating that RecQ4 may participate in the repair of DNA damage caused by UV irradiation. In agreement with this observation, it has been previously reported that cells from different RTS patients have a NER defect (25). RTS patients also have NER deficiency-related symptoms. 1) Manifestations of photosensitivity, including erythema and bullous eruption after exposure to sunlight, are seen in about 30% of patients (26). 2) Early occurrence of skin malignancies is another relevant feature of the syndrome (27). These observations together with our data strongly indicate that RecQ4 may be required for optimal NER process in cells.
Several RecQ4 interaction partners have been identified. Specifically, it interacts with two DNA repair proteins, Rad51 (8) and PARP-1 (11), suggesting its role in the repair of DNA damage. Here we have demonstrated that RecQ4 could interact with XPA both physically and functionally. XPA is a DNA-binding protein and a unique factor for NER, indispensable for both subpathways of NER in cells (22). XPA recruits many other NER factors, such as RPA, XPG, and XPF/ERCC1, to the DNA damage sites through protein-protein interactions (28). We propose that RecQ4 may also be recruited by XPA to the DNA damage sites after UV irradiation. The possible evidences we observed are as follows. 1) The interaction between RecQ4 and XPA was increased upon exposure to UV irradiation (Fig. 5, A and B). 2) The RecQ4 nuclear foci could colocalize with XPA nuclear foci after UV irradiation (Fig. 5C). 3) XPA null cells lost RecQ4 foci after UV irradiation (Fig. 5D). 4) Both RecQ4 and XPA protein bound more tightly to chromatin after UV treatment (Fig. 6).
However, it remains to be determined how RecQ4 affects NER pathway in cells. Several possible mechanisms can be considered. Firstly, RecQ4 has been reported to be important for the initiation of DNA replication (6); therefore, RecQ4 might help DNA resynthesis after the excision of DNA lesions, which is the late stage of NER process. Our finding that RecQ4 nuclear foci formed predominantly at 4 h after UV irradiation supports this postulation. Secondly, RecQ4 possesses a DNA-dependent ATPase activity (4, 5), and NER is a complicated process that requires ATP in multiple steps (29). It is possible that RecQ4 facilitates NER efficiency by promoting the hydrolysis of ATP. Nevertheless, further study is required to clarify the exact mechanism of RecQ4 functioning in UV-induced DNA damage repair.
Acknowledgments
We thank Dr. Yue Zou for the gift of XPA plasmid and XPA purified protein. We also thank Drs. Patrick Sung and Ashok R Venkitaraman for the gift of RecQ4 plasmids. We thank Dr. Peter Newburger for providing us with the slot blot apparatus. We thank the Nucleic Acid Facility of University of Massachusetts Medical School for sequencing the plasmids. We also thank Xiaoyan Shi and Kai Li for the technical support.
This work was supported, in whole or in part, by National Institutes of Health Grant R01AG026534 (to J. L.) through the NIA. This work was also supported in part by grants from Worcester Foundation Annual Research Award. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: RTS, Rothmund-Thomson syndrome; XPA, xeroderma pigmentosum group A; RPA, replication protein A; CPD, cyclobutane pyrimidine dimer; 4NQO, 4-nitroquinoline 1-oxide; NER, nucleotide excision repair; GG-NER, global genome NER; TC-NER, transcription-coupled NER; GST, glutathione S-transferase; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; siRNA, small interfering RNA.
References
- 1.Bachrati, C. Z., and Hickson, I. D. (2003) Biochem. J. 374 577–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindor, N. M., Furuichi, Y., Kitao, S., Shimamoto, A., Arndt, C., and Jalal, S. (2000) Am J. Med. Genet. 90 223–228 [DOI] [PubMed] [Google Scholar]
- 3.Kitao, S., Ohsugi, I., Ichikawa, K., Goto, M., Furuichi, Y., and Shimamoto, A. (1998) Genomics 54 443–452 [DOI] [PubMed] [Google Scholar]
- 4.Yin, J., Kwon, Y. T., Varshavsky, A., and Wang, W. (2004) Hum. Mol. Genet. 13 2421–2430 [DOI] [PubMed] [Google Scholar]
- 5.Macris, M. A., Krejci, L., Bussen, W., Shimamoto, A., and Sung, P. (2006) DNA Repair (Amst.) 5 172–180 [DOI] [PubMed] [Google Scholar]
- 6.Sangrithi, M. N., Bernal, J. A., Madine, M., Philpott, A., Lee, J., Dunphy, W. G., and Venkitaraman, A. R. (2005) Cell 121 887–898 [DOI] [PubMed] [Google Scholar]
- 7.Matsuno, K., Kumano, M., Kubota, Y., Hashimoto, Y., and Takisawa, H. (2006) Mol. Cell Biol. 26 4843–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petkovic, M., Dietschy, T., Freire, R., Jiao, R., and Stagljar, I. (2005) J. Cell Sci. 118 4261–4269 [DOI] [PubMed] [Google Scholar]
- 9.Vennos, E. M., and James, W. D. (1995) Dermatol. Clin. 13 143–150 [PubMed] [Google Scholar]
- 10.Kumata, Y., Tada, S., Yamanada, Y., Tsuyama, T., Kobayashi, T., Dong, Y. P., Ikegami, K., Murofushi, H., Seki, M., and Enomoto, T. (2007) Biochim. Biophys. Acta 1773 556–564 [DOI] [PubMed] [Google Scholar]
- 11.Woo, L. L., Futami, K., Shimamoto, A., Furuichi, Y., and Frank, K. M. (2006) Exp. Cell Res. 312 3443–3457 [DOI] [PubMed] [Google Scholar]
- 12.Werner, S. R., Prahalad, A. K., Yang, J., and Hock, J. M. (2006) Biochem. Biophys. Res. Commun. 345 403–409 [DOI] [PubMed] [Google Scholar]
- 13.Park, S. J., Lee, Y. J., Beck, B. D., and Lee, S. H. (2006) DNA Cell Biol. 25 696–703 [DOI] [PubMed] [Google Scholar]
- 14.Wood, R. D. (1996) Annu. Rev. Biochem. 65 135–167 [DOI] [PubMed] [Google Scholar]
- 15.de Laat, W. L., Jaspers, N. G., and Hoeijmakers, J. H. (1999) Genes Dev. 13 768–785 [DOI] [PubMed] [Google Scholar]
- 16.Kitao, S., Lindor, N. M., Shiratori, M., Furuichi, Y., and Shimamoto, A. (1999) Genomics 61 268–276 [DOI] [PubMed] [Google Scholar]
- 17.He, Z., Henricksen, L. A., Wold, M. S., and Ingles, C. J. (1995) Nature 374 566–569 [DOI] [PubMed] [Google Scholar]
- 18.Wu, X., Shell, S. M., Liu, Y., and Zou, Y. (2007) Oncogene 26 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hickson, I. D. (2003) Nat. Rev. Cancer 3 169–178 [DOI] [PubMed] [Google Scholar]
- 20.Ababou, M., Dumaire, V., Lecluse, Y., and Amor-Gueret, M. (2002) Oncogene 21 2079–2088 [DOI] [PubMed] [Google Scholar]
- 21.Harrigan, J. A., Wilson, D. M., Prasad, R., Opresko, P. L., Beck, G., May, A., Wilson, S. H., and Bohr, V. A. (2006) Nucleic Acids Res. 34 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu, X., Shell, S. M., Yang, Z., and Zou, Y. (2006) Cancer Res. 66 2997–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costa, R. M., Chigancas, V., Galhardo Rda, S., Carvalho, H., and Menck, C. F. (2003) Biochimie (Paris) 85 1083–1099 [DOI] [PubMed] [Google Scholar]
- 24.Li, G., and Ho, V. C. (1998) Br. J. Dermatol. 139 3–10 [PubMed] [Google Scholar]
- 25.Vasseur, F., Delaporte, E., Zabot, M. T., Sturque, M. N., Barrut, D., Savary, J. B., Thomas, L., and Thomas, P. (1999) Acta Dermato-Venereol. 79 150–152 [DOI] [PubMed] [Google Scholar]
- 26.Berg, E., Chuang, T. Y., and Cripps, D. (1987) J. Am. Acad. Dermatol. 17 332–338 [PubMed] [Google Scholar]
- 27.Vennos, E. M., Collins, M., and James, W. D. (1992) J. Am. Acad. Dermatol. 27 750–762 [DOI] [PubMed] [Google Scholar]
- 28.Sancar, A., Lindsey-Boltz, L. A., Unsal-Kacmaz, K., and Linn, S. (2004) Annu. Rev. Biochem. 73 39–85 [DOI] [PubMed] [Google Scholar]
- 29.Riedl, T., Hanaoka, F., and Egly, J. M. (2003) EMBO J. 22 5293–5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu, X., Shell, S. M., and Zou, Y. (2005) Oncogene 24 4728–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]