Abstract
Trehalose dimycolate (TDM), also known as cord factor, is a major surface glycolipid of the cell wall of mycobacteria. Because of its potent biological functions in models of infection, adjuvancy, and immunotherapy, it is important to determine how its biosynthesis is regulated. Here we show that glucose, a host-derived product that is not readily available in the environment, causes Mycobacterium avium to down-regulate TDM expression while up-regulating production of another major glycolipid with immunological roles in T cell activation, glucose monomycolate (GMM). In vitro, the mechanism of reciprocal regulation of TDM and GMM involves competitive substrate selection by antigen 85A. The switch from TDM to GMM biosynthesis occurs near the physiological concentration of glucose present in mammalian hosts. We further demonstrate that GMM is produced in vivo by mycobacteria growing in mouse lung. These results establish an enzymatic pathway for GMM production. More generally, these observations provide a specific enzymatic mechanism for dynamic alterations of cell wall glycolipid remodeling in response to the transition from noncellular to cellular growth environments, including factors that are monitored by the host immune system.
Mycobacterium avium complex (MAC)2 includes a group of acid-fast bacteria that distribute widely in natural environments, including soil, water, aerosols, and dust (1). Although less virulent than Mycobacterium tuberculosis, these environmental mycobacteria occasionally infect humans, especially patients infected with human immunodeficiency virus type 1, where they represent a major cause of morbidity. The incidence of clinically overt MAC infection has increased significantly in recent years, and because of the multidrug resistance evolved by the microbes, MAC infection is difficult to clear with chemotherapeutic agents. Thus, M. tuberculosis and MAC are now the two major groups of mycobacteria species that require further efforts for prevention and treatment. Unlike M. tuberculosis, which transmits primarily from individuals with active disease, epidemiologic evidence suggests that such transmission pathways are unlikely for MAC. Rather, MAC infection appears to occur when susceptible individuals are exposed to environmental MAC. These observations predict that, upon infection, environmental MAC should undergo significant adaptive changes to allow its survival and replication within the host.
Mycobacteria possess highly lipid-rich cell walls that are critical not simply for their acid-fast properties but also for their survival and replication. The cell wall contains mycolic acids, an α-alkyl-β-hydroxy fatty acid with extremely long carbon chains (∼C80), which are densely aligned in covalent association with the 6-position of arabinose termini of the underlying arabinogalactan sugar layer or exist as free molecules complexed to sugars, either glucose or trehalose. Arabinogalactan-linked mycolates are proposed to extend outward and interact noncovalently with carbon chains of the so-called surface-exposed glycolipids, including trehalose 6-monomycolate (TMM), trehalose 6,6′-dimycolate (TDM), and glucose 6-monomycolate (GMM), thereby forming the hydrophobic cell wall architecture that is essential for protection against chemical attack, such as reactive oxygen intermediates and hydrolytic enzymes derived from the host cells. Among the most abundant surface-exposed glycolipids is TDM that is biosynthesized from its precursor, TMM, by the mycolyltransferase activity of antigen 85 (Ag85) (2). Many biological functions have been assigned to TDM (3) that may impact on survival of mycobacteria within the host and possibly their virulence. Therefore, it is important to determine how biosynthesis of TDM and other mycolic acid-containing glycolipids is regulated by external factors. GMM exists at varied levels in the mycobacterial cell wall (4, 5). In addition to its role in cell wall barrier functions, GMM is a granuloma-forming agent in mice (6) as well as a CD1b presented antigen in humans (7).
Here we identify Ag85A as an enzyme that produces GMM by transfer of mycolate to glucose. Furthermore, mechanistic studies show that glucose present in its growth environment regulates the spectrum of mycolylglycolipids made by MAC, and glucose from the host influences GMM production in vivo during infection of mice. Mechanistic studies showed that glucose and trehalose compete as substrates for Ag85A, linking the biosynthesis pathways of GMM and TDM.
EXPERIMENTAL PROCEDURES
Reagents and Bacteria—Chemical reagents were purchased from Nacalai Tesque (Kyoto, Japan) unless otherwise indicated. M. avium ATCC 35767 (serovar 4) was obtained from American Type Culture Collection (Manassas, VA). The bacteria were maintained on a plate of Middlebrook 7H10 media supplemented with 10% oleic acid/albumin/dextrose/catalase (BD Biosciences). For extraction of the total lipid fraction, the bacteria were cultured in Middlebrook 7H9 broth media (containing 0.05% Tween 80 but not glycerol) supplemented with 10% albumin/dextrose/catalase (BD Biosciences). The log phase culture was diluted with 20 volumes of 7H9 media containing various concentrations of glucose, and the culture was continued for another 5–7 days until the absorbance at 600 nm reached ∼1. In some experiments, bacteria were grown in media containing either 0.01 or 0.1% glucose, and the media were replaced every day with fresh media containing the same concentrations of glucose. After 5 days, the bacteria were harvested for lipid extraction. To monitor early GMM production, bacteria were grown either in 7H9 media containing 0.01 or 0.1% glucose or in human serum and were harvested after 2, 4, 8, 18, and 24 h of culture.
Preparation of Mycolylglycolipids from MAC—Total lipids from mycobacteria were prepared as described previously (8). The total lipids were then dissolved in chloroform/methanol (C/M, 2:1, v/v), and 20 volumes of ice-cold acetone were added. After 30 min of incubation on ice, the suspension was subjected to centrifugation at 1,500 × g for 15 min at 1 °C, and the supernatant was carefully removed. The pellet was then washed with ice-cold acetone, and the residue was dissolved in C/M (2:1) and fractionated by TLC using an Analtech TLC plate (Newark, DE) with a solvent system of chloroform/methanol/acetone/acetic acid (90:10:10:1, v/v). GMM, TDM and TMM fractions were extracted with C/M (2:1) from the silica gels. For GMM and TDM purification, the fractions were further fractionated by TLC with a solvent system of chloroform/acetone/methanol/water (50:60:2.5:0.6, v/v). Finally, the GMM, TDM and TMM fractions were extracted with C/M (2:1), dried, and rinsed several times with methanol at room temperature to remove any residual contamination of glycopeptidolipids and phospholipids.
Matrix-assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS)—MALDI-TOF MS analyses of glycolipids were carried out according to the method described previously (9). Briefly, MALDI-TOF MS spectra were acquired on a Voyager DE-STR MALDI-TOF mass spectrometer (Applied Biosystems) with a pulse laser emitting at 337 nm. Samples were analyzed in the reflectron mode with an accelerating voltage operating in positive ion mode of 20 kV. As the matrix, 2,5-dihydroxybenzoic acid was used.
Gas Chromatography-Mass Spectrometry (GC-MS)—GC-MS analysis of the sugar moiety of GMM was carried out according to the method described previously (9). Briefly, GMM was hydrolyzed with 2 m trifluoroacetic acid at 120 °C for 2 h. The aqueous phase was dried, reduced with 10 mg/ml solution of NaBD4 (1 m NH4OH/C2H5OH, 1:1, v/v) at room temperature for 2 h, and then acetylated with acetic anhydride/pyridine (1:1, v/v) at 100 °C for 1 h. The resulting alditol acetate derivatives were analyzed by GC-MS with GCMS-QP2010 plus (Shimazu Co., Ltd., Kyoto, Japan), using a fused silica capillary column (SP-2380, 30 m × 0.25 mm inner diameter; Supelco Inc.). GC oven was operated at 50 °C for 0.5 min, and then the temperature was increased to 235 °C at a rate of 65 °C/s. The temperature was then kept at 235 °C for 12 min. Flow rate of helium gas was 44.4 cm/min.
Isolation of the Antigen 85A Gene from MAC, Preparation of the Recombinant Enzyme and Its Enzymatic Assay—The genomic DNA was isolated from the MAC strain using the Isoplant kit according to the manufacturer's instruction (Wako Pure Chemical Co. Ltd., Osaka, Japan). The gene that encoded the mature Ag85A lacking the signal sequence was amplified by PCR, using a specific primer set as follows: 5′-gga att cca tat gtt ctc gcg ccc cgg tct gcc-3′ (a sense primer, in which the NdeI restriction site is underlined) and 5′-ccg ctc gag ggt gcc ctgg ccg ttc ccg g-3′ (an antisense primer, in which the XhoI restriction site is underlined). PCR was carried out using a Takara LA-TaqDNA polymerase (Takara Co. Ltd., Tokyo, Japan), and the cycling conditions for PCR amplification were as follows: 94 °C, 2 min, followed by 30 cycles of 98 °C, 20 s and 72 °C, 1.5 min, and a final extension step of 72 °C, 3 min. The amplified PCR products were digested with NdeI and XhoI and ligated to a NdeI-XhoI-digested pET-21c plasmid vector (Merck). The nucleotide sequences of the Ag85A gene were determined for four isolated clones. Escherichia coli BL21 (DE3) was transformed with the Ag85A gene in pET-21c, and induction of protein expression was performed according to a method of Kremer et al. (10).
The bacteria expressing the His-tagged mature Ag85A were harvested and disrupted by sonication in ice-cold 20 mm Tris-HCl buffer (pH 7.9) containing 0.5 m NaCl and 60 mm imidazole (sonication buffer). The sonicate was centrifuged at 10,000 × g for 30 min at 4 °C to remove insoluble materials, and then the supernatant was applied onto a Ni2+-resin column equilibrated with the sonication buffer at 4 °C. After washing the column with the sonication buffer, the recombinant Ag85A was eluted with 20 mm Tris-HCl buffer (pH 7.9) containing 0.5 m NaCl and 0.5 m imidazole. The eluate was concentrated and dialyzed against 50 mm Tris-HCl buffer (pH 7.4) containing 10% glycerol overnight at 4 °C. Protein concentration of the recombinant Ag85A preparation was determined by the Quick Start Bradford protein assay kit (Bio-Rad). Purity of the preparation was determined by SDS-PAGE and Coomassie staining.
Mycolyltransferase assays were carried out by modification of a method of Kremer et al. (10). Twenty μg of purified TMM was dispersed by sonication in 150 μl of 50 mm sodium phosphate buffer (pH 7.4) in the presence or absence of indicated concentration of d-glucose. The reaction was started by the addition of 50 μl of the enzyme preparation containing 50 μgof protein. After 1 h of incubation at 37 °C, the reaction was stopped by the addition of 2 ml of C/M (2:1) and 0.3 ml of distilled water. The lipids were extracted by the method of Kremer et al. (10) and analyzed by silica gel TLC. The lipids on the TLC plate were visualized by spraying 50% sulfuric acid and baking.
GMM Detection in Vivo—Mouse infections were carried out via the aerosol route with 102 M. tuberculosis Erdman strain with mice sacrificed after ∼21 days of infection. Lungs were homogenized with beads and centrifuged at 2000 × g for 30 min at room temperature. The bacterial pellet was treated with 2% NaOH to disperse phospholipid bilayers, neutralized with 0.27 m phosphoric acid in phosphate buffered saline, and centrifuged at 2000 × g for 30 min to recover bacteria. Lipids were extracted from this mixture with three serial extractions in C/M (2:1, 1:1, and 1:2), evaporated to dryness under nitrogen, and resuspended in 1:1 C/M. These lipids were further fractionated by cold acetone precipitation to enrich for lipids that were analyzed by normal phase chromatography on a diol column. Solvent A was methanol, and solvent B was 60:40 (v/v) hexane/2-propanol. Both solvents contained 0.1% (v/v) formic acid and 0.05% (v/v) ammonium hydroxide. A binary gradient was used beginning at 5% solvent A for 3 min, linearly increasing to 40% solvent A over 5 min, holding at 40% solvent A for 6 min, linearly increasing to 100% solvent A over 2.2 min, holding at 100% solvent A for 3 min, linearly decreasing to 5% solvent A over 3.6 min, and finally holding at 5% solvent A for 3.2 min. Compounds matching the expected mass for GMM were detected at after 3.6–3.9 min of elution under these conditions. The accurate mass experiment was carried out with an Agilent 6520 Accurate Mass QTOFLC-MS operated in the positive mode with an Agilent Technologies 1200 Series high pressure liquid chromatography system. CID-MS was carried out with a ThermoLCQ Advantage Ion Trap mass spectrometer with nano-electrospray ionization in comparison with GMM derived from Mycobacterium fallax (11).
GMM-specific T Cell Assays—The T cell receptor (TCR)-deficient Jurkat cells (J.RT3) reconstituted by transfection with GMM-specific, CD1b-restricted TCRs have been described previously (12). The T cells (5 × 104/well) were cocultured in 96-well microtiter plates with the C1R human B-lymphoblastoid cells (1 × 105/well) stably transfected either with CD1b (C1R/CD1b) or with empty vector alone (C1R/mock) (13) in the presence of phorbol 12-myristate 13-acetate (10 ng/ml) and indicated concentrations of lipid preparations. In some experiments, monocyte-derived dendritic cells were used as antigen-presenting cells. After 20 h, aliquots of the culture supernatants were collected, and the amount of interleukin-2 (IL-2) released into the supernatants was measured by the IL-2 ELISA kit (BD Biosciences).
RESULTS
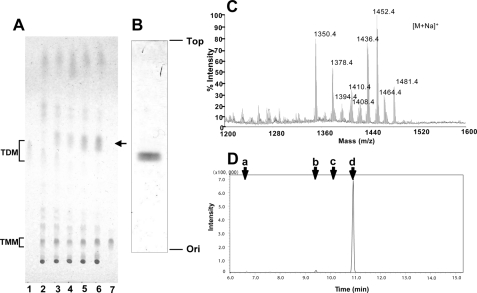
Reciprocal Production of TDM and GMM by MAC in Response to Glucose—Glucose is an essential nutrient to living organisms, which is utilized as a source not only for energy production but also for biosynthesis of glycosylated constituents of cellular architecture. Unlike other hexose sugars, glucose is maintained at high levels in the blood and tissues of mammalian hosts. Therefore, we predicted that, upon infection into the host, MAC grown in glucose-limited environments might undergo significant alterations in glycolipid biosynthesis by exposure to host-derived glucose. To gain insights into the impact of exogenous glucose on glycolipid composition in mycobacteria, we first monitored glycolipid production by M. avium strain (serovar 4) that was harvested after cultivation in liquid media supplemented with different concentrations of glucose. The total lipid fraction was obtained by extracting each bacterial preparation with chloroform and methanol. The lipids were then analyzed on a TLC plate developed with a solvent system suitable for separation of chemically diverse glycolipid species (Fig. 1A). When grown in the presence of a trace amount of glucose (0.01%, w/v), mycobacteria produced high levels of TDM and TMM (Fig. 1A, lane 2, shown with brackets). As the glucose concentrations present in media were increased, TDM production decreased, whereas the amount of TMM remained constant (Fig. 1A, lanes 2–6). Also, an increase in a discrete, unknown lipid species with a retardation factor (Rf) slightly greater than that of TDM was noted (Fig. 1A, lanes 3–6, indicated with an arrow). To determine the molecular identity of the unknown lipid, it was purified and subjected to TLC and MS analyses. The purified lipid was resolved as doublet bands on a TLC plate developed with a solvent system of C/M (9:1, v/v) (Fig. 1B). MALDI-TOF MS analysis revealed that the mass numbers of given ions were matched with those of sodium adducts of hexose monomycolate (Fig. 1C). Within the limits of error of the method of detection, the masses matched both in terms of the expected m/z of the dominant ions, the range of mass variation expected of individual molecular species of mycolate derivatives, and the absolute mass differences among the major ions, which can be accounted for by differences in carbon chain length and substitution of R groups (14). For example, m/z 1452.4 corresponds to the expected mass of sodium adduct of hexose monomycolate with C85 fatty acid and a wax ester-type R group on the meromycolate chain (Fig. 1C). GC-MS analysis of an alditol acetate derivative of the sugar moiety derived from the purified lipid identified glucose as the hexose group attached to mycolates (Fig. 1D). The doublet bands observed on a TLC plate were thus likely to represent two stereoisomers of mycolates as described previously (5, 15). Finally, the production of GMM in response to added glucose is expected based on the ability of mycobacteria to couple abundant hexose sugars at mycolyl esters (5). These results detected a reciprocal production of TDM and GMM by MAC in response to exogenous glucose without apparent alterations in the steady state levels of TMM. This experiment, carried out in live bacteria, raised the possibility that mycolyltransferases might compete for carbohydrate substrates.
FIGURE 1.
A reciprocal production of TDM and GMM in MAC in response to glucose. A, MAC was cultured in media containing 0.01% (w/v, lane 2), 1% (w/v, lane 3), 2% (w/v, lane 4), 5% (w/v, lane 5), and 10% glucose (w/v, lane 6), and the total lipid fractions (50 μg each) were analyzed on a TLC plate that was developed with chloroform/methanol/acetone/acetic acid (90:10:10:1, v/v). Purified TDM (lane 1) and TMM (lane 7) were used as references. Glucose dose-dependent production of a lipid species (indicated with an arrow) was detected. B, lipid species was purified and analyzed on a silica gel TLC plate that was developed with chloroform/methanol (9:1, v/v). C, MALDI-TOF MS profiles of the purified lipid species. D, GC-MS analysis of the sugar moiety of the purified lipid species. Arrows indicate retention times for the alditol acetate derivatives of arabinose (a), mannose (b), galactose (c)/ and glucose (d). Ion chromatogram of m/z 290 is shown. The retention time of the major ion corresponded with that of a glucose alditol acetate derivative.
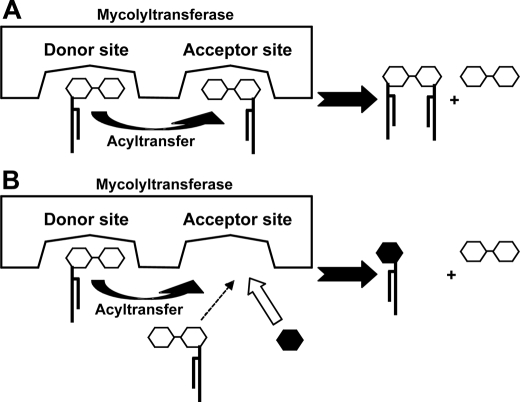
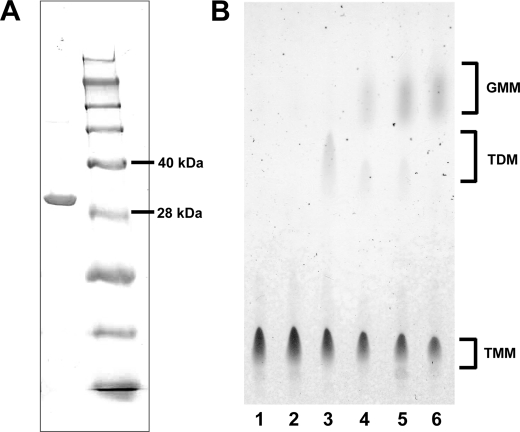
Ag85 Utilized Glucose for GMM Biosynthesis—Mycobacteria-derived mycolyltransferases, known also as Ag85, catalyze the final step of TDM biosynthesis, using TMM as a substrate. Current models of the Ag85-catalyzed reaction predicted that two molecules of TMM are captured in the two substrate-binding pockets of the enzyme, and the mycolyl acyl group of the TMM substrate bound in one substrate-binding pocket (donor site) is transferred to the other TMM substrate bound in the other pocket (acceptor site), resulting in generation of one molecule of TDM and one molecule of trehalose (Fig. 2A) (2, 16). Although GMM can be an abundant structure in the cell wall and functions to activate T cells and form granulomas, its mechanism of synthesis was unknown. We hypothesized that GMM biosynthesis could be catalyzed by Ag85 if glucose, instead of TMM, occupied the acceptor site (Fig. 2B). To address this possibility, we made recombinant Ag85A enzyme from the M. avium strain (serovar 4), and we performed in vitro enzymatic reaction experiments. To accomplish this, we first carried out PCR from the genome of the MAC strain as a template, and isolated the Ag85A gene that encoded the mature protein lacking the signal sequence. DNA sequencing of the isolated gene revealed that three nucleotides were altered as compared with the previously reported Ag85A gene derived from M. avium serovar 1 strain (17), but the deduced amino acid sequences were identical in both strains. We then constructed an expression plasmid in which the initiation codon was placed at the 5′-end and the sequence encoding a His tag was attached in frame at the 3′-end of the isolated Ag85A gene. The His-tagged enzyme was expressed in E. coli and affinity-purified by Ni2+-charged resin column chromatography. The purified material was resolved as a single band with an apparent molecular mass of ∼33 kDa on a Coomassie-stained SDS-polyacrylamide gel, consistent with its being the Ag85A protein (Fig. 3A). Incubation of TMM in vitro in the presence of this enzyme preparation resulted in generation of TDM (Fig. 3B, lane 3), confirming the mycolyltransferase activity exerted by the recombinant protein. Strikingly, addition of glucose to this reaction condition resulted in decreased TDM production in a dose-dependent manner, which was associated with an increase in GMM (Fig. 3B, lanes 3–6). GMM synthesis was completely abrogated when heat-inactivated enzyme was used (Fig. 3B, lane 2). This further confirmed that GMM was produced enzymatically by the mycolyltransferase activity of Ag85A but not as a result of nonenzymatic hydrolysis. These results indicate that Ag85A mediates synthesis of GMM. In this molecular model, we propose that TMM and glucose compete for access to the acceptor site of the Ag85A, and the enzyme preferentially catalyzes biosynthesis of GMM, rather than TDM, when glucose is readily available (Fig. 2B). The substrate selection by the mycolyltransferase would likely provide a molecular basis for the glucose-dependent TDM-GMM exchange detected in cultured MAC.
FIGURE 2.
Proposed scheme for TDM (A) and GMM (B) production catalyzed by mycolyltransferase. In model A, both the donor site and the acceptor site of the enzyme interact with TMM, resulting in TDM formation. In model B, a glucose substrate competes against a TMM substrate for access to the acceptor site. When glucose is readily available, glucose rather than TMM preferentially gain access to the site, resulting in production of GMM.
FIGURE 3.
TDM-GMM exchange mediated by recombinant Ag85A. A, purified MAC Ag85A (left lane) and a size marker (right lane) were resolved on a Coomassie-stained SDS-polyacrylamide gel. Positions for the 40- and 28-kDa marker proteins are indicated. B, enzymatic reactions were performed at 37 °C at conditions indicated below, and the lipids were extracted from the reaction mixtures, followed by analysis on a TLC plate. Lane 1, Ag85A and TMM with 5% glucose (w/v), 0 h of incubation; lane 2, heat-inactivated (100 °C, 3 min) Ag85A and TMM with 5% glucose (w/v), 1 h of incubation; lanes 3–6, Ag85A and TMM either with 0.2% (w/v) glucose (lane 4), 1% glucose (w/v) (lane 5), and 5% (w/v) glucose (lane 6) or without glucose (lane 3), 1 h of incubation.
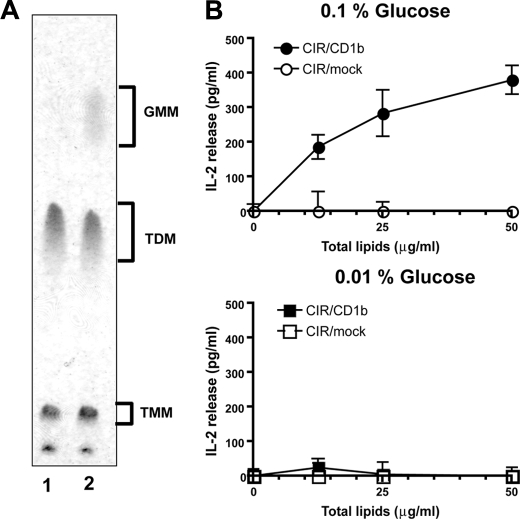
GMM Production Occurs at a Physiological Glucose Concentration—The observations made above have established an enzymatic pathway for GMM production in live mycobacteria that are grown in the presence of high levels of exogenous glucose. However, it remains to be addressed whether mycobacteria can produce GMM under physiological concentrations of glucose present in mammalian hosts, which is maintained at ∼100 mg/dl (0.1% w/v). To address this issue, we measured GMM production by mycobacteria cultured in liquid media with a glucose concentration comparable with that in the host. The MAC culture was started in the presence of either 0.01 or 0.1% glucose, and every 24 h, the culture media were replaced with fresh media to maintain the glucose concentrations at constant levels. After 5 days of culture, the bacteria were harvested, and the total lipids were extracted. Subsequently, methanol-insoluble lipids were isolated from these total lipids, followed by separation on TLC plates (Fig. 4A). Although TDM production was readily detected in both cultures, GMM production was detected only in the presence of 0.1% glucose (Fig. 4A, lane 2) but not in the presence of 0.01% glucose (lane 1). This was also confirmed by T cell-based assays (Fig. 4B) in which Jurkat T cells expressing specific TCRs recognizing GMM in the context of CD1b molecules were used. Incubation of the T cells with CD1b-expressing cells (C1R/CD1b) in the presence of the total lipids from the 0.1% glucose-containing culture resulted in dose-dependent IL-2 production by the T cells, demonstrating high levels of antigenicity when growing at physiological glucose concentrations (Fig. 4B, upper panel). The specific response was not observed when CD1b-negative cells (C1R/mock) were used as antigen-presenting cells, supporting that the response was CD1b-restricted.
FIGURE 4.
GMM production by mycobacteria cultured at a physiological glucose concentration. A, MAC was cultured in liquid media containing either 0.01 or 0.1% glucose, and the culture media were replaced with fresh media every day to maintain the glucose concentrations. After 5 days, the bacteria were harvested, and the total lipids were extracted. The methanol-insoluble fraction was then obtained from 100 μg of each total lipid preparation and analyzed by TLC. B, GMM-specific, CD1b-restricted TCR-expressing Jurkat T cells were cocultured with either C1R/CD1b or C1R/mock in the presence of different concentrations of the total lipids derived from the 0.1% glucose-containing (upper panel) and the 0.01% glucose-containing (lower panel) cultures. The T cell response was assessed by measuring IL-2 released into the media.
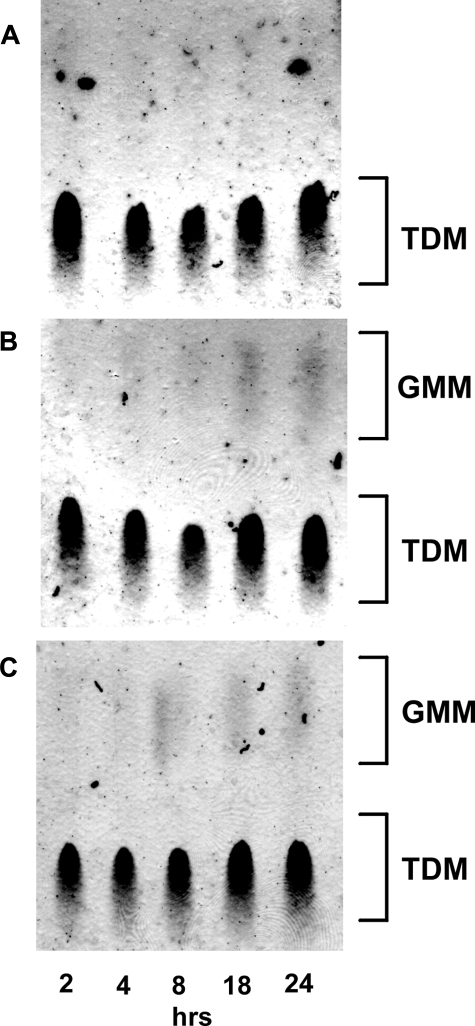
We then addressed how quickly induction of GMM production occurred after exposure to 0.1% glucose. MAC was cultured either in liquid media containing 0.01% (Fig. 5A) or 0.1% (B) or in human serum (C), and the bacteria were harvested at 2, 4, 8, 18, and 24 h. GMM production was observed as early as 8 h after the start of the culture both in 0.1% glucose-containing media and in human serum but not in media containing 0.01% glucose. These observations suggest that GMM production can occur quickly after exposure to high levels of glucose presumably as a result of competitive substrate selection by preexisting mycolyltransferases.
FIGURE 5.
GMM production by mycobacteria during early phases of culture. MAC was cultured either in liquid media containing 0.01% (A) or 0.1% (B) glucose or in human serum (C). At indicated time points, the bacteria were harvested, and the total lipids were analyzed by TLC.
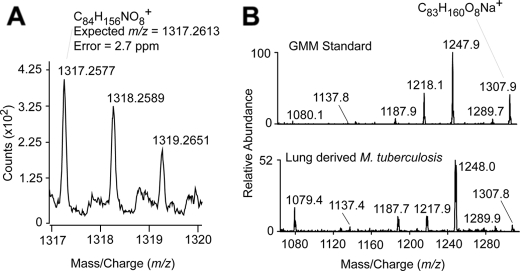
GMM Production Occurs in Mycobacteria-infected Tissues—A previous study detected GMM comigrating lipids can be derived from Mycobacterium leprae, raising the possibility that GMM is produced by mycobacteria in tissues (5). However, the chemical structures of such candidate glycosyl mycolates could not be directly determined, and it remained unknown whether M. tuberculosis produces GMM during infection. Therefore, we infected CH3 mice with M. tuberculosis Erdman strain and isolated mycobacteria directly from the lungs after ∼3 weeks of infection. Bacteria were enriched from lung preparations by centrifugation and treatment with weak base to disperse lung tissue. The resulting preparations contained predominantly mycobacterial lipids when analyzed by LC-MS (data not shown). By comparing total M. tuberculosis lipids from lung with an M. fallax GMM standard in LC-MS experiments, we analyzed the in vivo derived lipids that nearly copurified with the GMM standard. Mass measurements with an Accurate Mass QTOF capable of mass resolution of 10 ppm detected an ion at 1317.2577 in lung-derived lipids (Fig. 6A). Both the absolute m/z and the isotope ratios matched the predicted masses of an ammonium adduct of a GMM carrying a C78 α-mycolic acid within expected error (C84,H162O8,C78 GMM, expected m/z 1317.2613). Further supporting the identification of this ion as GMM, mycolic acid derivatives are characteristically synthesized as a series of molecules that differ from one another by mass increments corresponding to C2H2, and the spectrum of the lung-derived lipids contained two additional ions (m/z 1345.3033, 1289.2304) corresponding to the expected masses of C80 and C76 GMM (data not shown). Finally, a separate CID-MS experiment, carried out with an M. fallax GMM standard and the lung-derived lipids, showed nearly identical product ions, including ions with mass intervals corresponding to the loss of 60, 90, and 120 units (m/z 1248.0, 1217.9, and 1187.7), which likely represent the loss of C2H4O2, C3H6O3, and C4H8O4, which are products expected from cleavage through the hexose sugar (Fig. 6B). These data provide strong evidence that GMM is made in the host in vivo during an experimental infection.
FIGURE 6.
GMM production in vivo by M. tuberculosis. A, among the mixture of lipids extracted from M. tuberculosis derived from mouse lungs, lipids that copurified with a GMM standard were analyzed in the positive mode on an Accurate Mass QTOF MS. The detected mass of m/z 1317.2577 corresponds to the predicted mass of an ammonium adduct of C78 GMM (m/z 1317.2613). B, positive mode CID-MS analysis in ion trapping mass spectrometry of M. fallax GMM and the lung-derived candidate GMM molecule detected as sodium adducts show a similar pattern of product ions.
DISCUSSION
MAC represents a group of environmental mycobacteria that have evolved the capacity to adapt to low nutrition environments. In fact, MAC can survive and replicate in water supply systems (1), where aggregates of the microbes exist in close association with the surface area. This biofilm formation confers significant resistance to a variety of physical and chemical stresses, such as exposure to disinfectants and antibiotics, and thus is an important strategy for the environmental mycobacteria to maintain their life cycles safely in natural environments. These environmental mycobacteria stably express TDM on the surface of their cell wall but fail to biosynthesize GMM because of highly limited availability of glucose. This study argues that, upon entry into the host, this glycolipid phenotype would be modified enzymatically by utilizing the host-derived glucose as a substrate. All three functional mycolyltransferases (Ag85A, Ag85B, and Ag85C) identified in M. tuberculosis (18) appear capable of catalyzing TDM synthesis from TMM in vitro, and the corresponding Ag85 isoforms have been found also in M. avium. Although these enzymes have certain overlapping functions, their differential transcription patterns have been noted in mycobacteria grown under distinct conditions. Ag85A is an isoform that is preferentially expressed in macrophage-resident mycobacteria (19, 20), and thus, its catalytic potential for the TDM-GMM exchange could have an impact on macrophage functions if TDM and GMM have differential ability to activate the cells. Indeed, we recently found that interferon-γ-primed macrophages produced only a marginal level of nitric oxides when stimulated with GMM, which contrasted sharply with those stimulated with TDM that were capable of mounting robust nitric oxide responses (data not shown). Therefore, the TDM-GMM exchange may be valuable in minimizing the nitric oxide response by the host macrophages. Prior to this report, the identity of any mycolyltransferase that could produce GMM was unknown, so these results establish that Ag85A has this function.
Once pathogens break the frontline defense mediated by the host innate immunity, they are then challenged by specific T lymphocytes that belong to the acquired immunity. Glycolipid-specific T lymphocyte reactions are elicited in humans and guinea pigs infected with mycobacteria, and activation of these T cells is restricted not by the classical major histocompatibility complex-encoded class I and class II molecules but rather by nonmajor histocompatibility complex-encoded group 1 CD1 molecules (CD1a, CD1b, and CD1c in humans) (21). These CD1 molecules are expressed in activated macrophages as well as dendritic cells, the two major cell types for mycobacterial infection. Notably, infiltration of GMM-specific, CD1b-restricted T cells is detected in human skin infected with M. leprae, and the human T cell lines (7) and polyclonal T cells (22) exhibit cytotoxic effects, suggesting that the CD1-restricted T cell response directed against GMM could potentially function to clear infection. Taken together, these results raise an interesting possibility that GMM generated as a result of TDM-GMM exchange functions to reduce the innate immune response, but provides the host with a new opportunity to monitor live mycobacteria and eliminate them in the subsequent phases of the acquired immunity. This may represent an example of how the immune system has been constructed during the long processes of evolution to fight efficiently against pathogens.
Despite the fact that GMM is produced by pathogenic mycobacteria and structurally related to the well studied TDM, it has not been the target for focused investigation until recently. Presumably, this is partly because only a tiny amount of GMM, as compared with TDM, is synthesized by pathogenic slow growing mycobacteria, such as M. tuberculosis and M. avium, when cultured in the Middlebrook “standard” media formulations. Ironically, the standard media used for cultivation of fast growing saprophytic bacteria, such as Rhodococcus ruber, contain 1% glucose, and thus, the nonpathogenic bacteria cultured in such a medium produce GMM abundantly, and its structure and biological activities have been studied extensively (6, 23). In many previous studies, the composition, structure, and function of mycobacterial lipids were determined by using bacteria grown in standard media, but the present study suggests that the “lipid world” that is constructed by the bacteria grown in standard culture conditions is substantially different from the lipid world constructed as a result of interaction with the continuously changing host environments.
Acknowledgments
We thank Dr. Seiko Mizuno (Soai University, Osaka, Japan) for use of the GC-MS facility and Dr. Tan-Yun Cheng (Brigham and Women's Hospital, Harvard Medical School, Boston, MA) for reagents and advice.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 07155 (NIAID) (to D. B. M.). This work was also supported by grants-in-aid from scientific research on priority areas from the Ministry of Education, Culture, Sports, Science and Technology (to M. S.), grants-in-aid for scientific research B (to M. S.) and C (to I. M.) from the Japan Society for the Promotion of Science, grants from the Ministry of Health, Labour, and Welfare Research on Emerging and Re-emerging Infectious Diseases (to M. S.), and by the Burroughs Wellcome Fund (to D. B. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBank™/EBI Data Bank with accession number(s) AB325677.
Footnotes
The abbreviations used are: MAC, Mycobacterium avium complex; Ag85, antigen 85; GC-MS, gas chromatography-mass spectrometry; GMM, glucose 6-monomycolate; IL-2, interleukin-2; MALDI-TOF MS, matrix-assisted laser desorption ionization-time of flight mass spectrometry; TCR, T cell receptor; TDM, trehalose 6,6′-dimycolate; TMM, trehalose 6-monomycolate; LC-MS, liquid chromatography-mass spectrometry.
References
- 1.Primm, T. P., Lucero, C. A., and Falkinham, J. O., III (2004) Clin. Microbiol. Rev. 17 98–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sathyamoorthy, N., and Takayama, K. (1987) J. Biol. Chem. 262 13417–13423 [PubMed] [Google Scholar]
- 3.Ryll, R., Kumazawa, Y., and Yano, I. (2001) Microbiol. Immunol. 45 801–811 [DOI] [PubMed] [Google Scholar]
- 4.Brennan, P., and Ballou, C. E. (1967) J. Biol. Chem. 242 3046–3056 [PubMed] [Google Scholar]
- 5.Moody, D. B., Guy, M. R., Grant, E., Cheng, T. Y., Brenner, M. B., Besra, G. S., and Porcelli, S. A. (2000) J. Exp. Med. 192 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsunaga, I., Oka, S., Inoue, T., and Yano, I. (1990) FEMS Microbiol. Lett. 55 49–53 [DOI] [PubMed] [Google Scholar]
- 7.Moody, D. B., Reinhold, B. B., Guy, M. R., Beckman, E. M., Frederique, D. E., Furlong, S. T., Ye, S., Reinhold, V. N., Sieling, P. A., Modlin, R. L., Besra, G. S., and Porcelli, S. A. (1997) Science 278 283–286 [DOI] [PubMed] [Google Scholar]
- 8.Matsunaga, I., Bhatt, A., Young, D. C., Cheng, T. Y., Eyles, S. J., Besra, G. S., Briken, V., Porcelli, S. A., Costello, C. E., Jacobs, W. R., Jr., and Moody, D. B. (2004) J. Exp. Med. 200 1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enomoto, Y., Sugita, M., Matsunaga, I., Naka, T., Sato, A., Kawashima, T., Shimizu, K., Takahashi, H., Norose, Y., and Yano, I. (2005) Biochem. Biophys. Res. Commun. 337 452–456 [DOI] [PubMed] [Google Scholar]
- 10.Kremer, L., Maughan, W. N., Wilson, R. A., Dover, L. G., and Besra, G. S. (2002) Lett. Appl. Microbiol. 34 233–237 [DOI] [PubMed] [Google Scholar]
- 11.Cheng, T. Y., Relloso, M., Van Rhijn, I., Young, D. C., Besra, G. S., Briken, V., Zajonc, D. M., Wilson, I. A., Porcelli, S., and Moody, D. B. (2006) EMBO J. 25 2989–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant, E. P., Degano, M., Rosat, J. P., Stenger, S., Modlin, R. L., Wilson, I. A., Porcelli, S. A., and Brenner, M. B. (1999) J. Exp. Med. 189 195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugita, M., Porcelli, S. A., and Brenner, M. B. (1997) J. Immunol. 159 2358–2365 [PubMed] [Google Scholar]
- 14.Barry, C. E., III, Lee, R. E., Mdluli, K., Sampson, A. E., Schroeder, B. G., Slayden, R. A., and Yuan, Y. (1998) Prog. Lipid. Res. 37 143–179 [DOI] [PubMed] [Google Scholar]
- 15.Natsuhara, Y., Oka, S., Kaneda, K., Kato, Y., and Yano, I. (1990) Cancer Immunol. Immunother. 31 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson, D. H., Harth, G., Horwitz, M. A., and Eisenberg, D. (2001) J. Mol. Biol. 307 671–681 [DOI] [PubMed] [Google Scholar]
- 17.Ohara, N., Matsuo, K., Yamaguchi, R., Yamazaki, A., Tasaka, H., and Yamada, T. (1993) Infect. Immun. 61 1173–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belisle, J. T., Vissa, V. D., Sievert, T., Takayama, K., Brennan, P. J., and Besra, G. S. (1997) Science 276 1420–1422 [DOI] [PubMed] [Google Scholar]
- 19.Hou, J. Y., Graham, J. E., and Clark-Curtiss, J. E. (2002) Infect. Immun. 70 3714–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mariani, F., Cappelli, G., Riccardi, G., and Colizzi, V. (2000) Gene (Amst.) 253 281–291 [DOI] [PubMed] [Google Scholar]
- 21.Matsunaga, I., and Sugita, M. (2007) Curr. Immunol. Rev. 3 145–150 [Google Scholar]
- 22.Ulrichs, T., Moody, D. B., Grant, E., Kaufmann, S. H., and Porcelli, S. A. (2003) Infect. Immun. 71 3076–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsunaga, I., Oka, S., Fujiwara, N., and Yano, I. (1996) J. Biochem. (Tokyo) 120 663–670 [DOI] [PubMed] [Google Scholar]