Abstract
We previously demonstrated that a portion, or perhaps all, of the residues between 931 and 1000 of apolipoprotein (apo) B100 are required for the initiation of apoB-containing particle assembly. Based on our structural model of the first 1000 residues of apoB (designated as apoB:1000), we hypothesized that this domain folds into a three-sided lipovitellin-like “lipid pocket” via a hairpin-bridge mechanism. We proposed that salt bridges are formed between four tandem charged residues 717–720 in the turn of the hairpin bridge and four tandem complementary residues 997–1000 located at the C-terminal end of the model. To identify the specific motif within residues 931 and 1000 that is critical for apoB particle assembly, apoB:956 and apoB:986 were produced. To test the hairpin-bridge hypothesis, the following mutations were made: 1) residues 997–1000 deletion (apoB:996), 2) residues 717–720 deletion (apoB:1000Δ717–720), and 3) substitution of charged residues 997–1000 with alanines (apoB:996 + 4Ala). Characterization of particles secreted by stable transformants of McA-RH7777 cells demonstrated the following. 1) ApoB:956 did not form stable particles and was secreted as large lipid-rich aggregates. 2) ApoB:986 formed both a lipidated particle that was denser than HDL3 and large lipid-rich aggregates. 3) Compared with wild-type apoB:1000, apoB:1000Δ717–720 displayed the following: (i) significantly diminished capacity to form intact lipidated particles and (ii) increased propensity to form large lipid-rich aggregates. 4) In striking contrast to wild-type apoB:1000, (i) apoB:996 and apoB:996 + 4Ala were highly susceptible to intracellular degradation, (ii) only a small proportion of the secreted proteins formed stable HDL3-like lipoproteins, and (iii) a majority of the secreted proteins formed large lipid-rich aggregates. We conclude that the first 1000 amino acid residues of human apoB100 are required for the initiation of nascent apoB-containing lipoprotein assembly, and residues 717–720 and 997–1000 play key roles in this process, perhaps via a hairpin-bridge mechanism.
Apolipoprotein (apo)2 B100 is one of the largest monomeric proteins known. It is composed of 4536 amino acid residues and has a molecular mass of 550 kDa (1–5). ApoB100 is synthesized primarily in mammalian liver; it is an essential structural component for the formation and secretion of very low density lipoproteins and serves as a ligand for receptor-mediated uptake of low density lipoproteins by a variety of cells (4, 5). ApoB is present as a single molecule per lipoprotein particle (6), and therefore, its concentration in the plasma approximates the number of potential atherogenic lipoprotein particles. ApoB is highly insoluble in aqueous solutions and thus remains with the lipoprotein particle throughout its metabolism (7). Because of the size and insoluble nature of apoB, it has been difficult to confirm the structural motifs responsible for the lipid-associating properties of this nonexchangeable apolipoprotein.
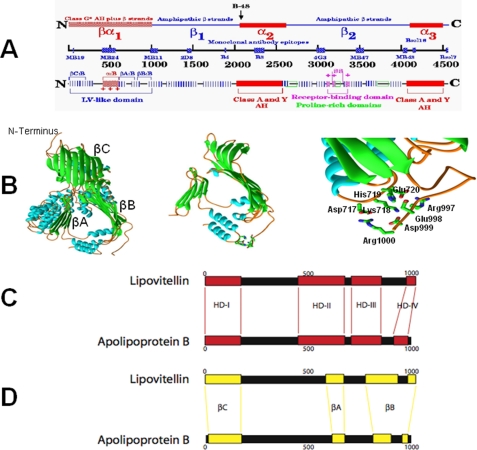
Early studies using calorimetry and x-ray diffraction (8–10), and more recently electron and cryoelectron microscopy (11–15), indicated that apoB is located on the surface of a spherical lipoprotein particle. Analysis of lipid-associating domains of apoB100 with a computer program called LOCATE confirmed the presence of two regions of amphipathic β-strands alternating with two regions of amphipathic α-helices reported previously (2, 9, 16–18) and suggested the presence of a third N-terminal globular domain, giving apoB100 the currently accepted pentapartite model NH2-βα1-β1-α2-β2-α3-COOH (19) (Fig. 1A). The β domains contain multiple amphipathic β-strands, and the α domains contain multiple α-helices (20, 21); the globular βα1 domain is a mixture of amphipathic β-strands and α-helices (21). A recent study on the structure of solubilized human apoB100 (22) is consistent with the proposed pentapartite model for apoB100 (19). The βα1 domain is the N-terminal 1000 amino acid residues of the mature apoB100 without the signal peptide and is referred to as apoB:1000 throughout.
FIGURE 1.
A, schematic diagram of the pentapartite structural model, NH2-βα1-β1-α2-β2-α3-COOH, for apoB100. B, ribbons diagram of the apoB:1000 model, including the modeled helix-loop-helix region (residues 700–744) and the four putative salt bridges. C, schematic diagram of local sequence homology between the first 1000 residues of APOB and the first 1020 residues of lamprey LV using the alignment program MACAW. The red boxes represent local blocks of sequence conservation. D, schematic diagram of structural conservation between the crystal structure of lamprey LV and the predicted secondary structure of APOB using the program LOCATE. Areas that have amphipathic structural conservation are highlighted by the yellow boxes.
Sequence analyses and intron/exon boundary identification of apoB indicated that the N-terminal βα1 domain of apoB is homologous to the lipovitellins (23, 24). Lipovitellins are major lipid transport proteins of egg-laying species, transporting lipids from the liver to the oocytes. The crystal structure of silver lamprey lipovitellin (LV) has been determined at 2.8 Å resolution and reveals a β-barrel structure (βC-region) on the apex of a triangular-shaped cavity (25–27). The cavity is bounded by three β-sheets (βA, βB, and βD) that are proposed to accommodate lipids within the cavity or lipid pocket. Based on the homology between LV and the βα1 domain of apoB100, we proposed (28, 29) that this domain might be a lipid-binding pocket for apoB (28). The apoB “lipid pocket” model (Fig. 1B) is based upon the highly significant sequence homology/similarity/identity (20.1% identity and 39.6% similarity) between the βα1 domain of apoB and the N-terminal region of lipovitellin (30). Any z-score (probability) <0.1 is strong evidence of homology, and all homologous proteins share regions of similar tertiary structure. Because human apoB has a z-score of 8.6 × 10-10 when compared with lamprey LV (28), it is almost certain that both proteins share major structural similarities within these homologous regions. Because the homology of apoB did not extend to the βD-sheet of LV, it was originally suggested on the basis of the presence of a pronounced cluster of amphipathic β-strands identified by the program LOCATE that microsomal triglyceride transfer protein (MTP) might function as the surrogate βD domain, closing the base of the lipid pocket (29). MTP has been proposed to act as a lipid transfer protein and is required for the secretion of apoB-containing lipoproteins (31). Alternatively, the initiation of particle assembly was proposed to occur when the βα1 domain folds into a three-sided LV-like lipid-binding cavity (29).
Subsequent experimentally derived results (32) demonstrated the following. (i) A portion, or perhaps all, of the amino acid residues between 931 and 1000 of apoB100 are necessary for the formation of a stable, lipid-containing nascent particle. (ii) The lipid pocket created by the first 1000 residues of apoB100 is PL-rich and has a maximum capacity on the order of 50 molecules of PL, 11 molecules of TAG, and 7 molecules of cholesteryl ester for a surface to core ratio of ∼ 3:1, a number similar to that observed for LV lipid pocket (27). These studies (32) also failed to show any measurable MTP in isolated apoB: 1000-containing particles and indicated that the first 1000 amino acid residues of apoB100 are competent to complete the lipid pocket without a structural requirement for MTP. Further support for the above results was provided by a structural model of apoB:1000 (30). The overall topology of the model (Fig. 1B) showed three unique structural domains in the first 1000 residues, i.e. β-barrel, α-helical bundle, and two β-sheets. Analysis of the model suggested a tandem series of four charged residues (Arg997, Glu998, Asp999, and Arg1000) located at the C-terminal end of the model form a portion of the base of the βB-sheet (30). On further analysis of the apoB:1000 sequence, a complementary tandem series of four charges (Glu720, His719, Lys718, and Asp717) was found in the unmodeled loop region (residues 670–745) located in the middle of the βA-sheet (30) (Fig. 1B). When these regions are modeled, four salt bridges can be formed from Arg997–Glu720, Glu998–His719, Asp999–Lys718, and Arg1000–Asp717 (Fig. 1B). We proposed that the salt bridges serve as a template to guide closure of the third side of the asymmetric pyramidal lipid pocket by the creation of a hairpin bridge that completes the nascent step of lipoprotein particle assembly (30).
The above studies raised two new questions. (i) What amino acid residues between 931 and 1000 of apoB100 are critical for the formation of a stable, lipid-containing nascent lipoprotein particle? (ii) Are the putative complementary salt bridges, formed between charged amino acid residues 717–720 and 997–1000, involved in the formation of a stable, lipid-containing lipoprotein particle? To address these questions, we truncated apoB between residues 1000 and 931 to produce apoB: 956, apoB:986, and apoB:996. Characterization of particles secreted by stable transformants of rat hepatoma McA-RH7777 cells demonstrated the following. (i) All of the amino acid residues between 931 and 1000 of apoB100 are necessary for the initiation of apoB-containing particle assembly and formation of stable HDL3-like lipoproteins, and (ii) amino acid residues 997–1000 may be important in this process.
EXPERIMENTAL PROCEDURES
Materials—Horse serum, antibiotic-antimycotic, and Tris-glycine gels were obtained from Invitrogen. Dulbecco's modified Eagle's medium (DMEM), minimum essential medium, trypsin, and G418 were purchased from Mediatech, Inc. (Herndon, VA). Fetal bovine serum, sodium deoxycholate, Triton X-100, benzamidine, phenylmethylsulfonyl fluoride, leupeptin, aprotinin, pepstatin A, brefeldin A (BFA), lactacystin, N-acetyl-leucyl-leucyl-norleucinal (ALLN), dimethyl sulfoxide (Me2SO), and fatty acid-free bovine serum albumin (BSA) were from Sigma. Protein G-Sepharose CL-4B, [3H]glycerol, [14C]oleic acid, and Amplify were from Amersham Biosciences. Tran35S-label, [35S]methionine/cysteine ([35S]Met/Cys), was from MP Biomedicals (Irvine, CA). Immobilon polyvinylidene difluoride transfer membrane and Centriprep centrifugal filter device YM-30 was purchased from Millipore Corp. (Bedford, MA). Affinity-purified polyclonal antibody to human apoB100 was prepared in our laboratory and was biotinylated at Brookwood Biomedical (Birmingham, AL). The apoB100 cDNA was a gift from Dr. Zemin Yao (University of Ottawa Heart Institute, Ottawa, Ontario, Canada).
Construction of Truncated ApoB Expression Plasmid—The wild-type truncated forms of apoB, including apoB:800, apoB: 931, apoB:956, apoB:986, apoB:996, and apoB:1000, denoting amino acid residues 1–800, 1–931, 1–956, 1–986, 1–996, and 1–1000, respectively, of the mature protein lacking the signal peptide were generated as described previously in detail (33). Truncated apoB cDNAs spanning nucleotides 1–2481 (apoB: 800), 1–2874 (apoB:931), and 1–3081 (apoB:1000) of the full-length apoB100 cDNA were prepared from pB100L-L (34) as a PCR template and appropriate C terminus primers as described previously in detail (33). The new truncated apoB cDNAs spanning nucleotides 1–2949 (apoB:956), 1–3039 (apoB:986), and 1–3069 (apoB:996) of the full-length apoB100 cDNA were prepared using the following C-terminal primers: apoB:956 (5′-CTAGTTGGAGTAAGCGCCTGAGGTGCAG-3′), apoB:986 (5′-CTACTGCTCAATCTCTCCTGTAGGCCTC-3′), and apoB:996 (5′-CTACTGGAGCTCATAGGTTGCGCTGAC-3′) as described previously (33). Standard cloning procedures were used to identify clones with 100% correct sequence as described previously (33). The apoB fragments were ligated into the mammalian expression vector, the Moloney murine leukemia virus-based retrovirus LNCX (35), and expression vectors were used for transformation. Clones harboring plasmids containing apoB gene with the correct orientation were identified by restriction enzyme digestion and confirmed by nucleotide sequencing and used to transfect McA-RH7777 cells as described previously (33).
Site-directed Mutagenesis—To test the hairpin-bridge hypothesis further, we made the following two mutants in addition to apoB:996: 1) apoB:1000 with charged amino acid residues 717–720 deletion (designated as apoB:1000Δ717–720) and 2) apoB:1000 with alanine substitution for charged amino acid residues 997–1000 (designated as apoB:1000 R997A, E998A, D999A, R1000A); for simplicity, we will refer to this mutant as apoB:996 + 4Ala. Amino acid additions and deletions were introduced by Quick Change® XL site-directed mutagenesis kit (Stratagene, catalogue number 200516) according to the manufacturer's protocol using pLNB:1000 and pLNB:996 as templates and the following primers. The primers for generation of apoB:1000Δ717–720 mutant were as follows: sense strand (5′-TCTTAGTGGACCACTTTGGCTATACCAAAGATCAGGATATGGTAAATGG-3′) and antisense strand (5′-CCATTTACCATATCCTGATCTTTGGTATAGCCAAAGTGGTCCACTAAGA-3′). The primers for generation of apoB:996 + 4Ala were as follows: sense strand (5′-CAACCTATGAGCTCCAGGCAGCAGCAGCATAGAAGGGCGAATTAGC-3′) and antisense strand (5′-GCTAATTCGCCCTTCTATGCTGCTGCTGCCTGGAGCTCATAGGTTG-3′). The entire open reading frames of apoB inserts were sequenced to confirm the occurrence of the desired mutations and the fidelity of the rest of the sequence. The apoB expression vectors, pLNCB:1000 Δ717–720 and pLNCB:996 + 4Ala with 100% correct sequence, were used to transfect McA-RH7777 cells as described previously (33).
Cell Culture and Transfection—Rat hepatoma McA-RH7777 (referred to as McA-RH herein) cells were obtained from American Type Culture Collection (Manassas, VA). Clonal stable transformants of McA-RH cells expressing apoB:800, apoB: 931, apoB:956, apoB:986, apoB:996, apoB:1000, apoB: 1000Δ717–720, and apoB:996 + 4Ala were generated as described previously in detail (33). Cells were grown in DMEM containing 20% horse serum, 5% fetal bovine serum, and 0.2 mg/ml G418n and medium was changed every 48 h. All experiments were conducted with 3–4-day-old cells as described previously (33).
De Novo Synthesis and Secretion of Truncated Forms of ApoB—Clonal stable transformants of McA-RH cells expressing apoB: 800, apoB:931, apoB:956, apoB:986, apoB:996, apoB:1000, apoB:1000Δ717–720, and apoB:996 + 4Ala were grown for 3–4 days in 6-well dishes. At the start of the experiments, maintenance media were removed, and monolayers were washed twice with phosphate-buffered saline (PBS), and serum-, methionine- and cysteine-free DMEM was added. The incorporation of [35S]Met/Cys (50 μCi/ml of medium) into newly synthesized truncated forms of apoB was determined after 3.5 h or overnight incubation, as indicated in each experiment. After the indicated incubation time, the 35S-labeled conditioned medium was collected, and preservative mixture at a final concentration of 500 units/ml penicillin-G, 50 μg/ml streptomycin sulfate, 20 μg/ml chloramphenicol, 1.3 mg/ml ε-aminocaproic acid, 1 mg/ml EDTA, 1 mm benzamidine, and 1 mm phenylmethylsulfonyl fluoride was added to prevent oxidative and proteolytic damage. The medium was centrifuged at 2,000 rpm for 30 min at 4 °C to remove broken cells and debris. Cell monolayer was washed with cold PBS, and lysis buffer containing preservative mixture described above plus leupeptin (50 μg/ml), pepstatin A (50 μg/ml), and aprotinin (100 kallikrein-inactivating units/ml) were added, and cells were processed as described previously (33). The 35S-labeled apoB in the conditioned medium and cell lysate was isolated by immunoprecipitation as described below.
Pulse-Chase Analysis and Immunoprecipitation—Cells were grown in 35-mm dishes for 4 days under conditions described above. At the start of each experiment, maintenance medium was removed, and cells were preincubated with serum- and methionine/cysteine-free DMEM for 1 h. Cells were then pulsed for 10 min in fresh preincubation medium containing [35S]Met/Cys (100 μCi/ml). Chase was initiated by removing the pulse medium followed by washing the cell monolayer twice with cold PBS and incubation in serum-free DMEM containing 10 mm unlabeled methionine and cysteine. At 0, 20, 40, 60, 90, 120, 150, and 180 min of chase, media were collected and supplemented with preservative mixture described above. Cell monolayer was washed with cold PBS, and lysis buffer containing preservatives was added as above, and cell lysate and medium were processed as described above.
In experiments designed to determine the role of proteasomal pathway in the intracellular degradation of truncated forms of apoB, lactacystin, ALLN, and brefeldin A were dissolved in water, dimethyl sulfoxide (Me2SO), and ethanol, respectively. Final concentrations of Me2SO and ethanol in all control and experimental dishes were 1 and 0.1%, respectively. Aliquots of medium and cell lysate were immunoprecipitated using monospecific polyclonal antibody to human apoB100 coupled to protein G-Sepharose CL-4B as described previously (32, 36). The 35S-labeled proteins were extracted from protein G (32, 36) and resolved on 4–12% SDS-PAGE (37). After electrophoresis, the gels were analyzed by autoradiography or immunoblotting as described below and in the figure legends. The relative intensities of the bands corresponding to truncated proteins were determined by computer-assisted image processing.
Metabolic Labeling of the Lipid Component of ApoB-containing Lipoprotein Particles—Cells were grown for 4 days under culture conditions described above. At the start of experiments, maintenance medium was removed; cell monolayer was washed twice with PBS, and serum-free DMEM containing [3H]glycerol (7 μCi/ml of medium) was added. In experiments where the radiolabeled lipids associated with the particles was determined by autoradiography, cells were labeled with both [3H]glycerol and 0.4 mm [14C]oleic acid bound to 0.75% BSA to enhance the signal. After overnight (17–20 h) incubation, labeled conditioned medium was collected and processed as described above. The incorporation of [3H]glycerol into various lipid moieties of secreted apoB-containing lipoproteins was determined by nondenaturing gradient gel electrophoresis (NDGGE) of concentrated labeled conditioned medium as described below. Cell monolayer was washed with cold PBS, scraped off the plate in PBS, and sonicated for determination of protein content by the method of Lowry et al. (38).
Lipoprotein Isolation—After overnight (17–20 h) incubation of cells with unlabeled serum-free DMEM (6 ml per 100-mm dish) or serum-free DMEM containing either [35S]Met/Cys or [3H]glycerol/[14C]oleate, conditioned medium was collected and processed as described above. The density of the conditioned medium was adjusted to either 1.23 or 1.25 g/ml using solid KBr, and lipoproteins (d < 1.23 g/ml or d < 1.25 g/ml) were isolated by centrifugation for 45 h at 50,000 rpm. The lipoprotein fractions and infranatants (d > 1.23 g/ml or d > 1.25 g/ml) were dialyzed against PBS (supplemented with preservative mixture described above) and were concentrated 5- and 10-fold, respectively, using Centriprep YM-30. The concentrated conditioned medium and lipoprotein fractions were analyzed by NDGGE, immunoblotting, and lipid analysis.
Density Gradient Ultracentrifugation—After overnight incubation with serum-free DMEM, conditioned medium from 10 100-mm dishes (for apoB:931, apoB:986, and apoB:1000) and 20 100-mm dishes (for apoB:956 and apoB:996) was collected and concentrated 10- and 20-fold, respectively. The density of 10 ml of concentrated conditioned medium was adjusted to 1.36 g/ml with solid KBr, overlayered with 12 ml of d = 1.26 g/ml and 18 ml of d = 1.06 g/ml KBr solutions containing preservative mixture and centrifuged for 6 h at 70,000 rpm at 7 °C (39). Forty fractions of 1.0 ml each were collected from the bottom of the centrifuge tube, and after measuring their densities were analyzed by NDGGE and immunoblotting.
NDGGE—ApoB-containing lipoprotein particles in the conditioned medium and lipoprotein fractions were run on 4–20% NDGGE; gels were stained and the bands corresponding to truncated forms of apoB, identified by immunoblotting of a duplicate gel, were excised and analyzed for proteins and lipids as described below. The incorporation of 3H-labeled glycerol and [14C]oleate into the total lipid content of intact apoB-containing lipoproteins was also determined by NDGGE of the labeled conditioned medium and lipoproteins isolated therefrom.
Lipid Analysis of Isolated Truncated ApoB-containing Particles—The bands corresponding to labeled truncated apoB-containing particles were excised from NDGGE, and lipids were extracted with chloroform/methanol (2:1) as described previously in detail (32); complete extraction was assessed by liquid scintillation counting the final gel homogenate. Total labeled lipids associated with gel-isolated apoB-containing lipoproteins were washed by the Folch method (40) and applied to thin layer chromatography (TLC) plates as described previously (41). The bands corresponding to PL, diacylglycerols (DAG), and TAG, identified by comparison to known standards, were visualized with iodine; each band was scraped off the plate, placed in a counting vial, and quantified by liquid scintillation counting.
Immunoblot Analysis—The isolated apoB-containing particles were run on 4–12% SDS-PAGE (37) or on 4–20% NDGGE. After electrophoresis, proteins were detected by Western blot analysis (42) using biotinylated antibody to human apoB100 as described previously (33).
Calculation of Number of Lipid Molecules per Particle—Calculations were made essentially as described by Carraway et al. (43) and reported previously (32).
RESULTS
Strategy for Testing the Relationship of the Local Sequence Homologies of Residues 931–1000 of ApoB and LV and ApoB Particle Assembly—Previously, a homology search using BLASTp indicated that the first 1000 residues of human apoB (hAPOB) are homologous to the first 1075 amino acids of lamprey LV (30). Two alignment programs, ClustalW and MACAW, were employed to create the best alignment possible (30). ClustalW (default settings), which provided an alignment over entire sequences of apoB and LV, showed that the alignment was highly conserved over the first 620 residues of apoB (30). MACAW analysis proved pivotal in determining blocks of locally conserved regions between sequences with homology gaps (28, 30). Multiple local alignments of sequence blocks between hAPOB and vertebrate LV precursors (vLV), performed using the program MACAW (28), identified four homologue domains (HD) for vLV precursors and hAPOB (vLV:B): HD-vLV:B-I (residues 1–180), HD-vLV:B-II (residues 450–680), HD-vLV:B-III (residues 710–860), and HD-vLV: B-IV (residues 980–1020 for vLV and residues 925–986 for hAPOB) (28). A schematic diagram of these homologue domains is shown in Fig. 1C.
We then used the results of the multiple alignments to map the known secondary structure of lamprey LV onto the amphipathic secondary structure predicted for hAPOB (28) as follows. HD I:1–180 maps to the following: (i) the βC:LV antiparallel β-sheet region of lamprey LV, residues 1–183, and (ii) a corresponding cluster of amphipathic β-strands in hAPOB (residues 16–186, βC:B). HD II:450–680, maps in its C-terminal portion to the following: (i) the first of the two separate sequences that make up the βA:LV antiparallel β-sheet domain of lamprey LV, residues 584–672, and (ii) a corresponding well defined cluster of amphipathic β-strands in hAPOB (residues 615–676, βA:B). HD III:710–860, maps to the following: (i) the first of the two separate sequences that make up the βB:LV antiparallel β-sheet domain of lamprey LV, residues 774–932, and (ii) a corresponding, very well defined cluster of amphipathic β-strands in hAPOB (residues 808–897, βB:B). Finally, HD IV:980–1020 maps to the following: (i) the second of the two separate sequences that make up the βB:LV antiparallel β-sheet domain of lamprey LV, residues 989–1057, and (ii) a corresponding cluster of amphipathic β-strands in hAPOB (residues 967–996, βB:B) (28). A schematic diagram of these homologue domains is shown in Fig. 1D.
To test the relationship of the local sequence homologies between βB:LV and human apoB (βB:B) (Fig. 1A) and the formation of a stable lipid-containing particle, apoB was truncated between residues 1000 and 900 to produce apoB:986 and apoB: 956 (half-way between residues 925 and 986). Expression plasmids encoding apoB:956 (N-terminal 21.08% of apoB100) and apoB:986 (N-terminal 21.74% of apoB100) (Fig. 2A) were used to generate clonal stable transformants of McA-RH cells as described previously (32). The properties of secreted particles containing the above truncated forms of apoB and the previously established (32) apoB:931 (N-terminal 20.52% of apoB100) were compared with apoB:1000 (N-terminal 22.05% of apoB100)-containing particles to test the potential role of short sequences between residues 931 and 1000 in the formation of apoB lipid pocket.
FIGURE 2.
Location and stable expression of truncated apoB constructs terminating at the junction of βα1 and β1 domains. A, schematic diagram showing the location of the C-terminal truncation sites relative to the LV homologue domains. The domains of apoB homologues to LV, βC:B, α:B, βA:B, and βB:B are shown as solid black boxes. The sequences outside the boxes (e.g. residues 300–450) may have structural similarity to LV but are not homologous. The location of the LV domains, βC:LV, α:LV, βA:LV, and βB:LV, is indicated below the sequence. The regions between the two separate βA:B and βB:B domains are not seen in the crystal structure and are presumably mobile. Expression plasmids encoding truncated forms of apoB were constructed as described under “Experimental Procedures.” ApoB:800, apoB:931, apoB:956, apoB:986, apoB:996, and apoB:1000 denote amino acid residues 1–800, 1–931, 1–956, 1–986, 1–996, and 1–1000, respectively, of the mature protein lacking the signal sequence. Clonal cell lines of McA-RH stably expressing the above truncated forms of apoB were incubated for 3 h in serum-, methionine-, and cysteine-free DMEM containing [35S]Met/Cys (50 μCi/ml of medium). The 35S-labeled truncated apoB proteins in the cell lysate and secreted into the conditioned medium (B) of LNCX (neo)-transfected cells (lane 1), apoB:800- (lane 2), apoB:931- (lane 3), apoB:956- (lane 4), apoB:986- (lane 5), apoB:996- (lane 6), and apoB:1000- (lane 7) expressing cells were immunoprecipitated with anti-human apoB100. Extracted proteins were resolved on 4–12% SDS-PAGE and visualized by autoradiography. C, the intact 35S-labeled apoB-containing particles in the conditioned medium were separated by 4–20% NDGGE for 48 h at 4 °C; gels were dried, and intact particles were visualized by autoradiography. Results are representative of five separate experiments.
Strategy for Testing the Importance of Four Putative Salt Bridges in the Formation of Stable ApoB Lipid Pocket—The structural model for apoB lipid pocket suggests a hairpin-bridge mechanism for lipid pocket completion without the structural requirement for MTP (30). We have made the conjecture that salt bridges are formed between the four tandem charged residues 717–720 in the turn of the hairpin bridge and four tandem complementary residues 997–1000 located at the C-terminal end of the model (30). To test the potential role of these putative salt bridges in the formation of the lipid pocket, residues 997–1000 were deleted to produce apoB:996. As a more stringent test of the hairpin-bridge hypothesis, we made the following additional mutants: 1) apoB:1000 with charged amino acid residues 717–720 deletion (designated as apoB:1000Δ717–720) and 2) apoB:1000 with alanine substitution for charged amino acid residues 997–1000 (designated as apoB:1000 R997A, E998A, D999A, R1000A); for simplicity, we will refer to this mutant as apoB:996 + 4Ala. Expression plasmids encoding apoB:996 (N-terminal 21.96% of apoB100), apoB:1000Δ717–720, and apoB:996 + 4Ala were used to generate clonal stable transformants of McA-RH cells as described previously (32). The properties of secreted particles containing the above mutated forms of apoB truncates were compared with those of apoB:1000-containing particles.
Expression and Secretion of C-terminally Truncated Forms of ApoB—Stable transformants of McA-RH cells were incubated for 3.5 h with serum-, methionine-, and cysteine-free DMEM containing [35S]Met/Cys (50 μCi/ml of medium). Aliquots of cell lysate and conditioned medium were immunoprecipitated using monospecific polyclonal antibody to human apoB-100, and 35S-labeled proteins were separated by 4–12% SDS-PAGE and were analyzed by autoradiography. Results demonstrated the presence of 35S-labeled apoB in the cell lysate and conditioned medium (Fig. 2B) with the expected molecular mass of 92,000 Da for apoB:800 (lane 2), 107,065 Da for apoB:931 (lane 3), 109,940 Da for apoB:956 (lane 4), 113,390 Da for apoB:986 (lane 5), 114,540 Da for apoB:996 (lane 6), and 115,000 Da for apoB:1000 (lane 7). Consistent with our previous studies (32, 33), apoB:800 (Fig. 2B, lane 2) was expressed and secreted at a significantly lower rate compared with the other five constructs. Interestingly, apoB:996 secretion (Fig. 2B, lane 6) was also markedly lower than that of apoB:986 (Fig. 2B, lane 5) and apoB:1000 (Fig. 2B, lane 7). Analysis of 35S-labeled conditioned medium by NDGGE and autoradiography demonstrated that both apoB:931 (Fig. 2C, lane 3) and apoB:956 (Fig. 2C, lane 4) had very low capacity to form intact 35S-labeled particles with an Sd of ∼110 Å. In comparison, a substantially larger proportion of secreted apoB:986 was in the form of particles with an Sd of ∼110 Å (Fig. 2C, lane 5). The capacity of apoB:996 to form particles with a Sd of ∼112 Å (Fig. 2C, lane 6) was markedly lower than that of apoB:1000 (Fig. 2C, lane 7).
All Amino Acid Residues between 931 and 1000 of ApoB100 Are Necessary for the Formation of a Lipidated Particle within the HDL3 Density Range, and Residues 997–1000 Appear to Be Important for This Process—To determine the effect of residues 931–1000 on the density of the secreted apoB-containing particles, unlabeled conditioned medium was collected and concentrated. The density of the concentrated conditioned medium was adjusted to either d = 1.23 g/ml or d = 1.25 g/ml, and d < 1.23 g/ml, d < 1.25 g/ml, d > 1.23 g/ml, and d > 1.25 g/ml fractions were isolated by ultracentrifugation as described under “Experimental Procedures.” All fractions were analyzed by NDGGE and immunoblotted with anti-human apoB100 as described previously (32). Significant differences were observed in the floatation properties of the secreted apoB-containing particles. As shown in Fig. 3, only a minor fraction of the major particles (indicated by an arrow) formed by apoB:931, apoB: 956, and apoB:986 floated at d = 1.23 g/ml (lane T, top fraction), and the majority was recovered in d > 1.23 g/ml (lane B, bottom fraction) indicating that they are lipid-poor. In sharp contrast, a significant proportion of the major apoB:1000-containing particles (Sd of ∼112 Å) floated at d = 1.23 g/ml (Fig. 3) indicating that these particles are relatively lipid-rich. Compared with apoB:1000, after normalization for the volumes of the fractions, a significantly smaller proportion of apoB:996-containing particles floated at d = 1.23 g/ml. When the density of the conditioned medium was adjusted to 1.25 g/ml, a larger proportion of apoB:931-, apoB:956-, apoB:986-, and apoB:996-containing particles floated (Fig. 3).
FIGURE 3.
Floatation properties of the secreted apoB-containing particles. Stable transformants of McA-RH cells expressing various truncated forms of apoB were incubated with serum-free DMEM for 20 h. The density of 10 ml of conditioned medium of apoB:931-, apoB:986-, and apoB:1000-expressing cells and 10 ml of 2-fold concentrated conditioned medium of apoB:956- and apoB:996-expressing cells was adjusted to either 1.23 or 1.25 g/ml using solid KBr, and lipoprotein fractions (d < 1.23 g/ml or d < 1.25 g/ml) were isolated by centrifugation for 45 h at 50,000 rpm. The lipoprotein fractions and infranatants (d > 1.23 g/ml or d > 1.25 g/ml) were dialyzed against PBS and were concentrated 5- and 10-fold, respectively. The concentrated conditioned medium (TM), lipoprotein fractions (T), and infranatants (B) were analyzed by 4–20% NDGGE and immunoblotting using polyclonal antibody to human apoB100. Results are representative of four separate experiments.
The density gradient distribution of the particles containing various truncated forms of apoB is shown in Fig. 4. All 40 fractions isolated by density gradient ultracentrifugation of unlabeled concentrated conditioned media were subjected to NDGGE and analyzed by immunoblotting with anti-human apoB100. We previously demonstrated (33) that the major apoB:931-containing particle (with a Sd of 110 Å) had a peak hydrated density of 1.25 g/ml, which is denser than the traditional HDL3, whereas apoB:1000 formed HDL3-like particles with a peak hydrated density of 1.208 g/ml (also shown in Fig. 4). Here the effect of sequential addition of local sequences in the second βB domain of apoB (residues 925–986) on the ability of the truncated apoB to form stable particles with HDL3-like hydrated density was determined. The larger particle (Sd ∼ 110 Å) formed by apoB:956 had a peak hydrated density of 1.24 g/ml (Fig. 4). ApoB:986 formed two major particles, the larger particle (Sd ∼ 111 Å) had a peak hydrated density of 1.23 g/ml, and the smaller particle had a peak hydrated density of 1.28 g/ml (Fig. 4).
FIGURE 4.
Density gradient ultracentrifugation distribution of apoB-containing particles secreted by McA-RH cells. Stable transformants of McA-RH cells expressing various truncated forms of apoB were incubated with serum-free DMEM for 20 h. Medium from 10 100-mm dishes (10 ml medium/dish) for apoB:986 and apoB:1000 and 20 dishes for apoB:956 and apoB:996 were collected; preservative mixture was added, and medium was concentrated 10-fold for apoB:986 and apoB:1000 and 20-fold for apoB:956 and apoB:996. The density of 10 ml of concentrated conditioned medium was adjusted to 1.36 g/ml with solid KBr, overlayered with 12 ml of d = 1.26 g/ml and 18 ml of d = 1.06 g/ml KBr solutions, and centrifuged for 6 h at 70,000 rpm at 7 °C (39). Forty fractions of 1.0 ml each were collected from the bottom of the centrifuge tube, and after measuring their densities were analyzed by NDGGE and immunoblotting with polyclonal antibody to human apoB100. Results are representative of three separate experiments.
Similarly to apoB:1000, the large particle (Fig. 4, indicated by an arrow) formed by apoB:996 had a diameter that remained relatively constant at ∼112 Å across a wide range of densities and a peak hydrated density of 1.21 g/ml. Interestingly, lipoproteins formed by apoB:996 appeared to have a wider density range than those formed by apoB:986 and apoB:1000 (Fig. 4). We have provided a plausible explanation for this observation under the “Discussion.” The above results indicate the following: (i) apoB:931, apoB:956, and apoB:986 lack the structural elements necessary for the formation of a stable particle in the traditional HDL3 density range of 1.125–1.21 g/ml, (ii) deletion of residues 997–1000 diminishes the ability of the majority of the resulting apoB:996 to form monodisperse stable particles in the HDL3-like density range.
Analysis of Metabolically Labeled Lipids Associated with Secreted Particles Containing Truncated Forms of ApoB100 Suggests That Amino Acid Residues 997–1000 Might Be Critical for the Assembly of Stable Lipidated Nascent Particles and Prevention of the Formation of Large Lipid-rich Aggregates—Cells were metabolically labeled with [3H]glycerol/[14C]oleic acid (0.4 mm bound to 0.75% BSA), and aliquots of concentrated conditioned medium, normalized for the apoB expression level, were subjected to 4–20% NDGGE. Western blot analysis demonstrated that apoB:931 (Fig. 5A, lane 1) and apoB:956 (Fig. 5A, lane 2) had very low capacity to form stable particles (p). Neither apoB:931-containing particles (Fig. 5B, lane 1) nor apoB: 956-containing particles (Fig. 5B, lane 2) contained any detectable lipids. Instead, nearly all the lipid lipids were associated with the large aggregates (a). ApoB:986 formed both a distinct particle of ∼110 Å (Fig. 5A, lane 3), which contained lipids (Fig. 5B, lane 3), and large lipid-rich aggregates. In sharp contrast to apoB-931, apoB:956, and apoB:986, apoB:1000 had a very high capacity to form intact particles (Fig. 5A, lane 5) which also contained nearly all the labeled lipids (Fig. 5B, lane 5). The lipid to protein ratios of intact particles (p) and large lipid-rich aggregates (a) on top of each lane are described below.
FIGURE 5.
Amino acid residues 997–1000 are critical for the formation of a stable lipid-containing lipoprotein particle. Stable transformants of McA-RH cells expressing apoB:931 (lane 1), apoB:956 (lane 2), apoB:986 (lane 3), apoB:996 (lane 4), and apoB:1000 (lane 5) were incubated for 20 h in serum-free DMEM containing [3H]glycerol/[14C]oleic acid (0.4 mm bound to 0.75% BSA). Aliquots of concentrated conditioned medium, normalized for the apoB expression level, were subjected to 4–20% NDGGE. Gels were run in duplicate; proteins were detected by immunoblotting with polyclonal antibody to human apoB100 (A), and a duplicate gel was analyzed for labeled lipids in apoB-containing lipoproteins by autoradiography (B). C, cells were incubated for 20 h in serum-free DMEM containing [35S]Met/Cys (100 μCi/ml of medium). The labeled conditioned medium was concentrated and analyzed by 4–20% NDGGE; gels were run in duplicate. One gel was subjected to autoradiography (C); particles (p) and aggregates (a) were excised from the second stained gel. 35S-Labeled proteins were extracted from particles and aggregates as described under “Experimental Procedures” and resolved by SDS-PAGE (D). Proteins and lipids were visualized by autoradiography of the gels, and the intensities of the labeled proteins and lipids were determined by computer-assisted image processing. Results are representative of five separate experiments.
Based on the apoB:1000 model (30), four salt bridges can be formed from the following: Arg997–Glu720, Glu998–His719, Asp999–Lys718, and Arg1000–Asp717 (Fig. 1B). We proposed that the salt bridges serve as a template to guide closure of the third side of the asymmetric pyramidal lipid pocket by the creation of a hairpin bridge that completes the nascent step of lipoprotein particle assembly (30). If this model is correct, deletion of amino acid residues 997–1000 should retard the complete lipidation of the lipid pocket and induce instability in the nascent particle. To test this hypothesis, the incorporation of [3H]glycerol/[14C]oleic acid (0.4 mm bound to 0.75% BSA) into the lipids associated with secreted apoB:996-containing intact particles was determined. Two distinct differences were observed between apoB:996 and apoB:1000 as follows. 1) Compared with apoB:1000 (Fig. 5, A and B, lane 5), apoB:996 formed considerably less distinct lipid-containing particles with similar Sd (Fig. 5, A and B, lane 4). 2) In contrast to apoB:1000 (Fig. 5B, lane 5), a substantial band of radioactivity was detected at the top of the gel for apoB:996 (Fig. 5B, lane 4) suggestive of lipid-rich aggregate formation. Thus, a unique feature of apoB:1000 is that it does not have the propensity to form large lipid-rich aggregates, and labeled lipids are detected essentially only in the particles with Sd ∼ 112 Å, and that this property is lost by deleting amino acid residues 997–1000.
Determination of Relative Distribution of Truncated Forms of ApoB between Intact Particle and Large Aggregate Reveals a Unique Property of ApoB:1000—To determine the relative concentration of the truncated apoB in the particles and aggregates, cells were metabolically labeled with [35S]Met/Cys. The labeled conditioned medium was concentrated and subjected to 4–20% NDGGE immediately as described above and analyzed by autoradiography (Fig. 5C). The bands corresponding to intact particles (p) and the aggregates (a) on top of the gel were excised from a duplicate gel. Proteins were extracted from the gels as described previously (32), resolved by SDS-PAGE, and visualized by autoradiography (Fig. 5D). Results shown in Fig. 5D demonstrated that in both apoB:931 (Fig. 5C, lane 1) and apoB:956 (Fig. 5C, lane 2), the majority of the protein was associated with the large lipid-rich aggregates. In apoB:986, a large proportion of protein was associated with the particle (p) and a relatively smaller proportion was in the large lipid-rich aggregate (Fig. 5D). In apoB:996, the protein appeared to be almost equally distributed between the particles and the large aggregate (Fig. 5D). It is noteworthy that when the concentrated 35S-labeled conditioned medium of apoB:996-expressing cells was subjected to NDGGE 2 days after collection, a larger proportion of the protein was recovered in the aggregate (data not shown). ApoB:1000 was clearly different from all other truncated forms of apoB in that essentially all the apoB:1000 protein was recovered in the major particle (Fig. 5D), which also contained nearly all the labeled lipids (Fig. 5B, lane 5). No major changes were observed in the properties of apoB:1000 upon storage at 4 °C for up to 2 weeks.
Lipid Composition of Secreted Particles Containing Truncated Forms of ApoB—From the above results it would appear that apoB:986 has the capacity to form a stable lipidated particle, similar to that of apoB:1000, in addition to a large lipid-rich aggregate. To evaluate the difference between apoB:986 and apoB:1000, labeled conditioned medium was analyzed by NDGGE immediately after collection. The intensities of 3H-/14C-labeled lipids and 35S-labeled proteins in the major particles with Sd of ∼110–112 Å, shown in Fig. 5, B and C, respectively, were determined by computer-assisted image processing of the autoradiogram. In three separate experiments, the ratio of 35S-labeled apoB:986 to apoB:1000 was 0.65 and 35S-labeled apoB:996 to apoB:1000 was 0.076. The corresponding 3H-/14C-lipids were 0.49 and 0.075. These results indicate the following: 1) each apoB:986-containing particle contains ∼25% less lipids than the apoB:1000-containing particle, and 2) when analyzed immediately after secretion, the small proportion apoB:996 that forms an intact particle with an Sd of ∼112 Å has similar amount of lipids to that in apoB:1000-containing particles.
To determine the stoichiometry of the lipid component of the truncated apoB-containing particles, cells were incubated overnight with serum-free DMEM containing 3H-labeled glycerol with or without 0.4 mm unlabeled oleic acid bound to 0.75% BSA. The density of the conditioned media was adjusted to d = 1.25 g/ml with KBr, and the d < 1.25 g/ml fraction was isolated by centrifugation for 45 h at 50,000 rpm. The lipoprotein fractions were dialyzed, concentrated, and subjected to NDGGE as described under “Experimental Procedures.” Bands corresponding to individual apoB-containing particles, identified by immunoblotting of a duplicate gel and their Sd, were excised, and labeled lipids were extracted as described previously (32). In the absence of oleic acid, apoB:986-containing particles contained 84% PL, 5% DAG, and 10% TAG, and this composition was not altered by oleic acid (Table 1). The calculated stoichiometries of PL, DAG, and TAG per apoB:986 were 50, 4, and 6, respectively, for the total of 60 lipid molecules per particle (Table 1). In case of apoB:996, PL, DAG, and TAG accounted for 76, 12, and 12%, respectively, of the total labeled lipids associated with the secreted particles, and this composition was not significantly altered by oleic acid (Table 1). The calculated numbers of PL, DAG, and TAG per apoB:996-containing particle were 50, 10, and 7, respectively. Thus, the lipid composition and stoichiometry of a small proportion of apoB:996 that formed intact particles were similar to that obtained for apoB:1000-containing particles shown in Table 1 and reported previously (32), provided that analysis was performed on fresh conditioned medium of apoB:996-expressing McA-RH cells.
TABLE 1.
Composition of [3H]glycerol-labeled lipids associated with apoB:986- and apoB:996-containing lipoproteins secreted by McA-RH7777 cells and isolated by nondenaturing gradient gel electrophoresis Cells were incubated with serum-free DMEM with or without 0.4 mm oleic acid bound to 0.75% BSA, and [3H]glycerol-secreted particles were isolated by NDGGE. Values are means ± S.E. of seven separate experiments.
| Truncated ApoB | Addition | PL | DAG | TAG | Total lipid molecules | |
|---|---|---|---|---|---|---|
| % of total lipids | ||||||
| ApoB:986 | Control | 84.4 ± 1.4 | 5.2 ± 0.7 | 10.4 ± 0.9 | ||
| ApoB:986 | Oleic acid | 77.7 ± 2.2 | 7.5 ± 0.9 | 14.7 ± 1.6 | ||
| ApoB:996 | Control | 76.0 ± 2.8 | 11.7 ± 1.5 | 11.9 ± 1.4 | ||
| ApoB:996 | Oleic acid | 71.0 ± 2.2 | 11.7 ± 2.4 | 17.4 ± 1.6 | ||
| ApoB:1000 | Control | 74.0 ± 1.0 | 8.0 ± 0.4 | 18.0 ± 0.8 | ||
| ApoB:1000 | Oleic acid | 70.0 ± 3.0 | 8.0 ± 0.7 | 22.0 ± 3.0 | ||
| calculated no. lipid molecules/particle | ||||||
| ApoB:986 | Control | 51.0 ± 1.2 | 4.0 ± 0.6 | 6.0 ± 0.5 | 61.0 ± 0.4 | |
| ApoB:986 | Oleic acid | 46.0 ± 1.9 | 6.0 ± 0.6 | 8.0 ± 0.8 | 60.0 ± 0.6 | |
| ApoB:996 | Control | 51.0 ± 2.6 | 10.0 ± 1.1 | 7.0 ± 0.7 | 68.0 ± 0.8 | |
| ApoB:996 | Oleic acid | 45. ± 2.0 | 10.0 ± 1.9 | 10.0 ± 0.8 | 65.0 ± 0.7 | |
| ApoB:1000 | Control | 50.0 ± 1.0 | 7.0 ± 0.4 | 11.0 ± 2.0 | 69.0 ± 0.3 | |
| ApoB:1000 | Oleic acid | 48.0 ± 2.0 | 6.0 ± 2.0 | 14.3 ± 1.5 | 8.0 ± 1.3 | |
Deletion of Amino Acid Residues 997–1000 Renders the Resulting ApoB:996 Susceptible to Intracellular Degradation—ApoB is a constitutively synthesized protein (44). Regulation of apoB production is thought to occur largely by intracellular degradation (45–49), at both co- and post-translational levels (50). This regulatory mechanism of apoB production has also been observed in McA-RH cells (51, 52). Therefore, we performed pulse-chase experiments to determine whether low efficiency of apoB:996 secretion (Fig. 2B, lane 6), in comparison with apoB:1000 (Fig. 2B, lane 7), was due to its intracellular degradation. Because of the observed differences in the properties of apoB:931 and apoB:986 compared with those of apoB: 1000, their intracellular stability and secretion kinetics were also assessed for comparison. Cells were pulse-labeled with [35S]Met/Cys for 10 min, and the disappearance of labeled apoB from the cells and their subsequent accumulation in the conditioned medium were monitored during a 20–180-min chase. ApoB:931 (Fig. 6A) and apoB:986 (Fig. 6B) did not appear to undergo intracellular degradation because 85–90% of the labeled proteins that was lost from the cells at 120–150 min of chase was recovered in the medium. However, an interesting and consistent observation was a marked (∼45%) decrease in the recovery of 35S-labeled apoB:931 in the medium between 150 and 180 min of chase (Fig. 6A), which was not affected by the presence of oleic acid during the pulse-chase period (data not shown).
FIGURE 6.
Intracellular stability of apoB:931, apoB:986, and apoB:1000. Stable transformants of McA-RH cells expressing apoB:931 (A), apoB:986 (B), or apoB:1000 (C and D) were pulse-labeled for 10 min with serum-, methionine-, and cysteine-free DMEM containing [35S]Met/Cys (100 μCi/ml of medium). The pulse medium was removed, and cell monolayers were washed with cold PBS, and serum-free DMEM containing 10 mm each of unlabeled methionine and cysteine was added. At the indicated chase period, medium and cell lysate were collected, and 35S-labeled apoB:931, apoB:986, and apoB:1000 were isolated by immunoprecipitation with polyclonal antibody to human apoB100. Proteins were resolved by 4–12% SDS-PAGE and visualized by autoradiography. The intracellular stability of apoB:1000 was determined in the absence (C) or presence (D) of 0.4 mm oleic acid bound to 0.75% BSA, which was present during both the pulse and chase periods. The intensities of the labeled proteins were determined by computer-assisted image processing. The autoradiogram is representative of six samples from three separate experiments.
ApoB:1000 was highly stable, and the newly synthesized protein that disappeared from the cells at 180 min of chase was recovered almost quantitatively in the medium; only a small proportion (∼10%) was degraded intracellularly both in the absence (Fig. 6C) or presence (Fig. 6D) of oleic acid. In striking contrast, apoB:996 was highly susceptible to intracellular degradation. At 120 min of chase, 80% of 35S-labeled apoB:996 that disappeared from the cells was not recovered in the medium (Fig. 7A). Addition of oleic acid during both pulse and chase periods, which has been shown to attenuate the degradation of apoB (53, 54), failed to rescue apoB:996 from intracellular degradation (Fig. 7B).
FIGURE 7.
ApoB:996 is susceptible to intracellular degradation. Stable transformants of McA-RH expressing apoB:996 were pulse-labeled for 10 min with serum-, methionine-, and cysteine-free DMEM containing [35S]Met/Cys (100 μCi/ml of medium) in the absence (A) or presence (B) of 0.4 mm oleic acid bound to 0.75% BSA. The pulse medium was removed, cell monolayers were washed with cold PBS, serum-free DMEM containing 10 mm each of unlabeled methionine and cysteine was added, and incubation was continued in the absence (A) or presence (B) of oleic acid. At the indicated chase period, medium and cell lysates were collected, and 35S-labeled apoB:996 was isolated by immunoprecipitation with polyclonal antibody to human apoB100. Proteins were resolved by 4–12% SDS-PAGE and visualized by autoradiography. The intensities of the labeled proteins were determined by computer-assisted image processing. The autoradiogram is representative of six samples from three separate experiments. C, the effects of ALLN (100 μm), lactacystin (10 μm), and BFA (2 μg/ml) on the recovery of 35S-labeled apoB:996 in cells and medium was determined after a 3-h steady state incubation with 35S-labeled Met/Cys (100 μCi/ml of medium). Labeled apoB:996 in cells and medium was isolated by immunoprecipitation with polyclonal antibody to human apoB100. Proteins were resolved by 4–12% SDS-PAGE and visualized by autoradiography. The intensities of the labeled proteins were determined by computer-assisted image processing. Values are mean ± S.E. of three dishes representative of three separate experiments.
The known pathways involved in apoB degradation are ER-associated degradation, post-ER presecretory proteolysis, and receptor-mediated re-uptake from the cell surface (55, 56). The ER-associated degradation of apoB occurs through both ubiquitin-proteasome, which has been established in both HepG2 and McA-RH cells (57–61), and nonproteasomal pathways (61). Proteins that have abnormal conformation are retained in the ER (62) and could be targeted to the nonproteasomal pathway (61). To assess the mechanism of apoB:996 degradation, cells were incubated with [35S]Met/Cys for 3 h in the presence or absence of ALLN (100 μm) or lactacystin (10 μm), known inhibitors of proteasomal degradation (63). The 35S-labeled apoB:996 in media and cell lysates was determined by immunoprecipitation as described above. ALLN and lactacystin were not effective in protecting apoB:996 from intracellular degradation (Fig. 7C) indicating that the proteasomal pathway was not involved. To test if the nonproteasomal degradation of apoB:996 was ER-associated, pulse-chase experiments were carried out in the presence of BFA (2 μg/ml), a known inhibitor of ER to Golgi protein trafficking. If degradation occurred in the Golgi, then the total recovery of apoB: 996 (in the cells plus the medium) after 3 h of incubation should be higher in the presence of BFA. This was not the case, as the total level of 35S-labeled apoB:996 was actually 12% lower in BFA-treated cells compared with that in control cells (Fig. 7C). Furthermore, in the presence of both lactacystin and BFA, the cellular content of 35S-labeled apoB:996 was the same as with BFA alone (Fig. 7C). These results indicate that intracellular degradation of apoB:996 occurred through a nonproteasomal ER-associated pathway.
Deletion of Charged Residues 717–720 and Substitution of Residues Arg997, Glu998, Asp999, and Arg1000 with Four Alanines Drastically Alter the Intracellular Stability and Properties of ApoB:1000-containing Particles—The above results strongly suggested that the charged amino acid residues 997–1000 are important for the formation of stable lipidated pre-nascent apoB-containing particles. As a more stringent test of the hairpin-bridge hypothesis, we made two additional mutants as follows: 1) apoB:1000 with charged amino acid residues 717–720 deletion (designated as apoB:1000Δ717–720), and 2) apoB: 1000 with alanine substitution for charged amino acid residues 997–1000 (designated as apoB:1000 R997A, E998A, D999A, R1000A); for simplicity, we will refer to this mutant as apoB:996 + 4Ala. Expression plasmids encoding apoB: 1000Δ717–720 and apoB:996 + 4Ala were used to generate clonal stable transformants of McA-RH cells as described previously (32). Stable transformants of McA-RH cells expressing the wild-type and mutated forms of apoB:1000 were incubated with either [35S]Met/Cys or [3H]/[14C]glycerol/oleic acid. Metabolic labeling experiments and pulse-chase studies were carried out exactly as described above and in the figure legends. To accurately evaluate potential differences between the properties of the secreted intact apoB-containing particles, the volumes of the labeled conditioned medium from McA-RH cell lines expressing wild-type or mutated apoB:1000 were adjusted based on the expression rate of each apo B truncate so that equal amounts of proteins were used for analysis by NDGGE described below.
Metabolic labeling studies demonstrated that deletion of residues 717–720 in apoB:1000 significantly altered the properties of the secreted intact particles. The level of 35S-labeled apoB: 1000Δ717–720 in the conditioned medium was ∼50% lower than that of apoB:1000, after normalization for cell protein (Fig. 8A). Deletion of residues 717–720 also diminished (by ∼60%) the ability of the mutated protein to form a stable intact particle (p) with Sd of ∼112 Å, as determined by Western blot analysis (Fig. 8B) and autoradiography (Fig. 8C) when compared with wild-type apoB:1000. Furthermore, metabolic labeling with [3H]glycerol/[14C]oleic acid demonstrated that compared with wild-type apoB:1000, the intact apoB:1000Δ717–720-containing particles (p) contained 85–90% lower content of 3H-/14C-labeled lipids (Fig. 8D). An interesting and consistent observation was increased propensity of apoB:1000Δ717–720 to form large lipidated aggregates when compared with wild-type apoB:1000 (Fig. 8D). In sharp contrast to apoB:1000, more than 60% of the total 3H-/14C-labeled lipids were associated with the aggregate formed by apoB:1000Δ717–720 (Fig. 8D).
FIGURE 8.
Deletion of residues 717–720 and substitution of charged residues 997–1000 with four alanines in apoB:1000 drastically changes the properties of the protein. Stable transformants of McA-RH cells expressing apoB:1000, apoB:1000Δ717–720, apoB:996 + 4Ala, or apoB:996 were incubated for 20 h in serum-free DMEM containing either [35S]Met/Cys (100 μCi/ml of medium) (A, B, C, E, and F) or [3H]glycerol/[14C]oleic acid (0.4 mm bound to 0.75% BSA) (D and G). The 35S-labeled truncated forms of apoB in cell lysate and conditioned medium were isolated by immunoprecipitation with polyclonal antibody to human apoB100; proteins were resolved by 4–12% SDS-PAGE and visualized by autoradiography (A and E). B and C, aliquots of 35S-labeled concentrated conditioned medium, normalized for the expression levels of apoB:1000 and apoB: 1000Δ717–720 (based on the results shown in A), were subjected to 4–20% NDGGE. Gels were run in duplicate; intact apoB-containing lipoproteins were detected by immunoblotting with polyclonal antibody to human apoB100 (B), and a duplicate gel was analyzed for 35S-labeled intact particles by autoradiography (C). F, aliquots of 35S-labeled concentrated conditioned medium, normalized for the expression levels of apoB:1000 and apoB:996 + 4Ala (based on the results shown in E), were subjected to 4–20% NDGGE. Gels were run in duplicate; 35S-labeled intact apoB-containing lipoproteins were detected by autoradiography (F); particles (p) and aggregates (a) were excised from the second stained gel. 35S-Labeled proteins were extracted from particles and aggregates as described under “Experimental Procedures” and resolved by SDS-PAGE (H). D and G, aliquots of concentrated 3H-/14C-labeled conditioned medium, normalized for apoB expression levels, were analyzed by 4–20% NDGGE, and labeled lipids were visualized by autoradiography. The intensities of the labeled proteins and lipids were determined by computer-assisted image processing. Results are representative of three separate experiments.
Substitution of residues Arg997, Glu998, Asp999, and Arg1000 with four alanines (designated as apoB:996 + 4Ala) drastically altered the properties of the secreted intact particles containing the mutated apoB:1000. Metabolic labeling studies demonstrated a marked decrease in the de novo synthesis and secretion of apoB:996 + 4Ala when compared with wild-type apoB: 1000, albeit to a lesser extent than that observed for apoB:996 (Fig. 8E). Relative to apoB:1000, the levels of 35S-labeled apoB: 996 + 4Ala in the cells and conditioned medium were decreased by 80 and 55%, respectively (Fig. 8E). Metabolic labeling studies demonstrated that compared with apoB:1000, apoB:996 + 4Ala had the following: (i) a markedly lower capacity to form intact 35S-labeled particles (p) with Sd of ∼112 Å (Fig. 8F), (ii) formed considerably less intact 3H-/14C-lipid-containing particles (Fig. 8G), and (iii) had a much greater propensity to form large lipidated aggregates (Fig. 8G). In three separate experiments, more than 50% of the 3H-/14C-labeled lipids in apoB:996 + 4Ala were associated with the aggregates as opposed to less than 10% detected in the same position in the wild-type apoB:1000 (Fig. 8G).
Relative distribution of apoB:996 + 4Ala between intact particle and large aggregate revealed further differences between this mutant and apoB:1000. The bands shown in Fig. 8F, corresponding to truncated apoB-containing particles (P) and the aggregates (a) on top of the gel were excised, and proteins were extracted from the gels and separated as described in detail for Fig. 5. Consistent with studies shown in Fig. 5, essentially all (greater than 90%) of apoB:1000 protein was recovered in the major ∼112-Å particle (Fig. 8H). In apoB:996 + 4Ala, the distribution of protein between the major ∼112 Å particle and large lipid-rich aggregate was 70 and 30%, respectively (Fig. 8H). The above results indicate that charged residues 997–1000 play a critical role in the formation of a stable lipidated pre-nascent apoB-containing particle. Results of pulse-chase studies demonstrated that, in sharp contrast to wild-type apoB:1000 (Fig. 6, C and D), apoB:996 + 4Ala was susceptible to intracellular degradation (Fig. 9). At 120-min chase, 80% of 35S-labeled apoB:996 + 4Ala that disappeared from the cells was not recovered in the medium (Fig. 9A). Addition of oleic acid during both pulse and chase periods had a modest protective effect but did not rescue apoB:996 + 4Ala from degradation. At 120 min of chase, 55–60% of 35S-labeled apoB:996 + 4Ala that disappeared from the cells was not recovered in the medium (Fig. 9B). Thus, the intracellular processing and the lipid-binding properties of apoB:996 + 4Ala closely resembled those of apoB:996.
FIGURE 9.
Substitution of charged residues 997–1000 with four alanines renders the resulting apoB truncate susceptible to intracellular degradation. Stable transformants of McA-RH expressing apoB:996 + 4Ala were pulse-labeled for 10 min with serum-, methionine-, and cysteine-free DMEM containing [35S]Met/Cys (100 μCi/ml of medium) in the absence (A) or presence (B) of 0.4 mm oleic acid bound to 0.75% BSA. The pulse medium was removed, cell monolayers were washed with cold PBS, serum-free DMEM containing 10 mm each of unlabeled methionine and cysteine was added, and incubation was continued in the absence (A) or presence (B) of oleic acid. At the indicated chase period, medium and cell lysate were collected, and 35S-labeled apoB:996 + 4Ala was isolated by immunoprecipitation with polyclonal antibody to human apoB100. Proteins were resolved by 4–12% SDS-PAGE and visualized by autoradiography. The intensities of the labeled proteins were determined by computer-assisted image processing. The autoradiogram is representative of six samples from three separate experiments.
DISCUSSION
The N-terminal 1000 residues (apoB:1000) of the mature apoB compose the βα1 superdomain of apoB100. The βα1 domain is a mixture of amphipathic β-strands and amphipathic α-helices related to its structural homology to lamprey lipovitellin (LV) (28). Because the homology of the βα1 domain did not extend to the βD-sheet of LV, the question of how the apoB lipid pocket is completed remained to be answered. Previously, based on our experimentally derived results (32) and molecular modeling of apoB:1000 (30), we proposed the following model. (i) ApoB:1000 is competent to complete the lipid pocket without a structural requirement for MTP. (ii) Four salt bridges can be formed between a tandem series of four charged residues (Arg997, Glu998, Asp999, and Arg1000) located at the C-terminal end of the model, with a complementary tandem series of four charges (Glu720, His719, Lys718, and Asp717) in the unmodeled loop region (residues 670–745) located in the middle of the βA-sheet (shown in Fig. 1B). (iii) These putative salt bridges might serve as a template to guide closure of the third side of the asymmetric pyramidal lipid pocket by the creation of a hairpin bridge that completes the nascent step of lipoprotein particle assembly (30).
In a recent study (64), we demonstrated that the addition of PL to apoB:1000 and initiation of apoB-containing lipoprotein assembly in stable transformants of McA-RH cells occur independently of MTP lipid transfer activity. In the present study, we set out to identify the narrow domain between amino acid residues 931 and 1000 of apoB that might be critical for the formation of a stable HDL3-like lipoprotein and to ascertain the potential role of the four putative salt bridges in the formation of a pre-nascent apoB-containing particle. Results consistently and reproducibly demonstrated that apoB:931 and apoB:956 did not have the capacity to form stable lipid-containing particles. Thus, residues 1–956 of apoB100 are not competent to direct particle assembly and instead form large highly lipidated aggregates. Although apoB:986 had the capacity to form a lipidated particle with an Sd ∼ 112 Å, in comparison with apoB: 1000, these particles were denser than the traditional HDL3, contained ∼25% fewer lipid molecules, and had a high propensity to form large lipid-rich aggregates. Thus, amino acid residues 925–986 do not contain the complete structural elements necessary for the formation of a stable lipid-containing HDL3-like particle. These results are in disagreement with studies by other investigators who have concluded that the translation of the first ∼884 amino acids of apoB completes the domain capable of initiating the assembly of nascent lipoprotein (65). One plausible reason for the inconsistency in the results is the use of cell lines with distinctly different tissue origins. In this study, we used lipoprotein producing rat hepatoma McA-RH cells, whereas the other investigators (65) used COS cells, transformed African green monkey kidney fibroblast cells, which normally do not produce lipoproteins. Considering the complexity of apoB-containing lipoprotein assembly, COS cells may not have the full complement of numerous factors and chaperones known to be necessary for the formation of nascent apoB-containing particle (5). This possibility is supported by studies demonstrating that nonhepatic cell lines synthesize apoB but cannot process it into mature lipoproteins (66).
The above results allowed us to refine the motif that could potentially be critical for the formation of a stable HDL3-like apoB-containing particle to amino acid residues between 986 and 1000. Referring to our structural model of apoB:1000 (30), we wondered if the tandem series of four charged residues (Arg997, Arg1000) at the C terminus of apoB:1000, which we have proposed to form salt bridges with the complementary tandem series of four charged amino acids, Glu720, His719, Lys718, Asp717, in the turn of the hairpin-bridge (30), could be the critical residues. Our experimental approach, which involved side by side comparison of apoB:996, apoB:996 + 4Ala, and apoB: 1000Δ717–720 properties with those of wild-type apoB:1000, generated crucial information in support of our hairpin-bridge hypothesis.
Several distinct differences between the properties of apoB: 996, apoB:996 + 4Ala, and apoB:1000Δ717–720 and those of wild-type apoB:1000 were observed. ApoB:1000 was expressed and secreted at a high rate and was recovered predominantly as monodisperse lipidated particle and had HDL3-like hydrated density; only a negligible proportion of secreted apoB:1000 formed large lipid-rich aggregates. ApoB:1000 also exhibited a very high intracellular stability. Considering the very small difference in the number of amino acid residues between apoB: 996 and apoB:1000, it would be anticipated that both polypeptides undergo similar intracellular processing. Contrary to this expectation and in striking contrast to apoB:1000, we found that apoB:996 was secreted at a markedly lower rate and had a very high propensity to form large lipid-rich aggregates. The low secretion rate of apoB:996 was because of its enhanced intracellular degradation, irrespective of the presence or absence of exogenous oleic acid. Thus, the supply of lipid that has been shown to inhibit intracellular degradation of the full-length apoB100 (53) did not rescue apoB:996 from intracellular degradation. The combined results of studies using lactacystin, ALLN, and BFA indicated that the degradation of apoB:996 occurred in the ER via a nonproteasomal pathway due, most likely, to its abnormal conformation.
Of great significance to our hairpin-bridge hypothesis were the observations that substitution of charged residues 997–1000 with four alanines and deletion of charged residues 717–720 significantly altered the properties of the mutated proteins when compared with wild-type apoB:1000. The properties of apoB:996 + 4Ala more closely resembled those of apoB:996 than those of apoB:1000. ApoB:996 + 4Ala had a lower secretion rate, enhanced intracellular degradation, diminished ability to form intact lipidated particles, and increased propensity to form large lipid-rich aggregates. If our hypothesis is correct, charged residues 717–720 should also play a critical role in the formation of the proposed hairpin-bridge mechanism. This was, in fact, the case because deletion of residues 717–720 in apoB:1000 significantly altered the properties of the mutated protein when compared with the wild-type apoB:1000. The secretion rate and the capacity of apoB:1000Δ717–720 to form intact stable particles were significantly reduced; these particles also had 85–90% lower content of labeled lipids compared with wild-type apoB:1000. Significantly, apoB:1000Δ717–720 had increased propensity to form large lipid-rich aggregates. These results provided further validation for our hairpin-bridge hypothesis.
What might be the reason(s) for the rapid intracellular degradation of apoB:996 and apoB:996 + 4Ala not observed for either apoB:986 or apoB:1000? There are at least two structurally related possibilities as follows: 1) residues 986–995 may induce conformational changes that would render the protein susceptible to intracellular degradation; and/or 2) deletion of residues 997–1000 or their substitution with alanine might disrupt the formation of the putative salt bridges and cause misfolding of the resulting apoB:996 and apoB:996 + 4Ala channeling them into the nonproteasomal ER degradation pathway. Computer analysis of the βα1 domain of apoB100 with the program LOCATE indicated a unique characteristic of the domain between residues 986 and 995, i.e. alternating hydrophobic and hydrophilic residues (QYSVSATYEL), suggesting that this region is an amphipathic β-strand (21, 28–30). The model further suggests that it is also likely that region 895–900 is a strand, and that the two strands are hydrogen-bonded together to form a small β-sheet (30). This putative β-sheet might play a role in the degradation of apoB. This possibility is based on the studies showing that the translocation efficiency of apoB and its susceptibility to proteasomal degradation are determined by the presence of β-sheets in the β1 domain of full-length protein (67). However, our studies did not provide any evidence for proteasomal degradation of apoB:996. Furthermore, the 986–995 sequence is also present in apoB:1000 and yet does not cause its intracellular degradation. Therefore, there is a unique structural feature in apoB:1000, not present in either apoB:996 or apoB:996 + 4Ala, that protects it from intracellular degradation. The present results suggest that intracellular degradation of apoB is not a simple function of its size. Rather, it might be determined, at least in part, by the sequence-specific folding (presumably in the presence of a lipid core) of the polypeptide and the structure of the protein on the particle.
The observed vast differences in the tendencies of apoB:986, apoB:996, apoB:996 + 4Ala, apoB:1000Δ717–720, and apoB: 1000 to form large lipid-rich aggregates warrant scrutiny. We offer the following plausible, albeit speculative, explanations. In the model of apoB:1000 (30), it was proposed that residues 700–744 form a helix-loop-helix and residues 717–720 in the loop region form salt bridges with residues 997–1000. We made the conjecture that this hairpin bridge guides the closure of the third side of the asymmetric pyramidal apoB lipid pocket to complete the nascent step of lipoprotein particle assembly (30). If the helix-loop-helix in apoB:986 (shown in Fig. 10) was tilted and rotated toward the C-terminal Gln986, it could be able to bond to the C terminus of the truncate. In this conformation, Gln986 could form a hydrogen bond with Asp717 and Glu985 could form a salt bridge with Lys718, without stearic hindrance from the β-sheet. Although this arrangement could help stabilize the lipid binding pocket, it would not be as energetically stable as apoB:1000 so it could form both a major stable lipidated particle and large lipid-rich protein aggregates (shown in Fig. 5). This conformation would result in the loss of volume in the lipid binding pocket formed by apoB:986 and fewer lipid molecules. Our experimentally derived results (Table 1) support this possibility.
FIGURE 10.
The apoB1000 model (30) was used to create models of the C-terminal truncates of apoB:986 (left image) and apoB:996 (right image). The critical residues in the helix-loop-helix are labeled, along with those residues that might be important in the C terminus for lipoprotein stability. The white arrow in apoB:996 model indicates the small helix connecting the βA- and βB-sheets. The figure was generated using the visual molecular dynamics program (68).
In apoB:996, the helix-loop-helix is directly adjacent to the small helix (indicated by the white arrow) connecting the βA- and βB-sheets (Fig. 10) limiting its movement and stability. One possible bond that could form is a hydrogen bond between Gln996 and Glu720, but Asp717 and Lys718 would not form stabilizing bonds. This lack of bonds could seriously undermine the ability of the helix-loop-helix to form a stable base for the lipid pocket formed by apoB:996 resulting in destabilization of particle and prevalence of lipid-rich aggregates.
We believe that the hairpin bridge is a transient intermediate in lipoprotein assembly with two distinct forms as follows: (i) a locked hairpin bridge that closes the pocket to allow the formation of a stable nascent lipoprotein particle with defined lipid content observed in apoB:1000, and (ii) an unlocked form that allows an increase in the particle size and decrease in the particle density through addition of lipid as more residues in the β1 domain (beyond residue 1000) are translated. Based on our results, we propose that the intracellular stability and particle assembly properties of apoB:1000 are disrupted upon removal of the locked hairpin bridge by either deletion of key charged residues 997–1000 or their substitution with four alanines. In this conformation, a small proportion of apoB:996 can temporarily hold lipids and is secreted as lipidated particles. However, the majority of the particles is unstable and is released from the ER membrane before significant lipidation occurs leading to the formation of particles with abnormal conformation. These particles are mostly channeled to nonproteasomal degradation pathway and if secreted can form large lipid-rich aggregates. This structurally based characteristic might be the reason for the observed wide heterogeneity in apoB:996-containing particle size shown in Fig. 4.
The differences in the known structure of LV lipid pocket (25–27) and our proposed mechanism for apoB lipid pocket formation warrant rationalization. There are two unique structural features in apoB that are not present in LV. First, there is small loop-helix at the apex of the two-sided pyramid (residues 764–779) that connects the two β-sheets (Fig. 1, A and B) (30). There is a large undefined loop joining two adjacent strands in the βA-sheet (residues 666–746) that is not homologous to any sequence in lipovitellin or any other sequence in the GenBank™ data base. LOCATE analysis of residues 670–745 showed the presence of two well defined amphipathic helices 702–716 and 721–738, predicted to be strongly lipid-associating (30). Significantly, the complementary tandem charges 717–720 are located precisely between the two putative amphipathic helices, suggesting a helix-turn-helix motif (30) (Fig. 1B). Second, as described above under the “Results” and in Fig. 1D, HD IV:980–1020 maps to the following: (i) the second of the two separate sequences that make up the βB:LV antiparallel β-sheet domain of lamprey LV, residues 989–1057, and (ii) a corresponding cluster of amphipathic β-strands in hAPOB (residues 967–996, βB:B) (28). In the absence of the βD-domain, which serves to close the third side of three-sided pyramid of LV lipid pocket, charged residues 717–720 and 997–1000 in apoB could appear as a compensatory mechanism for the completion of the lipid pocket by forming the proposed hairpin bridge.
In summary, the major findings of this study are as follows: (i) the entire 1000 amino acid residues of the βα1 domain of apoB100 are necessary for the formation of a stable particle in the HDL3-like density range and the initiation of apoB particle assembly; and (ii) the charged amino acid residues 997–1000 may play a key role in this process by forming salt bridges with a tandem series of four complementary charged residues 717–720, in the following arrangement: Arg997–Glu720, Glu998–His719, Asp999–Lys718, and Arg1000–Asp717. Results strongly support our proposal that the salt bridges serve as a lock to close the third side of the asymmetric pyramidal apoB lipid pocket by the creation of a hairpin bridge that completes the first step in the formation of the primordial apoB-containing lipoprotein particle.
This work was supported, in whole or in part, by National Institutes of Health Grants HL084685 and PO1 HL34343. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: apo, apolipoprotein; apoB:1000, N-terminal 22.05% (residues 1–1000) of the mature protein; ALLN, N-acetyl-leucyl-leucyl-norleucinal; BFA, brefeldin A; BSA, bovine serum albumin; DAG, diacylglycerol; DMEM, Dulbecco's modified Eagle's medium; DMSO, dimethyl sulfoxide; ER, endoplasmic reticulum; hAPOB, human apolipoprotein B; HD, homologue domain; HDL, high density lipoprotein; KBr, potassium bromide; LV, lipovitellin; Me2SO, dimethyl sulfoxide; MTP, microsomal triglyceride transfer protein; NDGGE, nondenaturing gradient gel electrophoresis; PL, phospholipids; PBS, phosphate-buffered saline; Sd, Stokes diameter; TAG, triacylglycerol; vLV, vertebrate lipovitellin.
References
- 1.Knott, T. J., Pease, R. J., Powell, L. M., Wallis, S. C., Rall, S. C., Jr., Innerarity, T. L., Blackhart, B., Taylor, W. H., Marcel, Y., Milne, R., Johnson, D., Fuller, M., Lusis, A. J., McCarthy, B. J., Mahley, R. W., Levy-Wilson, B., and Scott, J. (1986) Nature 323 734-738 [DOI] [PubMed] [Google Scholar]
- 2.Cladaras, C., Hadzopoulou-Cladaras, M., Nolte, R. T., Atkinson, D., and Zannis, V. I. (1986) EMBO J. 5 3495-3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, S. H., Yang, C. Y., Chen, P. F., Setzer, D., Tanimura, M., Li, W. H., Gotto, A. M., Jr., and Chan, L. (1986) J. Biol. Chem. 261 12918-12921 [PubMed] [Google Scholar]
- 4.Schumaker, V. N., Phillips, M. L., and Chatterton, J. E. (1994) Adv. Protein Chem. 45 205-248 [DOI] [PubMed] [Google Scholar]
- 5.Fisher, E. A., and Ginsberg, H. N. (2002) J. Biol. Chem. 277 17377-17380 [DOI] [PubMed] [Google Scholar]
- 6.Elovson, J., Chatterton, J. E., Bell, G. T., Schumaker, V. N., Reuben, M. A., Puppione, D. L., Reeve, J. R., Jr., and Young, N. L. (1988) J. Lipid Res. 29 1461-1473 [PubMed] [Google Scholar]
- 7.Kane, J. P. (1983) Annu. Rev. Physiol. 45 637-650 [DOI] [PubMed] [Google Scholar]
- 8.Deckelbaum, R. J., Shipley, G. G., Small, D. M., Lees, R. S., and George, P. K. (1975) Science 190 392-394 [DOI] [PubMed] [Google Scholar]
- 9.Atkinson, D., Deckelbaum, R. J., Small, D. M., and Shipley, G. G. (1977) Proc. Natl. Acad. Sci. U. S. A. 74 1042-1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deckelbaum, R. J., Shipley, G. G., and Small, D. M. (1977) J. Biol. Chem. 252 744-754 [PubMed] [Google Scholar]
- 11.Chatterton, J. E., Phillips, M. L., Curtiss, L. K., Milne, R., Fruchart, J. C., and Schumaker, V. N. (1995) J. Lipid Res. 36 2027-2037 [PubMed] [Google Scholar]
- 12.Spin, J. M., and Atkinson, D. (1995) Biophys. J. 68 2115-2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orlova, E. V., Sherman, M. B., Chiu, W., Mowri, H., Smith, L. C., and Gotto, A. M., Jr. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 8420-8425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Antwerpen, R., La Belle, M., Navratilova, E., and Krauss, R. M. (1999) J. Lipid Res. 40 1827-1836 [PubMed] [Google Scholar]
- 15.Gantz, D. L., Walsh, M. T., and Small, D. M. (2000) J. Lipid Res. 41 1464-1472 [PubMed] [Google Scholar]
- 16.De Loof, H., Rosseneu, M., Yang, C. Y., Li, W. H., Gotto, A. M., Jr., and Chan, L. (1987) J. Lipid Res. 28 1455-1465 [PubMed] [Google Scholar]
- 17.Chen, G. C., Hardman, D. A., Hamilton, R. L., Mendel, C. M., Schilling, J. W., Zhu, S., Lau, K., Wong, J. S., and Kane, J. P. (1989) Biochemistry 28 2477-2484 [DOI] [PubMed] [Google Scholar]
- 18.Yang, C. Y., Gu, Z. W., Weng, S. A., Kim, T. W., Chen, S. H., Pownall, H. J., Sharp, P. M., Liu, S. W., Li, W. H., and Gotto, A. M., Jr. (1989) Arteriosclerosis 9 96-108 [DOI] [PubMed] [Google Scholar]
- 19.Segrest, J. P., Jones, M. K., Mishra, V. K., Anantharamaiah, G. M., and Garber, D. W. (1994) Arterioscler. Thromb. 14 1674-1685 [DOI] [PubMed] [Google Scholar]
- 20.Segrest, J. P., Garber, D. W., Brouillette, C. G., Harvey, S. C., and Anantharamaiah, G. M. (1994) Adv. Protein Chem. 45 303-369 [DOI] [PubMed] [Google Scholar]
- 21.Segrest, J. P., Jones, M. K., Mishra, V. K., Pierotti, V., Young, S. H., Boren, J., Innerarity, T. L., and Dashti, N. (1998) J. Lipid Res. 39 85-102 [PubMed] [Google Scholar]
- 22.Johs, A., Hammel, M., Waldner, I., May, R. P., Laggner, P., and Prassl, R. (2006) J. Biol. Chem. 281 19732-19739 [DOI] [PubMed] [Google Scholar]
- 23.Baker, M. E. (1988) Biochem. J. 255 1057-1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shoulders, C. C., Narcisi, T. M., Read, J., Chester, A., Brett, D. J., Scott, J., Anderson, T. A., Levitt, D. G., and Banaszak, L. J. (1994) Nat. Struct. Biol. 1 285-286 [DOI] [PubMed] [Google Scholar]
- 25.Raag, R., Appelt, K., Xuong, N. H., and Banaszak, L. (1988) J. Mol. Biol. 200 553-569 [DOI] [PubMed] [Google Scholar]
- 26.Sharrock, W. J., Rosenwasser, T. A., Gould, J., Knott, J., Hussey, D., Gordon, J. I., and Banaszak, L. (1992) J. Mol. Biol. 226 903-907 [DOI] [PubMed] [Google Scholar]
- 27.Anderson, T. A., Levitt, D. G., and Banaszak, L. J. (1998) Structure (Lond.) 6 895-909 [DOI] [PubMed] [Google Scholar]
- 28.Segrest, J. P., Jones, M. K., and Dashti, N. (1999) J. Lipid Res. 40 1401-1416 [PubMed] [Google Scholar]
- 29.Segrest, J. P., Jones, M. K., De Loof, H., and Dashti, N. (2001) J. Lipid Res. 42 1346-1367 [PubMed] [Google Scholar]
- 30.Richardson, P. E., Manchekar, M., Dashti, N., Jones, M. K., Beigneux, A., Young, S. G., Harvey, S. C., and Segrest, J. P. (2005) Biophys. J. 88 2789-2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon, D. A. (1997) Curr. Opin. Lipidol. 8 131-137 [DOI] [PubMed] [Google Scholar]
- 32.Manchekar, M., Richardson, P. E., Forte, T. M., Datta, G., Segrest, J. P., and Dashti, N. (2004) J. Biol. Chem. 279 39757-39766 [DOI] [PubMed] [Google Scholar]
- 33.Dashti, N., Gandhi, M., Liu, X., Lin, X., and Segrest, J. P. (2002) Biochemistry 41 6978-6987 [DOI] [PubMed] [Google Scholar]
- 34.Yao, Z. M., Blackhart, B. D., Johnson, D. F., Taylor, S. M., Haubold, K. W., and McCarthy, B. J. (1992) J. Biol. Chem. 267 1175-1182 [PubMed] [Google Scholar]
- 35.Miller, A. D., Miller, D. G., Garcia, J. V., and Lynch, C. M. (1993) Methods Enzymol. 217 581-599 [DOI] [PubMed] [Google Scholar]
- 36.Dashti, N. (1992) J. Biol. Chem. 267 7160-7169 [PubMed] [Google Scholar]
- 37.Laemmli, U. K. (1970) Nature 227 680-685 [DOI] [PubMed] [Google Scholar]
- 38.Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951) J. Biol. Chem. 193 265-275 [PubMed] [Google Scholar]
- 39.Chung, B. H., Segrest, J. P., Ray, M. J., Brunzell, J. D., Hokanson, J. E., Krauss, R. M., Beaudrie, K., and Cone, J. T. (1986) Methods Enzymol. 128 181-209 [DOI] [PubMed] [Google Scholar]
- 40.Folch, J., Lees, M., and Sloane Stanley, G. H. (1957) J. Biol. Chem. 226 497-509 [PubMed] [Google Scholar]
- 41.Dashti, N., Feng, Q., Freeman, M. R., Gandhi, M., and Franklin, F. A. (2002) J. Nutr. 132 2651-2659 [DOI] [PubMed] [Google Scholar]
- 42.Burnette, W. N. (1981) Anal. Biochem. 112 195-203 [DOI] [PubMed] [Google Scholar]
- 43.Carraway, M., Herscovitz, H., Zannis, V., and Small, D. M. (2000) Biochemistry 39 9737-9745 [DOI] [PubMed] [Google Scholar]
- 44.Pullinger, C. R., North, J. D., Teng, B. B., Rifici, V. A., Ronhild de Brito, A. E., and Scott, J. (1989) J. Lipid Res. 30 1065-1077 [PubMed] [Google Scholar]
- 45.Bostrom, K., Wettesten, M., Boren, J., Bondjers, G., Wiklund, O., and Olofsson, S. O. (1986) J. Biol. Chem. 261 13800-13806 [PubMed] [Google Scholar]
- 46.Borchardt, R. A., and Davis, R. A. (1987) J. Biol. Chem. 262 16394-16402 [PubMed] [Google Scholar]
- 47.Sparks, J. D., and Sparks, C. E. (1990) J. Biol. Chem. 265 8854-8862 [PubMed] [Google Scholar]
- 48.Dixon, J. L., and Ginsberg, H. N. (1993) J. Lipid Res. 34 167-179 [PubMed] [Google Scholar]
- 49.Wang, C. N., Hobman, T. C., and Brindley, D. N. (1995) J. Biol. Chem. 270 24924-24931 [DOI] [PubMed] [Google Scholar]
- 50.Liao, W., Yeung, S. C., and Chan, L. (1998) J. Biol. Chem. 273 27225-27230 [DOI] [PubMed] [Google Scholar]
- 51.White, A. L., Graham, D. L., LeGros, J., Pease, R. J., and Scott, J. (1992) J. Biol. Chem. 267 15657-15664 [PubMed] [Google Scholar]
- 52.Cavallo, D., McLeod, R. S., Rudy, D., Aiton, A., Yao, Z., and Adeli, K. (1998) J. Biol. Chem. 273 33397-33405 [DOI] [PubMed] [Google Scholar]
- 53.Dixon, J. L., Furukawa, S., and Ginsberg, H. N. (1991) J. Biol. Chem. 266 5080-5086 [PubMed] [Google Scholar]
- 54.Mitchell, D. M., Zhou, M., Pariyarath, R., Wang, H., Aitchison, J. D., Ginsberg, H. N., and Fisher, E. A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 14733-14738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fisher, E. A., Pan, M., Chen, X., Wu, X., Wang, H., Jamil, H., Sparks, J. D., and Williams, K. J. (2001) J. Biol. Chem. 276 27855-27863 [DOI] [PubMed] [Google Scholar]
- 56.Pan, M., Cederbaum, A. I., Zhang, Y. L., Ginsberg, H. N., Williams, K. J., and Fisher, E. A. (2004) J. Clin. Investig. 113 1277-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeung, S. J., Chen, S. H., and Chan, L. (1996) Biochemistry 35 13843-13848 [DOI] [PubMed] [Google Scholar]
- 58.Fisher, E. A., Zhou, M., Mitchell, D. M., Wu, X., Omura, S., Wang, H., Goldberg, A. L., and Ginsberg, H. N. (1997) J. Biol. Chem. 272 20427-20434 [DOI] [PubMed] [Google Scholar]
- 59.Zhou, M., Fisher, E. A., and Ginsberg, H. N. (1998) J. Biol. Chem. 273 24649-24653 [DOI] [PubMed] [Google Scholar]
- 60.Cavallo, D., Rudy, D., Mohammadi, A., Macri, J., and Adeli, K. (1999) J. Biol. Chem. 274 23135-23143 [DOI] [PubMed] [Google Scholar]
- 61.Cardozo, C., Wu, X., Pan, M., Wang, H., and Fisher, E. A. (2002) Biochemistry 41 10105-10114 [DOI] [PubMed] [Google Scholar]
- 62.Ellgaard, L., Molinari, M., and Helenius, A. (1999) Science 286 1882-1888 [DOI] [PubMed] [Google Scholar]
- 63.Coux, O., Tanaka, K., and Goldberg, A. L. (1996) Annu. Rev. Biochem. 65 801-847 [DOI] [PubMed] [Google Scholar]
- 64.Dashti, N., Manchekar, M., Liu, Y., Sun, Z., and Segrest, J. P. (2007) J. Biol. Chem. 282 28597-28608 [DOI] [PubMed] [Google Scholar]
- 65.Shelness, G. S., Hou, L., Ledford, A. S., Parks, J. S., and Weinberg, R. B. (2003) J. Biol. Chem. 278 44702-44707 [DOI] [PubMed] [Google Scholar]
- 66.Dixon, J. L., Biddle, J., Lo, C. M., Stoops, J. D., Li, H., Sakata, N., and Phillips, T. E. (2002) J. Histochem. Cytochem. 50 629-640 [DOI] [PubMed] [Google Scholar]
- 67.Yamaguchi, J., Conlon, D. M., Liang, J. J., Fisher, E. A., and Ginsberg, H. N. (2006) J. Biol. Chem. 281 27063-27071 [DOI] [PubMed] [Google Scholar]
- 68.Humphrey, W., Dalke, A., and Schulten, K. (1996) J. Mol. Graphics 14 27-38 [DOI] [PubMed] [Google Scholar]