Abstract
The ubiquitin-proteasome pathway (UPP) regulates synaptic function, but little is known about specific UPP targets and mechanisms in mammalian synapses. We report here that the SCFβ-TRCP complex, a multisubunit E3 ubiquitin ligase, targets the postsynaptic spine-associated Rap GTPase activating protein (SPAR) for degradation in neurons. SPAR degradation by SCFβ-TRCP depended on the activity-inducible protein kinase Polo-like kinase 2 (Plk2). In the presence of Plk2, SPAR physically associated with the SCFβ-TRCP complex through a canonical phosphodegron. In hippocampal neurons, disruption of the SCFβ-TRCP complex by overexpression of dominant interfering β-TRCP or Cul1 constructs prevented Plk2-dependent degradation of SPAR. Our results identify a specific E3 ubiquitin ligase that mediates degradation of a key postsynaptic regulator of synaptic morphology and function.
Dendritic spines are tiny, actin-rich, dynamic protrusions radiating from the dendritic shaft of principal neurons and comprise the postsynaptic compartment of most glutamatergic synapses of the mammalian brain. The size and morphology of dendritic spines are correlated with their function. Thus, large mushroom-shaped spines tend to be more stable than small thin spines, contain more α-amino-3-hydroxy-5-methylisoxazole-4-proprionic acid receptors, and mediate stronger synaptic connections (1, 2). The morphology of spines, which changes during development and in response to synaptic activity, is influenced by multiple signaling pathways that emanate from postsynaptic glutamate receptors and act upon the actin cytoskeleton and associated proteins in the postsynaptic density (PSD)5 (2-4).
SPAR (spine-associated Rap GTPase activating protein (GAP)) is a PSD protein that regulates spine morphogenesis and forms a complex with the scaffold protein PSD-95 and N-methyl-d-aspartate-type glutamate receptors (5). Overexpression of SPAR results in enlargement of spine heads, an effect dependent upon the ability of SPAR to rearrange actin and function as a RapGAP. In contrast, dominant-negative SPAR produces long and thin spines (5). SPAR, in turn, is regulated by polo-like kinase 2 (Plk2; also known as serum-inducible serine/threonine kinase (SNK)) (6). Synaptic activity induces Plk2 expression (6-8), leading to degradation of SPAR, a mechanism recently shown to be critical in activity-dependent synaptic scaling (a principal form of homeostatic plasticity) (9, 43).
SPAR turnover depends upon the ubiquitin-proteasome pathway (UPP) since ubiquitinated SPAR accumulates in the presence of active Plk2 when proteasomes are inhibited (6). This necessitates the involvement of at least one E3 ubiquitin (Ub) ligase targeting SPAR. More generally, the UPP is known to play an important role in the activity-dependent turnover of several proteins in the PSD (10), a process that likely involves activity-driven redistribution of proteasomes into spines (11). However, as is the case for many neuronal processes in which the UPP has been implicated, the molecular and regulatory components upstream of the proteasome responsible for targeting the ubiquitination of SPAR and other PSD proteins are unknown.
Skp1/Cul1/F-box protein (SCF) complexes are one of the best understood E3 Ub-ligases with regard to mechanism of substrate recognition (12, 13). By recruiting substrates to the SCF complex, F-box proteins dramatically increase the specificity and rate of ubiquitin transfer to substrates. The F-box protein β-TRCP has been shown to target critical signaling proteins for degradation, including IκBα, β-catenin, and Cdc25A (14-18). Its function in neurons, however, has remained unexplored. Here, we identifyβ-TRCP as the F-box protein that targets Plk2-phosphorylated SPAR for degradation in neurons. Biochemical studies revealed that SPAR interacted with the SCFβ-TRCP complex through a canonical β-TRCP phosphodegron. Induction of Plk2 activity led to SPAR turnover, and this was prevented by dominant negative disruption of the SCFβ-TRCP complex or point mutations in the phosphodegron of SPAR preventing SPAR from interacting with β-TRCP.
EXPERIMENTAL PROCEDURES
DNA Plasmids—Expression plasmids expressing cDNA with a CMV promoter for myc-SPAR, HA-Plk2, and HA-Plk2K108M were previously described (6). pCMV-HA-Plk2D201A was created from pCMV-HA-Plk2 using PCR-based mutagenesis. cDNA for full-length SPAR as well as SPAR fragments were amplified from pGW1-myc-SPAR (5) and cloned into pENTR-6, compatible with the GATEway cloning system (Invitrogen). These were used for in vitro LR clonase reactions into pDEST-53 (for GFP-SPAR) or pDEST-N-myc (for myc-SPAR fragments). Point mutations in the β-TRCP phosphodegron were generated by PCR-based mutagenesis using pCMV-myc-SPAR or pDEST-myc-Act2 as templates. pCMV-myc-CKIε and pCMV-GSK3β were previously described (19). F-box proteins previously cloned from cDNA pools (14) were re-cloned into pENTR-6, and in vitro LR clonase reactions were performed with pDEST-27 to generate GST-fused F-box proteins. DNCul1 (residues 1-452) was previously described (14); DNCul3 and DNCul4 were prepared by cloning sequences encoding residues 1-418 of Cul3 and 1-440 of Cul4A into pcDNA3 (Invitrogen), respectively. β-TRCP1ΔF, Fbw7ΔF, and Skp2ΔF plasmids were previously described (14, 20, 21), and β-TRCP2ΔF was a gift from N. Khidekel (MIT). pRetroSuper (pRS)-shβ-TRCP and control shGFP plasmids were previously published (14).
HEK293T Transfections, Binding, Abundance, and Turnover Assays—HEK293T cells were grown in Dulbecco's modified Eagle's medium + 10% serum and seeded (1 × 106 cells/well of a 6-well dish) 16 h before transfection with Lipofectamine 2000 (Invitrogen). Cells were typically harvested ∼24-30 h post-transfection, except for RNAi experiments where cells were harvested 96 h post-transfection after a 48-h selection with puromycin (1 μg/ml) to enrich for a transfected population. Treatment of transfected cells with cycloheximide (25 μg/ml) began 96 h post-transfection.
For assays examining myc-SPAR abundance and turnover, cell pellets were lysed in 50 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.5% deoxycholate, 1% Nonidet P-40, 0.1% SDS, and cleared lysates were resolved on 4-12% gradient Tris-glycine SDS-PAGE gels. Resolved proteins were then transferred onto nitrocellulose (250 mA, 2 h) and immunoblotted with c-Myc 9E10 (sc-40, Santa-Cruz), HA F-7 (sc-7392, Santa Cruz), Cdk2 M2 (sc-163, Santa Cruz), β-TRCP1 (Cell Signaling), Cul1 (71-8700, Invitrogen/Zymed Laboratories Inc.), and Cdc25A Ab-3 (MS-640-P0, NeoMarkers), as indicated. Promega horseradish peroxidase-conjugate anti-mouse IgG (W402B) and anti-rabbit IgG (W401B) were used for secondary detection.
For single-cell immunofluorescence RNAi experiments, HEK293T cells were seeded onto 18-mm glass coverslips coated with poly-d-lysine (30 μg/ml) and laminin (2 μg/ml) and transfected at 20-30% confluency using calcium phosphate. Cells were fixed 96 h post-transfection for 10 min at room temperature using 4% paraformaldehyde and 4% sucrose in phosphate-buffered saline, and the GFP-SPAR signal was amplified using anti-GFP (A-11122, Invitrogen) at 1:1000 in GDB buffer (1% gelatin, 5% Triton X-100, 50 mm phosphate buffer, pH 7.4, 2 m NaCl) (6).
Binding and ubiquitination detection assays were performed with cleared whole cell lysates from transfected HEK293T cells in 50 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.5% deoxycholate, and 1% Nonidet P-40. In the case of ubiquitination detection assays, cells were treated with 25 μm MG-132 before harvesting, and 10 mm N-ethylmaleimide was added to the lysis buffer. GSH-Sepharose or c-Myc 9E10-agarose beads (10 μl per condition) were pre-washed in lysis buffer and incubated with 400 μg of lysate for 2 h at 4 °C while rocking. Beads were then washed 3-4 times in lysis buffer, and bound protein was eluded in 2× Laemmli protein loading buffer containing SDS. Both GSH-bound samples and crude extract were resolved on 4-12% gradient SDS-PAGE gels. Myc-bound samples were resolved on 6% Tris-Glycine SDS-PAGE gels. Samples were then transferred from the SDS-PAGE onto nitrocellulose and immunoblotted with anti-Myc, anti-HA, anti-FLAG (F3165, Sigma), or anti-GST (26H1, Cell Signaling), as indicated.
Neuron Cultures and Immunostaining—Medium-density dissociated hippocampal cultures were prepared and cultured from E19 Long Evans rat hippocampi as previously described (5). Neurons were transfected at DIV16, “super-infected” at DIV18, and fixed ∼18 h post-infection (DIV19) in 1% paraformaldehyde for 2 min at room temperature followed by -20 °C methanol for 10 min. Immunostaining was performed in GDB buffer (6) using rabbit SPAR polyclonal antibodies (5) and anti-FLAG M2 (Sigma).
Microscopy and Quantification—Fixed neurons and HEK293T cells were imaged with an LSM510 confocal system (Zeiss). A 40× oil immersion lens was used for confocal microscopy, and each image was comprised of 0.5-μm z-stacks projected into a single plane. SPAR immunostaining analysis was performed with MetaMorph Software and carried out blinded with respect to the experimental conditions. Quantification of SPAR puncta involved subjecting images stained for endogenous SPAR to threshold and was carried out from somatic and proximal dendritic regions from transfected, infected, or transfected plus superinfected cells. All SPAR intensity measurements represent integrated SPAR immunostaining intensity per area and are normalized to neighboring uninfected and untransfected cells.
Statistical Analysis—Statistical Methods are described in the figures legends.
RESULTS
Plk2-dependent SPAR Degradation Requires a Cul1-based E3 Ub-ligase—SPAR turnover in neurons is controlled through the UPP in a manner that requires active Plk2 protein kinase (6). Such regulation is reminiscent of the mechanism employed by most SCF E3 Ub-ligases, where substrate recognition depends upon upstream kinase signaling cascades. A central subunit of the SCF complex is the Cul1 scaffold protein. To explore the idea that SPAR turnover is regulated by an SCF E3 Ub-ligase, we made use of a previously reported dominant-negative version of Cul1 (DNCul1). This C-terminal-truncated protein binds substrates but fails to associate with ubiquitin-charged E2 ubiquitin conjugating enzymes, thereby preventing substrate turnover (14, 19, 22).
Cultured hippocampal neurons (16 days in vitro (DIV16)) were transfected with DNCul1 and then super-infected 2 days later with Sindbis virus driving expression of FLAG-tagged Plk2 for ∼18 h to promote degradation of endogenous SPAR (6). We infected at a titer that resulted in ∼10% infection rate of cells already transfected with DNCul1 (Fig. 1, A-C). The option of co-transfecting plasmids driving Plk2 expression was precluded by the low Plk2 expression achievable using this method.6
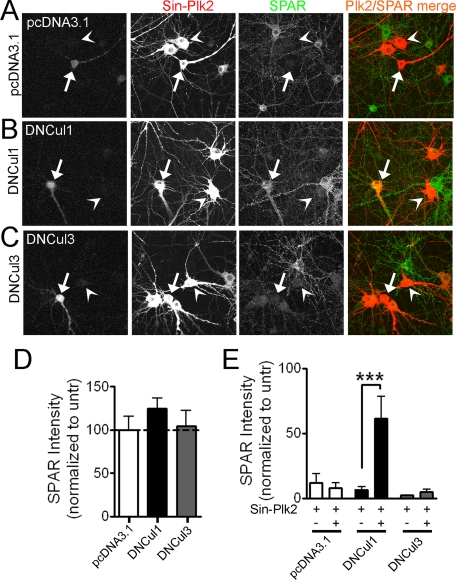
FIGURE 1.
Plk2-induced SPAR degradation requires a Cul1-based E3 Ubligase. A-C, dominant negative Cul1 constructs block Plk2-dependent loss of SPAR in hippocampal neurons. Dissociated rat hippocampal neurons (DIV16) were transfected with dominant negative Cullin plasmids (B and C) or a control plasmid (A) and super-infected 2 days later with FLAG-tagged Plk2 driven by Sindbis virus (Sin-Plk2). Neurons were fixed ∼18 h post-infection and immunostained for endogenous SPAR and infected Plk2. Transfected cells were identified during image acquisition by the presence of a co-transfected “fill” protein (GFP, seen in the first column of images). SPAR (green) and Plk2 (red) were pseudo-colored for illustrative purposes after image analysis. Arrows point to cells that are both transfected and infected; arrowheads point to cells that are infected only. Yellow indicates the presence of both SPAR and Plk2 staining. D and E, quantification of SPAR immunostaining in somatic and proximal dendritic regions as integrated immunofluorescence intensity per area in cells transfected with indicated plasmids and/or infected with Plk2 Sindbis virus (Sin-Plk2), normalized to nearby untransfected (untr) cells. Values represent the mean ± S.E., n > 17 cells for all conditions, ***, p < 0.001, Mann-Whitney test (E).
In the absence of Plk2 infection, transfection of control empty vector (pcDNA3.1) or a control dominant-negative Cullin 3 (DNCul3), which disrupts structurally related but functionally distinct Cul3-based complexes (23), had no effect on endogenous SPAR levels relative to nearby untransfected and uninfected cells as assessed by quantitative immunostaining (Fig. 1, A and C, quantified in D). Uninfected cells overexpressing DNCul1 displayed a trend toward increased SPAR levels compared with nearby untransfected cells, but did not reach statistical significance (p = 0.13; Fig. 1B, quantified in D).
In Plk2-infected but otherwise untransfected neurons, endogenous SPAR levels were close to undetectable (Fig. 1, A-C, arrowheads; quantified in E), in agreement with our previous findings (6). In Plk2-infected cells that had been previously transfected with control empty vector or DNCul3, SPAR levels fell to the same extent as in cells only infected with Plk2 (Fig. 1, A and C, compare SPAR staining in cells marked by arrowheads (infected) and arrows (transfected and infected); quantified in E). However, in cells transfected with DNCul1, infection with Plk2 Sindbis virus failed to reduce SPAR levels (Fig. 1B, notice the yellow color in cells marked by arrows, indicating the presence of both SPAR (green) and Plk2 (red); quantified in E). Thus, Plk2-driven SPAR degradation in neurons depended upon a Cul1-based SCF complex but not any of the structurally related Cul3-based Ub-ligases.
SPAR Physically Associates with the SCFβ-TRCP Complex—To further explore the idea that SPAR turnover is regulated by an SCF E3 Ub-ligase, we established a system in cultured HEK293T cells that recapitulates Plk2-dependent SPAR degradation. This system facilitated biochemical studies that were otherwise limited by the physical properties of dendritic spines and allowed the use of molecular reagents previously developed for study of the human SCF pathway (24). We found that expression of SPAR alone (as an Myc-tagged fusion protein) led to its accumulation in HEK293T cells (Fig. 2A, lane 3). In contrast, co-expression with Plk2 (but not a catalytically inactive mutant Plk2D201A) promoted the degradation of myc-SPAR (Fig. 2, A, lane 2 compared with lanes 4-6, and B, lanes 1 and 2). Co-expression of SPAR with wild type Plk2 correlated with the appearance of a slower mobility form of SPAR (presumably phosphorylated) that is sensitive to Plk2-induced degradation (Fig. 2A, myc-SPAR-P; see also Fig. 2, B and C). Our recapitulation of Plk2-dependent SPAR turnover in HEK293T cells provides a convenient system in which to search for components of the Plk2-dependent degradation pathway.
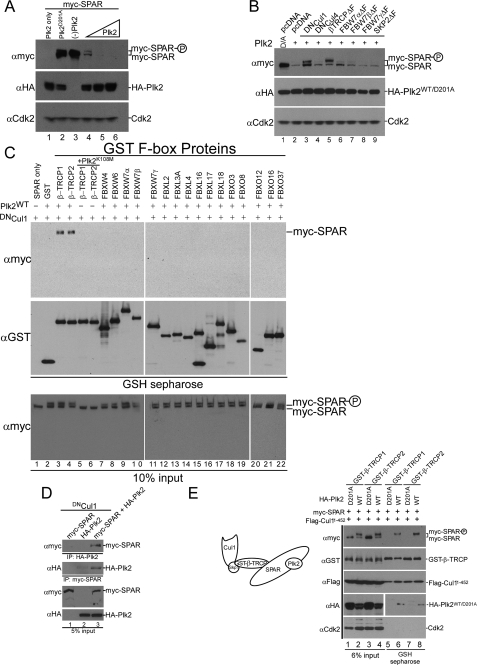
FIGURE 2.
SPAR physically associates with the SCFβ-TRCP complex. A, Plk2-dependent loss of SPAR. HEK293T cells were transfected with 1 μg of pCMV-HA-Plk2 (lane 1), 1 μg of pCMV-myc-SPAR (lane 3), or 1 μg of pCMV-myc-SPAR together with either 1 μg of catalytically inactive pCMV-HA-Plk2D201A (lane 2) or increasing amounts of pCMV-HA-Plk2WT (0.3, 1, or 2 μg) (lanes 4-6). The total amount of transfected DNA was kept constant among all conditions with use of empty vector. Whole cell lysates were immunoblotted with Myc antibody to assess myc-SPAR levels. B, dominant negative versions of Cul1 and β-TRCP stabilize SPAR. pCMV-myc-SPAR (0.5 μg) and pCMV-HA-Plk2D201A/WT (1 μg) (catalytically inactive, lane 1; wild type, lanes 2-9) were co-expressed in HEK293T cells with 2.5 μg of either empty vector, dominant negative Cullins, or dominant negative F-box proteins. Changes in the abundance of myc-SPAR were determined by immunoblotting with anti-Myc antibody. C, F-box protein interaction screen. pCMV-myc-SPAR (0.6 μg), pCMV-HA-Plk2WT/K108M (0.6 μg), and pCMV-DNCul1 (2 μg) were co-expressed as shown with pCMV-GST (lane 2) or the indicated F-box proteins as GST fusions (0.6 μg) (lanes 3-22) in HEK293T cells seeded in 6-well plates. After 24 h, cell extracts were used for GSH-Sepharose pull-down assays, and proteins were immunoblotted with anti-GST and anti-Myc antibodies. Crude lysates were blotted as an input control. D, coimmunoprecipitation of SPAR and Plk2. Extracts of HEK293T cells transfected with pCMV-DNCul1 and pCMV-myc-SPAR (lane 1), pCMV-HA-Plk2 (lane 2), or both (lane 3) were immunoprecipitated using anti-Myc or anti-HA antibodies as indicated. E, formation of a SPAR·Plk2·β-TRCP·Cul1 complex with active Plk2. Lysates from cells transfected with pCMV-myc-SPAR (0.5 μg), pCMV-HA-Plk2WT/D201A (0.5 μg), pCMV-GST-β-TRCP (0.5 μg), and pCMV-FLAG-Cul11-452 (2 μg), as indicated, were incubated with GSH-Sepharose and immunoblotted with anti-Myc, anti-HA, anti-GST, and anti-FLAG antibodies as shown. Lanes 1-4 show lysates (6% of input of the GSH-Sepharose binding reactions).
Consistent with the idea that SPAR is a target of one of the SCF complexes, overexpression of dominant-negative Cul1 (DNCul1) in HEK293T stabilized myc-SPAR despite cotransfection of active Plk2 (Fig. 2B, lane 3), in agreement with our findings in neurons (see Fig. 1, B and E). Moreover, these cells accumulated the slower migrating form of myc-SPAR that presumably corresponds to phosphorylated SPAR. In contrast, a dominant-negative form of the related Cullin4 (DNCul4) had no effect on SPAR turnover (lane 4), indicating that the effect of DNCul1 was specific.
F-box proteins serve as substrate receptors in Cul1-based E3s. To uncover candidate F-box proteins for SPAR, we performed a cell-based interaction screen in HEK293T cells between SPAR and a panel of co-expressed GST-tagged F-box proteins. Potential complexes between SPAR and GST-tagged F-box proteins were isolated by incubating cell lysates with GSH-Sepharose beads. Previous studies had indicated that the interaction of F-box proteins with substrates can be detected in tissue culture cells that co-express the F-box protein and substrate in the presence of DNCul1, which blocks substrate degradation (14, 19). Among a panel of F-box proteins individually co-transfected with myc-SPAR and HA-Plk2, we identified an interaction between myc-SPAR and both GST-β-TRCP1 and GST-β-TRCP2 (Fig. 2C, lanes 3 and 4). Our ability to assay for endogenous interactions was precluded by the unavailability of suitable antibodies for immunoprecipitation. Although produced from different genes, β-TRCP1 and β-TRCP2 are ∼85% identical and are believed to have largely redundant functions (25). Importantly, the binding of β-TRCP1 and β-TRCP2 to myc-SPAR required co-transfection of active (wild type (WT)) Plk2 and was undetectable in the presence of catalytically defective Plk2K108M (Fig. 2C, lanes 5 and 6). Together with the Plk2-dependent slower gel mobility of SPAR (Fig. 2, A-C), this suggests that SPAR associates with β-TRCP in a phosphorylation-dependent manner, as is the case for all other β-TRCP substrates identified to date (13). SPAR did not associate with any of 16 other F-box proteins tested (Fig. 2C, lanes 7-22), including 5 F-box proteins, which like β-TRCP, bind substrates through their WD40 repeats (Fig. 2C, lanes 7-11). This indicates a high degree of specificity in the interaction between SPAR and β-TRCP.
Consistent with previous studies that showed interaction between Plk2 and SPAR (6), we found that Plk2 associated with SPAR in a coimmunoprecipitation assay upon co-expression of DNCul1 (Fig. 2D), further validating our heterologous cell system. Interestingly, we also discovered that the associations of Plk2 and β-TRCP with SPAR were not mutually exclusive, as we were able to detect a ternary interaction with Plk2 that was not precluded by the β-TRCP-SPAR interaction (Fig. 2E, lanes 6 and 8). GST-β-TRCP1 (and GST-β-TRCP2) associated with SPAR, Plk2, and the N terminus of Cullin 1 (Cul11-452/DNCul1), whereas catalytically inactive Plk2D201A did not support assembly of the full complex (Fig. 2E, lanes 5-8). Moreover, the Plk2-dependent slower mobility form of SPAR was enriched in the complex compared with the faster mobility form of SPAR (Fig. 2E, compare lanes 2 and 4 with 6 and 8).
SCFβ-TRCP Regulates SPAR Abundance and Turnover and Promotes Its Ubiquitination—To validate a role for the SCFβ-TRCP complex in Plk2-dependent SPAR turnover, we next examined SPAR abundance in HEK293T cells after expression of dominant-negative β-TRCP (β-TRCPΔF) (Fig. 2B). The ΔF-box construct is unable to assemble with Cul1 due to absence of the F-box motif, but it maintains its ability to interact with substrates of both β-TRCP1 and -2 and can thereby sequester substrates and block their turnover (14, 21, 24). Expression of β-TRCPΔF resulted in increased levels of SPAR, especially of the slower migrating form of SPAR that is dependent upon Plk2 activity (Fig. 2B, lane 5). In contrast, ΔF-box dominant-negative versions of other F-box proteins Fbw7α/β/γ (containing WD40 repeats) and Skp2 (containing leucine-rich repeats) failed to promote an increase in the steady state abundance of SPAR (Fig. 2B, lanes 6-9).
To directly examine whether β-TRCP proteins are required for Plk2-dependent SPAR turnover, we took advantage of a shRNA vector (shβ-TRCP) that is capable of suppressing protein expression of both human β-TRCP1 and β-TRCP2. This hairpin sequence and this particular shRNA vector have been validated for numerous β-TRCP substrates (14, 19, 26). Cells were transfected with expression constructs for GFP-SPAR and red fluorescent protein (RFP) to mark transfected cells and simultaneously transfected with shβ-TRCP or control vector (pRSP) in the presence of active Plk2 or a catalytically inactive version, Plk2D201A (Fig. 3A). We subsequently visualized cells for the presence of GFP-SPAR in RFP-positive cells (Fig. 3A). RFP-positive cells expressing Plk2, but not those expressing catalytically inactive Plk2D201A, displayed very low levels of GFP-SPAR in the presence of the control shRNA vector pRSP (Fig. 3A, quantified in B). In contrast, RFP-positive cells transfected with shβ-TRCP contained high levels of GFP-SPAR despite the co-transfection of active Plk2 (Fig. 3A, quantified in B). Thus, depletion of β-TRCP by RNAi substantially protected GFP-SPAR from Plk2-dependent degradation. Immunoblotting of transfected cell lysates confirmed that Plk2-induced degradation of myc-SPAR requires β-TRCP (Fig. 3C). As expected, Plk2 promoted loss of myc-SPAR in cells expressing a control shRNA that targets GFP (shGFP) (lanes 1 and 2). In contrast, myc-SPAR was not efficiently degraded in cells depleted of β-TRCP despite the presence of active Plk2 (lane 4). Furthermore, depletion of β-TRCP led to accumulation of Cdc25A, a known target of SCFβ-TRCP used here as a positive control (14, 18).
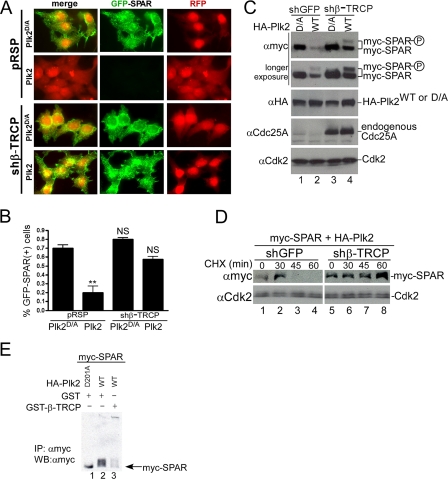
FIGURE 3.
SCFβ-TRCP regulates Plk2-dependent SPAR abundance, turnover, and promotes its ubiquitination. A and B, depletion of β-TRCP by RNAi protects GFP-SPAR from degradation in individual HEK293T cells. The indicated plasmids were transfected with pCMV-RFP as a co-transfection marker into HEK293T cells. After 96 h, cells were fixed and imaged for GFP-SPAR and RFP expression (A) and quantified as the percentage of RFP expressing cells that also expressed GFP-SPAR (B). Values represent the mean ± S.E. from three independent experiments, derived from analysis of 400 cells per experiment per condition (n = 1200 cells per condition). **, p < 0.01; NS, not significant; one-way analysis of variance, compared with control condition (pRSP+Plk2D/A). C and D, depletion of β-TRCP by RNAi stabilizes SPAR abundance and turnover in the presence of Plk2 activity. HEK293T cells were transfected with vectors expressing myc-SPAR (0.5 μg), WT, or catalytically inactive HA-Plk2 (D/A) (1 μg), and the indicated shRNA vector carrying a puromycin resistance selection marker (2.5 μg). 36 h post-transfection, cells were incubated with media containing 1 μg/ml puromycin to enrich for shRNA expression. Extracts were subsequently examined by immunoblotting 96 h post-transfection as indicated. Endogenous Cdc25A was probed to control for successful knockdown of β-TRCP. In panel D, HEK293T cells were transfected in an identical fashion to panel C, except that 24 h post-transfection the transfected cells were split among 5 wells in puromycin-containing media. After 48 h of puromycin selection, cells were treated with 25 μg/ml cycloheximide (CHX) and harvested at the indicated times before immunoblotting. E, expression of β-TRCP promotes Plk2-mediated SPAR ubiquitination. HEK293T cells were transfected with myc-SPAR (1 μg), His-Ub (1 μg), HA-Plk2 (1.5 μg, wild type or D201A), and GST or GST-β-TRCP (4.5 μg). Twenty hours post-transfection cells were treated with 25 μm MG-132 for 5 h and lysed in buffer containing 10 mm N-ethylmaleimide. Myc-SPAR was purified with c-Myc 9E10-agarose, resolved on 6% Tris-glycine SDS-PAGE gel, and immunoblotted (IB) with anti-Myc antibodies. IP, immunoprecipitates.
To directly examine whether β-TRCP is required for SPAR turnover, we performed a cycloheximide-chase experiment in cells expressing myc-SPAR and Plk2 in the presence of shGFP or shβ-TRCP (Fig. 3D). Myc-SPAR levels persisted after 45-60 min of cycloheximide treatment under conditions of β-TRCP depletion (shβ-TRCP) but disappeared after the same time period in control cells (shGFP) (Fig. 3D, lanes 7-8 compared with 3 and 4). Taken together, these data indicate that SCFβ-TRCP is critical for Plk2-dependent degradation of SPAR in heterologous cells.
To explore whether the SCFβ-TRCP complex promoted ubiquitination of SPAR, we immunoprecipitated myc-SPAR from MG-132-treated HEK293T cells and immunoblotted for modified myc-SPAR (Fig. 3E). Expression of active Plk2 resulted in detection of ubiquitinated SPAR, and the ubiquitination reaction was further driven in cells by ectopic expression of GST-β-TRCP (lanes 2 and 3). Together, this suggests that the Plk2-dependent turnover of myc-SPAR occurs through ubiquitination of SPAR.
Plk2-dependent Recognition of SPAR by β-TRCP Involves a Canonical Phosphodegron—SPAR is a large protein of 1804 amino acids containing two actin binding domains (Act1 and Act2), a RapGAP domain, a PDZ domain, and a guanylate kinase binding domain (GKBD) (Fig. 4A). Within the Act2 domain, we identified a candidate β-TRCP recognition motif (DSGIDT, residues 1304-1309) based upon the consensus β-TRCP recognition motif found in many of its targets (DpSGΦX(pS/T); Φ= hydrophobic residue, X = any residue, pS or pS/T = phosphoserine or threonine) (Fig. 4A). Initially, we surveyed three fragments of SPAR spanning the C terminus (C-1, C-2, C-3, Fig. 4A) for their ability to interact with co-transfected GST-β-TRCP1 in HEK293T cells in the presence of Plk2 and DNCul1. Fragments C-1 and C-2 (which contain the candidate phosphodegron) bound to GST-β-TRCP, whereas the C-3 construct lacking the phosphodegron failed to do so (Fig. 4B). N-terminal SPAR fragments that lack the DSGIDT motif also could not bind to β-TRCP (data not shown).
FIGURE 4.
A candidateβ-TRCP phosphodegron in SPAR. A, schematic representation of SPAR domains and SPAR fragments: actin-binding domains (Act1 and Act2), RapGAP, PDZ, and guanylate kinase binding (GKBD) domain. The candidate DSGIDT phosphodegron motif (residues 1304-1309) identified in the Act2 domain of SPAR closely resembles the consensus β-TRCP phosphodegron (inset). Boundaries of generated C-terminal fragments of SPAR are depicted; C-1, C-2, and Act2 fragments contain the putative phosphodegron motif, whereas C-3 does not. B and C, SPAR fragments spanning the β-TRCP phosphodegron bind to β-TRCP. HEK293T cells were transfected with vectors expressing SPAR fragments C-1, C-2, C-3 (B) or Act2 (C) (0.6 μg) either alone or with pCMV-GST or pCMV-GST-β-TRCP (0.6 μg). In addition, pCMV-HA-Plk2WT/K108M (0.6 μg) and DNCul1 (2 μg) were co-transfected as indicated. After 24 h, cell extracts were used for GSH-Sepharose pulldown assays, and proteins were immunoblotted with Myc antibodies. Crude lysates were blotted as an input control. D and E, phosphodegron-dependent binding to β-TRCP. Constructs expressing point mutations (S1305A, T1309A) in full-length SPAR (E) and the Act2 fragment (D) (myc-SPARAA or myc-Act2AA, 0.6 μg) were transfected into HEK293T cells along with pCMV-HA-Plk2WT/D201A (0.6 μg), pCMV-DNCul1 (2 μg), and pCMV-GST or pCMV-GST-β-TRCP (0.6 μg) as indicated. Cell lysates were incubated with GSH-Sepharose and immunoblotted with Myc antibodies. Crude extracts were resolved to control for input. Wild type constructs expressing myc-SPAR and myc-Act2 were used for comparison as a positive control for interaction with GST-β-TRCP. F, SPAR associates with endogenous SCFβ-TRCP1 complex in the presence of active Plk2 and dependent upon its phosphodegron. Constructs expressing full-length myc-SPAR (WT or AA) were co-expressed in HEK293T cells with active (WT) or catalytically inactive Plk2 (D201A). Before lysis and immunoprecipitation with 9E10-agarose, cells were treated with proteasome inhibitor MG-132 (25 μm) for 5 h. Proteins bound to 9E10-agarose were analyzed via immunoblotting using Myc antibodies and antibodies that recognized endogenous β-TRCP1 and Cul1. Myc-SPAR-C3, a fragment of SPAR that does not contain the phosphodegron served as a negative control, and myc-Cdc25A, a known target of SCFβ-TRCP1, served as a positive control. G, phosphodegron-dependent degradation of SPAR by Plk2 and β-TRCP. Myc-SPARWT and myc-SPARAA abundance was compared by immunoblotting with the indicated antibodies in HEK293T cells in the absence and presence of pCMV-HA-Plk2 in the background of empty vector or β-TRCPΔF.
A fourth fragment of SPAR spanning the Act2 domain was sufficient to interact with β-TRCP, and its association depended upon expression of active Plk2 (Fig. 4C, compare lanes 3 and 4 with 5 and 6). Mutation of Ser-1305 and Thr-1309 to alanines within the Act2 domain fragment alone (Act2AA) (Fig. 4D) or within full-length SPAR (SPARAA) (Fig. 4E) resulted in greatly diminished binding to β-TRCP compared with wild type SPAR constructs. Moreover, only in the presence of catalytically active Plk2 was full-length SPAR able to associate with endogenous β-TRCP and its associated Cul1 protein (Fig. 4F, compare lanes 2 and 3). Importantly, the C-3 fragment of SPAR that does not encompass the phosphodegron as well as full-length SPAR that carries point mutations of Ser-1305 and Thr-1309 did not support interaction with components of the endogenous SCFβ-TRCP complex (Fig. 4F, lanes 1 and 4). If this DSGIDT motif in SPAR is important for SPAR turnover, then mutation of this candidate phosphodegron should render SPAR resistant to Plk2-mediated turnover. Indeed, Plk2 efficiently promoted degradation of wild type SPAR but not the SPARAA mutant (Fig. 4G, lanes 1-4). Importantly, the accumulation of SPARAA seen in the presence of Plk2 was unchanged by the additional cotransfection of β-TRCPΔF (compare lanes 4 and 8), indicating that the mutation is specifically protecting turnover via the SCFβ-TRCP pathway. We further noted that SPARAA continued to migrate as multiple bands upon SDS-PAGE analysis of extracts from cells expressing Plk2 (Fig. 4E, input lanes 1, 2, and 4, Fig. 4G). This implies that the mobility shift seen with Plk2 expression does not solely rely on phosphorylation of residues Ser-1305 and Thr-1309.
SCFβ-TRCP Controls SPAR Abundance in Hippocampal Neurons—To investigate the role of the SCFβ-TRCP complex in post-mitotic neurons of the central nervous system, we first examined whether β-TRCP is present in the adult rat brain. mRNAs encoding β-TRCP1, β-TRCP2, and Cul1 are all present at high levels in rat brain, specifically in the CA1-CA3 regions of the hippocampus, the dentate gyrus, and cerebral cortex (Allen Brain Atlas).
Does the SCFβ-TRCP complex regulate turnover of SPAR in neurons? Unlike for the human mRNAs, we have yet to identify an shRNA sequence that can target both β-TRCP1 and β-TRCP2 transcripts efficiently in rat neurons. Co-transfection of different shRNAs that individually target β-TRCP1 and β-TRCP2 has not yielded satisfactory knockdown of both proteins, possibly due to low efficiency of cotransfection of both shRNA vectors in the same neuron. Therefore, we opted to disrupt endogenous SCFβ-TRCP function by overexpression of dominant-negative β-TRCP constructs β-TRCP1ΔF or β-TRCP2ΔF. As in Fig. 1, DIV16 cultured hippocampal neurons were transfected with either β-TRCP1ΔFor β-TRCP2ΔF and then super-infected 2 days later with Sindbis virus to drive expression of FLAG-tagged Plk2 and induce SPAR degradation (Fig. 5).
FIGURE 5.
Regulation of SPAR degradation in hippocampal neurons through the SCFβ-TRCP complex. A-C, dominant negative β-TRCP constructs block Plk2-dependent loss of SPAR in hippocampal neurons. As in Fig. 1, DIV16 dissociated rat hippocampal neurons were transfected with dominant negative β-TRCP plasmids (A and B) or control SkpΔF plasmid (C) and superinfected 2 days later with FLAG-tagged Plk2 driven by Sindbis virus (Sin-Plk2). Arrows point to cells that are both transfected and infected; arrowheads point to cells that are infected only. Yellow indicates the presence of both SPAR and Plk2 staining. D and E, quantification of SPAR immunostaining in somatic and proximal dendritic regions, as in Fig. 1. Values represent the mean ± S.E. n = 25-37 cells for all constructs in panel D; *, p < 0.05 indicates significant difference from theoretical mean of 100%, Student's t test (D). For panel E, n = 19-37 cells (infected only), n = 13-28 cells (infected and transfected), ***, p < 0.001, Mann Whitney test.
Transfection of control dominant-negative Skp2 F-box construct (Skp2ΔF) in the absence of Plk2 infection had no effect on endogenous SPAR levels relative to nearby untransfected and uninfected cells (Fig. 5C, quantified in D). In contrast, uninfected cells overexpressing β-TRCP1ΔF displayed higher levels of SPAR compared with nearby untransfected cells (p < 0.05, Fig. 5D). In neurons overexpressing β-TRCP2ΔF, SPAR levels trended upwards, but this did not reach statistical significance (p = 0.11, Fig. 5D). These results suggest that there may be some SCFβ-TRCP complex-dependent turnover of endogenous SPAR in hippocampal neurons cultured under basal conditions.
As expected, endogenous SPAR levels were close to undetectable in Plk2-infected neurons that were otherwise untransfected (Fig. 5, A-C, arrowheads, and E) as well as in Plk2-infected cells that had been previously transfected with Skp2ΔF (Fig. 5C, compare SPAR staining in cells marked by arrowheads (infected) and arrows (transfected and infected); quantified in E). In contrast, cells transfected with β-TRCP1ΔF or β-TRCP2ΔF showed no reduction in SPAR levels after infection with Plk2 Sindbis virus (Fig. 5, B and C, quantified in E). These results are consistent with disruption of the SCF complex through expression of DNCul1, which also prevented Plk2-dependent loss of SPAR (see Fig. 1), and together they demonstrate that the SCFβ-TRCP pathway is required for Plk2-induced SPAR degradation in neurons.
Because changes in SPAR expression can affect spine morphology (5) and synaptic strength (43), we wondered if blocking the SCFβ-TRCP pathway might also affect these aspects of neuronal function. Infection of DIV16 neurons with Plk2 led to a significant decrease in the size and number of spines, consistent with previous results (5). The additional overexpression of β-TRCPΔF in cells infected with Plk2, however, did not prevent the decrease in size and number of spines (supplemental Fig. S1, A-C). Similarly, blocking β-TRCP function had no effect on synaptic strength as assessed by recording of miniature excitatory postsynaptic currents (mEPSCs) (supplemental Fig. S1, D and E). These results are perhaps not surprising given that SCFβ-TRCP is likely to regulate additional synaptic substrates besides SPAR. Although we have clearly established the Plk2-SCFβ-TRCP-SPAR degradative pathway, its function in neurons remains to be resolved.
DISCUSSION
The UPP has received increasing attention in neurobiology as an important regulator of synapse development, synaptic transmission and plasticity, and the dynamic turnover of PSD proteins (27-31). Aberrant UPP function has been implicated in the pathogenesis of certain neurodegenerative disorders, e.g. Parkinson, Alzheimer, Huntington, and prion diseases, and amyotrophic lateral sclerosis (32, 33). To date most E3 substrate pairs in neurobiology have been identified in Caenorhabditis elegans and Drosophila melanogaster (34-42), and relatively little is known about the specific E3s that regulate synaptic proteins in the mammalian brain.
In this study we have uncovered a role for the E3 Ub-ligase SCFβ-TRCP in the degradation of a key postsynaptic scaffolding protein and enzyme (the RapGAP SPAR) that regulates the morphology of dendritic spines and the strength of synaptic transmission in hippocampal neurons (5, 43). Mammalian SCFβ-TRCP has been implicated in the ubiquitin-dependent turnover of several cell cycle and other signaling proteins, including IκBα, β-catenin, Cdc25A, Emi1, and the Period protein (14-17, 19). Moreover, it has been connected to the differentiation of neural progenitors through its targeting of REST transcription repressor (44, 45). Although we have defined the residues that are critical for SCFβ-TRCP-mediated degradation of SPAR, it is possible that SPAR degradation is also regulated by additional E3s, possibly in response to other signals and involving different determinants in the SPAR protein.
Previous studies have demonstrated that β-TRCP interacts with its substrates in a phosphorylation-dependent manner. The majority of β-TRCP substrates identified to date contain a DpSGΦXpS sequence (where pS is phosphoserine) that functions as a phosphodegron and directly binds to the WD40 repeats of β-TRCP (46). A variety of kinases have been implicated in the phosphorylation of β-TRCP phosphodegrons, and in many cases, multiple kinases collaborate to generate the phosphodegron, e.g. GSK3β/CKI (for β-catenin), Chk1 and another unknown kinase (for Cdc25A), and Cdc2/Plk1 (for Emi1) (14, 15, 47). Interestingly, Emi1 is one of several cell cycle regulators, in addition to Wee1, Emi2, and Claspin, that is phosphorylated by Plk family member Plk1 for degradation by the SCFβ-TRCP complex (47-55). Thus, Plk family members seem to collaborate with β-TRCP to regulate the degradation of a variety of substrates in different cellular contexts.
We found that SPAR contains a canonical β-TRCP phosphodegron sequence (1304DSGIDT) in its Act2 domain. Point mutations in Ser-1305 and Thr-1309 within SPAR inhibited Plk2-dependent SPAR degradation and its binding to β-TRCP, providing evidence that this sequence functions as a phosphodegron.
Currently, the identity of the kinase that phosphorylates 1304DSGIDT in SPAR is not definitively established. A direct demonstration that Plk2 phosphorylates Ser-1305 and Thr-1309 is currently lacking due, in part, to difficulty in purifying modified full-length SPAR. Thus, further studies are required to definitively correlate that these candidate sites are phosphorylated by Plk2. Nevertheless, because Plk2 promotes the SPAR-β-TRCP interaction and can phosphorylate SPAR in vitro (6), we favor the idea that Plk2 is the kinase responsible for phosphorylation of Ser-1305 and Thr-1309 in this phosphodegron. Supporting this idea is that Plk2 also binds to the Act2 domain of SPAR (6), so the kinase would be recruited to the vicinity of the phosphodegron. However, we cannot rule out the possibilities that Plk2 activity is required but not sufficient for phosphorylation of Ser-1305 and Thr-1309, that additional kinases are involved, or that additional phosphodegrons exist in SPAR. Plk2 is thus far the only kinase reported to promote SPAR degradation in neurons (6), although it is possible that other kinases could contribute as well. During early Xenopus and zebrafish development, casein kinase Iε has been implicated in the decrease of SPAR protein through Wnt signaling (56).
SPAR levels influence spine shape and size (5, 6), and SPAR degradation is involved in activity-dependent synaptic scaling (43). Our results here show that the SCFβ-TRCP complex targets SPAR for turnover in response to Plk2 activity. Nevertheless, blocking Plk2-dependent SPAR degradation through overexpression of the dominant-negative β-TRCP construct did not prevent Plk2-dependent changes in spine morphology. This suggests that the exact role of the Plk2-SCFβ-TRCP-SPAR pathway in neurons is still unclear. Plk2 may regulate the function or expression of additional proteins besides SPAR that are unaffected by disruption of the β-TRCP degradation pathway. Similarly, by globally disrupting the SCFβ-TRCP complex in the neuron, additional β-TRCP substrates may accumulate that offset the effects of accumulated SPAR protein. In C. elegans, the β-TRCP ortholog Lin-23 targets β-catenin/bar-1 for destruction in the ventral nerve cord, leading to altered glutamate receptor density (35).
Endogenous Plk2 expression is induced during periods of elevated neuronal activity and causes degradation of SPAR protein as well as thinning and elongation of spines (6). The accumulation of SPAR in spines upon inhibition of the SCFβ-TRCP pathway as well as the recent report that proteasomes redistribute into spines with synaptic stimulation (11) is consistent with the possibility that phosphorylation of SPAR by Plk2 leads to local interaction with β-TRCP and degradation of SPAR at postsynaptic sites. Because SPAR is a large scaffolding protein in the PSD that forms complexes with N-methyl-d-aspartate receptors and PSD-95 (5), regulation of SPAR degradation through the SCFβ-TRCP complex may be predicted to play a significant role in regulating the composition of proteins in the spine. Given the critical role of the proteasome in turning over many PSD proteins (10), it is likely that additional specific E3s will be identified that regulate synaptic composition.
Supplementary Material
Acknowledgments
We thank Azad Bonni (Harvard Medical School) for critical reading of the manuscript and Nelly Khidekel (Massachusetts of Technology) for the β-TRCP2ΔF construct.
This work was supported, in whole or in part, by National Institutes of Health Grants AG11085 and GM54137 (to J. W. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: PSD, postsynaptic density; Ub, ubiquitin; GAP, GTPase activating protein; SPAR, spine-associated Rap GTPase activating protein; Plk2, Polo-like kinase 2; UPP, ubiquitin-proteasome pathway; CMV, cytomegalovirus; HA, hemagglutinin; GFP, green fluorescent protein; SCF, Skp1/Cul1/F-box protein; WT, wild type; RFP, red fluorescent protein; GST, glutathione S-transferase; RNAi, RNA-mediated interference; DIV, days in vitro; shRNA, short hairpin RNA; E3, ubiquitin-protein isopeptide ligase; β-TRCP, β-transducin repeat-containing protein.
D. P. Seeburg, unpublished observation.
References
- 1.Kasai, H., Matsuzaki, M., Noguchi, J., Yasumatsu, N., and Nakahara, H. (2003) Trends Neurosci. 26 360-368 [DOI] [PubMed] [Google Scholar]
- 2.Tada, T., and Sheng, M. (2006) Curr. Opin. Neurobiol. 16 95-101 [DOI] [PubMed] [Google Scholar]
- 3.Bonhoeffer, T., and Yuste, R. (2002) Neuron 35 1019-1027 [DOI] [PubMed] [Google Scholar]
- 4.Carlisle, H. J., and Kennedy, M. B. (2005) Trends Neurosci. 28 182-187 [DOI] [PubMed] [Google Scholar]
- 5.Pak, D. T., Yang, S., Rudolph-Correia, S., Kim, E., and Sheng, M. (2001) Neuron 31 289-303 [DOI] [PubMed] [Google Scholar]
- 6.Pak, D. T., and Sheng, M. (2003) Science 302 1368-1373 [DOI] [PubMed] [Google Scholar]
- 7.Kauselmann, G., Weiler, M., Wulff, P., Jessberger, S., Konietzko, U., Scafidi, J., Staubli, U., Bereiter-Hahn, J., Strebhardt, K., and Kuhl, D. (1999) EMBO J. 18 5528-5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newton, S. S., Collier, E. F., Hunsberger, J., Adams, D., Terwilliger, R., Selvanayagam, E., and Duman, R. S. (2003) J. Neurosci. 23 10841-10851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turrigiano, G. (2007) Curr. Opin. Neurobiol. 17 318-324 [DOI] [PubMed] [Google Scholar]
- 10.Ehlers, M. D. (2003) Nat. Neurosci. 6 231-242 [DOI] [PubMed] [Google Scholar]
- 11.Bingol, B., and Schuman, E. M. (2006) Nature 441 1144-1148 [DOI] [PubMed] [Google Scholar]
- 12.Cardozo, T., and Pagano, M. (2004) Nat. Rev. Mol. Cell Biol. 5 739-751 [DOI] [PubMed] [Google Scholar]
- 13.Petroski, M. D., and Deshaies, R. J. (2005) Nat. Rev. Mol. Cell Biol. 6 9-20 [DOI] [PubMed] [Google Scholar]
- 14.Jin, J., Shirogane, T., Xu, L., Nalepa, G., Qin, J., Elledge, S. J., and Harper, J. W. (2003) Genes Dev. 17 3062-3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, C., Li, Y., Semenov, M., Han, C., Baeg, G. H., Tan, Y., Zhang, Z., Lin, X., and He, X. (2002) Cell 108 837-847 [DOI] [PubMed] [Google Scholar]
- 16.Margottin-Goguet, F., Hsu, J. Y., Loktev, A., Hsieh, H. M., Reimann, J. D., and Jackson, P. K. (2003) Dev. Cell 4 813-826 [DOI] [PubMed] [Google Scholar]
- 17.Winston, J. T., Strack, P., Beer-Romero, P., Chu, C. Y., Elledge, S. J., and Harper, J. W. (1999) Genes Dev. 13 270-283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Busino, L., Donzelli, M., Chiesa, M., Guardavaccaro, D., Ganoth, D., Dorrello, N. V., Hershko, A., Pagano, M., and Draetta, G. F. (2003) Nature 426 87-91 [DOI] [PubMed] [Google Scholar]
- 19.Shirogane, T., Jin, J., Ang, X. L., and Harper, J. W. (2005) J. Biol. Chem. 280 26863-26872 [DOI] [PubMed] [Google Scholar]
- 20.Bashir, T., Dorrello, N. V., Amador, V., Guardavaccaro, D., and Pagano, M. (2004) Nature 428 190-193 [DOI] [PubMed] [Google Scholar]
- 21.Welcker, M., Orian, A., Jin, J., Grim, J. E., Harper, J. W., Eisenman, R. N., and Clurman, B. E. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 9085-9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piva, R., Liu, J., Chiarle, R., Podda, A., Pagano, M., and Inghirami, G. (2002) Mol. Cell. Biol. 22 8375-8387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cullinan, S. B., Gordan, J. D., Jin, J., Harper, J. W., and Diehl, J. A. (2004) Mol. Cell. Biol. 24 8477-8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin, J., Ang, X. L., Shirogane, T., and Wade Harper, J. (2005) Methods Enzymol. 399 287-309 [DOI] [PubMed] [Google Scholar]
- 25.Guardavaccaro, D., Kudo, Y., Boulaire, J., Barchi, M., Busino, L., Donzelli, M., Margottin-Goguet, F., Jackson, P. K., Yamasaki, L., and Pagano, M. (2003) Dev. Cell 4 799-812 [DOI] [PubMed] [Google Scholar]
- 26.Fong, A., and Sun, S. C. (2002) J. Biol. Chem. 277 22111-22114 [DOI] [PubMed] [Google Scholar]
- 27.Patrick, G. N. (2006) Curr. Opin. Neurobiol. 16 90-94 [DOI] [PubMed] [Google Scholar]
- 28.Bingol, B., and Schuman, E. M. (2005) Curr. Opin. Neurobiol. 15 536-541 [DOI] [PubMed] [Google Scholar]
- 29.Yi, J. J., and Ehlers, M. D. (2005) Neuron 47 629-632 [DOI] [PubMed] [Google Scholar]
- 30.Cline, H. (2003) Curr. Biol. 13 514-516 [Google Scholar]
- 31.Jurd, R., Thornton, C., Wang, J., Luong, K., Phamluong, K., Kharazia, V., Gibb, S. L., and Ron, D. (2008) J. Biol. Chem. 283 301-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciechanover, A., and Brundin, P. (2003) Neuron 40 427-446 [DOI] [PubMed] [Google Scholar]
- 33.Petrucelli, L., and Dawson, T. M. (2004) Ann. Med. 36 315-320 [DOI] [PubMed] [Google Scholar]
- 34.Collins, C. A., Wairkar, Y. P., Johnson, S. L., and DiAntonio, A. (2006) Neuron 51 57-69 [DOI] [PubMed] [Google Scholar]
- 35.Dreier, L., Burbea, M., and Kaplan, J. M. (2005) Neuron 46 51-64 [DOI] [PubMed] [Google Scholar]
- 36.Juo, P., and Kaplan, J. M. (2004) Curr. Biol. 14 2057-2062 [DOI] [PubMed] [Google Scholar]
- 37.Kato, A., Rouach, N., Nicoll, R. A., and Bredt, D. S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 5600-5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao, E. H., Hung, W., Abrams, B., and Zhen, M. (2004) Nature 430 345-350 [DOI] [PubMed] [Google Scholar]
- 39.Myat, A., Henry, P., McCabe, V., Flintoft, L., Rotin, D., and Tear, G. (2002) Neuron 35 447-459 [DOI] [PubMed] [Google Scholar]
- 40.Nakata, K., Abrams, B., Grill, B., Goncharov, A., Huang, X., Chisholm, A. D., and Jin, Y. (2005) Cell 120 407-420 [DOI] [PubMed] [Google Scholar]
- 41.Stegmuller, J., Konishi, Y., Huynh, M. A., Yuan, Z., Dibacco, S., and Bonni, A. (2006) Neuron 50 389-400 [DOI] [PubMed] [Google Scholar]
- 42.van Roessel, P., Elliott, D. A., Robinson, I. M., Prokop, A., and Brand, A. H. (2004) Cell 119 707-718 [DOI] [PubMed] [Google Scholar]
- 43.Seeburg, D. P., Feliu-Mojer, M., Gaiottino, J., Pak, D. T., and Sheng, M. (2008) Neuron 58 571-583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guardavaccaro, D., Frescas, D., Dorrello, N. V., Peschiaroli, A., Multani, A. S., Cardozo, T., Lasorella, A., Iavarone, A., Chang, S., Hernando, E., and Pagano, M. (2008) Nature 452 365-369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westbrook, T. F., Hu, G., Ang, X. L., Mulligan, P., Pavlova, N. N., Liang, A., Leng, Y., Maehr, R., Shi, Y., Harper, J. W., and Elledge, S. J. (2008) Nature 452 370-374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, G., Xu, G., Schulman, B. A., Jeffrey, P. D., Harper, J. W., and Pavletich, N. P. (2003) Mol. Cell 11 1445-1456 [DOI] [PubMed] [Google Scholar]
- 47.Hansen, D. V., Loktev, A. V., Ban, K. H., and Jackson, P. K. (2004) Mol. Biol. Cell 15 5623-5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hansen, D. V., Tung, J. J., and Jackson, P. K. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 608-613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu, J., and Maller, J. L. (2005) Curr. Biol. 15 1458-1468 [DOI] [PubMed] [Google Scholar]
- 50.Mailand, N., Bekker-Jensen, S., Bartek, J., and Lukas, J. (2006) Mol. Cell 23 307-318 [DOI] [PubMed] [Google Scholar]
- 51.Mamely, I., van Vugt, M. A., Smits, V. A., Semple, J. I., Lemmens, B., Perrakis, A., Medema, R. H., and Freire, R. (2006) Curr. Biol. 16 1950-1955 [DOI] [PubMed] [Google Scholar]
- 52.Moshe, Y., Boulaire, J., Pagano, M., and Hershko, A. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 7937-7942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peschiaroli, A., Dorrello, N. V., Guardavaccaro, D., Venere, M., Halazonetis, T., Sherman, N. E., and Pagano, M. (2006) Mol. Cell 23 319-329 [DOI] [PubMed] [Google Scholar]
- 54.Rauh, N. R., Schmidt, A., Bormann, J., Nigg, E. A., and Mayer, T. U. (2005) Nature 437 1048-1052 [DOI] [PubMed] [Google Scholar]
- 55.Watanabe, N., Arai, H., Nishihara, Y., Taniguchi, M., Hunter, T., and Osada, H. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 4419-4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsai, I. C., Amack, J. D., Gao, Z. H., Band, V., Yost, H. J., and Virshup, D. M. (2007) Dev. Cell 12 335-347 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.