Abstract
SecA is an obligatory component of the Escherichia coli general secretion pathway. However, the oligomeric structure of SecA and SecA conformational changes during translocation processes are still unclear. Here we obtained the three-dimensional structure of E. coli wild-type full-length SecA in solution by single particle cryo-electron microscopy and determined its oligomeric organization. In this structure, SecA occurs as a dimer in which the two protomers are arranged in an antiparallel mode, with a novel electrostatic interface, and both protomers are in closed conformation. The system developed here may provide a promising technique for studying dynamic structural changes in SecA.
In bacteria, most extracytoplasmic proteins are translocated across the bacterial inner membrane via the general secretion pathway (the Sec pathway). SecA, along with SecYEG and other Sec proteins (1–6), is an intrinsic component of the Sec apparatus and has multiple functions (7). It interacts with most of the other components involved in protein translocation, including unfolded preprotein, acidic phospholipids, the SecYEG complex, and SecB (8–10). The SecA oligomeric state in the translocation process is still under debate. Some research groups have provided evidence that SecA functions as a dimer, whereas others have raised the possibility that SecA may be active as a monomer (11–13). In addition to controversy over the oligomeric state, the ternary conformational change in SecA coupled with preprotein translocation is not fully understood. To elucidate the mechanism of SecA function during preprotein translocation, the structure of SecA in the apo state and in complexes with other ligands should be investigated.
Since the first SecA structure was solved in 2002 (14) (1M6N), a total of six SecA structures from different species have become available. These structures provide detailed maps of the SecA domain organization, the SecA dimerization interface, and possible interaction sites with other components (14–19). Although all structures of SecA protomers are similar, two different conformations of full-length Bacillus subtilis SecA (bsSecA)3 have been found, i.e. open state and closed state (14, 16) (1TF5), and may provide hints on how the SecA conformation changes in the translocation process. However, which conformation SecA adopts in the ground state is not yet clear. The other remaining area to be elucidated is the oligomeric structure of SecA. Most of the SecA x-ray structure shows dimer packing (14, 17–19), whereas some structures are only present as monomers (16). Even in dimer SecAs, the dimerization interfaces differs among these structures. The full-length bsSecA structure shows dimeric SecA with two antiparallel SecA protomers related by crystallographic symmetry (14), and the dimerization interface is formed by N-terminal residues 1–10, the NBD1, and parts of the helical scaffold (14). The N-terminal 780-residue truncated fragment of the bsSecA (841 residues in total) was crystallized as a dimer in which two SecA protomers interact at the prominent groove formed by NBD2 and the preprotein interaction domain (18) (2BIM). Thermus thermophilus SecA has a parallel head-to-head dimerization mode (17) (2IPC). The Escherichia coli SecA fragment (9–861) has been crystallized as a dimer in which the dimerization of two antiparallel SecA protomers is mediated exclusively by DEAD motor residues (from the NBD and IRA2 domains) (19) (2FSF). These contradictory data suggest that the SecA structure may be highly flexible and may also diverge among species. The data also indicate that x-ray crystallography itself may not be able to fully solve the SecA structure. Researchers have used various methodologies to study SecA structures, including electron microscopy (EM) (20, 21) and NMR (22). A combination of structural studies using different methods may help to better understand SecA structures and functions.
In the present work, we used cryo-EM to study the ligand-free E. coli SecA (ecSecA) structure in solution, which is the ground state of SecA. Our EM results show that ecSecA adopts antiparallel dimeric packing in solution. This dimer interface mainly involves electrostatic interactions and differs from the ecSecA x-ray structure. The SecA structure also has a closed conformation.
MATERIALS AND METHODS
Protein Purification—SecA was purified as described (23) by stepwise elution on a Amersham Biosciences S-Sepharose column followed by gel filtration chromatography on a Superdex 200 column.
Specimen Preparation and Electron Microscopy—To prepare cryo-samples of SecA in solution, holey grids were covered with a layer of thin carbon film (5–10 nm) and glow-discharged for 30 s before use. Immediately before freezing, 3.5 μl of protein solution (100 μg/ml SecA, 20 mm Tris-Ac, pH 7.5, 1 mm DTT) was applied to the grid. The grid was then blotted and frozen in liquid ethane. The specimens were examined in a Phillips CM120 microscope operated at 100 kV. Films were developed in full strength D19 for 12 min. Images were digitized on Nikon Coolscan 9000ED scanner at a step size of 12.7 μm/pixel.
Three-dimensional Reconstruction of SecA—The three-dimensional reconstruction was mainly performed using the EMAN package (24). About 8,510 particles were manually selected. Due to the low resolution of current reconstruction, the particle images were only phase-flipped, and the amplitude was not corrected by contrast transfer function. Particle images were band pass-filtered (1–20 nm) and then centered and rotationally aligned using the CENALIGNINT command. About 10% of particles were discarded after this step due to bad alignment. The remaining 7,765 particles were classified into about 100 groups by the multivariance statistical analysis method. Eight typical class averages from 100 resulting classes were selected, and an initial model was obtained by angular reconstitution. Back projections of this initial model were computed at 10.1° intervals, and a projection match was performed on the data set of 7,765 particles for eight cycles. The final map was calculated from 6,249 particles with two-fold symmetry imposed. The resolution of the final map was estimated by both the Fourier shell correlation and the differential phase residual methods. The final three-dimensional map was low pass-filtered at 1.7 nm and was visualized by using the VMD software (25), and the isosurface threshold was selected to fit the molecular mass of 204 kDa assuming the protein density of 1.35 g/ml. The initial model was obtained by the EMAN package, and the refine procedure was performed using both SPIDER (26) and EMAN. There is no difference between models calculating by these two packages.
Docking Technique—The docking of SecA x-ray structure into the low resolution EM map was carried out using the Colores algorithm in the SITUS package (27).
RESULTS AND DISCUSSION
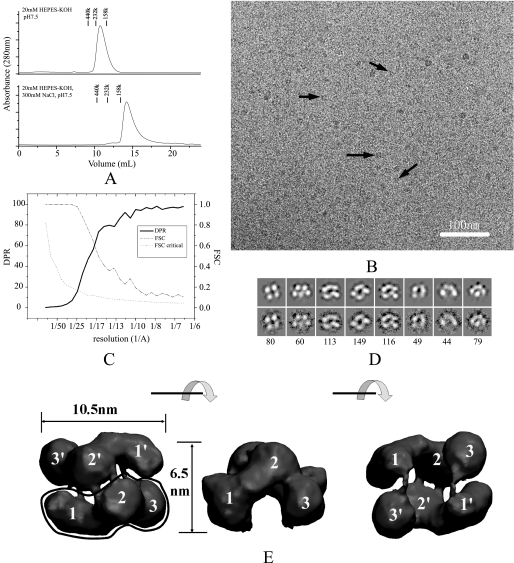
SecA Is a Distinct Antiparallel Dimer in Solution—It is generally accepted that most SecAs occur in dimeric form in the cytosol (12, 28). Previous studies have established that SecA undergoes monomer-dimer equilibrium in solution with a Kd of ∼1 μm that is sensitive to temperature and ionic strength (29). In our cryo-EM study, the protein concentration used was ∼1 μm, which favors the appropriate distribution density of single particles. To ensure that the majority of SecA was in the dimeric state, we kept the protein in low ionic strength buffer (10–20 mm Tris-Ac or HEPES-KOH, pH 7.5, 1 mm DTT). Under those conditions, SecA exhibited a major dimeric peak, with a molecular mass of ∼200 kDa, although it was still in a dimer-monomer equilibrium, as reflected by the asymmetric peak. In contrast, the major peak of SecA in high salt conditions had a monomer size (Fig. 1A).
FIGURE 1.
The E. coli SecA structure in solution. A, size-exclusion chromatography of SecA at 4 °C. A 125-μg sample of SecA was injected to a Superdex 200 column and eluted in a buffer of 20 mm HEPES-KOH, pH 7.5, or 20 mm HEPES-KOH, 300 mm KCl, pH 7.5. Marker proteins are: Ferritin (440 kDa), bovine catalase (232 kDa), and aldolase (158 kDa). B, cryo-EM image of 100 μg/ml SecA in buffer (20 mm Tris-Ac, pH 7.5, 1 mm DTT). Some individual particles are indicated by black arrows. The bar is 100 nm. C, resolution curves of the three-dimensional reconstruction. The resolutions calculated from two methods are shown: Fourier shell correlation (FSC) function (dashed line) and differential phase residual (DPR) function (solid line). The dotted line is the critical Fourier shell correlation function 3δ. D, distinct views of SecA cryo-EM samples. The top panel is the projection map of the reconstructed model; the bottom panel is the average map of all particles in this class; the number of particles in this class is indicated in the bottom panel. The box is 18.4 nm. E, surface representation of the three-dimensional reconstruction. The surface was rendered and displayed using the VMD software. Three views are shown: top view, side view, and bottom view. Each view was obtained after 90° rotation around the horizontal axis, as shown between the views. The three domains in the two subunits are designated as 1, 2, and 3, and 1′, 2′, and 3′.
Purified ecSecA molecules were frozen-hydrated, and images were recorded by cryo-EM in the low dose mode. Intact particles were apparent, but at low contrast, because of the relatively low molecular mass of SecA dimers (204 kDa; Fig. 1B). The protein particles on grids have no preferred orientation (supplemental Figs. S1 and S2). An angular reconstruction algorithm was used to produce the initial model, which was then refined by projection matching. Because the initial model showed two-fold symmetry, p2 symmetry was imposed in the refinement. A stable three-dimensional model was obtained from 6,249 cryo-particles with two-fold symmetry imposed. There was no difference between the symmetry-imposed model and the model constructed without any symmetry (supplemental Fig. S3). The resolution was estimated by comparing two independent data sets and gave a value of 1.7 nm for the Fourier shell correlation criterion (> 0.5) and 2.0 nm for the differential phase residual criterion (< 45°; Fig. 1C). The accuracy of the structure was assessed by the comparison of class averages with projections from the EM map at representative orientations (Fig. 1D and supplemental Fig. S4). The overall shape and dimensions of the reconstructed model are similar to a model deduced from a negative stained sample (30), but our model has more structural detail (Fig. 1E). The model shows a wedge-shaped structure composed of two antiparallel subunits. Each subunit has a prominent comma-shaped structure with three domains, designated domain 1, domain 2, and domain 3. These domains represent the distinct structural domains of the SecA subunits. In a SecA dimer, the two protomers have extensive contact through domain 2 as well as a connection between the subunits through domain 1 and domain 3. There is a deep cavity in the base surrounded by domain 3 and domain 1 from the two separate protomers.
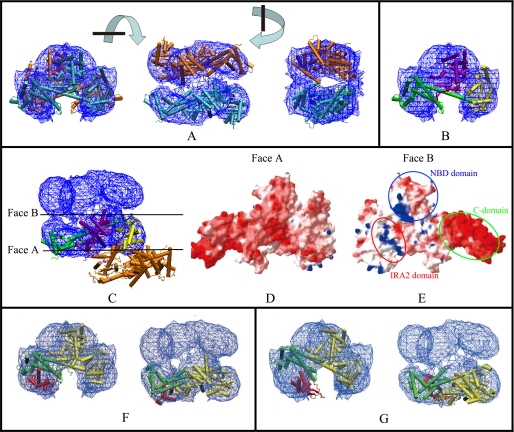
To identify the three domains, we docked the x-ray structure of ecSecA (2FSF, chain A) (14) into the cryo-EM map using the SITUS package. The Colores docking algorithm (correlation-based low resolution docking) placed two SecA molecules in the EM density map, and the x-ray structures fitted the EM density map well (Fig. 2A). We gave different colors to the different domains identified in the ecSecA structure so that we could locate the approximate domains in the EM map. Similarly, as assigned in the work of Papanikolau et al. (19), ecSecA was divided into four parts: the NBD domain (Fig. 2B, purple); Var, IRA2, and joint domain (Fig. 2B, yellow); PBD domain (Fig. 2B, red); and C-terminal 30-kDa domain comprising the IRA1, WD (wing subdomain), SD (scaffold subdomain), and CTD (C-terminal subdomain) subdomains (Fig. 2B, green). Domain 1 corresponds to the PBD domain and C-terminal domain; domain 2 corresponds to the NBD domain; and domain 3 corresponds to the domain comprising Var, IRA2, and the joint subdomain (Fig. 2B). The main disparity between the crystal structure (2FSF, chain A) and the EM map was located in the C-terminal domain and the PBD domain (domain 1), which are regions of high mobility (19).
FIGURE 2.
Docking of the cryo-EM three-dimensional map with the ecSecA crystal structure and comparison of the structures from cryo-EM and X-rays. A, chain A in the ecSecA (PDB number: 2FSF) was used for docking. The blue mesh represents the 1.5 σ contour of the electron density map. There are two molecules in the cryo-EM map, colored cyan and orange. Three views are shown. Each view is obtained after 90° rotation around the perpendicular or horizontal axis, as shown between the views. B, the docked crystal structure is rendered in different colors for different domains. For clarity, only one SecA protomer is shown. Similar to the assignment by Papanikolau et al. (19), ecSecA is divided into four parts: the NBD domain (purple); the Var, IRA2, and joint domains (yellow); the PBD domain (red); the C-terminal 30-kDa domain comprised of the IRA1, WD, SD, and CTD subdomains (green). As noted above, only a small fraction of PBD structure is seen in the refined x-ray structure. C, manual docking of one protomer (in different domain colors) in the x-ray dimeric structure (PDB number: 2FSF) into the electron density map. The colors in different domains were assigned as in B. The other protomer (orange) is obviously displaced in the EM map. The two side surfaces of the SecA protomer are designated Face A and Face B. D, view of Face A of the SecA protomer after electrostatic surface calculation. Electrostatic potential is represented as a tricolor gradient from blue (+1.8 kT (k, the constant of boltzmann; T, Thermodynamic temperature)) through white (neutral) to red (–5.8 kT), as represented using the Swiss-PDBviewer and calculated by DELPHY. E, view of Face B of the SecA protomer after electrostatic surface calculation. The patches of the C-domain, the IRA2 domain, and the NBD domain are indicated. The two protomers interact with each other through this interface. F and G, docking of the cryo-EM map with bsSecA x-ray structure in the closed (PDB number: 1M6N) (F) or open (PDB number: 1TF5) (G) state. For clarity, only one SecA protomer is shown. The PBD domain is red, and the C-domain is green. Both the side and the top view are shown.
SecA Adopts a Novel Dimerization Interface—Although two SecA protomers can be fitted into our EM density map, the resulting dimer differs from the reported ecSecA dimeric x-ray structure. We attempted to dock one protomer of the dimeric ecSecA into the EM map. However, when one molecule was fitted into the EM map, the other molecule was obviously outside the EM map (Fig. 2C). In the x-ray structure, SecA dimerization is exclusively mediated by DEAD motor residues. The contacts between the two protomers are primarily hydrophobic and are further stabilized by hydrogen bonds (19). Our cryo-EM map showed another dimerization interface. The two protomers are aligned antiparallel and have extensive interaction throughout all domains. The NBD domain from each protomer forms the major contact interface. In addition, the C-terminal domain of one protomer interacts with the IRA2 domain from another protomer. We calculated the electrostatic surface of ecSecA according to the x-ray structure. Face A, which is the solvent-exposed face, is exclusively negatively charged (Fig. 2D). Face B, which is the buried face of the two protomers, has alternatively negatively and positively charged patches (Fig. 2E). Specifically, SecA C-domain (660–901) is highly negative charged as shown by the strong red region in the figure. The IRA2 domain (420–590) is rich in positive residues, as shown by the blue patch in the figure. The NBD domain has separated positively and negatively charged patches. When the two protomers are aligned together in an antiparallel fashion through Face B, the negatively charged area of one protomer could interact with the positively charged area of the other. This interaction mode may explain why the electrostatic force is important to the dimerization of SecA and is consistent with results of salt concentration effects on the oligomeric state of SecA (29).
We directly demonstrated that dimeric SecA is in an antiparallel state in solution, which is consistent with previous experiments using fluorescence resonance energy transfer (31). However, the arrangement of the SecA protomers in SecA dimers differs from those found in any of the dimeric x-ray structures. This difference may come from the different constructs studied or from different buffer conditions used in EM analysis and x-ray crystallography. Further structural studies are required to determine the actual physiological structures, especially the SecA conformational changes that are coupled with the translocation process.
SecA Is in the Closed Dimerization Form—Previous studies have shown that there are two different conformations for SecA in the x-ray structures (16). In the so-called closed conformation, the PBD of SecA interacts extensively with the C-terminal domain to form a compact domain. In contrast, in the open conformation, PBD departs from the C domain and exposes most of the PBD surface to the solvent. We investigated whether the structure derived from cryo-EM is an open or closed structure. As noted above, only a small fraction of the PBD domain was visible in the refined ecSecA x-ray structure. Although the ecSecA PBD structure can be modeled from the bsSecA-PBD structure, it may not be accurate for discriminating the open or closed state. Because bsSecA and ecSecA are highly similar (52% identical in sequence) and both the open and the closed structures of bsSecA are available, we attempted to dock the two different x-ray structures of bsSecA (closed state, PDB number 1M6N; open state, PDB number 1TF5) into the EM map. Both x-ray structures can be docked into the EM map nicely, apart from the PBD part (Fig. 2, F and G). Only PBD in the closed conformation fits well into the EM map (Fig. 2F). This indicates that the ecSecA structure in solution revealed by cryo-EM is in a closed conformation.
Our results are consistent with a previous fluorescein maleimide labeling experiment (32), indicating that in solution alone, the ground state of SecA is in a closed conformation. A recent NMR study using methyl group labeling suggested that the PBD of SecA is in the open conformation in solution, with minimal interaction with the C-terminal tail (Fig. 2G), and that the equilibrium of the open and closed states is ∼90:10 (22). It is not known whether the labeling used in NMR studies favors one conformation over the other or whether the open and closed forms represent the monomer and dimer states, respectively. However, our data directly show that the SecA dimers in solution are in a closed state. Thus, this ground state is probably the physiological state before the SecA dimers in solution interact with the natural ligands, SecB, and the precursors, prior to interacting with SecYEG in membranes.
Supplementary Material
Acknowledgments
We thank W. Jiang for maintenance of the EM.
This work was supported by research grants from the National Nature Science Foundation of China and the National Basic Research Program 973 of China (Grant 2004 CB720005). This work was also supported in part by National Institutes of Health Grant 34766 (to P. C. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains three supplemental figures.
Footnotes
The abbreviations used are: bsSecA, B. subtilis SecA protein; ecSecA, E. coli SecA protein; EM, electron microscope; NBD, nucleotide binding domain; IRA1, intramolecular regulator of ATPase 1 subdomain; IRA2, intramolecular regulator of ATPase 2 domain; PBD, preprotein binding domain; C-domain, carboxyl domain; Var, the variable region; DTT, dithiothreitol; PDB, Protein Data Bank.
References
- 1.Cabelli, R. J., Chen, L., Tai, P. C., and Oliver, D. B. (1988) Cell 55 683–692 [DOI] [PubMed] [Google Scholar]
- 2.Cunningham, K., Lill, R., Crooke, E., Rice, M., Moore, K., Wickner, W., and Oliver, D. (1989) EMBO J. 8 955–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliver, D. B., and Beckwith, J. (1981) Cell 25 765–772 [DOI] [PubMed] [Google Scholar]
- 4.Manting, E. H., and Driessen, A. J. (2000) Mol. Microbiol. 37 226–238 [DOI] [PubMed] [Google Scholar]
- 5.Wickner, W., and Leonard, M. R. (1996) J. Biol. Chem. 271 29514–29516 [DOI] [PubMed] [Google Scholar]
- 6.Watanabe, M., and Blobel, G. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 9011–9015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Keyzer, J., van der Does, C., and Driessen, A. J. (2003) CMLS Cell Mol. Life Sci. 60 2034–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartl, F. U., Lecker, S., Schiebel, E., Hendrick, J. P., and Wickner, W. (1990) Cell 63 269–279 [DOI] [PubMed] [Google Scholar]
- 9.Hendrick, J. P., and Wickner, W. (1991) J. Biol. Chem. 266 24596–24600 [PubMed] [Google Scholar]
- 10.Woodbury, R. L., Topping, T. B., Diamond, D. L., Suciu, D., Kumamoto, C. A., Hardy, S. J., and Randall, L. L. (2000) J. Biol. Chem. 275 24191–24198 [DOI] [PubMed] [Google Scholar]
- 11.Or, E., Navon, A., and Rapoport, T. (2002) EMBO J. 21 4470–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Driessen, A. J. (1993) Biochemistry 32 13190–13197 [DOI] [PubMed] [Google Scholar]
- 13.Benach, J., Chou, Y. T., Fak, J. J., Itkin, A., Nicolae, D. D., Smith, P. C., Wittrock, G., Floyd, D. L., Golsaz, C. M., Gierasch, L. M., and Hunt, J. F. (2003) J. Biol. Chem. 278 3628–3638 [DOI] [PubMed] [Google Scholar]
- 14.Hunt, J. F., Weinkauf, S., Henry, L., Fak, J. J., McNicholas, P., Oliver, D. B., and Deisenhofer, J. (2002) Science 297 2018–2026 [DOI] [PubMed] [Google Scholar]
- 15.Sharma, V., Arockiasamy, A., Ronning, D. R., Savva, C. G., Holzenburg, A., Braunstein, M., Jacobs, W. R., Jr., and Sacchettini, J. C. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 2243–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osborne, A. R., Clemons, W. M., Jr., and Rapoport, T. A. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10937–10942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vassylyev, D. G., Mori, H., Vassylyeva, M. N., Tsukazaki, T., Kimura, Y., Tahirov, T. H., and Ito, K. (2006) J. Mol. Biol. 364 248–258 [DOI] [PubMed] [Google Scholar]
- 18.Zimmer, J., Li, W., and Rapoport, T. A. (2006) J. Mol. Biol. 364 259–265 [DOI] [PubMed] [Google Scholar]
- 19.Papanikolau, Y., Papadovasilaki, M., Ravelli, R. B., McCarthy, A. A., Cusack, S., Economou, A., and Petratos, K. (2007) J. Mol. Biol. 366 1545–1557 [DOI] [PubMed] [Google Scholar]
- 20.Wang, H. W., Chen, Y., Yang, H., Chen, X., Duan, M. X., Tai, P. C., and Sui, S. F. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 4221–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen, Y., Tai, P. C., and Sui, S. F. (2007) J. Struct. Biol. 159 149–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gelis, I., Bonvin, A. M., Keramisanou, D., Koukaki, M., Gouridis, G., Karamanou, S., Economou, A., and Kalodimos, C. G. (2007) Cell 131 756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen, X., Xu, H., and Tai, P. C. (1996) J. Biol. Chem. 271 29698–29706 [DOI] [PubMed] [Google Scholar]
- 24.Ludtke, S. J., Baldwin, P. R., and Chiu, W. (1999) J. Struct. Biol. 128 82–97 [DOI] [PubMed] [Google Scholar]
- 25.Humphrey, W., Dalke, A., and Schulten, K. (1996) J. Mol. Graph. 14 33–38 [DOI] [PubMed] [Google Scholar]
- 26.Frank, J., Radermacher, M., Penczek, P., Zhu, J., Li, Y., Ladjadj, M., and Leith, A. (1996) J. Struct. Biol. 116 190–199 [DOI] [PubMed] [Google Scholar]
- 27.Wriggers, W., and Chacon, P. (2001) Structure 9 779–788 [DOI] [PubMed] [Google Scholar]
- 28.Akita, M., Shinkai, A., Matsuyama, S., and Mizushima, S. (1991) Biochem. Biophys. Res. Commun. 174 211–216 [DOI] [PubMed] [Google Scholar]
- 29.Woodbury, R. L., Hardy, S. J., and Randall, L. L. (2002) Protein Sci. 11 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan, X. J., and Sui, S. (2005) Tsinghua Sci. Technol. 10 445–448 [Google Scholar]
- 31.Ding, H., Hunt, J. F., Mukerji, I., and Oliver, D. (2003) Biochemistry 42 8729–8738 [DOI] [PubMed] [Google Scholar]
- 32.van der Wolk, J. P., de Wit, J. G., and Driessen, A. J. (1997) EMBO J. 16 7297–7304 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.