Abstract
2,4-Dihydroxyquinoline (DHQ) is an abundant extracellular metabolite of the opportunistic pathogen Pseudomonas aeruginosa that is secreted into growth medium in stationary phase to concentrations comparable with those of the Pseudomonas quinolone signal. Using a combination of biochemical and genetic approaches, we show that PqsD, a condensing enzyme in the pqs operon that is essential for Pseudomonas quinolone signal synthesis, accounts for DHQ formation in vivo. First, the anthraniloyl moiety is transferred to the active-site Cys of PqsD to form an anthraniloyl-PqsD intermediate, which then condenses with either malonyl-CoA or malonyl-acyl carrier protein to produce 3-(2-aminophenyl)-3-oxopropanoyl-CoA. This short-lived intermediate undergoes an intramolecular rearrangement to form DHQ. DHQ was produced by Escherichia coli coexpressing PqsA and PqsD, illustrating that these two proteins are the only factors necessary for DHQ synthesis. Thus, PqsD is responsible for the production of DHQ in P. aeruginosa.
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that is a highly interactive organism secreting a variety of chemical signals collectively known as quorum-sensing molecules (1, 2). One intercellular signaling system unique to Pseudomonas and Burkholderia species is the production of hydrophobic quinolines (1, 3–5). These quinoline derivatives are 4-hydroxy-2-heptylquinoline (HHQ),2 3,4-dihydroxy-2-heptylquinoline (Pseudomonas quinolone signal (PQS)), and 2,4-dihydroxyquinoline (DHQ) (Scheme 1).
SCHEME 1.
PQS and HHQ are established quorum-sensing molecules that are potent regulators of gene expression, including many that are important determinants of virulence (1, 6, 7), and the N-oxide of HHQ exhibits antibacterial activity against Gram-positive organisms (8). Anthranilic acid is the precursor to both PQS and HHQ and is produced by the phnAB operon (9). All of the gene products in the pqsABCD gene cluster are thought to be essential for the formation of PQS and HHQ (9, 10). The prediction that PqsA is a CoA ligase is borne out by recent in vitro experiments illustrating that PqsA is an anthraniloyl-CoA synthetase (11). The details of HHQ biosynthesis are unknown, but the anthraniloyl-CoA intermediate is predicted to react with a β-keto fatty acid via a “head-to-head” condensation in a series of steps requiring PqsB, PqsC, and PqsD to form HHQ (12), which is converted to PQS by the hydroxylation of HHQ catalyzed by PqsH (3). DHQ is a recently identified secondary metabolite of P. aeruginosa. Although only pqsA in the pqsABCD gene cluster is reported to be required for DHQ biosynthesis (13), the addition of two carbons to the anthraniloyl moiety in the DHQ structure indicates that a condensing enzyme-like reaction is probably involved. Despite that both DHQ and HHQ contain an anthraniloyl moiety and require PqsA for the synthesis, there is no evidence of DHQ being the precursor of HHQ and PQS.
The condensing enzymes play a central role in bacterial fatty acid synthesis by elongating the growing acyl chains by two carbons (14). These enzymes catalyze the Claisen condensation reaction to form a carbon–carbon bond via a ping-pong kinetic mechanism involving an acyl-enzyme intermediate (15). In the first step, the acyl-enzyme is formed by the transfer of an acyl chain from either acyl-CoA or acyl-acyl carrier protein (ACP) to the conserved active-site cysteine residue to form a thioester linkage. In the second step, malonyl-ACP is decarboxylated, forming a carbanion, which attacks the acyl-enzyme intermediate to form the β-ketoacyl-ACP product (14, 16). In addition to their essential functions in bacterial fatty acid synthesis, condensing enzymes are also important components for the formation of secondary metabolites such as polyketides (17). Although PqsB, PqsC, and PqsD are designated as condensing enzymes in the sequencing data bases, only PqsD (or FabH1) possesses the Cys-His-Asn triad of active-site residues present in bone fide β-ketoacyl-ACP synthase III (FabH) condensing enzymes. PqsC lacks the Asn, and PqsB lacks all three of the active-site resides, although it retains some overall similarity to the N-terminal domains in the FabH condensing enzyme superfamily. In this study, we used a combination of biochemical and genetic analyses to demonstrate that the condensing enzyme activity of PqsD is responsible for the synthesis of DHQ in P. aeruginosa.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Reagents—PAO1 (pqsA, pqsB, pqsC, and pqsD) strains were cultured at 37 °C in LB or tryptic soy broth (BD Biosciences) unless specified otherwise. The reagents used in this study were purchased from the following suppliers: Sigma, anthranilic acid and [ring-UL-14C]anthranilic acid (specific activity of 61.1 mCi/mmol), malonyl-CoA, ATP, and DHQ; Analtech, Silica Gel H plates; Promega, restriction enzymes; and BD Biosciences, Pseudomonas isolation agar. Malonyl-ACP was prepared using AcpS (ACP synthase) with apo-ACP and malonyl-CoA as described (18). Anthraniloyl-CoA was prepared by PqsA with anthranilic acid and CoA. Briefly, the reaction contained 82 μm [ring-UL-14C]anthranilic acid (specific activity of 61.1 mCi/mmol), 200 μm CoA, 2 mm ATP, 10 mm MgCl2, and 12.5 μg/ml purified PqsA protein in 100 mm Tris-HCl (pH 7.5). The reaction was incubated at 37 °C for 30 min before being subjected to centrifugation in an Amicon Ultra concentration filter (10 kDa) to get rid of the PqsA protein. The complete conversion of anthranilic acid to anthraniloyl-CoA in the filtrate was confirmed by TLC.
Plasmid Construction and Mutant Generation—The pqsB, pqsC, and pqsD mutants generated by transposon insertions were obtained from the P. aeruginosa mutant collection at the University of Washington Genome Center. The DNA sequence located between the loxP sites in the transposon mutant was excised by Cre-mediated recombination by conjugation of SM10λ/pCre1 donors with the transposon mutant recipients, leaving a 189-bp insertion at the location of the original transposon. The pqsA mutant was created by gene replacement technology as described previously (19, 20). Briefly, a DNA fragment containing the upstream sequence of pqsA and ∼200 bp of pqsA 5′-end, a gentamicin cassette, and a DNA fragment containing downstream sequence of pqsA were cloned into pEX18ApGW (19). The resultant plasmid was conjugated from Escherichia coli SM10λpir into strain PAO1 with selection of gentamicin and carbenicillin on Pseudomonas isolation agar plates (BD Biosciences). Merodiploids formed via a single crossover event were resolved through 5% sucrose selection in the presence of gentamicin. The gentamicin resistance cassette was subsequently removed by Flp recombinase, resulting in a 1.3-kb deletion in pqsA. The presence of the deletion was verified by PCR and sequencing.
Protein Purification—The gene fragment encoding PqsA or PqsD was subcloned into expression plasmid pET-28a (Novagen) between the NdeI and HindIII restriction sites. The resultant plasmids were transformed into the Rosetta(DE3) strain (Novagen) to express the proteins with an N-terminal His tag. Transformed cells were grown at 37 °C in LB broth in the presence of 30 μg/ml kanamycin and 34 μg/ml chloramphenicol to an A600 of 0.8, heat-shocked at 42 °C for 15 min, and induced with 1 mm isopropyl β-d-thiogalactopyranoside at room temperature for 3 h. The cells were harvested by centrifugation; resuspended in 20 mm Tris-HCl (pH 8), 500 mm NaCl, and 10% glycerol containing 1 mm phenylmethylsulfonyl fluoride; and lysed with a French press. His-tagged protein in the cell-free extract was purified by nickel affinity chromatography as described previously (21). The fractions containing the purified protein were pooled, concentrated, and stored in 50% glycerol at –20 °C.
PqsA Activity Assay—The conversion of anthranilic acid to anthraniloyl-CoA by PqsA was measured by TLC. Briefly, each 40-μl reaction contained 82 μm [ring-UL-14C]anthranilic acid (specific activity of 61.1 mCi/mmol), 200 μm CoA, 2 mm ATP, 10 mm MgCl2, and 250 ng of purified PqsA protein in 100 mm Tris-HCl (pH 7.5). The reaction was initiated by the addition of PqsA. After a 10-min incubation at 37 °C, 20 μl of the reaction mixture was spotted on an activated Silica Gel H plate, which was developed in butanol/acetic acid/water (5:2:4, volume ratio). The dried TLC plate was exposed to a storage PhosphorImager screen, and the autoradiograph was scanned using a Typhoon 9200. The radioactivity was quantitated using ImageQuant Version 5.1 software.
Activity Assay of PqsD—The activity of PqsD was analyzed by TLC using malonyl-CoA and [14C]anthraniloyl-CoA produced in situ by PqsA or prepared in advance. The PqsD reaction contained 82 μm [14C]anthraniloyl-CoA, 125 μm malonyl-CoA, and the indicated concentration of PqsD in 100 mm Tris-HCl (pH 7.5). After incubation at 37 °C for 30 min, 20 μl of the 40-μl reaction mixture was spotted on an activated Silica Gel H plate. The TLC plate was developed in chloroform/methanol/acetic acid (95:5:1, volume ratio), and the product was quantitated and analyzed as described above.
Liquid Chromatography and Mass Spectrometry—The PqsA or PqsD reaction products with non-radioactive substrates were analyzed on a Waters Alliance HT liquid chromatography-mass spectrometry (LC-MS) system (Waters 2795 separation module linked to a Waters 2996 photodiode array detector), which was controlled by MassLynx Version 4.1 software. A 30-μl aliquot was injected onto a Waters XBridge C18 column (3.5 μm, 4.6 × 50 mm) and eluted using a methanol gradient at a flow rate of 1 ml/min: 0% methanol for 1 min, linear gradient from 0 to 60% methanol over 1.5 min, linear gradient from 60 to 95% methanol over 4.5 min, and linear gradient from 95 to 0% methanol over 2 min. The samples from the separation module were injected onto a Waters Micromass ZQ mass spectrometer and analyzed using positive ion mode electrospray mass spectrometry with a cone voltage of 60 V, a source temperature of 130 °C, and a desolvation temperature of 350 °C.
Extraction of P. aeruginosa Medium Supernatant—Strains of P. aeruginosa were grown in 50 ml of tryptic soy broth to stationary phase (A600 = 4.0). The cells were removed by centrifugation and filtration. The resultant medium supernatant was extracted twice with an equal volume of acidified ethyl acetate in a separating funnel. The two extraction mixtures were combined, dried under nitrogen, and resuspended in 0.1 ml of ethyl acetate for LC-MS analysis.
[14C]Anthranilic Acid Metabolic Labeling—Strains were grown in LB broth supplemented with 0.005% tryptophan and 33 μm [ring-UL-14C]anthranilic acid (specific activity of 61.1 mCi/mmol) and incubated at 37 °C for 16 h. The cells were removed by centrifugation, and the supernatant was collected. To detect DHQ levels,10 μl of supernatant was loaded onto a Silica Gel H plate (pre-activated at 90 °C for 1 h) and developed with chloroform/methanol/acetic acid (95:5:1, volume ratio). The dried TLC plate was exposed to a PhosphorImager screen, which was scanned with a Typhoon 9200.
Viability of MLE-12 Cells—MLE-12 cells were maintained in 2% fetal bovine serum supplemented HITES medium modified with transferrin (10 μg/ml), HEPES (10 mm), and l-glutamine (2 mm in addition to that in the base medium). Monolayers of MLE-12 cells were treated with 0.25% trypsin/EDTA solution for 15 min and subcultured in 6-well plates at 5% density. After overnight incubation, fresh medium containing 150 μm DHQ was added to DHQ-treated wells, whereas medium with an equal volume of solvent dimethyl sulfoxide (DMSO) was added to control wells. Each treatment was performed in triplicate. Total cells and viable cells excluding trypan blue stain were counted at 0, 24, and 48 h after DHQ or DMSO was added. To determine the effects of different concentrations of DHQ, the MLE-12 cells were treated with 10, 25, 75, and 150 μm DHQ or DMSO for 48 h.
RESULTS AND DISCUSSION
Functional Predictions in the pqs Operon of P. aeruginosa—The addition of an acetate moiety to anthranilate is required to form the basic structure of DHQ. Such two-carbon additions are typically carried out by condensing enzymes that decarboxylate malonyl-CoA (or malonyl-ACP) in the process of forming the new carbon–carbon bond. The first protein encoded by the pqs operon PqsA is an anthraniloyl-CoA synthetase (11), suggesting that anthraniloyl-CoA is the primer for the condensing enzyme step. Condensation reactions that use acyl-CoA primers are carried out by the β-ketoacyl-ACP synthase III or FabH family of proteins (Pfam08545). Therefore, the predicted amino acid sequences of the pqs genes were compared with the E. coli initiation condensing enzyme (FabH) to identify potential gene products involved in the postulated condensation reaction in DHQ biosynthesis in P. aeruginosa. PqsB, PqsC, and PqsD all have some degree of overall sequence similarity to E. coli FabH. Multiple sequence alignment of these three proteins with E. coli FabH showed that they are most highly related in the three areas surrounding the location of the Cys-His-Asn active-site triad in E. coli FabH (Fig. 1). PqsD possesses all three predicted active-site residues and is the most highly related protein to E. coli FabH, leading to its annotation as FabH1 in the P. aeruginosa genome data base. These similarities strongly suggest that PqsD carries out a condensation reaction. PqsC is the second most related protein. The Cys and His residues are present; however, PqsC lacks the conserved Asn. All three active-site residues are missing in PqsB. Previous work suggests that all three active-site residues contribute to catalysis (22), indicating that PqsB and PqsC are not functional condensing enzymes, although their overall relatedness to E. coli FabH suggests that they may possess a similar overall fold. These functional predictions indicate that the most likely candidate condensing enzyme for a role in DHQ formation in the pqs operon is PqsD.
FIGURE 1.
Sequence alignment of the FabH homologs encoded by the pqs operon in P. aeruginosa. PqsD, PqsC, and PqsB have three blocks of homologous sequence regions surrounding the active-site residues of E. coli (Ec) FabH (indicated by asterisks). Residues identical to those found in E. coli FabH are highlighted in black, and conservative substitutions are highlighted in gray.
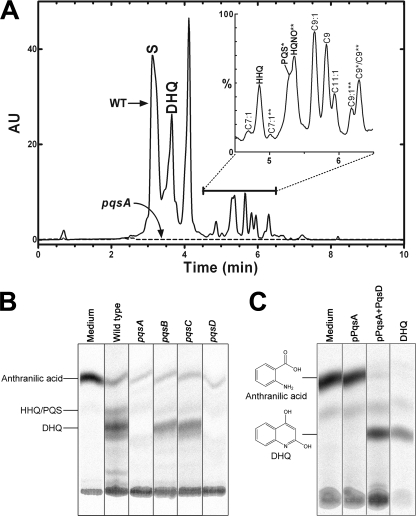
DHQ Production in the pqs Operon Mutants—To identify the functions of Pqs enzymes in DHQ production in vivo, pqsA, pqsB, pqsC, and pqsD mutant strains were tested for their abilities to accumulate DHQ in the growth medium by both LC-MS analysis and [14C]anthranilic acid labeling. Each of these mutant alleles knocked out the function of the transcribed protein by the insertion or deletion of DNA sequences and did not introduce elements that could potentially create a polar effect on the downstream genes in the operon. The efficiency of DHQ extraction from the growth medium by acidified ethyl acetate was evaluated using a DHQ standard. LC-MS result showed that, at 0.15 mg/ml, >90% of DHQ was extracted into ethyl acetate. Thus, ethyl acetate was used to extract the extracellular quinolines from the spent media of the wild-type and mutant strains. The wild-type PAO1 strain secreted DHQ into the medium in addition to HHQ, PQS, and 2-n-heptyl-4-hydroxyquinoline N-oxide species with different acyl chain modifications (Fig. 2, A and B). LC-MS analyses of the growth medium extracts showed that no detectable levels of DHQ or other quinolines were observed in the pqsA mutant strain, demonstrating a requirement of PqsA for the formation of DHQ and the quinoline signals (Fig. 2A). Of the three FabH homologs, only the PqsD function was required for DHQ production because both the pqsB and pqsC mutant strains produced DHQ, whereas the pqsD mutant strain did not secrete detectable DHQ (Fig. 2B). Consistent with these results using the Pseudomonas mutant strains, expression of pqsA plus pqsD in E. coli, an organism that does not produce DHQ, led to robust DHQ formation from extracellular [14C]anthranilic acid (Fig. 2C). These data show that PqsA and PqsD are both necessary and sufficient for DHQ biosynthesis. PqsA forms the anthraniloyl-CoA primer, which is converted to DHQ by PqsD.
FIGURE 2.
Extracellular metabolites produced by the wild-type or pqs mutant strains of P. aeruginosa. A, liquid chromatographs of the extracted spent media from the wild-type (WT; solid line) and pqsA (dashed line) strains. The solvent peak is indicated (S). The expanded mass trace of the wild-type extract between 4.5 and 6.5 min is shown in the inset. PQS, HHQ, and 2-n-heptyl-4-hydroxyquinoline N-oxide (HQNO) species with different acyl chains were identified by mass spectrometry. PQS species are indicated by single asterisks, and 2-n-heptyl-4-hydroxyquinoline N-oxide species are indicated by double asterisks; peaks without asterisks are HHQ species. AU, absorbance units. B, [14C]anthranilic acid metabolic labeling of the wild-type and pqs mutant strains of P. aeruginosa. C, [14C]anthranilic acid metabolic labeling of E. coli strains expressing Pqs proteins.
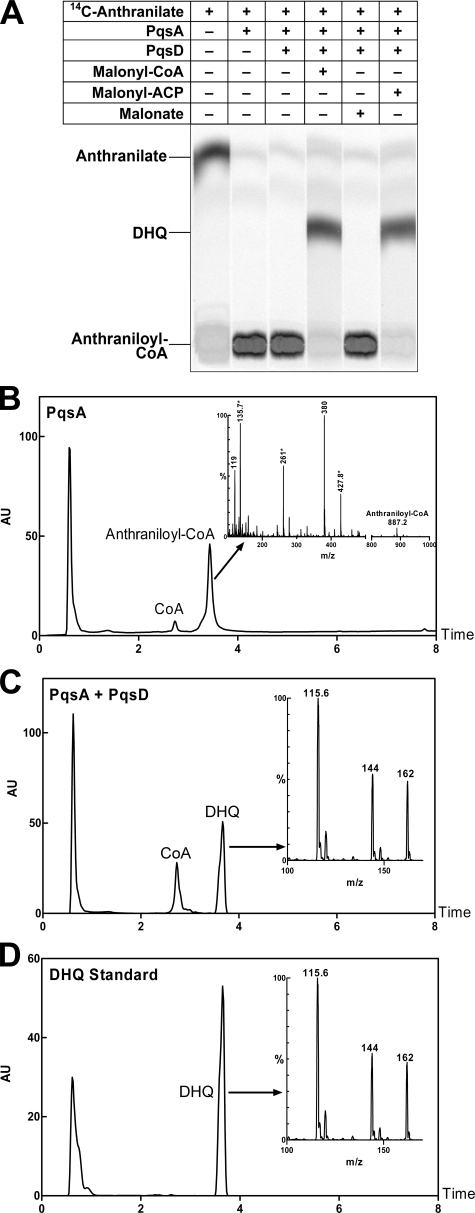
DHQ Formation by PqsD in Vitro—On the basis of the structure of DHQ, we hypothesized that DHQ is formed by the condensation of anthraniloyl-CoA with an activated form of a three-carbon acid, such as malonyl-CoA. Analyses of the pqs mutant strains demonstrated that PqsD was involved in DHQ formation in vivo; therefore, we purified recombinant PqsD protein to test this conclusion in vitro. Anthraniloyl-CoA, which was produced in situ by PqsA, was condensed with malonyl-CoA by PqsD to form a new product as shown by TLC (Fig. 3A). The identity of the product formed in the coupled reaction was confirmed by LC-MS to be DHQ, which exhibited the same elution time (3.67 min) and mass spectrum as the DHQ standard (Fig. 3, C and D). The conversion of anthraniloyl-CoA to DHQ was nearly complete in these experiments using 1 μg of each of the enzymes. As anticipated, anthraniloyl-CoA was the product in the presence of PqsA alone (Fig. 3B). Anthraniloyl-CoA was eluted at 3.43 min; the mass spectrum of the anthraniloyl-CoA product from the PqsA reaction revealed fragment peaks characteristic of both the anthraniloyl group and CoA (Fig. 3B). Malonyl-ACP was also a substrate for DHQ formation by PqsD, whereas malonic acid did not support DHQ formation (Fig. 3A).
FIGURE 3.
PqsD catalyzes the formation of DHQ in vitro. A, TLC analysis of PqsD reaction products showed that both malonyl-CoA and malonyl-ACP were substrates, whereas malonic acid was not. The reactants are indicated above each lane. The identity of the PqsD reaction product was determined by LC-MS analyses (B–D). (Each panel shows the liquid chromatograph with the mass spectrum of the indicated peak shown in the inset.) B, anthraniloyl-CoA was the product of PqsA. Anthraniloyl-CoA was eluted at 3.43 min, and the protonated molecular ion (M + H+) at m/z 887.2 was present. The ions indicated by asterisks were derived from CoA. The ion at m/z 119 was the anthraniloyl moiety; the ion at m/z 380 was the anthraniloyl moiety plus the CoA ion at m/z 261. AU, absorbance units. C, DHQ and CoA were the products of the PqsA-plus-PqsD reaction. D, shown are the results from LC-MS of the DHQ standard. DHQ was eluted at 3.67 min; the molecular ion of DHQ was at m/z 162. The fragment ions at m/z 144 and 115 are characteristic of DHQ (13).
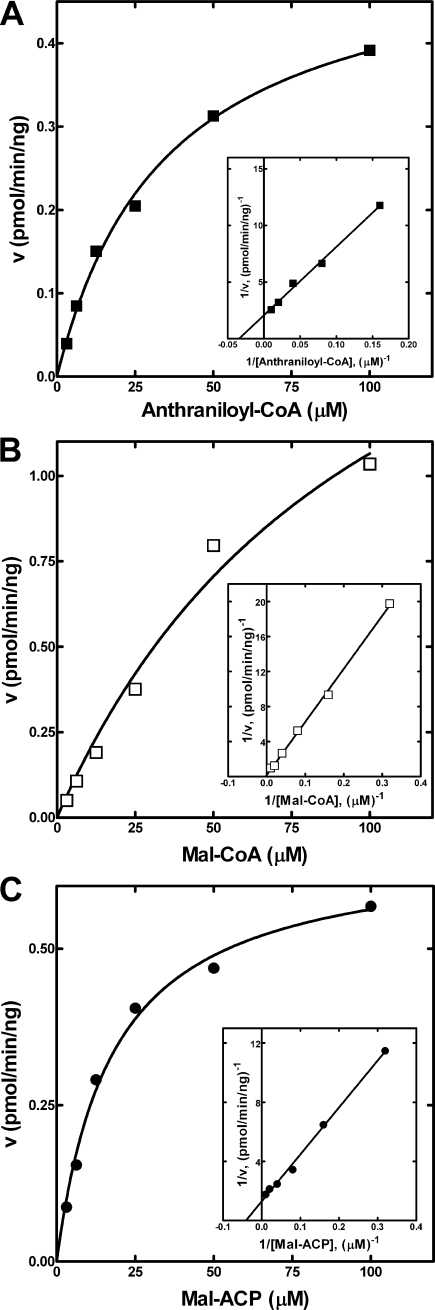
We determined the kinetic parameters of PqsD for different substrates. The apparent Km values of PqsD for anthraniloyl-CoA, malonyl-CoA, and malonyl-ACP were 35 ± 4, 104 ± 37, and 18 ± 2 μm, respectively (Fig. 4, A–C). PqsD exhibited a specific activity of 0.22 ± 0.03 pmol/min/ng for DHQ formation when assayed in the linear range. Consistent with the in vivo results, neither purified PqsB nor PqsC catalyzed DHQ synthesis in vitro.
FIGURE 4.
Kinetic parameters of PqsD. A, the apparent Km of PqsD for anthraniloyl-CoA was 35 ± 4 μm. B, the apparent Km of PqsD for malonyl (Mal)-CoA was 104 ± 37 μm. C, the apparent Km of PqsD for malonyl-ACP was 18 ± 2 μm. These kinetic parameters were determined by fitting the data with the Michaelis-Menten equation using Prism Version 5 software (GraphPad Software).
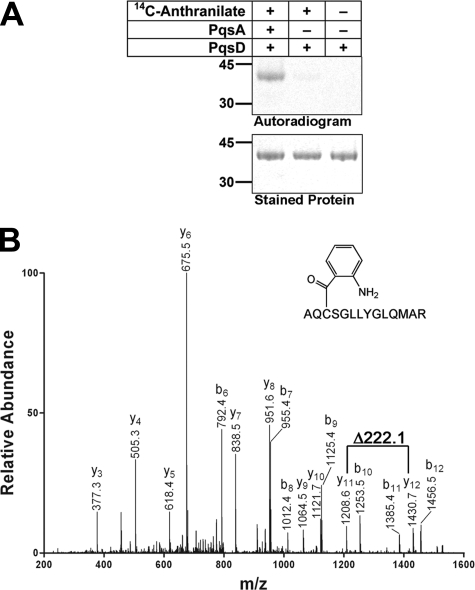
Formation of Anthraniloyl-enzyme Intermediate—A common property of condensing enzymes is the presence of a conserved active-site Cys that forms a thioester bond with an acyl primer. Our results showed that anthraniloyl-CoA was used as a primer to form an acyl-enzyme intermediate with PqsD. Incubation of PqsD with [14C]anthraniloyl-CoA produced by PqsA in situ led to the formation of a radioactive protein intermediate, indicating that the [14C]anthraniloyl group was transferred from CoA to the PqsD protein (Fig. 5A). [14C]Anthranilic acid was not a substrate for PqsD. LC-MS/MS analysis showed that the covalent modification of PqsD occurred at Cys144, which aligned with the active-site Cys of the E. coli FabH protein (Fig. 1). The increase in mass of 119 Da in peptide AQCSGLLYGLQMAR was consistent with the molecular mass of an attached anthraniloyl group (Fig. 5B). None of the other five Cys residues of PqsD was modified.
FIGURE 5.
Formation of the anthraniloyl-PqsD intermediate. A, formation of the anthraniloyl-PqsD intermediate. The upper panel is an autoradiograph of an SDS gel stained with SimplyBlue SafeStain (Invitrogen) shown in the lower panel. The composition is indicated above each lane. B, mass spectrum of a peptide from the anthraniloyl-PqsD sample that was eluted from an SDS gel and digested with trypsin protease. The deduced sequence of the peptide was AQCSGLLYGLQMAR; the cysteine in this peptide was predicted to be the active-site cysteine based on multiple sequence alignment (see Fig. 1). The mass difference between y11 and y12 is 222.1 Da, consistent with a cysteine residue (103.1 Da) covalently modified by an anthraniloyl moiety (119 Da).
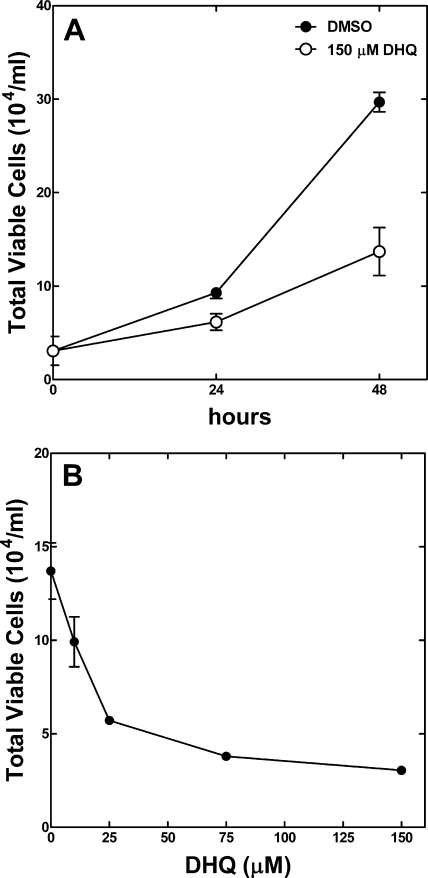
DHQ Inhibits MLE-12 Cell Viability—The lungs of cystic fibrosis patients are colonized by P. aeruginosa biofilms, where extracellular metabolites such as DHQ are accumulated. We tested the effect of DHQ on mouse lung epithelial MLE-12 cells in the presence of 150 μm DHQ, the concentration found in the growth medium of P. aeruginosa. The addition of DHQ to the cell culture medium significantly inhibited the growth of MLE-12 cells, particularly after 48 h of treatment (Fig. 6A). In addition to the reduced total cell numbers, the fraction of viable cells treated with DHQ was reduced to 30% compared with 85% of the DMSO-treated control cells. The inhibition of MLE-12 cells by DHQ was evident at 10 μm (Fig. 6B). The growth inhibitory effect of DHQ on the mouse lung epithelial cells suggests that DHQ contributes to the pathogenicity of P. aeruginosa infection. Further experiments are needed to determine how DHQ reduces cell viability and what the outcomes are from long-term exposure to DHQ.
FIGURE 6.
DHQ inhibits MLE-12 cell viability. A, MLE-12 cells were treated with either 150 μm DHQ (○) or an equal volume of DMSO (•) for 24 and 48 h. The addition of DHQ to the culture medium significantly inhibited MLE-12 cell viability. p < 0.05 at 24 h; p < 0.005 at 48 h. B, DHQ inhibited the viability of MLE-12 cells in a concentration-dependent manner. The MLE-12 cells were treated in the presence of different concentrations DHQ for 48 h. The error bars are means ± S.E. from three independent measurements.
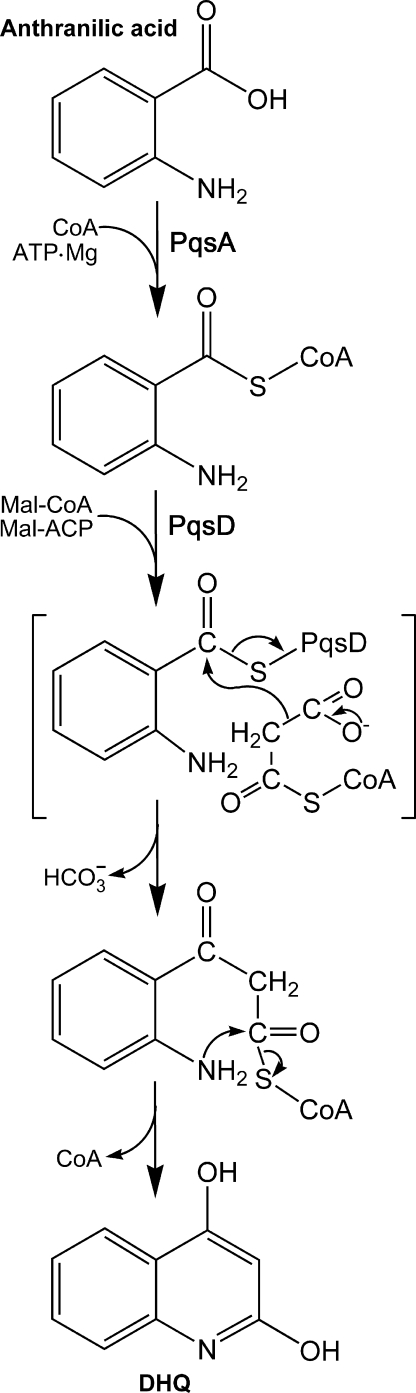
Conclusion—Our experiments led to the model for DHQ biosynthesis outlined in Fig. 7. The first step is the formation of anthraniloyl-CoA by PqsA. This intermediate acts as a primer to load the active-site cysteine of PqsD, forming an acyl-enzyme intermediate. Next, either malonyl-CoA or malonyl-ACP binds to the acylated enzyme, which catalyzes the condensation reaction with the release of bicarbonate and the formation of 3-(2-aminophenyl)-3-oxopropanoyl-CoA (or ACP). PqsD exhibited a slightly higher affinity for malonyl-ACP than for malonyl-CoA (Fig. 4, B and C) in vitro. However, without the information on the endogenous concentrations of these two substrates in P. aeruginosa, we cannot rule out one or the other as the real substrate for PqsD in vivo. In E. coli, malonyl thioesters compose typically 5% of both the CoA and ACP pools, but the total CoA concentration is 10-fold higher compared with ACP (23–26). These considerations suggest that malonyl-CoA provides the bulk of the two-carbon units to DHQ. The above-described intermediate then undergoes intramolecular ring closure to form DHQ. We postulate that this cyclization reaction may occur spontaneously, as is observed in the formation of triacetic acid lactone by the elongation condensing enzymes of fatty acid synthesis (18) and in the examples of spontaneous ring closure in polyketide synthases (27). However, the idea that the cyclization is promoted by the PqsD condensing enzyme cannot be ruled out because we have been unable to detect the postulated CoA thioester intermediate in any of our experiments.
FIGURE 7.
Proposed mechanism for DHQ synthesis. First, PqsA is an anthraniloyl-CoA synthetase that forms the intermediate that is used to prime the active-site cysteine of PqsD. This acyl-enzyme intermediate then reacts with malonyl (Mal)-CoA (or malonyl-ACP) to form 3-(2-aminophenyl)-3-oxopropanoyl-CoA (or ACP). This molecule undergoes a spontaneous intramolecular cyclization reaction to form DHQ.
The model illustrated in Fig. 7 is consistent with both the in vitro biochemical analyses of PqsD and the requirement for only PqsA and PqsD to produce DHQ both in P. aeruginosa and in the heterologous E. coli system. Our conclusion that PqsD is the condensing enzyme in DHQ synthesis is different from that reached by Lépine et al. (13). On the basis of the production of DHQ by a pqsB insertion mutant that they surmised would have a polar effect that would eliminate the expression of pqsC and pqsD, these investigators concluded that none of the pqs genes other than pqsA was involved in DHQ formation. However, they did not describe their mutant in sufficient detail to allow a comparison with our work, and their assumption that the pqsB gene disruption used in their study also disrupted transcription/translation of the downstream pqsCD genes was not experimentally verified. Although our data definitively establish PqsD as a condensing enzyme, its role in PQS biosynthesis remains ambiguous. It may catalyze the postulated head-to-head condensation of anthraniloyl-CoA with a β-keto acid, but it is equally likely that the condensing enzyme is responsible for the formation of the β-keto acid precursor.
Acknowledgments
We thank Ruobing Zhou for expert technical support and Raghu Chitta (Hartwell Center, St. Jude Children's Research Hospital) for the LC-MS/MS analyses of the PqsD peptides.
This work was supported, in whole or in part, by National Institutes of Health Grant GM 34496 and Cancer Center CORE Support Grant CA 21765. This work was also supported by the American Lebanese Syrian Associated Charities. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HHQ, 4-hydroxy-2-heptylquinoline; PQS, Pseudomonas quinolone signal; DHQ, 2,4-dihydroxyquinoline; ACP, acyl carrier protein; LC-MS, liquid chromatography-mass spectrometry; DMSO, dimethyl sulfoxide.
References
- 1.Diggle, S. P., Cornelis, P., Williams, P., and Camara, M. (2006) Int. J. Med. Microbiol. 296 83–91 [DOI] [PubMed] [Google Scholar]
- 2.Swift, S., Downie, J. A., Whitehead, N. A., Barnard, A. M., Salmond, G. P., and Williams, P. (2001) Adv. Microb. Physiol. 45 199–270 [DOI] [PubMed] [Google Scholar]
- 3.Deziel, E., Lépine, F., Milot, S., He, J., Mindrinos, M. N., Tompkins, R. G., and Rahme, L. G. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 1339–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pesci, E. C., Milbank, J. B., Pearson, J. P., McKnight, S., Kende, A. S., Greenberg, E. P., and Iglewski, B. H. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 11229–11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diggle, S. P., Lumjiaktase, P., Dipilato, F., Winzer, K., Kunakorn, M., Barrett, D. A., Chhabra, S. R., Camara, M., and Williams, P. (2006) Chem. Biol. 13 701–710 [DOI] [PubMed] [Google Scholar]
- 6.Diggle, S. P., Matthijs, S., Wright, V. J., Fletcher, M. P., Chhabra, S. R., Lamont, I. L., Kong, X., Hider, R. C., Cornelis, P., Camara, M., and Williams, P. (2007) Chem. Biol. 14 87–96 [DOI] [PubMed] [Google Scholar]
- 7.Dietrich, L. E., Price-Whelan, A., Petersen, A., Whiteley, M., and Newman, D. K. (2006) Mol. Microbiol. 61 1308–1321 [DOI] [PubMed] [Google Scholar]
- 8.Machan, Z. A., Taylor, G. W., Pitt, T. L., Cole, P. J., and Wilson, R. (1992) J. Antimicrob. Chemother. 30 615–623 [DOI] [PubMed] [Google Scholar]
- 9.Gallagher, L. A., McKnight, S. L., Kuznetsova, M. S., Pesci, E. C., and Manoil, C. (2002) J. Bacteriol. 184 6472–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Argenio, D. A., Calfee, M. W., Rainey, P. B., and Pesci, E. C. (2002) J. Bacteriol. 184 6481–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coleman, J. P., Hudson, L. L., McKnight, S. L., Farrow, J. M., III, Calfee, M. W., Lindsey, C. A., and Pesci, E. C. (2008) J. Bacteriol. 190 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bredenbruch, F., Nimtz, M., Wray, V., Morr, M., Muller, R., and Haussler, S. (2005) J. Bacteriol. 187 3630–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lépine, F., Dekimpe, V., Lesic, B., Milot, S., Lesimple, A., Mamer, O. A., Rahme, L. G., and Deziel, E. (2007) Biol. Chem. 388 839–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White, S. W., Zheng, J., Zhang, Y.-M., and Rock, C. O. (2005) Annu. Rev. Biochem. 74 791–831 [DOI] [PubMed] [Google Scholar]
- 15.Heath, R. J., and Rock, C. O. (2002) Nat. Prod. Rep. 19 581–596 [DOI] [PubMed] [Google Scholar]
- 16.Hirota, M., Kitagaki, M., Itagaki, H., and Aiba, S. (2006) J. Toxicol. Sci. 31 149–156 [DOI] [PubMed] [Google Scholar]
- 17.Hutchinson, C. R. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 3336–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, Y.-M., Hurlbert, J., White, S. W., and Rock, C. O. (2006) J. Biol. Chem. 281 17390–17399 [DOI] [PubMed] [Google Scholar]
- 19.Choi, K.-H., and Schweizer, H. P. (2005) BMC Microbiol. 5 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu, K., Choi, K.-H., Schweizer, H. P., Rock, C. O., and Zhang, Y.-M. (2006) Mol. Microbiol. 60 260–273 [DOI] [PubMed] [Google Scholar]
- 21.Heath, R. J., and Rock, C. O. (1996) J. Biol. Chem. 271 27795–27801 [DOI] [PubMed] [Google Scholar]
- 22.Davies, C., Heath, R. J., White, S. W., and Rock, C. O. (2000) Structure (Lond.) 8 185–195 [DOI] [PubMed] [Google Scholar]
- 23.Jackowski, S., and Rock, C. O. (1981) J. Bacteriol. 148 926–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackowski, S., and Rock, C. O. (1986) J. Bacteriol. 166 866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vallari, D. S., Jackowski, S., and Rock, C. O. (1987) J. Biol. Chem. 262 2468–2471 [PubMed] [Google Scholar]
- 26.Heath, R. J., and Rock, C. O. (1995) J. Biol. Chem. 270 15531–15538 [DOI] [PubMed] [Google Scholar]
- 27.Zhou, H., Zhan, J., Watanabe, K., Xie, X., and Tang, Y. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 6249–6254 [DOI] [PMC free article] [PubMed] [Google Scholar]