Abstract
Multiple pathways link expression of PTR2, the transporter of di- and tripeptides in the yeast Saccharomyces cerevisiae, to the availability and quality of nitrogen sources. Previous work has shown that induction of PTR2 by extracellular amino acids requires, in particular, SSY1 and PTR3. SSY1 is structurally similar to amino acid transporters but functions as a sensor of amino acids. PTR3 acts downstream of SSY1. Expression of the PTR2 peptide transporter is induced not only by amino acids but also by dipeptides with destabilizing N-terminal residues. These dipeptides bind to UBR1, the ubiquitin ligase of the N-end rule pathway, and allosterically accelerate the UBR1-dependent degradation of CUP9, a transcriptional repressor of PTR2. UBR1 targets CUP9 through its internal degron. Here we demonstrate that the repression of PTR2 by CUP9 requires TUP1 and SSN6, the corepressor proteins that form a complex with CUP9. We also show that the induction of PTR2 by amino acids is mediated by the UBR1-dependent acceleration of CUP9 degradation that requires both SSY1 and PTR3. The acceleration of CUP9 degradation is shown to be attained without increasing the activity of the N-end rule pathway toward substrates with destabilizing N-terminal residues. We also found that GAP1, a general amino acid transporter, strongly contributes to the induction of PTR2 by Trp. Although several aspects of this complex circuit remain to be understood, our findings establish new functional links between the amino acids-sensing SPS system, the CUP9-TUP1-SSN6 repressor complex, the PTR2 peptide transporter, and the UBR1-dependent N-end rule pathway.
Biological processes addressed by this study include the mechanisms and regulation of peptide import. Peptides can serve as a source of amino acids and nitrogen in all organisms. The import of di- and tripeptides (di/tripeptides) in the yeast Saccharomyces cerevisiae has been shown to be regulated by the N-end rule pathway, one proteolytic pathway of the ubiquitin (Ub)4-proteasome system (1–4). The N-end rule relates the in vivo half-life of a protein to the identity of its N-terminal residue (reviewed in Refs. 5–7). Although prokaryotes lack the Ub system, they still contain the N-end rule pathway, albeit Ub-independent versions of it (6, 8, 9). In eukaryotes, this pathway recognizes several kinds of degradation signals (degrons), including a set called N-degrons (5–7, 10). Specific N-degrons that are active (recognized) in a cell give rise to that cell's N-end rule. The determinants of an N-degron in a substrate protein are a destabilizing N-terminal residue that bears the unmodified N-terminal amino group, a substrate's internal Lys residue (the site of formation of a poly-Ub chain), and a nearby conformationally disordered region (11–13).
The N-end rule has a hierarchic structure. In eukaryotes, N-terminal Asn and Gln are tertiary destabilizing residues in that they function through their enzymatic deamidation, to yield the secondary destabilizing N-terminal residues Asp and Glu (Ref. 7, and references therein). Destabilizing activity of N-terminal Asp and Glu requires their conjugation to Arg, one of the primary destabilizing residues, by the ATE1-encoded Arg-tRNA-protein transferase (arginyl-transferase or R-transferase) (14–18). In mammals and other eukaryotes that produce nitric oxide (NO), the set of arginylated residues contains not only Asp and Glu but also N-terminal Cys, which is arginylated after its oxidation to Cys-sulfinate or Cys-sulfonate (14). The in vivo oxidation of N-terminal Cys requires NO, as well as oxygen (O2) or its derivatives (15, 18). The N-end rule pathway is thus a sensor of NO, through the ability of this pathway to destroy proteins with N-terminal Cys, at rates controlled by NO, O2, and their derivatives.
E3 Ub ligases of the N-end rule pathway are called N-recognins (7, 10, 19). They recognize (bind to) primary destabilizing N-terminal residues. (The term “Ub ligase” denotes either an E2–E3 holoenzyme or its E3 component.) At least four N-recognins, including UBR1, mediate the N-end rule pathway in mammals and other multicellular eukaryotes (7). The known N-recognins share a ∼70-residue motif called the UBR box. Mouse UBR1 and UBR2 are sequelogous (similar in sequence) 200-kDa RING-type E3 Ub ligases that are 47% identical. Several other mammalian N-recognins, either confirmed or putative ones, are HECT-type or SCF-type Ub ligases that share the UBR motif with the RING-type UBR1 and UBR2 but are largely nonsequelogous to them otherwise (20, 21).5
The functions of the N-end rule pathway include: (i) the sensing of heme, owing to inhibition, in both yeast and mammals, of the ATE1 R-transferase by hemin (Fe3+-heme), which also inhibits N-recognins, the latter at least in yeast; (ii) the sensing of NO and O2, and the resulting control of signaling by transmembrane receptors, through the conditional, NO/O2-mediated degradation of G-protein regulators RGS4, RGS5, and RGS16; (iii) regulation of import of short peptides, through the degradation, modulated by peptides, of CUP9, the repressor of import; (iv) fidelity of chromosome segregation, through degradation of a separase-produced cohesin fragment; (v) regulation of apoptosis, through degradation of a caspase-processed inhibitor of apoptosis; (vi) a multitude of processes mediated by the transcription factor c-FOS, a conditional substrate of the N-end rule pathway; (vii) regulation of the human immunodeficiency virus replication cycle, through degradation of human immunodeficiency virus integrase; and (viii) regulation of meiosis, spermatogenesis, neurogenesis, and cardiovascular development in mammals, and leaf senescence in plants (Refs. 2–4, 7, 14, 15, 17–19, 23–25, and references therein). Mutations in human UBR1, one of the E3s of the N-end rule pathway, are the cause of Johansson-Blizzard syndrome, which comprises mental retardation, physical malformations, and severe pancreatitis (26).
The N-end rule pathway of S. cerevisiae is mediated by a single N-recognin, UBR1, a 225-kDa sequelog of mammalian UBR1 and UBR2 (2, 10, 27). S. cerevisiae UBR1 contains at least three substrate-binding sites. The type-1 site is specific for basic N-terminal residues of polypeptides (Arg, Lys, and His). The type-2 site is specific for bulky hydrophobic N-terminal residues (Trp, Phe, Tyr, Leu, and Ile). The third binding site of UBR1 recognizes an internal (non-N-terminal) degron in target proteins. The third binding site of UBR1 is autoinhibited but can be allosterically activated through a conformational change that is caused by the binding of short peptides, such as dipeptides, to the other, type-1 and type-2, binding sites of UBR1. The known substrate of the third binding site of UBR1 is CUP9 (1–3), a homeodomain protein and transcriptional regulator, largely a repressor, of more than 30 genes in S. cerevisiae.6 The regulon of CUP9 includes PTR2, a gene encoding the main transporter of di/tripeptides (28, 29). The reversal of UBR1 autoinhibition by imported di/tripeptides with destabilizing N-terminal residues accelerates the UBR1-dependent ubiquitylation of CUP9, leads to its faster degradation, and thereby causes a derepression of PTR2. The resulting positive-feedback circuit allows S. cerevisiae to detect the presence of extracellular peptides and to react by increasing their uptake (2, 3).
The evolution of peptide-import circuits is under conflicting selective pressures, because the ability of a cell to import peptides confers both a benefit (utilization of peptides as food) and a vulnerability to toxins that resemble short peptides. Although PTR2 is the major transporter of di/tripeptides in S. cerevisiae, some di/tripeptides can also be imported (with a low efficacy) by DAL5, whose major function is the import of other nitrogen sources, such as allantoate and ureidosuccinate (4). Another set of S. cerevisiae peptide transporters comprises OPT1 (HGT1) and OPT2, which have partially overlapping functions, do not import di/tripeptides, but can import peptides of 4–5 residues. In addition, OPT1 (HGT1) is a high affinity importer of glutathione, a “noncanonical” tripeptide (Ref 30, and references therein). Similarly to the PTR2 transporter of di/tripeptides, the expression of OTP2 is down-regulated by CUP9, whereas the expression of OPT1 (HGT1) is independent of CUP9 (30). In addition to PTR2 and OPT2, the N-end rule pathway also controls the expression of DAL5, but in a manner opposite to that of the other two transporters: whereas CUP9 is a transcriptional repressor of PTR2 and OPT2, CUP9 apparently up-regulates the expression of DAL5 (28, 30). It is unknown whether CUP9 down-regulates a repressor of DAL5 or whether CUP9 acts, in the context of DAL5, as a transcriptional activator.
The induction of the PTR2 peptide transporter by di/tripeptides, a process controlled by the UBR1-CUP9 circuit (3), is just one of regulatory inputs that couple PTR2 expression to the availability and quality of nutrients. For example, PTR2 expression is down-regulated by certain nitrogen sources, including ammonia, but not by other nitrogen sources, such as urea and allantoin (31). The underlying systems, including the N-end rule pathway, ensure that a cell does not waste resources synthesizing large amounts of the PTR2 transporter in the absence of extracellular peptides, or when a more efficacious nitrogen source, such as ammonia, is present.
PTR2 is also induced by extracellular amino acids, particularly leucine or tryptophan (32). This response is likely to be adaptive in natural habitats, because the amino acids that S. cerevisiae (a scavenging heterotroph) encounters outside the laboratory tend to be the breakdown products of peptides and thus signify a likely presence of di/tripeptides. In addition, it has not been precluded that the S. cerevisiae PTR2 transporter might itself import amino acids, in addition to peptides. For example, AtPTR2 and AtCHL1, members of the PTR2 family in the plant Arabidopsis thaliana, transport histidine and nitrate, respectively (Ref. 33, and references therein). Either of the above possibilities may underlie the fact that amino acids regulate PTR2 expression.
Extracellular amino acids regulate PTR2 through the SPS (SSY1-PTR3-SSY5) pathway (34–40). SSY1, an integral membrane protein and a sensor of amino acids, is a sequelog of amino acid transporters but does not function as a transporter (36, 37, 41), a disposition that recurs with other nutrient sensors as well (37, 42). Both the inferred design of SSY1 and experimental evidence suggest that it is the concentration ratio of an amino acid across the plasma membrane, rather than the absolute concentration of extracellular amino acid that determines the signaling output by SSY1 (36). Activated SSY1 induces expression of a regulon that includes the PTR2 peptide transporter and amino acid transporters such as AGP1, BAP2, BAP3, TAT1, TAT2, and GNP1 (38). PTR3 and SSY5 are peripheral membrane proteins associated with SSY1 (41). SSY5 is a protease regulated, in particular, by PTR3. SSY5 can cleave, and thereby activate, the latent (conditionally cytosolic) transcriptional activators STP1 and STP2, leading to their import into the nucleus and the induction of genes that encode, in particular, amino acid transporters (34–37, 39, 40, 43).
In the present work, we show that an extracellular amino acid such as Trp acts via the SPS system to induce the PTR2-mediated import of di/tripeptides through the acceleration of degradation of CUP9 (the repressor of import) by the UBR1-dependent N-end rule pathway. The bulk of this effect of Trp on the rate of CUP9 degradation requires both SSY1 and PTR3. At the same time, no significant activation of the N-end rule pathway toward substrates with destabilizing N-terminal residues (i.e. toward substrates with N-degrons) was observed under these conditions, suggesting a differential regulation of three substrate-binding sites of the UBR1 Ub ligase. We also show that the repression of PTR2 by CUP9 requires the global corepressors TUP1 and SSN6, and that GAP1, a general amino acid transporter, strongly contributes to the induction of PTR2 by Trp. Although several aspects of this complex circuit remain to be understood, our findings establish new functional links between the amino acids-sensing SPS system, the CUP9-TUP1-SSN6 repressor complex, the PTR2 peptide transporter, and the UBR1-dependent N-end rule pathway.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Genetic Techniques—The S. cerevisiae strains used in this study are described in Table 1. AVY24 and AVY25 were constructed in the background of strain RJD347 (MATα ura3–52; a gift from Dr. R. Deshaies, California Institute of Technology). A PCR-based deletion strategy (44) was employed, using DNA fragments encoding the myc3-URA3-myc3 cassette to precisely delete SSY1 and PTR3 open reading frames (myc3 denotes 3 repeats of a nucleotide sequence encoding the myc epitope, flanked on either side by 50 bp of sequences identical to the sequences of either the SSY1 or the PTR3 gene, respectively). This technique was employed to produce deletions of the SSY1 and PTR3 open reading frames. We then selected for recombination within the myc3-URA3-myc3 module using 5-fluoroorotic acid, to produce strains AVY27 (MATα ura3–52 ssy1Δ::myc3) and AVY28 (MATα ura3–52 ptr3Δ::myc3), and verified the identity of these strains by Southern hybridization (data not shown). AVY26 (MATα ura3–52 ubr1Δ::HisG) was also a derivative of RJD347 (Table 1). It was constructed using the same PCR-based deletion strategy (44), except that a HisG-URA3-HisG cassette (see below) was used. Strains AVY30 (MATα leu2–3,112 ubr1Δ::LEU2), AVY31 (MATα leu2–3,112 cup9Δ::LEU2), and AVY32 (MATα LEU2) (Table 1) were derivatives of RJD350 (MATα leu2–3,112) (a gift from Dr. R. Deshaies).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotype | Reference |
|---|---|---|
| JD52 | MATaura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 | (56) |
| JD55 | MATaura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 ubr1-Δ1::HIS3 | (56) |
| AVY24 | MATα ura3-52 ssy1Δ::myc3-URA3-myc3 | This study |
| AVY25 | MATα ura3-52 ptr3Δ::myc3-URA3-myc3 | This study |
| AVY26 | MATα ura3-52 ubr1Δ::HisG | This study |
| AVY27 | MATα ura3-52 ssy1Δ::myc3 | This study |
| AVY28 | MATα ura3-52ptr3Δ::myc3 | This study |
| AVY30 | MATα leu2-3,112 ubr1Δ::LEU2 | (3) |
| AVY31 | MATα leu2-3,112 cup9Δ::LEU2 | (3) |
| AVY32 | MATα LEU2 | (3) |
| AVY50 | MATaura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 cup9Δ::LEU2 | CBY19 in Ref. 1 |
| AVY51 | MATaura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 ubr1Δ::HIS3 cup9Δ::LEU2 | CBY17 in Ref. 1 |
| AVY60 | MATaura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 ssn6Δ::HisG | This study |
| AVY61 | MATaura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 tup1Δ::HisG | This study |
| AVY62 | MATaura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 ubr1Δ::HIS3 ssn6Δ::HisG | This study |
| AVY63 | MATaura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 ubr1Δ::HIS3 tup1Δ::HisG | This study |
| AVY64 | MATaura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 cup9Δ::LEU2 ssn6Δ:::HisG | This study |
| AVY65 | MATaura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 cup9Δ::LEU2 tup1Δ::HisG | This study |
| AVY66 | MATaura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 ubr1Δ::HIS3 cup9Δ::LEU2 ssn6Δ::HisG | This study |
| AVY67 | MATaura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 ubr1Δ::HIS3 cup9Δ::LEU2 tup1Δ::HisG | This study |
| AVY107 | MATaura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 ubr1Δ::myc3 | (3) |
| RJD347 | MATα ura3-52 | A gift from Dr. R. Deshaies |
| RJD350 | MATα leu2-3,112 | A gift from Dr. R. Deshaies |
| M4272 | MATaSUC2 mal gal2 CUP1 ura3 Δ gap1 Δ stp1 Δ stp2 | (57) |
| M4270 | MATaSUC2 mal gal2 CUP1 ura3 Δ gap1 Δ stp2 | (57) |
| M4173 | MATaSUC2 mal gal2 CUP1 ura3 Δ gap1 Δ stp1 | (57) |
| M4054 | MATaSUC2 mal gal2 CUP1 ura3 Δ gap1 | (57) |
| M3750 | MATaSUC2 mal gal2 CUP1 ura3 | (58) |
| 23344c | MATα, ura3 | (59) |
| JA115 | MATα grr1Δ::kanMX2 ura3 | (59) |
PCR with S. cerevisiae genomic DNA was used to produce a fragment containing SSN6 flanked by 901 bp and 400 bp of upstream and downstream sequences, respectively. The fragment was ligated to NotI/EcoRI-cut pRS314 (45). The same procedure was used to construct a fragment containing TUP1 flanked by 459 bp and 400 bp of upstream and downstream sequences, respectively, followed by ligation to SpeI/ClaI-cut pRS416 (45). pSsn6Δ, which carried ssn6Δ::HisG-URA3-HisG, was constructed by subcloning the SSN6-containing NotI-EcoRI fragment into NotI/EcoRI-cut Bluescript (Stratagene), producing pBlueSSN6. Using PCR, a deletion (from 50 bp upstream of the SSN6 start codon to position 2,585 of its open reading frame) was introduced, while simultaneously creating an EclXI site at the upstream breakpoint of the deletion. The EclXI-PstI fragment of pAS315 (a gift from Dr. A. Sil, University of California, San Francisco), containing HisG-URA3-HisG, was then inserted. In vivo integration of the NotI-EcoRI fragment of the resulting plasmid (pSsn6Δ), carrying ssn6Δ::HisG-URA3-HisG, produced a deletion of SSN6 that spanned the region from –50 to 2585, relative to the start codon. pTup1Δ, which carried tup1Δ::HisG-URA3-HisG, was constructed similarly, using the SpeI-ClaI fragment containing TUP1 (from pTUP1), its subcloning into pBluescript (thus yielding pBlueTUP1), and PCR to produce a deletion allele of TUP1 (from 7 bp upstream of its start codon to position 2016 of the TUP1 open reading frame) that contained a (PCR-added) BamHI site at the deletion breakpoint. The BamHI-EcoRI fragment of pAS135 was then inserted into pBlueTUP1. In vivo integration of the SpeI-XhoI fragment of the resulting plasmid (carrying tup1Δ::HisG-URA3-HisG) produced a deletion of TUP1, from –7 to +2016, relative to the start codon. To select for recombination/excision within the integrated HisG-URA3-HisG cassette and thereby to produce ssn6Δ::HisG and tup1Δ::HisG alleles, 5-fluoroorotic acid-resistant yeast colonies were isolated, and the identities of strains were verified by Southern hybridization (data not shown). This procedure was performed in the JD52 background to construct strains AVY60 and AVY61; in the JD55 (ubr1Δ) background (Table 1) (23) to construct AVY62 and AVY63; in the AVY50 background (1) to construct AVY64 and AVY65; and in the AVY51 background (1) to construct AVY66 and AVY67 (Table 1).
SSN6 tagged at the C terminus with two copies of the myc epitope (SSN6myc2) and expressed from its own promoter on a high copy (2μ) plasmid was a gift from Dr. R. Zitomer (State University of New York, Albany). For plasmid-based complementation assays, plasmids encoding SSN6 (pSSN6) and TUP1 (pTUP1) were constructed using standard techniques. fDHFR-UbK48R-CUP9NSF was expressed from the PMET25 promoter in the plasmid pMET416UPRCUP9NSF, based on the low copy (CEN) vector p416MET25 (3). The previously described (27) pUb23-X plasmids expressed X-β-galactosidase reporter proteins (derived from the corresponding Ub-X-β-galactosidase fusions; X = Met, Ala, His, or Tyr) from the PGAL1 promoter in a high copy vector.
Coimmunoprecipitation, Pulse-chase, β-Galactosidase, and Northern Hybridization Assays—For CUP9-SSN6 coimmunoprecipitation assays, S. cerevisiae carrying the indicated plasmids were grown to A600 of ∼1 in SD medium (12) containing auxotrophic supplements. Cells from a 10-ml culture were harvested by centrifugation, washed in 1 ml of water, and resuspended in 0.8 ml of ice-cold buffer C (0.25 m NaCl, 1 mm EDTA, 50 mm HEPES, pH 7.5) containing ovalbumin at 1 mg/ml. Cells were disrupted by vortexing with 0.5 ml (packed volume) of 0.5-mm glass beads in the presence of protease inhibitors (Roche Applied Science). Extracts were prepared and immunoprecipitations were carried out as described (46), with two modifications. First, after incubation of an extract with antiFLAG beads (Sigma) for 45 min at 4 °C, the resulting immunoprecipitate was washed once with 1 ml of buffer C, and then twice more with 1 ml of buffer C lacking ovalbumin. Second, immunoprecipitates were eluted from anti-FLAG beads through a 15-min incubation, at room temperature, with the FLAG peptide at 0.5 mg/ml, in buffer C lacking ovalbumin. An equal volume of 2× SDS-PAGE sample buffer was added, followed by SDS-13% PAGE and immunoblotting with anti-myc antibody (Covance, Berkeley, CA), followed by a secondary horseradish peroxidase-conjugated goat anti-mouse antibody (Bio-Rad) at 1:1,000 dilution, and thereafter a detection of the latter antibody using ECL assay (GE Healthcare, Piscataway, NJ).
For pulse-chase assays, S. cerevisiae was grown to A600 of ∼0.8 in SHM medium (2% glucose, 0.1% allantoin, and 0.17% yeast nitrogen base without amino acids and without ammonium sulfate) that either contained or lacked tryptophan (98 μm, 20 μg/ml), an inducing amino acid (32). Cells were harvested, washed in 0.8 ml of SHM ± Trp, resuspended in 0.4 ml of the same medium, and labeled for 5 min at 30 °C with 0.16 mCi of 35S-EXPRESS (PerkinElmer Life Sciences). Thereafter cells were pelleted, resuspended in fresh SHM ± tryptophan containing 4 mm l-methionine and 2 mm l-cysteine, and incubated further at 30 °C. Samples of 0.1 ml were taken at indicated time points and transferred to chilled tubes, each containing 0.5 ml of 0.5-mm glass beads, 0.7 ml of ice-cold lysis buffer (1% Triton X-100, 0.15 m NaCl, 5 mm EDTA, 50 mm HEPES, pH 7.5), and a mixture of protease inhibitors (final concentrations: 1 mm freshly prepared phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, 0.5 μg/ml leupeptin, and 0.7 μg/ml pepstatin). Extracts were prepared, and immunoprecipitations were carried out as described previously (46), using anti-FLAG M2 beads (Sigma). Immunoprecipitated proteins were fractionated by SDS-13% PAGE, followed by autoradiography. Some of the pulse-chase assays were carried out in the presence of cycloheximide (at 0.2 mg/ml) during the chase.
For β-galactosidase (βgal) activity assays, S. cerevisiae carrying a pUB23-X plasmid (27) and expressing, from the PGAL1 promoter, an X-βgal test protein (derived from Ub-X-β-galactosidase, with X = Met, Ala, His, or Tyr) (47) were grown in SHM medium overnight. Cultures were diluted to A600 of ∼0.1 in SHM containing 2% galactose instead of glucose and either containing or lacking 98 μm Trp, followed by growth until A600 of ∼0.5. Cell extracts were thereafter prepared and processed for assays with o-nitrophenyl o-nitrophenyl-β-d-galactopyranoside as previously described (27). For Northern analyses of PTR2 induction by amino acids, cells were grown to A600 of ∼0.6 in SHM containing or lacking 98 μm (20 μg/ml) Trp. Total RNA was prepared as described previously (16), using an RNeasy Mini Kit (Qiagen, Valencia, CA). Samples containing 25 μg of RNA were electrophoresed in 1% formaldehyde-agarose gels, followed by blotting for Northern analysis (3). Northern assays of the effects of deletions of specific genes on PTR2 expression were carried out identically, in the absence of added Trp.
RESULTS AND DISCUSSION
Repression of PTR2 Transcription by CUP9 Involves the CUP9-TUP1-SSN6 Corepressor Complex—UBR1, the E3 Ub ligase of the S. cerevisiae N-end rule pathway, regulates the import of peptides by controlling, through degradation of the transcriptional repressor CUP9, the expression of the peptide transporter PTR2 (1). PTR2 mRNA was originally shown to be absent in ubr1Δ cells (1, 48), consistent with a mechanism in which a normally short-lived repressor of PTR2 becomes long-lived in the absence of UBR1, thereby increasing in concentration and shutting down PTR2 transcription. CUP9 was identified using a bypass screen for mutations that restored dipeptide uptake and PTR2 expression in ubr1Δ cells (1).
Further analysis, described below, of our collection of bypass mutants identified additional isolates that did not belong to the CUP9 complementation group. Many of these non-cup9 mutants (Fig. 1A) exhibited flocculation (aggregation during growth in liquid culture) and had a reduced mating ability. These phenotypes are characteristic of a defect in either TUP1 or SSN6, which encode two “global” corepressors that form a complex interfering with transcription initiation (reviewed in Ref. 49). The TUP1-SSN6 complex, which does not bind to DNA by itself, is recruited to individual promoters through interactions with specific DNA-binding proteins. One such protein is the mating-type regulator MATα2 (50). In the absence of either TUP1 or SSN6, the MATα2-mediated repression of MATa-specific genes is defective, resulting in mating-defective, pseudodiploid cells (49).
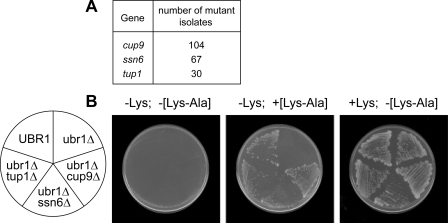
FIGURE 1.
Mutations in SSN6 or TUP1 rescue the block to dipeptide import in ubr1Δ S. cerevisiae, which lack the N-end rule pathway. A, a collection of 201 mutants in which the ability of a ubr1Δ strain to import dipeptides had been restored (cup9 members of that collection were examined in the earlier study (1)) were classed into complementation groups that included SSN6 and TUP1 using plasmid-based or mating-based complementation tests (1). The number of isolates in each complementation group, cup9, tup1, and ssn6, is shown. B, ssn6Δ and tup1Δ restore dipeptide import to a ubr1Δ strain. Strains JD52, AVY107, AVY51, AVY62, and AVY63 (Table 1), with relevant genotypes indicated on the plate diagram, were tested for their ability to use the dipeptide Lys-Ala as a source of essential nutrient, lysine. All strains were lys2 and thus auxotrophic for Lys. Strains were tested for growth on SD medium lacking Lys (left), or supplemented with Lys-Ala (center), or supplemented with Lys (right). Only strains capable of importing dipeptides could form colonies on the [–Lys; +[Lys-Ala]] plate. Deletion of either SSN6, TUP1, or (as shown previously (1)) CUP9 restored dipeptide import to a ubr1Δ strain (center plate).
To determine whether the above non-cup9 isolates (Fig. 1A) were either tup1 or ssn6 mutants, they were transformed with low copy plasmids that expressed either TUP1 or SSN6 from their natural promoters. These plasmids indeed suppressed the clumpy-growth and peptide-uptake phenotypes of all of the flocculating bypass mutants. The entire collection of our 201 bypass mutants (including cup9 mutants) was partitioned into complementation groups using a combination of the above plasmid-based complementation tests as well as mating-based complementation assays (the latter could not be comprehensive, owing to mating deficiency of some isolates). The 201 bypass suppressors were found to belong to three complementation groups, defined by CUP9, TUP1, and SSN6. Apart from genes required for cell viability, it is likely that our (nonconditional) screen was an exhaustive one, given the numbers of isolates in each of the three complementation groups thus identified: 104, 67, and 30 isolates of cup9, ssn6, and tup1 mutants, respectively (Fig. 1A).
To verify the role of TUP1 and SSN6 in regulating peptide uptake, we tested the effects of disrupting either one of these genes on the ability of a ubr1Δ strain to import dipeptides. Congenic (tup1Δ ubr1Δ) and (ssn6Δ ubr1Δ) double mutants were constructed using homologous recombination in the lysine-auxotrophic (lys2) genetic background, and the identities of mutants were verified by Southern hybridization (data not shown). The ability of these Lys-requiring double mutants to import dipeptides was then assayed by monitoring their growth on a minimal medium containing the dipeptide Lys-Ala as the sole source of Lys (Fig. 1B). A lys2 strain containing the intact UBR1 gene was able to import sufficient amounts of Lys-Ala dipeptide to support its growth on this medium, whereas a congenic (ubr1Δ lys2) strain could not grow under these conditions (it could be rescued by free Lys) (Fig. 1B). In contrast to the ubr1Δ lys2 strain, both tup1Δ ubr1Δ lys2 and ssn6Δ ubr1Δ lys2 triple mutants formed colonies on Lys-Ala plates, as did a cup9Δ ubr1Δ lys2 strain (Fig. 1B). Thus, a deletion of either CUP9, or TUP1, or SSN6 is each capable of restoring dipeptide uptake to a ubr1Δ strain. The slightly smaller colonies formed by the ssn6Δ ubr1Δ lys2 strain on Lys-Ala plates could be caused by a growth defect of ssn6Δ strains, a phenotype that we also observed with the ssn6Δ ubr1Δ lys2 strain on a control (Lys-supplemented) plate (Fig. 1B).
The fact that cup9, tup, and ssn6 mutants all bypassed the block to dipeptide uptake in a ubr1Δ strain suggested that CUP9 represses PTR2 expression by functioning as a part of a TUP1/SSN6-containing complex. To address this model, we compared the levels of PTR2 mRNA in ssn6Δ and tup1Δ mutants with those in other strains, in the absence of added PTR2 inducers such as dipeptides or amino acids. In a strain retaining all five of the studied genes, PTR2, UBR1, CUP9, SSN6, and TUP1, and also in a ubr1Δ strain, the level of PTR2 mRNA was low enough to be undetectable at the sensitivity of Northern assays used (Fig. 2A, lanes a and b; compare with lanes c–i). With ubr1Δ strains, no PTR2 mRNA could be detected even by higher sensitivity Northern blots (data not shown), whereas wild-type (UBR1) strains, in the absence of either added dipeptides or “inducing” amino acids, contained low but detectable levels of PTR2 mRNA, as shown previously (1, 3). In contrast, the levels of PTR2 mRNA were high in the absence of either CUP9, or SSN6, or TUP1, irrespective of the presence or absence of UBR1 (Fig. 2A, lanes c–i; compare with lanes a and b; see also Fig. 3A, lane c versus lane i). Thus, the tight block to dipeptide uptake in a ubr1Δ strain, caused by increased levels of the CUP9 repressor, could be bypassed not only through the removal of CUP9, but also through the removal of either SSN6 or TUP1 (Fig. 2A).
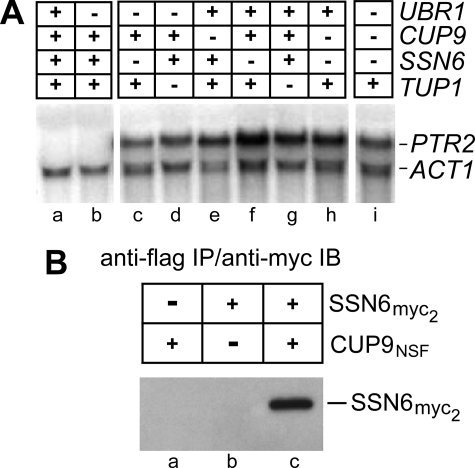
FIGURE 2.
CUP9 represses PTR2 transcription via SSN6 and TUP1. A, SSN6, TUP1, and CUP9 are required for repression of the peptide transporter gene PTR2. Northern analysis of PTR2 mRNA in the wild-type strain JD52, and in congenic ssn6Δ (AVY60), tup1Δ (AVY61), cup9Δ (AVY50), and ubr1Δ (AVY107) mutants, as well as in various combinations thereof (AVY51 and AVY62–AVY67) (Table 1). The status of each gene (–, deleted; +, wild-type allele) is shown in the upper diagram. PTR2 and ACT1 (actin) mRNA are indicated. S. cerevisiae strains were grown in SHM without PTR2-inducing amino acids or peptides. B, coimmunoprecipitation of myc-tagged SSN6 (SSN6myc2) with CUP9NSF, a FLAG-tagged CUP9 derivative (see “Results” and “Discussion”). Extracts from cells expressing the indicated combinations of SSN6myc2 and CUP9NSF were immunoprecipitated with a monoclonal anti-FLAG antibody, followed by SDS-PAGE and immunoblotting with a monoclonal anti-myc antibody (see “Experimental Procedures”).
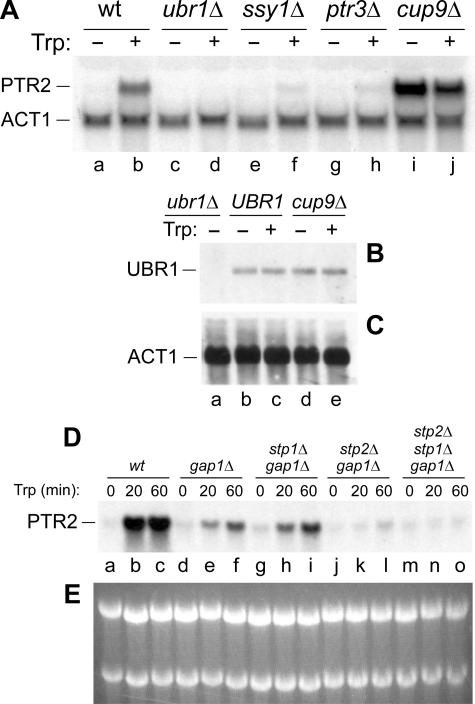
FIGURE 3.
Northern analyses of PTR2 and UBR1 under different conditions and in different genetic backgrounds. A, amino acid-inducible expression of PTR2 requires UBR1 and CUP9. Congenic strains of the indicated genotype (AVY32, AVY30, AVY24, AVY25, and AVY31) (Table 1) were grown in SHM medium containing or lacking 98 μm (20 μg/ml) tryptophan. Isolated total RNA was analyzed by Northern hybridization with PTR2 and ACT1 DNA probes, as indicated. B and C, levels of UBR1 expression do not change significantly in the presence versus absence of Trp. Total RNA from the strains AVY30-AVY32 (Table 1) was analyzed by Northern hybridization with UBR1 (B) and ACT1 (C). Lane a, RNA from ubr1Δ cells (negative control). Lanes b and c, RNA from wild-type (UBR1) cells either in the presence or absence of added Trp (98 μm). Lanes d and e, same as lanes b and c, but RNA from cup9Δ cells. D, Trp-mediated induction of PTR2 in wild-type and mutant S. cerevisiae. Total RNA was isolated from congenic strains of indicated genotypes either before the addition of 98 μm Trp, or 20 min thereafter, or 60 min thereafter, and the relative levels of PTR2 mRNA were determined by Northern hybridization. E, ethidium-stained rRNA (loading controls) in agarose gel before the transfer of RNA to hybridization membrane.
High levels of PTR2 mRNA in the absence of either CUP9, SSN6, or TUP1 (Figs. 2A and 3A) indicated that each of these proteins was essential for repression of the PTR2 gene. Both the demonstrated functional dependence of CUP9 on SSN6/TUP1 (Figs. 1 and 2A) and the close analogy between the homeodomain repressors CUP9 and MATα2, the latter of which is known to interact with SSN6/TUP1 (see above), suggested that CUP9 also interacted with SSN6/TUP1. To address this experimentally, we carried out a coimmunoprecipitation assay, using epitope-tagged SSN6 and CUP9 (Fig. 2B). Because overexpression of wild-type CUP9 inhibits cell growth, most likely owing to its interactions with DNA (data not shown), we employed a previously constructed nontoxic derivative of CUP9 (3). CUP9 interacts with DNA through its homeodomain motif. Based on the previously studied interactions of other homeodomain proteins with DNA, an Asn → Ser substitution at position 265, within the recognition helix of the CUP9 homeodomain, was predicted to strongly reduce the affinity of CUP9 for DNA without causing a significant structural perturbation. This nontoxic CUP9 derivative, tagged at the C terminus with the FLAG epitope, was termed CUP9NSF (3).
Extracts were prepared from S. cerevisiae stably expressing both CUP9NSF and SSN6 (SSN6myc2) (the latter C-terminally tagged with a double myc epitope), followed by immunoprecipitation with anti-FLAG antibody, SDS-PAGE, and immunoblotting with anti-myc antibody. SSN6myc2 was coimmunoprecipitated with CUP9NSF in this assay (Fig. 2B, lane c). No SSN6myc2 was detected in immunoprecipitates of extracts from cells that did not also express CUP9NSF (Fig. 2B, lane b), and the expression levels of CUP9NSF and SSN6myc2 were similar in cells that coexpressed the two proteins (data not shown). Thus, in agreement with the above expectation, these results (Fig. 2B) suggested that untagged SSN6 and untagged CUP9, at their (lower) natural expression levels in S. cerevisiae, would also form a complex that is analogous to the SSN6/MATα2 complex (49), a model that accounts for the observed functional interdependence among CUP9, SSN6, and TUP1 (Figs. 1B, 2A, and 3A). Although we did not examine the immunoprecipitated CUP9NSF-containing complexes for the presence of TUP1, extensive previous work has established that SSN6 and TUP1 form a stable complex both in vivo and as purified proteins (Ref. 49, and references therein), indicating that it is the complex of SSN6 and TUP1 that interacts with CUP9, similarly to the previously characterized interaction between SSN6/TUP1 and MATα2, another homeodomain repressor (49).
Both CUP9 and UBR1 Are Required for Inducibility of PTR2 by Amino Acids—When an amino acid, particularly a bulky hydrophobic one such as Leu or Trp, is added to a culture of S. cerevisiae growing on a poor nitrogen source such as, for example, allantoin, cells induce their PTR2 gene and thus increase their capacity for the import of di/tripeptides (32). Effective concentrations of extracellular Leu or Trp can be as low as 1 μm (32). Previous work has shown that an accelerated degradation of CUP9 by the UBR1-dependent N-end rule pathway in response to imported dipeptides with destabilizing N-terminal residues, strongly enhances PTR2 expression (3). This result led us to consider whether other signals that up-regulate PTR2 expression, e.g. amino acids, may also act through acceleration of CUP9 degradation.
To begin addressing this possibility, we examined the effects of Trp, a strongly inducing amino acid, on PTR2 mRNA levels in several S. cerevisiae strains, AVY32, AVY24, AVY25, AVY30, and AVY31, that were constructed to be completely prototrophic, and in particular did not require extracellular amino acids for viability (Table 1). These strains were grown in allantoin-based media (SHM) containing or lacking Trp. Allantoin is a nonrepressing nitrogen source (32), so the expression of PTR2 was not influenced by the nitrogen catabolite repression under these conditions. PTR2 mRNA levels were extremely low in wild-type UBR1 CUP9 cells grown in medium lacking amino acids, but in the presence of 98 μm (20 μg/ml) Trp the expression of PTR2 was greatly increased (Fig. 3A, lanes a and b). By contrast, in the absence of CUP9 (in a cup9Δ strain), PTR2 was highly expressed in either the presence or absence of Trp (Fig. 3A, lanes i and j; compare with lanes a and b). Finally, PTR2 mRNA was undetectable in a ubr1Δ strain grown in either the presence or absence of Trp (Fig. 3A, lanes c and d). Thus, both UBR1 and CUP9 are required for inducibility of PTR2 expression by an amino acid.
Three genes, SSY1, PTR3, and SSY5, are a part of the SPS amino acid-sensing pathway in S. cerevisiae (see introduction). We examined the influence of added Trp on the levels of PTR2 mRNA in ssy1Δ and ptr3Δ strains, comparing the consequences of deleting these genes with the effects of deleting either CUP9 or UBR1. The induction of PTR2 by Trp was dramatically reduced in both ssy1Δ and ptr3Δ strains (Fig. 3A, lanes e–h). However, a weak Trp-mediated increase in PTR2 expression was still observed in these strains, in contrast to the complete absence of PTR2 expression (and of Trp effect) in ubr1Δ cells, and in contrast to a high level of PTR2 mRNA in cup9Δ cells (Fig. 3A). Thus the bulk (but apparently not all) of the Trp effect on the expression of PTR2 requires SSY1 and PTR3.
We next examined whether the general amino acid permease GAP1 (37) was required for Trp-mediated induction of PTR2 mRNA. Expression of PTR2 in wild-type cells and a congenic gap1Δ strain was measured by Northern analysis as a function of time after the addition of Trp. Wild-type cells exhibited a strong induction of PTR2 by 20 min in the presence of Trp (Fig. 3, D and E, lanes a–c). Remarkably, the induction of PTR2 by Trp, while still occurring in the absence of GAP1, was found to be strongly decreased (Fig. 3, D and E, lanes d–f; compare with lanes a–c). To the best of our knowledge, this is the first indication of a major involvement of GAP1 in the amino acid-mediated induction of the PTR2 peptide transporter. Being an inducer of PTR2 and other transporter genes via the amino acid sensor SSY1, the extracellular Trp exerts its effect without its import across the plasma membrane (see introduction). Therefore the absence of GAP1 as an importer of Trp would not be expected, a priori, to influence the PTR2-inducing effect of Trp. Moreover, Trp can also be imported by the Trp-specific permease TAP2 (SCM2) (51). In sum, the observed decrease (but not elimination) of PTR2 induction in the absence of GAP1 (Fig. 3, D and E, lanes a–f) stems from a function of GAP1 that is separate from its role as a transporter of amino acids. Indeed, S. cerevisiae GAP1 is known to function as an amino acid sensor for activation of protein kinase A targets. The protein kinase A and SCH9 kinases have both overlapping and distinct functions in controlling the adaptation of S. cerevisiae to nutrient availability, and GAP1 plays a role at least in the protein kinase A-mediated part of this circuit (Ref. 52, and references therein). Thus, although the mechanistic understanding of involvement of GAP1 in the induction of PTR2 by Trp remains to be attained, the above finding (Fig. 3, D and E) revealed a new aspect of PTR2 control and yet another function of GAP1 as a nutritional regulator.
We also measured, in the gap1 background, the influence of STP1 and/or STP2 on the Trp-mediated induction of PTR2 mRNA. STP1 and STP2 are two sequelogous, conditionally active transcriptional activators that mediate the signaling by the SPS pathway (see introduction) (35, 39, 40, 43). Northern analyses of PTR2 expression as a function of time after addition of Trp showed that the Trp-mediated induction of PTR2 was largely retained in a gap1Δ stp1Δ strain (which lacked the STP1 transcriptional activator), in comparison to congenic gap1Δ strain, whereas the STP2 transcriptional activator was essential for PTR2 induction by Trp: both a gap1Δ stp2Δ mutant and a triple mutant gap1Δ stp1Δ stp2Δ did not exhibit a significant induction of PTR2 by Trp, in contrast to parental gap1Δ and gap1Δ stp1Δ strains (Fig. 3, D and E). Thus, as could be expected from the presence of STP1/2-recognized UASAA nucleotide sequence motifs (53) upstream of the PTR2 gene (data not shown), the induction of PTR2 proceeds both through its transcriptional up-regulation, in particular by the Trp-activated STP2 (Fig. 3, D and E), and through a decrease in its repression, owing to the Trp-induced acceleration of degradation of the CUP9 repressor by the N-end rule pathway, as described below (Fig. 4).
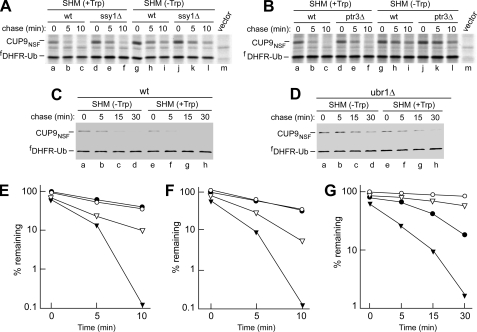
FIGURE 4.
The bulk of Trp-accelerated CUP9 degradation requires SSY1, PTR3, and UBR1. Pulse-chase analyses, using the UPR technique and fDHFR-UbK48R reference protein, of the CUP9 degradation. S. cerevisiae of the indicated genotypes were grown in SHM medium containing or lacking 98 μm Trp (20 μg/ml). [35S]Met/Cys labeling for 5 min, followed by chases for 5 and 10 min (A and B) or for 5, 15, and 30 min (C and D), the latter with cycloheximide at 0.2 mg/ml. E and F, PhosphorImager-based quantitation of pulse-chase patterns in A and B, respectively. Closed circles and triangles, wild-type cells, without and with added Trp, respectively. Open circles, either ssy1Δ cells (A) or ptr3Δ cells (B) in the absence of added Trp. Open triangles, same but with added Trp. G, quantitation of pulse-chase patterns in C and D. Closed circles and triangles, wild-type cells, without and with added Trp, respectively. Open circles and triangles, same but with ubr1Δ cells. Data in C, D, and G were from experiments independent of those in A, B, E, and F, and in addition were carried out with cycloheximide. See “Experimental Procedures” for additional details.
The SPS Pathway Induces PTR2 Expression through Acceleration of CUP9 Degradation—Some of the above results (Fig. 3A) suggested that the induction of PTR2 transporter by amino acids might occur through an increase in the rate of degradation of CUP9 repressor. A precedent for this possibility was the previously demonstrated acceleration of CUP9 degradation by dipeptides with destabilizing N-terminal residues (3). In the latter case, the bulk of CUP9 degradation, both before and after the addition of dipeptides to the medium, was carried out by the UBR1-dependent N-end rule pathway (3). To determine whether the rate of CUP9 degradation was altered by a PTR2-inducing amino acid such as Trp, we carried out pulse-chase assays with CUP9. These experiments utilized CUP9NSF, the above-described nontoxic, single-residue mutant of CUP9. CUP9NSF was expressed as an fDHFR-UbK48R-CUP9NSF fusion, where fDHFR is the N-terminally FLAG-tagged mouse dihydrofolate reductase. Deubiquitylating enzymes cotranslationally cleave this fusion at the UbK48R-CUP9NSF junction, yielding the long-lived reference protein fDHFR-UbK48R and the test protein CUP9NSF (3). The reference fDHFR-UbK48R served as a “built-in” internal control for variations in expression levels and immunoprecipitation efficiency. This generally applicable method, called the Ub-protein-reference (UPR) technique (Refs. 12, 47, and references therein), increases the accuracy of pulse-chase assays. The in vivo degradation of CUP9NSF was indistinguishable from that of wild-type CUP9 and was also not altered significantly by the incorporation of this protein into the above (cotranslationally processed) UPR-type fusion (3).
We expressed CUP9NSF from a URA3-bearing plasmid in S. cerevisiae strains that did not require extracellular amino acids for viability (RJD347, AVY26, AVY27, and AVY28; see Table 1). In wild-type (UBR1) cells grown in the SHM medium lacking amino acids, CUP9NSF was degraded with t½ of ∼10 min (Fig. 4, A, B, E, and F). Remarkably, the addition of Trp (to 98 μm, or 20 μg/ml) to the medium resulted in a strong acceleration of CUP9NSF degradation, with t½ of ∼2 min between 0 and 5 min of chase, and even <2 min between 5 and 10 min of chase (Fig. 4, A, B, E, and F). This effect of Trp on the rate of CUP9NSF degradation was reproducible in separate and independent pulse-chase assays, including a set of assays in which cycloheximide, a translation inhibitor, was present during chase (Fig. 4, C and G). (The other assays, in Fig. 4 (A, B, E, and F) did not involve cycloheximide.) Although in cycloheximide-based assays (Fig. 4, C and G) the CUP9NSF protein was degraded more slowly (t½ of ∼20 min and 3–4 min in the absence and presence of Trp, respectively) than in cycloheximide-free assays (Fig. 4, A, B, E, and F), the effect of Trp was comparably strong in both cases (Fig. 4). In addition to increasing the accuracy of pulse-chase assays, the UPR technique, specifically its built-in, long-lived reference protein, makes it possible to determine the relative level of a test protein (measured as the ratio of 35S in a test versus reference protein) at the beginning of chase, i.e. at the end of pulse labeling (12). In Fig. 4 (E–G), 100% was assigned, in each of three panels, to the relative amount of 35S (relative to the reference protein) of CUP9NSF at time “0” (the end of pulse labeling) in the absence of Trp in the medium. Thus, a value below 100% at time “0” reflects the relative extent of CUP9NSF degradation during the 5-min pulse. In all of pulse-chase assays (Fig. 4, E–G), these initial (time-zero) levels of CUP9NSF were found to correlate, consistently, with Trp-induced changes in the rates of CUP9NSF degradation that were measured during the chase.
The presence of SSY1 (Fig. 4, A and E) and also, independently, of PTR3 (Fig. 4, B and F) was found to be strongly but partially required for the Trp-accelerated degradation of CUP9NSF. Specifically, the bulk of enhanced degradation of CUP9NSF upon the addition of Trp was absent in ssy1Δ and ptr3Δ mutants (Fig. 4, A, B, E, and F). Interestingly, however, the degradation of CUP9NSF was still detectably accelerated by Trp in these mutant strains (Fig. 4, A, B, E, and F), in agreement with the weak but still detectable induction, by Trp, of PTR2 mRNA in ssy1Δ and ptr3Δ mutants (Fig. 3A). As we observed previously as well (3), CUP9NSF was greatly but not completely stabilized in ubr1Δ cells (Fig. 4G and data not shown). Interestingly, the addition of Trp to ubr1Δ cells resulted in a weak but still detectable Trp-induced destabilization of the (now long-lived) CUP9NSF, despite the absence of the N-end rule pathway (Fig. 4G, open circles versus open triangles). The “residual” (UBR1-independent) instability of CUP9NSF and the residual sensitivity of this degradation to Trp suggest that CUP9 may also be targeted, at a much lower rate, by a UBR1-independent proteolytic pathway. Our attempts to identify a relevant E3 Ub ligase, through the testing of non-UBR1 E3 mutants, have not been successful, thus far (data not shown).
The Levels of UBR1 mRNA Are Not Changed Significantly by a PTR2-inducing Amino Acid—Previous work has shown that the in vivo concentration of UBR1 is rate-limiting for degradation of N-end rule substrates by the S. cerevisiae N-end rule pathway (reviewed in Ref. 5). To determine whether the Trp-induced acceleration of CUP9 degradation (Fig. 4) was accompanied by an increased level of the UBR1 Ub ligase, we used Northern hybridization to compare the levels of UBR1 mRNA before and after the addition of Trp. Extracellular Trp did not cause a significant alteration in the levels of UBR1 mRNA (Fig. 3, B and C). Together with findings described in the next section (Fig. 5), these results (Fig. 3, B and C) strongly suggested that the observed acceleration of CUP9 degradation in the presence of extracellular Trp (Fig. 4) was not caused by increased levels of UBR1. In addition, the levels of UBR1 mRNA remained the same or nearly the same in the absence of CUP9 (Fig. 3, B and C), indicating that UBR1 is not a part of the CUP9 regulon.
FIGURE 5.
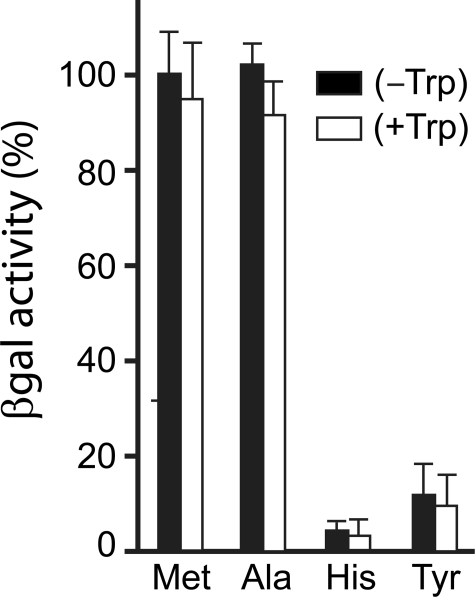
Relative metabolic stabilities of X-βgal test proteins in the absence or presence of added tryptophan. In vivo degradation of X-βgal proteins (X = Met, Ala, His, or Tyr) was assessed by comparing their steady-state levels in wild-type S. cerevisiae in the absence or presence of added Trp. In contrast to CUP9, which is targeted for UBR1-dependent degradation through its internal, C terminus-proximal degron, the short-lived His-βgal and Tyr-βgal (bearing, respectively, a type-1 and type-2 destabilizing N-terminal residue) are targeted by UBR1 via their respective N-degrons.
Degradation of Reporter Proteins Bearing N-degrons Is Not Changed Significantly by a PTR2-inducing Amino Acid—CUP9 lacks an N-degron and is recognized by the UBR1 Ub ligase through a C terminus-proximal degron (see introduction). To determine whether the acceleration of CUP9 degradation by Trp (Fig. 4) was accompanied by increased activity of the N-end rule pathway toward substrates with N-degrons, we employed X-βgal reporters. They were produced through cotranslational deubiquitylation of the corresponding Ub-X-βgal fusions, where X was a variable residue (47, 54). Previous work has shown that the enzymatic activity of βgal in extracts from cells that expressed an X-βgal reporter protein was a reliable measure of the reporter's metabolic stability in vivo (27). Of the four X-βgal reporters in Fig. 5, Met-βgal and Ala-βgal had stabilizing N-terminal residues, whereas His-βgal and Tyr-βgal had, respectively, a type-1 and type-2 primary destabilizing N-terminal residues. As would be expected from the UBR1-dependent degradation of His-βgal and Tyr-βgal (5), their levels in wild-type (UBR1) cells were much lower than the levels of long-lived Met-βgal and Ala-βgal under the same conditions (Fig. 5). We found that the addition of Trp, which led to a strong acceleration of the UBR1-dependent degradation of CUP9 (Fig. 4), did not result in significant alterations of the relative steady-state levels of His-βgal and Tyr-βgal reporters, in comparison to those of Met-βgal and Ala-βgal (Fig. 5). These findings were independently confirmed by carrying out pulse-chase assays with X-βgal reporter proteins (data not shown). Thus the activity of the N-end rule pathway toward substrates with N-degrons did not change significantly upon the addition of Trp.
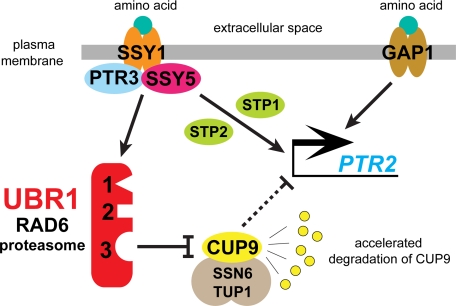
Concluding Remarks—Extracellular Trp does not increase the activity of the N-end rule pathway toward its N-degron-bearing substrates, whereas it does accelerate degradation of the CUP9 repressor by the same pathway (Figs. 4 and 5), thereby making possible the induction of the PTR2 peptide transporter. This dichotomy constrains possible interpretations of our findings. Yet another constraint stems from the fact that the bulk of the Trp-induced acceleration of CUP9 degradation by the N-end rule pathway requires the amino acid-sensing SPS pathway (Figs. 3A and 6). CUP9 is targeted for degradation via its internal degron, through its interaction with a third substrate-binding site of the UBR1 E3 Ub ligase, which functions as the UBR1-RAD6 holoenzyme (see introduction). The autoinhibited CUP9-binding site of UBR1 can be activated, allosterically, by dipeptides with destabilizing N-terminal residues that interact with the other two (type-1 and type-2) binding sites of UBR1 (2, 3). Conformational equilibria, in the in vivo ensembles that contain UBR1, would determine a fraction of UBR1 that is active toward CUP9. We suggest that such equilibria can be shifted toward CUP9-targeting conformations of UBR1 not only in response to UBR1-binding di/tripeptides, as demonstrated previously (2, 3), but also through other routes, for example UBR1 phosphorylation. Other studies have shown that signaling by the SSY1 sensor of amino acids that results in the PTR3/SSY5-dependent (proteolytic) activation of the latent transcription factors STP1 and STP2 is mediated, at least in part, by an increased phosphorylation of PTR3, in reactions mediated by the SSY1-associated YCK1/YCK2 kinases (35, 40, 55). In sum, a parsimonious, testable model that can account for our findings (Figs. 1, 2, 3, 4, 5) and is also consistent with other evidence is that the Trp-activated signaling by the SPS system (Fig. 6) may increase the rate of CUP9 degradation by the N-end rule pathway through a phosphorylation of the UBR1 Ub ligase. This phosphorylation may be mediated, at least in part, by the previously described SSY1-associated kinases that mediate the SSY1-induced phosphorylation of PTR3 (35, 40, 55). (As shown in Figs. 3 and 4, both SSY1 and PTR3 are required for the bulk of Trp-induced acceleration of CUP9 degradation.)
FIGURE 6.
The amino acid-sensing SPS pathway and its connections with the PTR2-CUP9-UBR1 circuit, including the amino acid-mediated acceleration of degradation of CUP9, a transcriptional repressor of the PTR2 peptide transporter. See the main text for details, including the evidence for involvement of a GAP1-mediated pathway.
Our recent findings6 indicate that UBR1 is a multiply phosphorylated protein. They also indicate that UBR1 phosphorylation involves a set of kinases that includes YCK1/YCK2, and that the resulting modifications of UBR1 regulate its functions.6 Work is underway to dissect UBR1 phosphorylation in vivo and to determine whether specific kinds of phosphorylated UBR1 underlie the Trp-induced acceleration of degradation of the CUP9 repressor (Fig. 4). Work is also underway to identify one or more putative E3 Ub ligases that appears to mediate the residual degradation of CUP9 in ubr1Δ cells, which lack the N-end rule pathway. Yet another aspect of these circuits that remains to be understood is a mechanism that underlies our finding (Fig. 3, D and E) that the general amino acid permease GAP1 contributes to the induction of PTR2 by Trp. Given recent studies of GAP1 as an amino acid sensor (Ref. 52 and references therein), its Trp-mediated effect on PTR2 is likely to involve a phosphorylation-based pathway. A better understanding of spatiotemporal aspects of CUP9 regulation would also be important. Because steady-state levels of (short-lived) CUP9 in wild-type cells are very low, and because overexpression of wild-type CUP9 is toxic, this and previous studies (2, 3) utilized CUP9NSF, a missense mutant that does not bind to cognate DNA sites that CUP9 normally recognizes. The Trp-induced acceleration of CUP9NSF degradation (Fig. 4) implies that in vivo interactions of CUP9 with DNA do not play a significant role in the Trp effect. Nevertheless, it would be important to determine whether wild-type CUP9 can be targeted for degradation by the N-end rule pathway directly at sites of its association with DNA in vivo, whether a modification of CUP9, e.g. its phosphorylation, might also be involved in the Trp-induced, UBR1-dependent acceleration of CUP9 degradation, and whether this degradation takes place in the context of CUP9-containing complexes such as CUP9-SSN6-TUP1. Some of the other transcriptional repressors, such as MATα2, are metabolically stabilized by their association with SSN6-TUP1 (50).
Acknowledgments
We thank R. J. Deshaies (California Institute of Technology, Pasadena, CA), B. André (Université Libre de Bruxelles, Belgium), M. C. Kielland-Brandt (Carlsberg Laboratory, Copenhagen, Denmark), and C. Wittenberg (Scripps Research Institute, La Jolla, CA) for S. cerevisiae strains, R. Zitomer (State University of New York, Albany, NY) for the SSN6-expressing plasmid, and A. Sil (University of California, San Francisco, CA) for the pAS315 plasmid. We are grateful to former and current members of the Varshavsky laboratory for helpful discussions and comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants DK39520 and GM31530 (to A. V.). This work was also supported by the Sandler Program for Asthma Research, and from the Ellison Medical Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Ub, ubiquitin; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; βgal, β-galactosidase; UPR, Ub-protein-reference.
A note on terminology: “sequelog” and “spalog” denote, respectively, a sequence that is similar, to a specified extent, to another sequence, and a three-dimensional structure that is similar, to a specified extent, to another three-dimensional structure (22). Besides their usefulness as separate terms for sequence and spatial similarities, the rigor-conferring advantage of sequelog and spalog is their evolutionary neutrality, in contrast to interpretation-laden terms such as “homolog,” “ortholog,” and “paralog.” The latter terms are compatible with the sequelog/spalog terminology and can be used to convey understanding about functions and common descent, if this (additional) information is available.
C. S. Hwang and A. Varshavsky, unpublished data.
References
- 1.Byrd, C., Turner, G. C., and Varshavsky, A. (1998) EMBO J. 17 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du, F., Navarro-Garcia, F., Xia, Z., Tasaki, T., and Varshavsky, A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 14110–14115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner, G. C., Du, F., and Varshavsky, A. (2000) Nature 405 579–583 [DOI] [PubMed] [Google Scholar]
- 4.Homann, O. R., Cai, H., Becker, J. M., and Lindquist, S. L. (2005) PLoS Genet. 1 e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varshavsky, A. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 12142–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mogk, A., Schmidt, R., and Bukau, B. (2007) Trends Cell Biol. 17 165–172 [DOI] [PubMed] [Google Scholar]
- 7.Tasaki, T., and Kwon, Y. T. (2007) Trends Biochem. Sci. 32 520–528 [DOI] [PubMed] [Google Scholar]
- 8.Graciet, E., Hu, R. G., Piatkov, K., Rhee, J. H., Schwarz, E. M., and Varshavsky, A. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 3078–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou, J. Y., Sauer, R. T., and Baker, T. A. (2008) Nat. Struct. Mol. Biol. 15 288–294 [DOI] [PubMed] [Google Scholar]
- 10.Xia, Z., Webster, A., Du, F., Piatkov, K., Ghislain, M., and Varshavsky, A. (2008) J. Biol. Chem. 283 24011–24028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachmair, A., and Varshavsky, A. (1989) Cell 56 1019–1032 [DOI] [PubMed] [Google Scholar]
- 12.Suzuki, T., and Varshavsky, A. (1999) EMBO J. 18 6017–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inobe, T., and Matouschek, A. (2008) Curr. Opin. Struct. Biol. 18 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon, Y. T., Kashina, A. S., Davydov, I. V., Hu, R.-G., An, J. Y., Seo, J. W., Du, F., and Varshavsky, A. (2002) Science 297 96–99 [DOI] [PubMed] [Google Scholar]
- 15.Hu, R.-G., Sheng, J., Xin, Q., Xu, Z., Takahashi, T. T., and Varshavsky, A. (2005) Nature 437 981–986 [DOI] [PubMed] [Google Scholar]
- 16.Hu, R.-G., Brower, C. S., Wang, H., Davydov, I. V., Sheng, J., Zhou, J., Kwon, Y. T., and Varshavsky, A. (2006) J. Biol. Chem. 281 32559–32573 [DOI] [PubMed] [Google Scholar]
- 17.Hu, R.-G., Wang, H., Xia, Z., and Varshavsky, A. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, M. J., Tasaki, T., Moroi, K., An, J. Y., Kimura, S., Davydov, I. V., and Kwon, Y. T. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 15030–15035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon, Y. T., Xia, Z. X., An, J. Y., Tasaki, T., Davydov, I. V., Seo, J. W., Xie, Y., and Varshavsky, A. (2003) Mol. Cell. Biol. 23 8255–8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tasaki, T., Mulder, L. C. F., Iwamatsu, A., Lee, M. J., Davydov, I. V., Varshavsky, A., Muesing, M., and Kwon, Y. T. (2005) Mol. Cell. Biol. 25 7120–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tasaki, T., Sohr, R., Xia, Z., Hellweg, R., Hörtnagl, H., Varshavsky, A., and Kwon, Y. T. (2007) J. Biol. Chem. 282 18510–18520 [DOI] [PubMed] [Google Scholar]
- 22.Varshavsky, A. (2004) Curr. Biol. 14 R181–R183 [DOI] [PubMed] [Google Scholar]
- 23.Rao, H., Uhlmann, F., Nasmyth, K., and Varshavsky, A. (2001) Nature 410 955–960 [DOI] [PubMed] [Google Scholar]
- 24.Ditzel, M., Wilson, R., Tenev, T., Zachariou, A., Paul, A., Deas, E., and Meier, P. (2003) Nat. Cell Biol. 5 467–473 [DOI] [PubMed] [Google Scholar]
- 25.Sasaki, T., Kojima, H., Kishimoto, R., Ikeda, A., Kunimoto, H., and Nakajima, K. (2006) Mol. Cell 24 63–75 [DOI] [PubMed] [Google Scholar]
- 26.Zenker, M., Mayerle, J., Lerch, M. M., Tagariello, A., Zerres, K., Durie, P. R., Beier, M., Hülskamp, G., Guzman, C., Rehder, H., Beemer, F. A., Hamel, B., Vanlieferinghen, P., Gershoni-Baruch, R., Vieira, M. W., Dumic, M., Auslender, R., Gil-da-Silva-Lopes, V. L., Steinlicht, S., Rauh, R., Shalev, S. A., Thiel, C., Winterpacht, A., Kwon, Y. T., Varshavsky, A., and Reis, A. (2005) Nat. Genet. 37 1345–1350 [DOI] [PubMed] [Google Scholar]
- 27.Xie, Y., and Varshavsky, A. (1999) EMBO J. 18 6832–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai, H., Hauser, M., Naider, F., and Becker, J. M. (2007) Eukaryot. Cell 6 1805–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai, H., Kauffman, S., Naider, F., and Becker, J. M. (2006) Genetics 172 1459–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiles, A. M., Cai, H., Naider, F., and Becker, J. M. (2006) Microbiology 152 3133–3145 [DOI] [PubMed] [Google Scholar]
- 31.Godard, P., Urrestarazu, A., Vissers, S., Kontos, K., Bontempi, G., van Helden, J., and André, B. (2007) Mol. Cell. Biol. 27 3065–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Island, M. D., Naider, F., and Becker, J. M. (1987) J. Bacteriol. 169 2132–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frommer, W. B., Hummel, S., and Rentsch, D. (1994) FEBS Lett. 347 185–189 [DOI] [PubMed] [Google Scholar]
- 34.Andréasson, C., Heessen, S., and Ljungdahl, P. O. (2006) Genes Dev. 20 1563–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abdel-Sater, F., Bakkoury, E. I., Urrestarazu, A., Vissers, S., and Andre, B. (2004) Mol. Cell. Biol. 24 9771–9785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, B., Ottow, K., Poulsen, P., Gaber, R. F., Albers, E., and Kielland-Brandt, M. C. (2006) J. Cell Biol. 173 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boles, E., and André, B. (2004) Top. Curr. Genet. 9 121–153 [Google Scholar]
- 38.Fosberg, H., and Ljungdahl, P. O. (2001) Curr. Genet. 40 91–109 [DOI] [PubMed] [Google Scholar]
- 39.Boban, M., and Ljungdahl, P. O. (2007) Genetics 176 2087–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, Z., Thornton, J., Spirek, M., and Butow, R. A. (2008) Mol. Cell. Biol. 28 551–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klasson, H., Fink, G. R., and Ljungdahl, P. O. (1999) Mol. Cell. Biol. 19 5405–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holsbeeks, I., Lagatie, O., Nuland, A. V., Van de Velde, S., and Thevelein, J. M. (2004) Trends Biochem. Sci. 29 556–564 [DOI] [PubMed] [Google Scholar]
- 43.Andréasson, C., and Ljungdahl, P. O. (2002) Genes Dev. 16 3158–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider, B. L., Seufert, W., Steiner, B., Yang, Q. H., and Futcher, B. (1995) Yeast 11 1265–1274 [DOI] [PubMed] [Google Scholar]
- 45.Sikorski, R. S., and Hieter, P. (1989) Genetics 122 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghislain, M., Dohmen, R. J., Levy, F., and Varshavsky, A. (1996) EMBO J. 15 4884–4899 [PMC free article] [PubMed] [Google Scholar]
- 47.Varshavsky, A. (2005) Methods Enzymol. 399 777–799 [DOI] [PubMed] [Google Scholar]
- 48.Alagramam, K., Naider, F., and Becker, J. M. (1995) Mol. Microbiol. 15 225–234 [DOI] [PubMed] [Google Scholar]
- 49.Malavé, T. M., and Dent, S. Y. (2006) Biochem. Cell Biol. 84 437–443 [DOI] [PubMed] [Google Scholar]
- 50.Laney, J. D., Mobley, E. F., and Hochstrasser, M. (2006) Mol. Cell. Biol. 26 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao, H. C., Xiao, Z., and Fitzgerald-Hayes, M. (1994) Mol. Gen. Genet. 244 260–268 [DOI] [PubMed] [Google Scholar]
- 52.Roosen, J., Engelen, K., Marchal, K., Mathys, J., Griffoen, G., Cameroni, E., Thevelein, J. M., De Vergilio, C., De Moor, B., and Winderickx, J. (2005) Mol. Microbiol. 55 862–880 [DOI] [PubMed] [Google Scholar]
- 53.Abdel-Sater, F., Iraqui, I., Urrestarazu, A., and André, B. (2004) Genetics 166 1727–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turner, G. C., and Varshavsky, A. (2000) Science 289 2117–2120 [DOI] [PubMed] [Google Scholar]
- 55.Spielewoy, N., Flick, K., Kalashnikova, T. I., Walker, J. R., and Wittenberg, C. (2004) Mol. Cell. Biol. 24 8994–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson, E. S., Ma, P. C., Ota, I. M., and Varshavsky, A. (1995) J. Biol. Chem. 270 17442–17456 [DOI] [PubMed] [Google Scholar]
- 57.Nielsen, P. S., van den Hazel, B., Didion, T., de Boer, M., Jørgensen, M., Planta, R. J., Kielland-Brandt, M. C., and Andersen, H. A. (2001) Mol. Gen. Genet. 264 613–622 [DOI] [PubMed] [Google Scholar]
- 58.Jorgensen, M. U., Gjermansen, C., Andersen, H. A., and Kielland-Brandt, M. C. (1997) Curr. Genet. 31 241–247 [DOI] [PubMed] [Google Scholar]
- 59.Bernard, F., and André, B. (2001) FEBS Lett. 496 81–85 [DOI] [PubMed] [Google Scholar]