Abstract
Microcephalin/MCPH1 is one of the causative genes responsible for the autosomal recessive disorder primary microcephaly. Patients with this disease present with mental retardation and dramatic reduction in head size, and cells derived from these patients contain abnormally condensed chromosomes. MCPH1 contains an N-terminal BRCT and tandem C-terminal BRCT domains. More recently, MCPH1 has been implicated in the cellular response to DNA damage; however, the exact mechanism remains unclear. Here, we report the identification Condensin II as a major MCPH1-interacting protein. MCPH1 and Condensin II interact in vivo, mediated by the CAPG2 subunit of Condensin II binding to a middle domain (residues 376-485) of MCPH1. Interestingly, while Condensin II is not required for the IR-induced G2/M checkpoint, Condensin II-depleted cells have a defect in HR repair, which is also present in MCPH1-/-MEFs. Moreover, the Condensin II binding region of MCPH1 is also required for HR function. Collectively, we have identified a novel function of MCPH1 to modulate HR repair through Condensin II, and thereby maintain genome integrity.
Microcephalin/MCPH1 was previously identified as one of the genes mutated in autosomal recessive primary microcephaly (1). This is a neurological disease with genetic heterogeneity as MCPH1 is one of six loci responsible for this disease. To date mutations in MCPH1, CDKRAP2, ASPM, CENPJ, and Pericentrin have been shown to cause primary microcephaly (2-6). Patients that have this disease present with a prominent reduction in head size, between -5 and -10 S.D. below the mean, and also mental retardation (1, 7).
So far, three mutations have been described for MCPH1, two premature stop codon mutations (S25X, 427insA) and one missense mutation in the N-terminal BRCT domain (T27R) (3, 8, 9). MCPH1 also possesses C-terminal tandem BRCT domains, which are known phosphoprotein binding domains implicated in cell cycle control, DNA damage checkpoints, and DNA repair (10, 11). Indeed, recent work has highlighted the function of MCPH1 in the DNA damage response (12-17). Moreover, LOH of MCPH1 was observed in tumor samples (14), implicating that it might act as a tumor suppressor. Paradoxically, patients with MCPH1 mutations do not present with dramatic sensitivity to DNA-damaging agents, premature aging, or cancer predisposition, which is often a characteristic of other cancer susceptibility syndromes caused by mutations in the DNA damage response pathway (18). One unique characteristic of cells derived from MCPH1 patients is the premature chromosome condensation (PCC)4 phenotype observed in a subset of these patient cells (∼15%) (3, 8). These cells display an aberrant morphology under routine chromosome spreads with highly compacted, condensed DNA. Consequently, MCPH1 mutations were also discovered in a family with PCC syndrome, which shared the dramatic reduction in head size seen in primary microcephaly (8). Despite these strong cell biology data, a mechanistic link between MCPH1 and chromosome condensation remains to be identified.
Chromosome condensation is an important step in the cell cycle especially when cells are preparing to enter mitosis. In humans and other eukaryotes, there are several protein complexes that regulate chromosome condensation and segregation. These include cohesin, SMC5-SMC6-NseI, Condensin I, and Condensin II (19). These protein complexes share many similarities, especially the core domains of these multiprotein subunit complexes, which consist of Structural Maintenance of Chromosome (SMC) proteins, which are large ATPases, and their unique subunits, which contain HEAT or kleisin protein domains. Cohesin consists of an SMC heterodimer composed of SMC1/SMC3 and two specific subunits Scc1 and Scc3, and functions in sister chromatid cohesion (19). The SMC5/6-NseI complex again shares a core SMC heterodimer and then 4 specific subunits (NseI, -II, -III, and -IV) and has been shown to function in DNA repair and cell cycle checkpoints (20). Condensin I and Condensin II have a common SMC heterodimer (SMC2/SMC4) that associate to form a characteristic V-shape and also three distinct specific subunits (21). Together, each SMC protein forms an arm through self-folding of anti-parallel coiled-coil sequences forming a hinge domain at one end that is responsible for the association of the two molecules, and at the other end an ATPase “head” of the N- and C-terminal sequences that can engage ATP, which when hydrolyzed allows for movement of DNA. However, the detailed mechanism is still unclear (21). Either Condensin I or II are large molecules, with each arm about ∼50 nm in size, which is equivalent to 150 bp of dsDNA. Conceivably, this distance is big enough to wrap around and access nucleosomes. However, the mechanism by which Condensin I or II regulates chromosome condensation is still largely unknown (21).
Condensin I and II share the SMC2/SMC4 core heterodimer and three other subunits, CAP-D2, CAP-G, and CAP-H in the Condensin I complex, and CAP-D3, CAP-G2 and CAP-H2 in Condensin II. CAP-D2, CAP-D3, CAP-G, and CAP-G2 proteins contain HEAT repeats while CAP-H and CAP-H2 are members of the kleisin family, γ and β, respectively. Recent in vitro reconstitution experiments demonstrated that each complex has an identical subunit organization; CAP-H and CAP-H2 bind to the SMC2/4 heterodimer through their N and C terminus; thus, forming a closed ring and also function as a platform for the association of the other HEAT repeat containing subunits (22). While having similar subunit and complex architecture, Condensin I and Condensin II have different cellular localization patterns. During interphase, Condensin I remains in the cytoplasm while Condensin II localizes in the nucleus (23). At the onset of mitosis and the breakdown of the nuclear envelope Condensin I is believed to act together with Condensin II and function in compacting chromosomes preparing for mitosis (21). This condensation ability has been demonstrated in vitro since Condensin I can form positive super-coils using a DNA substrate (24, 25). However, exactly how Condensin I or II works in vivo and what other processes it may function remains a mystery.
At present, MCPH1 appears to function in diverse pathways regulating cell cycle control, DNA damage and chromosome biology. To fully understand how MCPH1 participates in these events, we have purified MCPH1-containing complexes and identified several MCPH1 associated proteins. Surprisingly, the Condensin II complex was isolated as a major binding partner of MCPH1. Subsequent experiments indicate that both MCPH1 and Condensin II are involved in homologous recombination repair, providing the first evidence supporting a role of Condensin II in DNA damage repair.
EXPERIMENTAL PROCEDURES
Cell Culture, Plasmids, and Antibodies—HeLa and 293T cells were purchased from American Tissue Type Culture (Manassas, VA) and maintained in RPMI 1640 supplemented with 10% bovine serum and 1% penicillin/streptomycin at 37 C in 5% CO2 (v/v). Microcephalin/BRIT1 knock-out mouse embryonic fibroblasts (MEFs) were isolated from E14.5 embryos, and cultured in Dulbecco's modified Eagle's medium supplemented with 20% fetal bovine serum and 1% penicillin/streptomycin. MEFs were immortalized with SV40.
Deletion mutants of MCPH1 were generated using the QuikChange site-directed mutagenesis (Stratagene) using pDNR-MCPH1 as template and then cloned into an EF1A HA-FLAG vector. All deletions were verified by sequencing. Primer sequences are available upon request. Condensin II subunits hCAP-D3, hCAP-G2, and hCAP-H2 cDNA was obtained from OpenBiosystems, and cloned into the pDONOR201 gateway vector.
MCPH1 polyclonal antibody was generated by immunizing rabbits with MBP fusions of the middle and C-BRCT domains of MCPH1. The antibody was purified using the AminoLink Immobilization kit (Pierce). Antibodies specifically for pH2AX, 53BP1, MDC1 have been described previously (15). CAPG2, CAPD3 antibodies were purchased from Bethyl Laboratories, and anti-SMC2, SMC4 were purchased from Novus Biologicals. Anti-Myc monoclonal antibody and anti-FLAG M2 antibody were obtained, respectively, from Santa Cruz Biotechnology and Sigma.
Transfection and Infection—Plasmid transfection was carried out using Lipofectamine (Invitrogen) per the manufacturer's instructions. siRNA transfection was carried out using Oligofectamine (Invitrogen) per the manufacturer's instructions. The coding strand for the control was UCCAGUGAAUCCUUGAGGUdTdT, and for CAPD3 was CUGGAUUUCACAGAGACUGTTdTdT. All siRNA were purchased from Dharmacon.
Full-length MCPH1 or its deletions was cloned into pEF1A-HA/FLAG retroviral vector using the gateway system. pEF1A-MCPH1 and its deletions were co-transfected with pCL-Amphobac in BOSC23-packaging cell lines to produce virus. Viral particles were collected 48 and 72 h after transfection and subsequently used to infect MEF cell lines. 48 h after the last infection, MEFs were irradiated as indicated, and immunofluoresence staining was performed. Alternatively, cells were selected in puromycin-containing media for establishing stable clones.
Tandem Affinity Purification of MCPH1 and MS Analysis—293T cells were transfected with plasmids encoding SFB-tagged MCPH1. Forty-eight hours after transfection, cells were split at the 1:10 ratio and cultured in the medium containing puromycin (2 μg/ml) for 3 weeks. The individual puromycin-resistant colonies were isolated and screened by Western blotting. 293T cells stably expressing MCPH1 were lysed with NETN buffer on ice for 20 min. Crude lysates were cleared by centrifugation at 14,000 rpm at 4 °C for 10 min, and supernatants were incubated with 300 μl of streptavidin-conjugated beads (Amer-sham Biosciences). The immunocomplexes were washed three times with NETN buffer, and then bead-bound proteins were eluted with 1 ml of NETN buffer containing 1 mg/ml biotin (Sigma). The eluted supernatant was incubated with 80 μl of S protein beads (Novagen). The immunocomplexes were washed three times with NETN buffer and subjected to SDS-PAGE. Protein bands were excised, digested, and the peptides were analyzed by mass spectrometry at the Taplin Mass Spec Facility at Harvard.
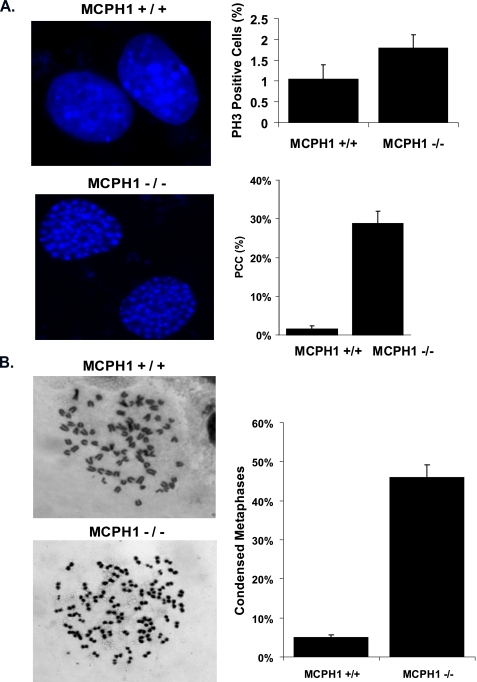
Immunofluoresence, Western Blotting, and FACS Analysis—Cells grown on coverslips were fixed in 3% paraformaldehyde solution for 20 min, rinsed 1× with PBS, and then permeabilized using 0.5% Triton X-100 for 5 min. Cells were then washed and incubated with primary antibodies diluted in 5% goat serum at 37 °C for 20 min. Cells were washed again 1× in PBS, and then secondary antibodies either fluorescein isothiocyanate-conjugated goat anti-mouse IgG or rhodamine-conjugated goat anti-rabbit IgG for 20 min at 37 °C. After washing, cells were counterstained with DAPI, washed with PBS, and then mounted onto slides with anti-Fade. Images were taken using a Nikon ECLIPSE E800 microscope. For PCC quantification using DAPI-staining positive cells were scored that had the representative condensed DNA seen in Fig. 3A, top left panel, bottom, and scored negative if the DAPI pattern was that of normal cells (Fig. 3A, top left panel, top).
FIGURE 3.
MCPH1-/- MEFs display prematurely condensed chromosomes. A, MCPH1 MEFs have abnormal condensed DAPI staining. Examples of DAPI staining of MCPH1+/+ (top) and MCPH1-/- (bottom) MEFs were presented. The top right panel shows number of mitotic cells in each cell line, MCPH1+/+ and MCPH1-/- cells were collected and analyzed by FACS for phospho-Histone H3 as a mitotic marker. An average of three experiments is presented. The bottom right panel displays quantification of percentage of cells with abnormal condensed DNA measured by DAPI staining. On average, three independent experiments were performed, and 500 cells were counted per experiment. Bars represent the S.D. B, MCPH1-/- MEFs have condensed chromosomes. Examples of metaphase spreads of MCPH1+/+ (top) and MCPH1-/- (bottom) cells were included. Three independent experiments were performed, and at least 100 metaphase spreads were counted per experiments. The average of percentages of cells with condensed chromosomes is shown. Bars indicate the S.D.
For Western blotting, cells were lysed with NETN (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40) on ice and then rocked for 10 min at 4 °C. Crude cell lysates were then centrifuged at 14,000 rpm for 10 min, and cleared lysates were collected. Samples were boiled in 2× Laemmli buffer and run on SDS-PAGE. Membranes were blocked in 5% milk-TBST and then probed with antibodies as indicated.
For cell cycle analysis, cells were trypsinized, washed twice in cold PBS, then resuspended in 300 μl of cold PBS and fixed with 700 μl of ice-cold 100% ethanol. Cells were incubated in RNase A in sodium citrate buffer (100 units/sample) for 30 min, before stained with propidium iodide (PI, 50 μg/ml) for 30 min. Cell cycle analysis was performed using FACScan.
Chromosome Spreads—Metaphase chromosome spreads were performed as described before (26). Briefly, cells were treated with colcemid for 4 h and then harvested. After treatment with 0.075 m KCl, cells were fixed in fresh methanol/acetone (3:1) solution and dropped from 10 cm onto coverslips. Cell were allowed to air dry on coverslips, stained with giemsa, and visualized under the microscope.
For quantification, 100 metaphase spreads were counted for each cell line in three independent experiments. For Fig. 3, representative spreads are shown for +/+ and -/- cells and were scored positive if they possessed condensed chromosomes. For Fig. 4, representative examples are shown in Fig. 4D, left panel. Spreads were scored positive if they possessed the condensed chromosome pattern as that of MCPH1-/- cells.
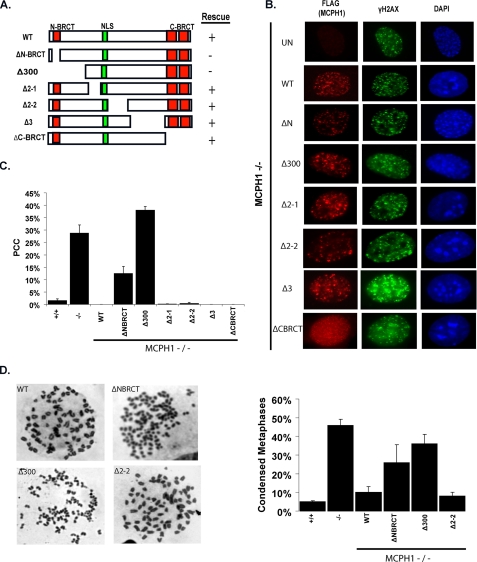
FIGURE 4.
Condensin II binding is not required to rescue the PCC defect in MCPH1-/- MEFs. A, diagrams of wild-type and deletion mutants of MCPH1 used in this study. B, N terminus of MCPH1 is important for the rescue of PCC in MCPH1-deficient cells. MCPH1-/- reconstituted cells were treated with 10 Gy, fixed, and stained 6 h later with the indicated antibodies. C, quantification of data presented in B. 500 cells were counted, the results of three independent experiments is shown. Bars represent S.D. D, loss of the N terminus but not the Condensin II binding region causes PCC. Chromosome spreads from MCPH1-/--reconstituted cells were counted. Shown on the left are examples of metaphase spreads seen in each cell line; on the right is quantification of condensed chromosomes in each cell line tested. Bars indicate the S.D.
Homologous Recombination Assays—The HR assay was done using the DR-GFP/I-sceI system as previously described (27). Briefly, after siRNA treatment, cells were electroporated with 16 μg of pCBASce. 24 h later, cells were collected and analyzed for GFP expression by FACS. All data are the average of three independent experiments and bars are S.D.
RESULTS
Isolation of Condensin II as an MCPH1-associated Protein—Because it is still uncertain as exactly how MCPH1 functions in DNA damage repair and cell cycle checkpoint control, we sought to identify MCPH1-associated proteins to further elucidate the function of MCPH1. To do this, we generated a 293T cell line stably expressing a triple-tagged (S-FLAG-strepavidin-binding peptide) MCPH1. Tandem affinity purification (TAP) was performed, and the purified complex underwent mass spectrometry analysis (Fig. 1, A and B). Our results revealed several MCPH1-associated proteins; however, the major associated proteins appeared to be members of the Condensin II complex, which is involved in chromosome condensation (19, 21, 28). This is exciting because a previous study has reported a possible link between the abnormal chromosome condensation observed in patients with MCPH1 mutations with Condensin II misregulation (29).
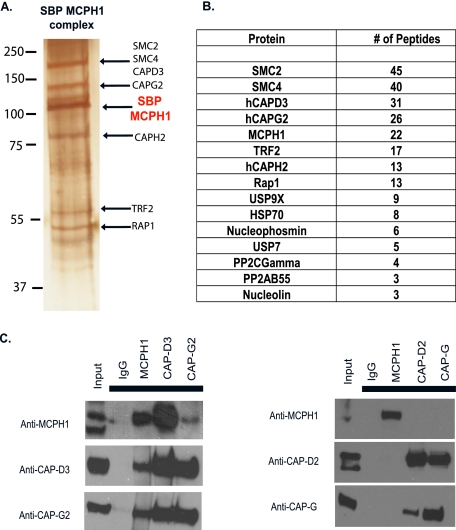
FIGURE 1.
Isolation of Condensin II as a MCPH1-associated protein. A, tandem affinity purification of MCPH1-containing complexes. 293T cells stably expressing MCPH1 underwent two rounds of affinity purification, and the final elution was run on SDS-PAGE and proteins visualized by silver stain. The pooled elution was sent for MALDI-TOF. B, list of peptides obtained from MS analysis. C, MCPH1 associates with Condensin II in vivo. Co-immunoprecipitation experiments were performed using the indicated antibodies.
MCPH1 Forms a Complex in Vivo with Condensin II—To explore this possible interaction further, we performed immunoprecipitation (IP) experiments. The CAP-D3 and CAP-G2 subunits were detected in the MCPH1 IP but not in a control IgG IP. In parallel, MCPH1 was detected in Condensin II IPs, but not in the immunoprecipitates of CAP-D2 or CAP-G IPs, which are members of the Condensin I complex (Fig. 1C). These data suggest that MCPH1 associates specifically with Condensin II but not Condensin I, which agrees with our purification data, because we did not recover any peptides derived from Condensin I-specific subunits.
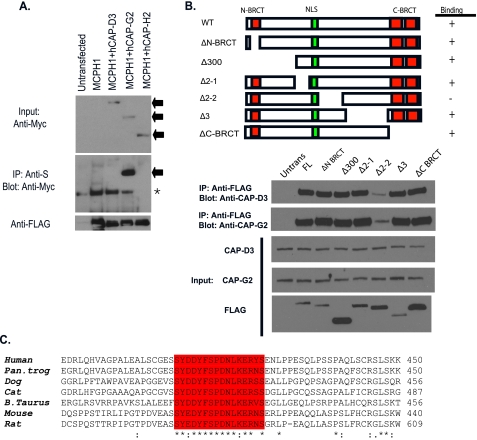
A Region within the Middle Domain (MD) of MCPH1 Is Required for Condensin II Binding—We co-overexpressed the Condensin II subunits together with MCPH1 and found a strong interaction between MCPH1 and CAP-G2 (Fig. 2A), implying that the interaction between MCPH1 and Condensin II may be mediated by this specific subunit of Condensin II. Next we wanted to determine which domain of MCPH1 mediates its interaction with Condensin II. MCPH1 contains 3 BRCT domains, which are known phosphoprotein binding domains. We generated a series of deletion mutations spanning the entire coding region of MCPH1. Surprisingly, the region required for MCPH1 binding was not the N- or C-terminal BRCT domains, but an uncharacterized region in the middle domain (residues 376-485) of MCPH1 (Fig. 2B). When we performed a data base search, we found that this region contains a highly conserved stretch of residues but does not have any similarity with known protein domains (Fig. 2C), indicating that the interaction between MCPH1 and Condensin II may be conserved and thus play an important role throughout evolution.
FIGURE 2.
A middle region of MCPH1 specifically mediates its interaction with Condensin II. A, CAP-G2 subunit of Condensin II associates with MCPH1. Immunoprecipitation reactions were performed using S beads and blotted with anti-Myc or anti-FLAG antibodies. Arrows show input bands for the Condensin II subunits. The asterisk denotes the nonspecific band. B, Condensin II binds to a middle region (residues 376-485) of MCPH1. A schematic representation of MCPH1 deletions used to map Condensin II binding region on MCPH1 is shown. Wild-type and deletion mutants of MCPH1 were ectopically expressed in 293T cells and immunoprecipitated with anti-FLAG antibodies and blotted with the indicated antibodies specifically for Condensin II subunits. C, Δ2-2 region contains a 20-amino acid conserved region across species. The ClustalW alignment of the conserved region among higher eukaryotes within the Condensin II binding domain of MCPH1 is presented.
MCPH1-/- MEFs Display PCC—Patients that harbor mutations within MCPH1 have primary microcephaly characterized by a drastic reduction in head size, and cell lines derived from these patients have a population of cells with abnormally condensed chromosomes. In order to generate a model for primary microcephaly and also confirm the cellular phenotypes observed in MCPH1-deficient patients, MCPH1 knock-out mice were generated.5 What is very noticeable in these MCPH1-/- mouse embryonic fibroblasts (MEFs) is that they also recapitulate a similar premature condensed chromosome phenotype present in MCPH1 mutant human cells (Fig. 3A). Wild-type MEFs stained with DAPI show normal nuclear DNA with several bright dots indicative of heterochromatin regions; however, in the MCPH1-/- MEFs we discovered a population of cells that displayed a pattern of intense DAPI staining, with unstained spaces in the nucleus (Fig. 3A). This was not due to an overabundance of mitotic cells; staining with phospho-H3 S10 as a mitotic marker showed only ∼2 to 2.5% of cells in mitosis in MCPH1-/- MEFs, slightly higher than in the MCPH1+/+ cells (Fig. 3A). The percentage of cells with this intense DAPI staining pattern was between 30 and 35% in MCPH1-deficient cells, but only 2 or 3% in wild-type cells (Fig. 3A).
To determine if this nuclear staining phenotype represented abnormally condensed chromosomes, we prepared metaphase chromosome spreads to determine their morphology. As shown in Fig. 3B, MCPH1+/+ cells showed normal chromosome architecture; however, chromosomes from MCPH1-/- cells were abnormally condensed. When scored under the microscope, ∼40% of MCPH1-deficient cells had condensed chromosomes. This number is similar to the percentage of cells with intense DAPI staining (Fig. 3A), suggesting that these two are related phenotypes, resulting from MCPH1 deficiency.
The N-BRCT, but Not the Condensin II Binding Region, Is Required to Rescue PCC—MCPH-/- cells recapitulate the chromosome condensation phenotype observed in cells derived from human patients. Our next question was what is the underlying mechanism responsible for this defect. We reconstituted MCPH1-/- MEFs with wild-type or MCPH1 deletion mutants to find which regions of MCPH1 were responsible for this defect. Reconstitution with wild-type MCPH1 fully rescued the chromosome condensation defect because we observed normal DAPI staining and chromosome spreads in these cells (Fig. 4). Surprisingly, when we tested the series of MCPH1 deletions, the region that is important for Condensin II binding (deleted in the Δ2-2 mutant) is not required for the rescue of the chromosome condensation defect, instead the N terminus of MCPH1 is (Fig. 4, A-C). Deletion of the N-BRCT domain alone or deletion of the N-BRCT domain plus a 150-amino acid region immediately after it failed to rescue the chromosome condensation defect as measured by DAPI staining (Fig. 4, A-C). We also performed metaphase spreads for wildtype, MCPH1-deficient, and reconstituted cells. In agreement with the DAPI assay, expression of wild-type MCPH1 rescued the chromosome condensation defect measured by this assay, the N-terminal deletion mutant failed to do so (Fig. 4D). Again, the Δ2-2 mutant, which cannot bind Condensin II, is still capable of rescuing the chromosome condensation defect as measured by metaphase spreads. Collectively, our data implicate that the N terminus including the N-BRCT domain is necessary for rescue of the PCC defect in MCPH1-/- cells; however the Condensin II binding domain appears to be dispensable for this function.
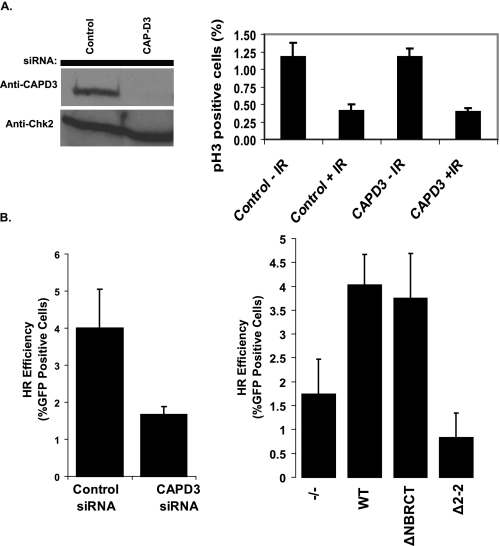
Condensin II Participates in Homologous Recombination Repair—If the interaction between MCPH1 and Condensin II is not essential for its function in regulating chromosome condensation, then what is the significance of this interaction? The only other known function of MCPH1 is its involvement in the DNA damage response. To test whether Condensin II would also take part in DNA damage checkpoint or DNA repair, we depleted CAP-D3, a subunit of the Condensin II complex, in HeLa cells using CAP-D3-specific siRNA. As shown in Fig. 5A, depletion of CAP-D3 had no effect on the G2/M checkpoint as the number of mitotic cells decreased similar to the control cells after IR treatment (Fig. 4A). Next, we investigated whether Condensin II may be important for DNA repair, as Condensin I has been shown to participate in nucleotide excision repair (30), and the SMC5/6 complex regulates DNA repair through recruitment of cohesin (31). We assayed for homologous recombination repair efficiency using a GFP reporter system (27). As shown in Fig. 5B, CAP-D3 depleted caused a 2.5-3-fold reduction in HR repair efficiency. Consistent with this observation, MCPH1-/- MEFs also displayed a 2.2-fold decrease in HR repair, which could be rescued by the expression of wild-type MCPH1 and the N-terminal BRCT domain deletion mutant of MCPH1 (ΔNBRCT) in these cells (Fig. 5B). However, the Δ2-2 mutant of MCPH1, which abrogates Condensin II binding, failed to rescue this HR defect. Taken together, our data suggest that not only MCPH1 and Condensin II, but also an interaction between MCPH1 and Condensin II are important for efficient HR repair.
FIGURE 5.
Condensin II is not required for G2/M checkpoint control, but plays a role in HR repair. A, G2/M checkpoint is active in Condensin II-depleted cells. HeLa cells were transfected with control or CAP-D3 siRNA. Cells were irradiated and harvested 1 h later, and mitotic cells were counted based on p-H3 staining. Average of three independent experiments with S.D. is shown. B, both Condensin II and MCPH1 are required for efficient HR repair. Left panel, U2OS-DRGFP was transfected with control or CAP-D3 siRNA, allowed to recover, and then transfected with 16 μg of pCBASce. 24 h later, cells were collected and analyzed for GFP expression by FACS. Right panel, MCPH1+/+ cells, MCPH1-/- cells, and MCPH1-/- cells reconstituted with indicated wild-type and deletion mutant of MCPH1 were assayed for HR capability by co-transfection of DR-GFP reporter plasmid with pCBASce. Cells were allowed to recover and analyzed 48 h later for GFP expression.
DISCUSSION
Despite intense scrutiny on how MCPH1 functions in cell cycle control and the DNA damage response, the exact role of this protein in these processes remains unclear. Here we describe the purification of Condensin II complex as MCPH1-associated proteins. In this study, we showed that this association is not required for checkpoint control, but participates in homologous recombination repair.
Mammalian cells contain two related Condensin complexes: Condensin I and II. It is possible that higher eukaryotes evolved more than one Condensin complex to perform functions other than chromosome condensation. Indeed, Condensin I and II have different localization patterns in interphase cells, with Condensin II mainly localizes in the nucleus and Condensin I confines in the cytoplasm (23). We speculate that this nuclear localization of Condensin II allows Condensin II to perform its role in HR repair with MCPH1 during interphase and then switch to its role in condensation with Condensin I during mitosis.
Exactly how Condensin II may function in HR repair is still unknown, but studies from bacterial Condensin provides some interesting hypotheses. A recent report has shown that MukB, the bacterial Condensin, forms clusters on DNA and that these clusters can interact and bring distant DNA ends together (32). It is therefore tempting to speculate that Condensin II in humans might also display this activity and function in a certain step in HR repair that would require this ability.
Besides the HR deficiency, we also observed abnormal chromosome condensation in MCPH1-/- MEFs, which can be rescued by the introduction of wild-type MCPH1. The condensation defect observed in MCPH1-/- MEFs recapitulates the defect observed in MCPH1 patient cell lines. Moreover, two recent reports on Drosophila MCPH1 also described these characteristics of the human syndrome with prematurely condensed DNA (33, 34). These and our study underline the importance of using genetic models to gain more insight into human disease. Interestingly, the MCPH1 mutant (Δ2-2), which is defective in Condensin II binding, is fully functional in restoring the chromosome condensation defect. On the other hand, the N terminus of MCPH1, especially the N-terminal BRCT domain of MCPH1, is required for rescuing this condensation defect. Given that BRCT domain is a protein-protein interaction domain, it is reasonable to speculate that the N terminus of MCPH1 may associate with another yet-to-be identified protein and thus participate in chromosome condensation. We believe that this function of MCPH1 may somehow be related to the function of Condensin II, but the detailed mechanism can only be worked out once this additional player is identified.
Acknowledgments
We thank all present and past members of the Chen laboratory, especially Ja-Eun Kim and Zhenkun Lou for helpful discussions and technical support.
This work was supported, in whole or in part, by National Institutes of Health Grants CA92312 and CA100109 (to J. C.) and R01CA109574-01A1 (to K. L.). This work was also supported by The Susan G. Komen Breast Cancer Foundation Grant BCTR0504162T (to K. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PCC, premature chromosome condensation; SMC, structural maintenance of chromosome; PBS, phosphate-buffered saline; DAPI, 4′,6-diamidino-2-phenylindole; MEF, mouse embryonic fibroblast; HR, homologous recombination; FACS, fluorescent-activated cell sorting; GFP, green fluorescent protein.
Y. Liang, H. Gao, S.-Y., Lin, P., Zhang, C., Zhu, O., Benjamin, A. S., Multani, S., Chang, J. A., Goss, F. C., Brunicardi, and K., Li, submitted manuscript.
References
- 1.Woods, C. G., Bond, J., and Enard, W. (2005) Am. J. Hum. Genet. 76 717-728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bond, J., Roberts, E., Springell, K., Lizarraga, S. B., Scott, S., Higgins, J., Hampshire, D. J., Morrison, E. E., Leal, G. F., Silva, E. O., Costa, S. M., Baralle, D., Raponi, M., Karbani, G., Rashid, Y., Jafri, H., Bennett, C., Corry, P., Walsh, C. A., and Woods, C. G. (2005) Nat. Genet. 37 353-355 [DOI] [PubMed] [Google Scholar]
- 3.Jackson, A. P., Eastwood, H., Bell, S. M., Adu, J., Toomes, C., Carr, I. M., Roberts, E., Hampshire, D. J., Crow, Y. J., Mighell, A. J., Karbani, G., Jafri, H., Rashid, Y., Mueller, R. F., Markham, A. F., and Woods, C. G. (2002) Am. J. Hum. Genet. 71 136-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffith, E., Walker, S., Martin, C. A., Vagnarelli, P., Stiff, T., Vernay, B., Al Sanna, N., Saggar, A., Hamel, B., Earnshaw, W. C., Jeggo, P. A., Jackson, A. P., and O'Driscoll, M. (2008) Nat. Genet. 40 232-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bond, J., Roberts, E., Mochida, G. H., Hampshire, D. J., Scott, S., Askham, J. M., Springell, K., Mahadevan, M., Crow, Y. J., Markham, A. F., Walsh, C. A., and Woods, C. G. (2002) Nat. Genet. 32 316-320 [DOI] [PubMed] [Google Scholar]
- 6.Rauch, A., Thiel, C. T., Schindler, D., Wick, U., Crow, Y. J., Ekici, A. B., van Essen, A. J., Goecke, T. O., Al-Gazali, L., Chrzanowska, K. H., Zweier, C., Brunner, H. G., Becker, K., Curry, C. J., Dallapiccola, B., Devriendt, K., Dorfler, A., Kinning, E., Megarbane, A., Meinecke, P., Semple, R. K., Spranger, S., Toutain, A., Trembath, R. C., Voss, E., Wilson, L., Hennekam, R., de Zegher, F., Dorr, H. G., and Reis, A. (2008) Science 319 816-819 [DOI] [PubMed] [Google Scholar]
- 7.Woods, C. G. (2004) Curr. Opin. Neurobiol. 14 112-117 [DOI] [PubMed] [Google Scholar]
- 8.Trimborn, M., Bell, S. M., Felix, C., Rashid, Y., Jafri, H., Griffiths, P. D., Neumann, L. M., Krebs, A., Reis, A., Sperling, K., Neitzel, H., and Jackson, A. P. (2004) Am. J. Hum. Genet. 75 261-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trimborn, M., Richter, R., Sternberg, N., Gavvovidis, I., Schindler, D., Jackson, A. P., Prott, E. C., Sperling, K., Gillessen-Kaesbach, G., and Neitzel, H. (2005) Hum. Mutat. 26 496. [DOI] [PubMed] [Google Scholar]
- 10.Yu, X., Chini, C. C., He, M., Mer, G., and Chen, J. (2003) Science 302 639-642 [DOI] [PubMed] [Google Scholar]
- 11.Manke, I. A., Lowery, D. M., Nguyen, A., and Yaffe, M. B. (2003) Science 302 636-639 [DOI] [PubMed] [Google Scholar]
- 12.Alderton, G. K., Galbiati, L., Griffith, E., Surinya, K. H., Neitzel, H., Jackson, A. P., Jeggo, P. A., and O'Driscoll, M. (2006) Nat. Cell Biol. 8 725-733 [DOI] [PubMed] [Google Scholar]
- 13.Lin, S. Y., Rai, R., Li, K., Xu, Z. X., and Elledge, S. J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 15105-15109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rai, R., Dai, H., Multani, A. S., Li, K., Chin, K., Gray, J., Lahad, J. P., Liang, J., Mills, G. B., Meric-Bernstam, F., and Lin, S. Y. (2006) Cancer Cell 10 145-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood, J. L., Singh, N., Mer, G., and Chen, J. (2007) J. Biol. Chem. 282 35416-35423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu, X., Lee, J., and Stern, D. F. (2004) J. Biol. Chem. 279 34091-34094 [DOI] [PubMed] [Google Scholar]
- 17.Jeffers, L. J., Coull, B. J., Stack, S. J., and Morrison, C. G. (2008) Oncogene 27 139-144 [DOI] [PubMed] [Google Scholar]
- 18.O'Driscoll, M., and Jeggo, P. A. (2006) Nat. Rev. Genet. 7 45-54 [DOI] [PubMed] [Google Scholar]
- 19.Hirano, T. (2006) Nat. Rev. Mol. Cell. Biol. 7 311-322 [DOI] [PubMed] [Google Scholar]
- 20.Strom, L., and Sjogren, C. (2007) Curr. Opin. Cell Biol. 19 344-349 [DOI] [PubMed] [Google Scholar]
- 21.Hirano, T. (2005) Curr. Biol. 15 R265-275 [DOI] [PubMed] [Google Scholar]
- 22.Onn, I., Aono, N., Hirano, M., and Hirano, T. (2007) EMBO J. 26 1024-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ono, T., Fang, Y., Spector, D. L., and Hirano, T. (2004) Mol. Biol. Cell 15 3296-3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takemoto, A., Kimura, K., Yokoyama, S., and Hanaoka, F. (2004) J. Biol. Chem. 279 4551-4559 [DOI] [PubMed] [Google Scholar]
- 25.Strick, T. R., Kawaguchi, T., and Hirano, T. (2004) Curr. Biol. 14 874-880 [DOI] [PubMed] [Google Scholar]
- 26.Lou, Z., Minter-Dykhouse, K., Franco, S., Gostissa, M., Rivera, M. A., Celeste, A., Manis, J. P., van Deursen, J., Nussenzweig, A., Paull, T. T., Alt, F. W., and Chen, J. (2006) Mol. Cell 21 187-200 [DOI] [PubMed] [Google Scholar]
- 27.Pierce, A. J., Johnson, R. D., Thompson, L. H., and Jasin, M. (1999) Genes Dev. 13 2633-2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirano, T., Kobayashi, R., and Hirano, M. (1997) Cell 89 511-521 [DOI] [PubMed] [Google Scholar]
- 29.Trimborn, M., Schindler, D., Neitzel, H., and Hirano, T. (2006) Cell Cycle 5 322-326 [DOI] [PubMed] [Google Scholar]
- 30.Heale, J. T., Ball, A. R., Jr., Schmiesing, J. A., Kim, J. S., Kong, X., Zhou, S., Hudson, D. F., Earnshaw, W. C., and Yokomori, K. (2006) Mol. Cell 21 837-848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potts, P. R., Porteus, M. H., and Yu, H. (2006) EMBO J. 25 3377-3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui, Y., Petrushenko, Z. M., and Rybenkov, V. V. (2008) Nat. Struct. Mol. Biol. 15 411-418 [DOI] [PubMed] [Google Scholar]
- 33.Brunk, K., Vernay, B., Griffith, E., Reynolds, N. L., Strutt, D., Ingham, P. W., and Jackson, A. P. (2007) J. Cell Sci. 120 3578-3588 [DOI] [PubMed] [Google Scholar]
- 34.Rickmyre, J. L., Dasgupta, S., Ooi, D. L., Keel, J., Lee, E., Kirschner, M. W., Waddell, S., and Lee, L. A. (2007) J. Cell Sci. 120 3565-3577 [DOI] [PubMed] [Google Scholar]