Abstract
PMEPA1 was identified originally as a highly androgen-inducible gene with prostate-abundant expression that was restricted to prostatic epithelial cells. PMEPA1 protein is a NEDD4 (ubiquitin-protein isopeptide ligase)-binding protein, which negatively regulates prostate cancer cell growth. In this study we establish that PMEPA1 is a direct transcriptional target of the androgen receptor (AR). We also demonstrate that PMEPA1 negatively regulates AR protein levels in different cell culture models. Transient expression of PMEPA1 down-regulates AR protein levels and AR transcriptional targets in prostate cancer cells. Conversely, knockdown of PMEPA1 leads to elevated levels of AR protein, AR transcriptional targets (prostate-specific antigen), and increased cell cycle S phase. We define that the PMEPA1-dependent down-regulation of AR is because of AR ubiquitination and proteasome-mediated degradation. The mutant PMEPA1 (PY1/2 motif mutation) that is impaired in NEDD4 recruitment shows attenuated AR ubiquitination and AR protein down-regulation. These data support the hypothesis that PMEPA1 negatively regulates the stability of AR protein by enhancing AR ubiquitination and proteasome-mediated degradation through NEDD4. The effect of PMEPA1 on AR ubiquitination and degradation appears to be MDM2-independent. Thus, the PMEPA1-AR degradation pathway may represent a new androgen-dependent mechanism for regulating AR levels in prostate epithelial cells. These findings underscore that the decreased PMEPA1 expression frequently noted in prostate cancers may lead to increased AR functions and strengthen the biological role of PMEPA1 in prostate cancers.
Androgen receptor (AR),3 the male hormone receptor and a nuclear transcription factor, plays a central role in the growth and differentiation of the prostate gland. AR dysfunctions may contribute to benign prostatic hyperplasia and prostate cancer (CaP) (1–3). Both non-nuclear and nuclear functions of AR have been described in prostate cancer cells (2). The mechanism of nuclear function of AR involves translocation of the dihydrotestosterone-bound AR to the nucleus, where it binds to AR-responsive elements (AREs) of target genes and regulates their transcription. AR transcriptional targets such as PSA, NKX3.1, ODC1, AMD1, CDK1–2, etc. carry out many of the downstream cellular functions of AR such as cell growth and differentiation in a context-specific manner (4, 5). Although the physiologic functions of PSA are involved in reproductive biology, PSA is a marker of luminal epithelial cell differentiation in the prostate, and higher serum PSA levels have been instrumental in the early detection of prostate cancer (6). NKX3.1, a prostate cancer suppressor gene, is involved in prostate development and differentiation, and loss of NKX3.1 contributes to the development of pre-neoplastic stages of prostate cancer (7, 8). In animal models it was shown that Nkx3.1 plays an important role in blocking prostate cancer initiation by PTEN loss (9). Because androgen-dependent AR signaling plays key roles in the growth and differentiation of the prostate gland, tight control of AR signaling may be critical in maintaining the homeostasis of the prostate gland (4, 10). Absence of physiologic levels of testosterone leads to degeneration of the prostate gland (11). In clinical practice, androgen ablation is the cornerstone for the treatment of advanced CaP (12).
Abnormal functions of AR, such as functional activation because of mutations, amplification, and cross-talk with other pro-cancer signaling pathways, are increasingly recognized in prostate cancer development and progression (13–15). Although AR protein synthesis has been extensively studied, only a few studies have addressed how AR protein turnover is regulated (16, 17). These studies show that in LNCaP cells that harbor T877A mutant AR, the half-life of AR is around 3 h. In contrast, in the androgen-independent LNCaP-C4-2 cells and CWR-R1 cells, the half-life of AR is between 6 and 12 h (18). It has been reported that AR is stabilized by S26 proteasome inhibitor MG-132, suggesting that AR is targeted for degradation to the ubiquitin-proteasome pathway (19). In addition, down-regulation of AR by proteasome degradation occurs during prostate cancer cell mitosis (15).
E3 ligases catalyze the ubiquitination of proteins. The abundance and specificity of the currently identified E3 ligases suggest that these molecules play central roles in determining the specificity of ubiquitination (20). There are four classes of proteins with ubiquitin E3 ligase activity as follows: the HECT domain proteins (homology to E6-AP C terminus), U-box proteins, PHD finger-type, and RING finger ligases. The RING finger-containing proteins such as BRCA-1, Snurf-1, and ARA54 associate with AR and modulate AR activity (21–23). Moreover, MDM2 has been shown to form a complex with Akt and AR, promoting phosphorylation-dependent AR ubiquitination and proteasomal degradation (16). This pathway requires the intact RING domain of MDM2 to function as a ubiquitin E3 ligase (16, 24). Nedd4 was originally identified as a developmentally regulated gene in mice (25–28). Nedd4 is a HECT domain-containing protein with E3 ubiquitin ligase activity harboring three WW domains composed of two highly conserved tryptophan residues with binding preference for proline-rich sequences (PPXY) known as the PY motif (29). NEDD4 (isoform 1) may also function as an oncogene by reducing cytoplasmic levels of PTEN in cancer cells (10, 30). Furthermore, NEDD4 plays a direct role in reducing the levels of RNA polymerase II during cellular damage responses (31). Therefore, NEDD4-interacting proteins may play significant roles in controlling the availability of NEDD4 in cancer cells.
PMEPA1 is a NEDD4-binding protein that was originally identified by our laboratory as a prostate-abundant, highly androgen-induced gene and was mapped to chromosome 20q13 (32, 33). Human PMEPA1 exhibits amino acid homology to the mouse Nedd4-binding protein Nedd4BP (33). PMEPA1 protein has two PY motifs (PPPY and PPTY), which are required for binding to WW domains of NEDD4 E3 ubiquitin ligase (34). We reported a decrease or loss of PMEPA1 mRNA expression in the tumor specimens of 62% of prostate cancer patients (33). Ectopic expression of PMEPA1 in prostate cancer cell lines exhibits cell growth inhibitory functions (33). Other studies have shown that PMEPA1 is a transforming growth factor-β-induced gene and a marker of terminal colonocyte differentiation (35). By using the combination of in silico and experimental approaches, we have defined androgen-responsive elements within the PMEPA1 promoter upstream sequences (36). Furthermore, we showed that DNA methylation may contribute to the down-regulation of PMEPA1 expression in prostate cancer cell culture models (44). Taken together, accumulating data suggest that PMEPA1 is an androgen-inducible negative regulator of prostate cancer cell growth. The observation that PMEPA1 is an androgen-regulated gene together with the regulatory functions of AR in prostate cancer led us to investigate whether PMEPA1 is involved in the regulation of AR protein turnover, through the ubiquitin-proteasome pathway.
In this study, we show that PMEPA1 is a direct AR transcriptional target and that PMEPA1 protein interacts with and mediates the down-regulation of AR protein. The down-regulation of AR is mediated by enhanced AR ubiquitination and degradation. Our findings also reveal a new MDM2-independent mechanism of AR ubiquitination and degradation pathway linked to the NEDD4 E3 ubiquitin ligase. Thus, AR and PMEPA1 form a feedback loop, and frequent loss of PMEPA1 expression noted in prostate tumors (33) at least in part may lead to the gain of AR functions contributing to prostate cancer.
EXPERIMENTAL PROCEDURES
Plasmid Constructs, RNAi, and PCR Primers—Mammalian expression vectors encoding wild-type (WT) and PY mutant PMEPA1-V5 and PMEPA1-GFP were described before (33). pcDNA-HA-ubiquitin was a kind gift from Dr. Dirk Bohmann (University of Rochester, NY). pCMV-AR (33, 37) and pCMV-NEDD4 (isoform 1) were described before (33). siRNAs, RNAi-1, and RNAi-2 targeting PMEPA1 and nontargeting control RNAi pool (D-001206-13-20) were purchased from Dharmacon (Lafayette, CO). The target sequences are RNAi-1-GCATCAGCGCCACGTGCTA and RNAi-2-GTTATCACCACGTTATATA.
Cell Cultures and Transfection—Prostate cell line, LNCaP, was purchased from American Type Culture Collection (Manassas, VA) and was grown in RPMI medium with fetal bovine serum (FBS) or with charcoal-stripped FBS (cFBS). A stable PMEPA1-GFP-Tet LNCaP transfectant was generated by using the pTet-Off expression system (Clontech). Expression of PMEPA1-GFP fusion protein in these cells was negatively regulated by tetracycline (Sigma). For this cell line 200 μg/ml G418 and 2 μg/ml puromycin were used to maintain cell growth. COS-7 cells were grown in DMEM with 10% of FBS. HEK 293 cells were purchased from ATCC. 2KO cells (p53–/–-MDM2–/–) were from Dr. Guillermina Lozano (MD Anderson Cancer Center). The transfections were performed by using Lipofectamine 2000 (Invitrogen).
Immunoprecipitation and Immunoblot Analysis—Cells were harvested in a lysis buffer containing 50 mm Tris-HCl, pH 8.0, 1 mm EDTA, 1% Nonidet P-40, 150 mm NaCl, 10% of glycerol, protease inhibitor mixture (Roche Applied Science), and phosphatase inhibitor mixtures I and II (Sigma) at 4 °C. After centrifugation at 10,000 × g for 3 min at 4 °C, the supernatant was collected, and protein concentration was measured by using the protein assay kit from Bio-Rad. Equal amounts of protein lysates were used for immunoprecipitations with protease A + G beads (Pierce). Washing buffer for the immunoprecipitated proteins on the beads contained 50 mm Tris-HCl, pH 8.0, 1 mm EDTA, 1% Nonidet P-40, 10% glycerol, and 250 mm NaCl. Immunoprecipitates solubilized in sample loading buffer according to the instructions (Invitrogen) were separated by electrophoresis on 4–12% of NuPAGE gel, and proteins were transferred onto polyvinylidene difluoride membranes (Invitrogen).
Antibodies and Chemicals—Antibodies used in this study were as follows: anti-AR (PG-21, 06-680, Upstate Biotechnology, Inc., Lake Placid, NY); anti-AR (H-280, sc-13062, Santa Cruz Biotechnology, Santa Cruz, CA); anti-AR monoclonal antibody (441, sc-7305, Santa Cruz Biotechnology); anti-AR polyclonal antibody (N-20, sc-816, Santa Cruz Biotechnology); anti-P27 antibody (sc-1641, Santa Cruz Biotechnology); antipol II antibody (sc-899, Santa Cruz Biotechnology); anti-human-PSA polyclonal antibody (A05662, DakoCytomation, Denmark); anti-GFP monoclonal antibody (632375, Clontech); anti-HA antibody (MMS-101P, Covance, Berkeley, CA); anti-acetyl histone H3 antibody (06-599, Upstate Biotechnology, Inc.); anti-actin antibody (A2668, Sigma); horseradish peroxidase-conjugated anti-rabbit and anti-mouse TrueBlot antibodies (18-8816, 18-8817-33 eBioscience, San Diego); and anti-PMEPA1 monoclonal antibody, 2A12 (H00056937-M01, ABNOVA, Taiwan). Tetracycline and proteasome inhibitor lactacystin (L6785) were purchased from Sigma. R1881 was obtained from PerkinElmer Life Sciences. Proteasome inhibitors MG132 (catalog number 474790), LLNL (catalog number 208719), and protease inhibitor E-64 (catalog number 324890) were purchased from Calbiochem. Anti-PMEPA1 polyclonal antibody corresponding to the PMEPA1 peptide sequence AIWSKEKDKQKGHPL was prepared by our laboratory. PMEPA1 antibody was immunoaffinity-purified by using the Sulfolink kit (Pierce).
Fluorescence-activated Cell Sorter Analysis—Equivalent numbers of LNCaP cells were grown in FBS-containing media and were transfected with RNAi molecules targeting PMEPA1. After 48 h of incubation the cells (floating or attached) were collected by centrifugation, fixed with 100% methanol, and stained with propidium iodide (Sigma) and analyzed by flow cytometry using modified LT software.
Quantitative RT-PCR—LNCaP cells were grown in cFBS-containing media for 5 days. Then R1881 synthetic androgen was added to the media to the indicated concentrations. After 24 h of incubation, cells were processed for total RNA extraction by using the TRIzol reagent (Invitrogen). RT-PCR was performed by using the SuperScript III first-strand (Invitrogen) kit with (dT)20. Quantitative gene expression analysis was performed by TaqMan-based quantitative reverse transcription-PCR on ABI 7700 (Applied Biosystems, Foster City, CA). Primers for detecting PMEPA1 (Locus ID GXL_128240) were as follows: forward 5′-CATGATCCCCGAGCTGCT-3′ and reverse 5′-TGATCTGAACAAACTCCAGCTCC-3′. The probe was 6FAM-AGGCGGACAGTCTCCTGCGAAA-TAMRA. For GAPDH (glyceraldehyde-3-phosphate dehydrogenase) amplification, primers (PN402869) were purchased from Applied Biosystems (Foster City, CA).
Ubiquitination Assay—HEK-293 cells or 2KO cells were cotransfected with pCMV-AR and WTPMEPA-V5 or PMEPA Py1+Py2 mutant-V5 or pcDNA3 vectors. After 24 h, cells were incubated with 5 μm MG132 proteasome inhibitor for 8 h. Cells were lysed with RIPA buffer. Cell lysates were immunoprecipitated with anti-AR antibody and analyzed by immunoblot assay.
Chromatin Immunoprecipitation Assay (ChIP)—LNCaP cells were grown in cFBS containing media for 5 days. Then R1881 was added to the cell cultures to indicated concentrations. After 24 h of incubation, cells were processed for ChIP assay as described before (36) using 1 μl of a 1:1 v/v mixture of anti-AR (H-280) and anti-AR (PG21) antibodies or anti-RNA polymerase II antibody or anti-acetyl histone H3 antibody. DNA fragments were amplified by PCR primers corresponding to the androgen receptor binding motifs of PMEPA1 5′ promoter upstream regions and PMEPA1 core promoter region. PCR primer sequences are as follows: to amplify the –2134 distal PMEPA1 ARE between positions –2251 and –2068, forward 5′-CCCTGGCACATCTAGGGTTA-3′ and reverse 5′-TGGACTGCCAGCACTCATAG-3′; PMEPA1 ARE –230 proximal between positions –284 and –156, forward 5′-CAGGGAGGGGAGGTCTCTTA-3′ and reverse 5′-TCAAAAGGGGTATGAGCAGG-3′; PMEPA1 core promoter between +81 and +199, forward 5′-AACTGAAGGCGGACAGTCTC-3′ and reverse 5′-TTCTGAGGAGCACAAGGTCC-3′; PMEPA1 low affinity ARE –629 between –721 and –493, forward 5′-TCACTTCCCAAATTCCAGC-3′ and reverse 5′-GTCACACAGTGGTGGAGCC-3′ primers were used.
RESULTS
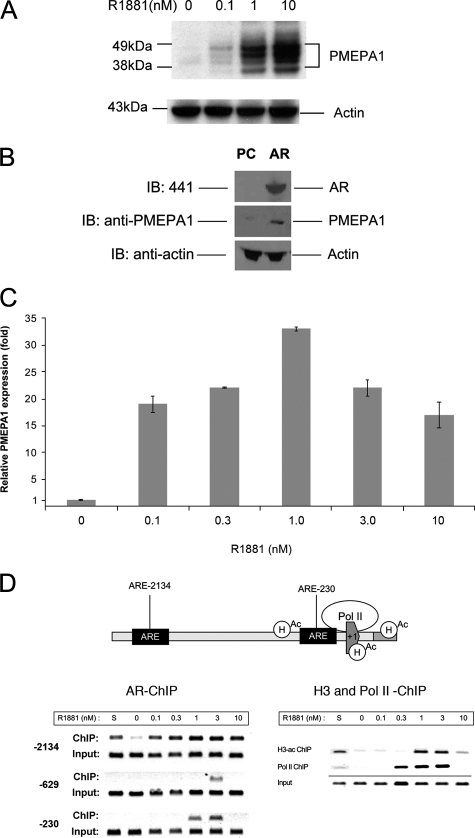
Androgen Receptor Activates PMEPA1—We have reported earlier that PMEPA1 was among the highly induced genes in the androgen-regulated transcriptome of LNCaP cells (32). Here we show a dose-dependent induction of the PMEPA1 protein in response to androgen (R1881) treatment of LNCaP cells (Fig. 1A). To further define if PMEPA1 induction is AR-dependent, we provide evidence that endogenous PMEPA1 protein is up-regulated by ectopic expression of AR in COS-7 cells that do not express endogenous AR (16, 18) (Fig. 1B). To establish AR as the direct transcriptional regulator of PMEPA1, we monitored the synthetic androgen, R1881, dose-dependent transcriptional activation of PMEPA1 relative to the expression levels in LNCaP cells grown in the absence of R1881 (Fig. 1C). Also, we monitored recruitment of AR to high affinity binding sites (–2134 and –230 relative to the transcription initiation site of PMEPA1) previously defined both by model prediction and by chromatin immunoprecipitation assays (Fig. 1D) (36). As a control, we compared AR recruitment to a low affinity AR-binding site (position –629) predicted by single matrix match. In semi-quantitative ChIP assays, androgen-dependent binding of AR to the PMEPA1 cognate sequences were highest in 1–3 nm R1881-treated LNCaP cells. Histone H3 acetylation and RNA polymerase II recruitment to these sites also peaked at the same R1881 concentration. Importantly, recruitment of AR, RNA polymerase II, histone H3 acetylation, and PMEPA1 expression were observed 24 h after R1881 induction at the same time point. These experiments suggest that PMEPA1 is a direct transcriptional target of AR, and androgen induction of the PMEPA1 mRNA is consistent with the expression of PMEPA1 protein.
FIGURE 1.
PMEPA1 expression is regulated by AR. A, LNCaP cells were maintained in hormone-depleted (cFBS) media for 5 days followed by the addition of indicated doses of R1881 synthetic androgen for 48 h. Thirty micrograms of cell lysates were subjected to 4–12% of NuPAGE and immunoblotted with anti-PMEPA1 monoclonal antibody 2A12 and anti-actin antibody. Molecular mass markers are shown in kDa. B, COS-7 cells were transfected with pcDNA3.1 (PC) or AR-pcDNA3.1 (AR) expression vectors under the control of cytomegalovirus promoter. After incubation for 24 h cell lysates were analyzed by immunoblot assays (IB) with anti-AR (441), anti-PMEPA1, and anti-actin antibodies visualizing AR (100 kDa), multiple forms of PMEPA1 (32–40 kDa) and actin (34 kDa) proteins, respectively. C and D, LNCaP cells were grown in hormone-depleted cFBS-containing media for 5 days followed either by the addition of 0, 0.1, 0.3, 1.0, 3.3, 10 nm of R1881 or by the addition of FBS containing media (S). Cells were incubated for 24 h, and the expression of PMEPA1 was analyzed by quantitative PCR. PMEPA1 expression fold change was normalized to the levels of the housekeeping gene GAPDH. PMEPA1 expression is shown relative to the levels in hormone-depleted (cFBS, 0 nm R1881) LNCaP cells (bars equal mean ± S.E. (n = 3)). In semi-quantitative chromatin immunoprecipitation assay anti-AR, anti-RNA polymerase II or anti-acetylated H3 histone antibodies were used. Specific primer pairs were selected for amplifying the –2134, –230, and the low affinity –629 AREs within the PMEPA1 promoter upstream sequences. To assay the core promoter for RNA polymerase II (polII) and acetyl histone H3 (H3-ac) recruitment the +81 and +199 were amplified. To achieve linear amplifications ranges the ChIP and input products were amplified with 38 and 33 cycles, respectively.
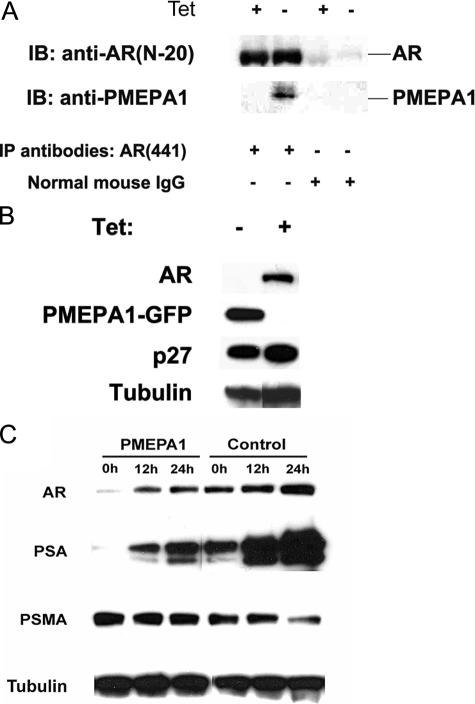
PMEPA1 Protein Binds to and Down-regulates AR Protein—To investigate the functions of PMEPA1 protein, we assessed interactions between PMEPA1 and AR. We generated a Tet-Off inducible PMEPA1-GFP-expressing LNCaP cell line. Using the anti-AR antibody 441 (Santa Cruz Biotechnology, Santa Cruz CA), we demonstrate that Tet-Off-induced PMEPA1 protein interacts with endogenous AR protein in LNCaP cells (Fig. 2A). These data support the hypothesis that PMEPA1 protein physically binds to the AR protein. When PMEPA1 expression was induced in this Tet-Off-PMEPA-LNCaP cell line, dramatic reduction in the AR protein levels was observed (Fig. 2B). In contrast, p27, a known E3 ligase-binding protein and proteasome target, did not show significant degradation in the presence of PMEPA1. This result suggests the selectivity of PMEPA1 for its target such as AR. Additionally, LNCaP cells with constitutive PMEPA1 overexpression also showed significant reduction of the newly synthesized AR as well as AR targets such as PSA (Fig. 2C).
FIGURE 2.
AR binds to PMEPA1 and PMEPA1 down-regulates AR protein. LNCaP-PMEPA1-GFP-tet stable transfectants, expressing PMEPA1-GFP fusion protein in response to tetracycline withdrawal (Tet-Off), were cultured in medium containing 2 μg/ml of tetracycline (+). PMEPA1-GFP expression was induced by changing to tetracycline-free media (–). AR, PMEPA1, PMEPA1-GFP (60 kDa), p27 (27 kDa), PSA (28 kDa), PSMA (100 kDa), and tubulin (58 kDa) proteins were detected by immunoblot assay. A, cell lysates were immunoprecipitated (IP) with either anti-AR monoclonal antibody (441) or by normal mouse IgG and immunoblotted (IB) with anti-AR polyclonal antibody (N-20) or by anti-PMEPA1 monoclonal antibody 2A12. Anti-rabbit and anti-mouse TrueBlot-horseradish peroxidase secondary antibodies were used to visualize the proteins. B, LNCaP-PMEPA1-GFP (Tet-Off system) cells were cultured with or without tetracycline for 10 days. Thirty μg of the cell lysates were analyzed by immunoblot assay with anti-AR (441), anti-GFP, anti-p27, and anti-tubulin antibodies. C, stable PMEPA1 expressing LNCaP cells were grown in hormone-depleted media (cFBS) for 5 days followed by the addition of 10 nm of R1881 and were incubated for 0, 12, or 24 h. Thirty μg of cell lysates were immunoblotted with the indicated antibodies.
AR, PSA, and Cell Cycle S Phase Are Increased in Response to PMEPA1 Expression Knockdown—Our earlier studies showed that ectopic expression of PMEPA1 in prostate cancer cell lines inhibited cell proliferation (34). To further elucidate the biological functions of PMEPA1, we knocked down the endogenous PMEPA1 expression by siRNA in LNCaP cells (Fig. 3, A and B). Decreased expression of PMEPA1 was associated with increased levels of the AR protein (Fig. 3C). We also observed a concomitant increase of PSA protein, a well known transcriptional target of AR (Fig. 3C). Furthermore, PMEPA1 knockdown was associated with modest but detectable increases in S phase of the cell cycle (Fig. 3D). This observation is consistent with the slow growth of androgen-responsive LNCaP cells. These data suggest a functional link between PMEPA1, AR, and cell growth.
FIGURE 3.
PMEPA1 knockdown increases AR protein levels, elevates PSA, and enhances S phase of cell cycle. A, LNCaP cells were transfected with nontargeting RNAi (NT-RNAi) or with PMEPA1 RNAi-1 or PMEPA1 RNAi-2. After 48 h of incubation, cells were lysed with TRIzol reagent for total RNA extraction and were processed for quantitative RT-PCR analysis. After normalization to GAPDH, changes in PMEPA1 transcript levels are shown as the percent of expression of the nontargeting RNAi treatment group. Bars equal mean ± S.E. (n = 4) from two independent experiments. B, LNCaP cells were grown in hormone-depleted (cFBS) media for 5 days followed by the transfection with 75 nm of either nontargeting RNAi (NT) or with PMEPA1 RNAi-1 or PMEPA1 RNAi-2 for 16 h. The cells were treated with 0.1 nm of synthetic androgen R1881 for 48 h. Forty five μg of cell lysates were analyzed on 4–12% NuPAGE by immunoblot (IB) assay with the indicated antibodies detecting AR, actin, and PSA (28 kDa) proteins. C, RNAi transfected LNCaP cells (the same condition as A) were incubated for 48 h, and 30 μg of cell lysates were separated on 4–12% NuPAGE and were immunoblotted with indicated antibodies. D, RNAi-transfected LNCaP cells (as in A and C) were incubated for 48 h followed by propidium iodide staining and subsequent flow cytometry analysis. Cell S phase was quantitated using the ModFit LT program. Percentage (%) of cells in S phase is shown on the graph. Bars represent mean ± S.E. (n = 3) from three independent experiments. Analysis of variance and Dunnett's t test were applied for statistic analysis, and SAS software was used.
PMEPA1 Down-regulates AR through the Ubiquitin-Proteasome Pathway—Polyubiquitinated form of AR is degraded by the MDM2-dependent ubiquitin-proteasome pathway (16). To address the question of whether the ubiquitin-proteasome pathway (16, 19) is involved in PMEPA1-mediated AR down-regulation, the S26 proteasome inhibitor MG132 was tested in prostate cancer cells. We demonstrate that Tet-Off-inducible PMEPA1 significantly down-regulated AR protein levels in LNCaP cells, and inhibition of the ubiquitin-proteasome pathway by MG132 partially rescued AR down-regulation by PMEPA1 (Fig. 4A). Moreover, we show that other proteasome inhibitors, lactacystin and LLnL but not E-64, a protease inhibitor, rescued the AR protein levels in PMEPA1 expressing LNCaP (Tet-Off) cells (Fig. 4B). Because proteasome degradation of AR requires AR ubiquitination (19), we tested the consequences of PMPEA1 expression on AR ubiquitination. In HEK 293 cells, HA-ubiquitin-dependent AR-ubiquitination was enhanced by the overexpression of the PMEPA1 protein (Fig. 4C). These data suggest that PMEPA1 down-regulates AR by enhancing AR polyubiquitination and by promoting proteasome-mediated degradation.
FIGURE 4.
PMEPA1 targets AR to the ubiquitin-proteasome degradation pathway. A, LNCaP-PMEPA1-GFP-tet (Tet-Off) cells were grown with or without tetracycline for 5 days. Fiftyμm of MG132 was added to the media, and the cells were further incubated for 8 h. Thirty μg of cell lysates were tested with the indicated antibodies in immunoblot (IB) assays to detect AR, PMEPA1, and tubulin proteins. B, LNCaP-PMEPA1-GFP Tet-off cells were grown with (+) or without (–) tetracycline for 11 days and followed by the addition of 30 μl of DMSO (control), 50 μm MG132, 50 μm LLnL, 10 μm lactacystin, and 50 μm E-64 for an additional 5 h. Thirty μg of cell lysates were separated with 4–12% of NuPAGE gel and immunoblotted to visualize AR, PMEPA1-GFP, and actin proteins with the indicated antibodies. C, HEK 293 cells were transfected with AR, hemagglutinin-ubiquitin (HA-Ub) and PMEPA1 expressing pcDNA3.1 vectors or with vector alone (MOCK). After 24 h of incubation, 10 nm of R1881 was added to the media, and the cells were further incubated for 16 h followed by the addition of 5 μm of MG132 proteasome inhibitor. After 8 h of incubation cells were lysed and immunoprecipitated (IP) with anti-AR antibody (441) and were immunoblotted with anti-HA antibody. Western blot analysis of PMEPA1 and tubulin proteins are shown in the upper panel.
PMEPA1 Enhances AR Ubiquitination and Degradation through NEDD4 in MDM2-independent Manner—We previously reported that PMEPA1 is an androgen-regulated NEDD4-binding protein (33). We have also shown (33) that two PY motifs (PPPY-PY1 and PPTY-PY2) are required for binding to the WW domain of NEDD4 ubiquitin E3 ligase (Fig. 5A). When COS-7 cells were co-transfected with the expression vectors of AR and wild-type (WT) PMEPA1 or with mutant PMEPA1 (PY1 and PY2, Y126A and Y197A, respectively), AR protein was down-regulated by WT PMEPA1, but AR down-regulation was appreciably inhibited by PY mutant PMEPA1 (Fig. 5B). Moreover, PY mutant PMEPA1 was less effective in enhancing AR polyubiquitination compared with WT PMEPA1 (Fig. 5C). These data indicate that down-regulation of AR by PMEPA1 is dependent on its NEDD4 binding activity in enhancing AR polyubiquitination and degradation. MDM2 is a known ubiquitin E3 ligase for AR ubiquitination and degradation. To test whether PMEPA1 enhanced AR polyubiquitination and degradation in the absence of MDM2, 2KO cells (p53–/– and MDM2–/–) were co-transfected with AR expression vector with increasing amounts of a WT PMEPA1 expressing construct (Fig. 6A). AR protein levels partially decreased with increasing amounts of PMEPA1 expression, indicating that PMEPA1 at least in part contributes to AR degradation in an MDM2-independent manner. These data also suggest that in addition to MDM2, NEDD4 is involved in regulating AR levels. Because PMEPA1 has neither RING domain nor HECT domain noted in ubiquitin E3 ligases, we propose that PMEPA1 recruits NEDD4 to AR by binding to WW motifs of NEDD4 for AR ubiquitination and degradation (Fig. 6B).
FIGURE 5.
Mutation of PY motifs impairs PMEPA1-mediated AR degradation. A, schematic representation of PY motifs within the PMEPA1 protein. aa, amino acids. B, COS-7 cells were co-transfected with AR and/or with pcDNA3.1 control (PC) or WT PMEPA1 (WT) or PY mutant PMEPA1 (PYmt) expressed by pcDNA3.1 vectors. Cells were incubated for 42 h. Thirty μgof cell lysates were subjected to immunoblot (IB) assay to detect AR, PMEPA1, and actin proteins with the indicated antibodies. C, double knock-out p53–/– and MDM2–/– 2KO cells were transfected with the combination of HA-tagged ubiquitin (HA-Ub), AR, NEDD4, PMEPA-V5 (PMEPA1), or PY mutant PMEPA1 (Pymut) or V5-pcDNA3.1 control vectors. After 24 h of incubation, cells were treated with 25 μm MG132 proteasome inhibitor for 5 h. Cells were lysed, and proteins were immunoprecipitated with anti-AR antibody (441) and were analyzed by Western blot with anti-AR or anti-HA antibodies. In the lower panel immunoblots with anti-PMEPA1 or anti-actin antibodies are shown.
FIGURE 6.
Down-regulation of AR by PMEPA1 is independent of MDM2 and p53. A, 2KO (p53–/–/MDM2–/–) cells were co-transfected with 0.5 μgof pCMV-AR and 0.2 μg of pcDNA3-GFP with 0, 1, 2, or 8 μg of PMEPA1 expressing vectors followed by incubation of cells for 42 h. Cells were lysed, and the proteins were separated by 4–12% of NuPAGE. Immunoblot (IB) assays were performed to visualize AR, PMEPA1, GFP (28 kDa), and actin proteins with the indicated antibodies. B, AR-PMEPA1 feedback loop model. AR is a transcriptional activator of PMEPA1. PMEPA1 negatively regulates AR protein levels by recruiting the NEDD4 ubiquitin E3 ligase, thus targeting AR to the ubiquitin-proteasome pathway. This mechanistic model also suggest that decreased PMEPA1 expression, frequently noted in prostate cancers, may lead to increased AR protein and functions in prostate tumors with lost or decreased PMEPA1 expression.
DISCUSSION
Deregulation of androgen signaling contributes to the continuum of prostate cancer progression. Consistently, AR alterations are more apparent at advanced stages. Numerous studies have addressed genomic or expression alterations of the AR, AR transcription factor dysfunctions, or cross-talk of AR with other pro-cancer signaling pathways in prostate cancer (2, 38). Compromised proteasome-dependent degradation of AR is known to play a role in prostate cancer progression. MDM2 is a RING finger-type of E3 ubiquitin ligase that has been shown to mediate AR degradation in an Akt-dependent manner (16). Here we provide evidence that PMEPA1 as an androgen-responsive gene functions as a negative regulator of AR by recruiting the NEDD4 ubiquitin E3 ligase.
We previously reported physical and functional interactions between PMEPA1 and NEDD4 E3 ubiquitin ligase, a HECT-domain containing member of the ubiquitin-proteasome pathway (33). Using LNCaP cells harboring Tet-Off-inducible PMEPA1-GFP, we demonstrate that AR protein levels decrease in response to elevated PMEPA1 expression. To rule out the nonspecific effects of PMEPA1 overexpression on protein turnover, we examined the levels of the p27 protein, known to be targeted by ubiquitin-proteasome pathway. No significant changes in p27 suggested selectivity for the PMEPA1-mediated decease in AR protein levels. Although the region(s) of contact between AR and PMEPA1 remains to be defined, our data establish that PMEPA1 protein physically binds to AR protein.
To address the question of whether or not the ubiquitin-proteasomal pathway is involved in PMEPA1-mediated AR degradation, we tested if S26 proteasome inhibitors, such as lactacystin, MG132, and LLnL, can rescue AR from PMEPA1-mediated depletion in prostate cancer cells. Indeed, inhibition of the ubiquitin-proteasomal pathway by lactacystin, MG132 and LLnL, partially rescued AR from PMEPA1-mediated down-regulation in prostate cancer cells. We also found that wild type but not the PY motif mutations (Y126A (PY1) and Y197A (PY2)) of PMEPA1 enhance AR ubiquitination and down-regulation. The PY motif mutation has been shown previously to abrogate the binding of PMEPA1 to NEDD4 (33). Our findings indicate that docking of NEDD4 E3 ubiquitin ligase to PMEPA1 was required for PMEPA-mediated AR degradation. Recent reports have established NEDD4 as a protooncogene by demonstrating that NEDD4-mediated monoubiquitination results in the reduction of cytoplasmic levels of PTEN tumor suppressor (39). Thus the role of PMEPA1 in other cancer pathways remains to be determined.
PMEPA1 is highly inducible by androgen, and AR activates PMEPA1 expression. Here we demonstrate that AR preferentially binds to upstream androgen-responsive elements (36) in a hormone dose-dependent manner, facilitating the recruitment of RNA polymerase II and acetyl histone H-3 to the PMEPA1 promoter. Although, similar to other hormone-regulated genes, PMEPA1 transcript levels may also be modulated by mRNA stabilization (40), our data suggest that AR is a direct transcriptional activator of PMEPA1. Taken together these observations reveal a tight feedback loop between AR and PMEPA1.
In cell culture models PMEPA1 exhibits cell growth inhibitory effects (33). Therefore, PMEPA1 may fall into the category of androgen-regulated genes that negatively regulate cell growth, such as NKX3.1, which is a tumor suppressor and is involved in prostate development and differentiation. Loss of Nkx3.1 in mouse prostate leads to prostate epithelial hyperplasia, resembling early stages of prostate cancer cooperating with decreased functions of PTEN in cancer progression (9, 41–43). Similar to PMEPA1, NKX3.1 is an androgen-inducible gene that can modulate downstream AR transcription targets (43). Although PMEPA1 functions remain to be defined in mouse models, our data in human prostate cancer specimens suggest that decrease or loss of PMEPA1 is associated with prostate cancer. Although the mechanism of reduced PMEPA1 expression in human tumors is under investigation, in cell culture models of prostate cancer inhibition of DNA methyltransferases can lead to increased PMEPA1 expression (1). In summary, our data suggest that AR is a transcriptional activator of PMEPA1, whereas PMEPA1 mediates the degradation of AR protein by recruiting NEDD4 E3 ubiquitin ligase. Therefore, we propose a negative feedback loop to describe this tight regulatory circuit between AR and PMEPA1. Although MDM2 has been shown to target AR to ubiquitin-proteasomal degradation in an Akt dependent manner, PMEPA1-dependent AR turnover may be significant in the context of androgen-dependent regulation of AR in prostate epithelial cells. Thus, understanding of AR protein turnover may provide important new clues about the role of AR and AR modulators such as PMEPA1 in prostate cancer, and such mechanistic studies may enhance the therapeutic strategies aiming to down-regulate AR levels in prostate tumor cells.
Acknowledgments
The excellent technical assistance of Soyon Oh and Karthik Dwarki are greatly appreciated. We are thankful to Stephen Doyle for the artwork and Anita Roundtree and Kara Leventhal for the preparation of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01CA106653 (NCI) (to S. S. and A. D.). This work was also supported by the Center for Prostate Disease Research Program through the Henry M. Jackson Foundation for the Advancement of Military Medicine under Contract HU001-04-C-1502 (2004) with the Uniformed Services University. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: AR, androgen receptor; ARE, AR-responsive element; CaP, prostate cancer; RNAi, RNA interference; FBS, fetal bovine serum; cFBS, charcoal-stripped FBS; HA, hemagglutinin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ChIP, chromatin immunoprecipitation; glyceraldehyde-3-phosphate dehydrogenase; E3, ubiquitin-protein isopeptide ligase; PSA, prostate-specific antigen.
References
- 1.Richter, E., Srivastava, S., and Dobi, A. (2007) Prostate Cancer Prostatic Dis. 10 114–118 [DOI] [PubMed] [Google Scholar]
- 2.Dehm, S. M., and Tindall, D. J. (2007) Mol. Endocrinol. 12 2855–2863 [DOI] [PubMed] [Google Scholar]
- 3.Dehm, S. M., and Tindall, D. J. (2006) J. Cell. Biochem. 99 333–344 [DOI] [PubMed] [Google Scholar]
- 4.Nieto, M., Finn, S., Loda, M., and Hahn, W. C. (2007) Int. J. Biochem. Cell Biol. 39 1562–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu, C. T., Altuwaijri, S., Ricke, W. A., Huang, S. P., Yeh, S., Zhang, C., Niu, Y., Tsai, M. Y., and Chang, C. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 12679–12684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson, I. M., and Ankerst, D. P. (2007) Can. Med. Assoc. J. 176 1853–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen, M. M., and Abate-Shen, C. (2003) Dev. Dyn. 228 767–778 [DOI] [PubMed] [Google Scholar]
- 8.Bieberich, C. J., Fujita, K., He, W. W., and Jay, G. (1996) J. Biol. Chem. 271 31779–31782 [DOI] [PubMed] [Google Scholar]
- 9.Lei, Q., Jiao, J., Xin, L., Chang, C. J., Wang, S., Gao, J., Gleave, M. E., Witte, O. N., Liu, X., and Wu, H. (2006) Cancer Cell 9 367–378 [DOI] [PubMed] [Google Scholar]
- 10.Wang, X., Trotman, L. C., Koppie, T., Alimonti, A., Chen, Z., Gao, Z., Wang, J., Erdjument-Bromage, H., Tempst, P., Cordon-Cardo, C., Pandolfi, P. P., and Jiang, X. (2007) Cell 128 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu, C. P., and Gu, F. L. (1991) Prog. Clin. Biol. Res. 370 249–255 [PubMed] [Google Scholar]
- 12.Mostaghel, E. A., Montgomery, R. B., and Lin, D. W. (2007) Curr. Urol. Rep. 8 224–232 [DOI] [PubMed] [Google Scholar]
- 13.Debes, J. D., and Tindall, D. J. (2004) N. Engl. J. Med. 351 1488–1490 [DOI] [PubMed] [Google Scholar]
- 14.Gelmann, E. P. (2002) J. Clin. Oncol. 20 3001–3015 [DOI] [PubMed] [Google Scholar]
- 15.Litvinov, I. V., Vander Griend, D. J., Antony, L., Dalrymple, S., De Marzo, A. M., Drake, C. G., and Isaacs, J. T. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 15085–15090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, H. K., Wang, L., Hu, Y. C., Altuwaijri, S., and Chang, C. (2002) EMBO J. 21 4037–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, D. K., and Chang, C. (2003) J. Clin. Endocrinol. Metab. 88 4043–4054 [DOI] [PubMed] [Google Scholar]
- 18.Gregory, C. W., Johnson, R. T., Jr., Mohler, J. L., French, F. S., and Wilson, E. M. (2001) Cancer Res. 61 2892–2898 [PubMed] [Google Scholar]
- 19.Sheflin, L., Keegan, B., Zhang, W., and Spaulding, S. W. (2000) Biochem. Biophys. Res. Commun. 276 144–150 [DOI] [PubMed] [Google Scholar]
- 20.Crosetto, N., Bienko, M., and Dikic, I. (2006) Mol. Cancer Res. 4 899–904 [DOI] [PubMed] [Google Scholar]
- 21.Kang, Z., Pirskanen, A., Janne, O. A., and Palvimo, J. J. (2002) J. Biol. Chem. 277 48366–48371 [DOI] [PubMed] [Google Scholar]
- 22.Poukka, H., Karvonen, U., Yoshikawa, N., Tanaka, H., Palvimo, J. J., and Janne, O. A. (2000) J. Cell Sci. 113 2991–3001 [DOI] [PubMed] [Google Scholar]
- 23.Yeh, S., Hu, Y. C., Rahman, M., Lin, H. K., Hsu, C. L., Ting, H. J., Kang, H. Y., and Chang, C. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 11256–11261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang, S., Jensen, J. P., Ludwig, R. L., Vousden, K. H., and Weissman, A. M. (2000) J. Biol. Chem. 275 8945–8951 [DOI] [PubMed] [Google Scholar]
- 25.Ingham, R. J., Gish, G., and Pawson, T. (2004) Oncogene 23 1972–1984 [DOI] [PubMed] [Google Scholar]
- 26.Harvey, K. F., Dinudom, A., Komwatana, P., Jolliffe, C. N., Day, M. L., Parasivam, G., Cook, D. I., and Kumar, S. (1999) J. Biol. Chem. 274 12525–12530 [DOI] [PubMed] [Google Scholar]
- 27.Anan, T., Nagata, Y., Koga, H., Honda, Y., Yabuki, N., Miyamoto, C., Kuwano, A., Matsuda, I., Endo, F., Saya, H., and Nakao, M. (1998) Genes Cells 3 751–763 [DOI] [PubMed] [Google Scholar]
- 28.Kumar, S., Harvey, K. F., Kinoshita, M., Copeland, N. G., Noda, M., and Jenkins, N. A. (1997) Genomics 40 435–443 [DOI] [PubMed] [Google Scholar]
- 29.Macias, M. J., Hyvonen, M., Baraldi, E., Schultz, J., Sudol, M., Saraste, M., and Oschkinat, H. (1996) Nature 382 646–649 [DOI] [PubMed] [Google Scholar]
- 30.Trotman, L. C., Wang, X., Alimonti, A., Chen, Z., Teruya-Feldstein, J., Yang, H., Pavletich, N. P., Carver, B. S., Cordon-Cardo, C., Erdjument-Bromage, H., Tempst, P., Chi, S. G., Kim, H. J., Misteli, T., Jiang, X., and Pandolfi, P. P. (2007) Cell 128 141–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anindya, R., Aygun, O., and Svejstrup, J. Q. (2007) Mol. Cell 28 386–397 [DOI] [PubMed] [Google Scholar]
- 32.Xu, L. L., Shanmugam, N., Segawa, T., Sesterhenn, I. A., McLeod, D. G., Moul, J. W., and Srivastava, S. (2000) Genomics 66 257–263 [DOI] [PubMed] [Google Scholar]
- 33.Xu, L. L., Shi, Y., Petrovics, G., Sun, C., Makarem, M., Zhang, W., Sesterhenn, I. A., McLeod, D. G., Sun, L., Moul, J. W., and Srivastava, S. (2003) Cancer Res. 63 4299–4304 [PubMed] [Google Scholar]
- 34.Jolliffe, C. N., Harvey, K. F., Haines, B. P., Parasivam, G., and Kumar, S. (2000) Biochem. J. 351 557–565 [PMC free article] [PubMed] [Google Scholar]
- 35.Brunschwig, E. B., Wilson, K., Mack, D., Dawson, D., Lawrence, E., Willson, J. K., Lu, S., Nosrati, A., Rerko, R. M., Swinler, S., Beard, L., Lutterbaugh, J. D., Willis, J., Platzer, P., and Markowitz, S. (2003) Cancer Res. 63 1568–1575 [PubMed] [Google Scholar]
- 36.Masuda, K., Werner, T., Maheshwari, S., Frisch, M., Oh, S., Petrovics, G., May, K., Srikantan, V., Srivastava, S., and Dobi, A. (2005) J. Mol. Biol. 353 763–771 [DOI] [PubMed] [Google Scholar]
- 37.Sun, C., Shi, Y., Xu, L. L., Nageswararao, C., Davis, L. D., Segawa, T., Dobi, A., McLeod, D. G., and Srivastava, S. (2006) Oncogene 25 3905–3913 [DOI] [PubMed] [Google Scholar]
- 38.Lange, C. A., Gioeli, D., Hammes, S. R., and Marker, P. C. (2007) Annu. Rev. Physiol. 69 171–199 [DOI] [PubMed] [Google Scholar]
- 39.Salmena, L., and Pandolfi, P. P. (2007) Nat. Rev. Cancer 7 409–413 [DOI] [PubMed] [Google Scholar]
- 40.Ing, N. H. (2005) Biol. Reprod. 72 1290–1296 [DOI] [PubMed] [Google Scholar]
- 41.Bowen, C., Bubendorf, L., Voeller, H. J., Slack, R., Willi, N., Sauter, G., Gasser, T. C., Koivisto, P., Lack, E. E., Kononen, J., Kallioniemi, O. P., and Gelmann, E. P. (2000) Cancer Res. 60 6111–6115 [PubMed] [Google Scholar]
- 42.Bhatia-Gaur, R., Donjacour, A. A., Sciavolino, P. J., Kim, M., Desai, N., Young, P., Norton, C. R., Gridley, T., Cardiff, R. D., Cunha, G. R., Abate-Shen, C., and Shen, M. M. (1999) Genes Dev. 13 966–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen, H., Nandi, A. K., Li, X., and Bieberich, C. J. (2002) Cancer Res. 62 338–340 [PubMed] [Google Scholar]
- 44.Richter, E., Masuda, K., Cook, C., Ehrich, M., Tadese, A. Y., Li, H., Owusu, A., Srivastava, S., and Dobi, A. (2007) Epigenetics 2 100–105 [DOI] [PubMed] [Google Scholar]